Abstract
The cis-regulatory systems that control developmental expression of two sea urchin genes have been subjected to detailed functional analysis. Both systems are modular in organization: specific, separable fragments of the cis-regulatory DNA each containing multiple transcription factor target sites execute particular regulatory subfunctions when associated with reporter genes and introduced into the embryo. The studies summarized here were carried out on the CyIIIa gene, expressed in the embryonic aboral ectoderm and on the Endo16 gene, expressed in the embryonic vegetal plate, archenteron, and then midgut. The regulatory systems of both genes include modules that control particular aspects of temporal and spatial expression, and in both the territorial boundaries of expression depend on a combination of negative and positive functions. In both genes different regulatory modules control early and late embryonic expression. Modular cis-regulatory organization is widespread in developmentally regulated genes, and we present a tabular summary that includes many examples from mouse and Drosophila. We regard cis-regulatory modules as units of developmental transcription control, and also of evolution, in the assembly of transcription control systems.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bienz M., Saari G., Tremml G., Müller J., Züst B., Lawrence P. A. Differential regulation of Ultrabithorax in two germ layers of Drosophila. Cell. 1988 May 20;53(4):567–576. doi: 10.1016/0092-8674(88)90573-9. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Davidson E. H. Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. Q Rev Biol. 1971 Jun;46(2):111–138. doi: 10.1086/406830. [DOI] [PubMed] [Google Scholar]
- Calzone F. J., Thézé N., Thiebaud P., Hill R. L., Britten R. J., Davidson E. H. Developmental appearance of factors that bind specifically to cis-regulatory sequences of a gene expressed in the sea urchin embryo. Genes Dev. 1988 Sep;2(9):1074–1088. doi: 10.1101/gad.2.9.1074. [DOI] [PubMed] [Google Scholar]
- Cameron R. A., Britten R. J., Davidson E. H. Expression of two actin genes during larval development in the sea urchin Strongylocentrotus purpuratus. Mol Reprod Dev. 1989;1(3):149–155. doi: 10.1002/mrd.1080010302. [DOI] [PubMed] [Google Scholar]
- Cameron R. A., Davidson E. H. Cell type specification during sea urchin development. Trends Genet. 1991 Jul;7(7):212–218. doi: 10.1016/0168-9525(91)90367-y. [DOI] [PubMed] [Google Scholar]
- Cameron R. A., Fraser S. E., Britten R. J., Davidson E. H. Segregation of oral from aboral ectoderm precursors is completed at fifth cleavage in the embryogenesis of Strongylocentrotus purpuratus. Dev Biol. 1990 Jan;137(1):77–85. doi: 10.1016/0012-1606(90)90009-8. [DOI] [PubMed] [Google Scholar]
- Cameron R. A., Hough-Evans B. R., Britten R. J., Davidson E. H. Lineage and fate of each blastomere of the eight-cell sea urchin embryo. Genes Dev. 1987 Mar;1(1):75–85. doi: 10.1101/gad.1.1.75. [DOI] [PubMed] [Google Scholar]
- Capovilla M., Brandt M., Botas J. Direct regulation of decapentaplegic by Ultrabithorax and its role in Drosophila midgut morphogenesis. Cell. 1994 Feb 11;76(3):461–475. doi: 10.1016/0092-8674(94)90111-2. [DOI] [PubMed] [Google Scholar]
- Charité J., de Graaff W., Vogels R., Meijlink F., Deschamps J. Regulation of the Hoxb-8 gene: synergism between multimerized cis-acting elements increases responsiveness to positional information. Dev Biol. 1995 Oct;171(2):294–305. doi: 10.1006/dbio.1995.1282. [DOI] [PubMed] [Google Scholar]
- Coffman J. A., Kirchhamer C. V., Harrington M. G., Davidson E. H. SpRunt-1, a new member of the runt domain family of transcription factors, is a positive regulator of the aboral ectoderm-specific CyIIIA gene in sea urchin embryos. Dev Biol. 1996 Feb 25;174(1):43–54. doi: 10.1006/dbio.1996.0050. [DOI] [PubMed] [Google Scholar]
- Cox K. H., Angerer L. M., Lee J. J., Davidson E. H., Angerer R. C. Cell lineage-specific programs of expression of multiple actin genes during sea urchin embryogenesis. J Mol Biol. 1986 Mar 20;188(2):159–172. doi: 10.1016/0022-2836(86)90301-3. [DOI] [PubMed] [Google Scholar]
- Davidson E. H. How embryos work: a comparative view of diverse modes of cell fate specification. Development. 1990 Mar;108(3):365–389. doi: 10.1242/dev.108.3.365. [DOI] [PubMed] [Google Scholar]
- Davidson E. H. Lineage-specific gene expression and the regulative capacities of the sea urchin embryo: a proposed mechanism. Development. 1989 Mar;105(3):421–445. doi: 10.1242/dev.105.3.421. [DOI] [PubMed] [Google Scholar]
- Davidson E. H. Spatial mechanisms of gene regulation in metazoan embryos. Development. 1991 Sep;113(1):1–26. doi: 10.1242/dev.113.1.1. [DOI] [PubMed] [Google Scholar]
- Dearolf C. R., Topol J., Parker C. S. Transcriptional control of Drosophila fushi tarazu zebra stripe expression. Genes Dev. 1989 Mar;3(3):384–398. doi: 10.1101/gad.3.3.384. [DOI] [PubMed] [Google Scholar]
- Doyle H. J., Kraut R., Levine M. Spatial regulation of zerknüllt: a dorsal-ventral patterning gene in Drosophila. Genes Dev. 1989 Oct;3(10):1518–1533. doi: 10.1101/gad.3.10.1518. [DOI] [PubMed] [Google Scholar]
- Emery J. F., Bier E. Specificity of CNS and PNS regulatory subelements comprising pan-neural enhancers of the deadpan and scratch genes is achieved by repression. Development. 1995 Nov;121(11):3549–3560. doi: 10.1242/dev.121.11.3549. [DOI] [PubMed] [Google Scholar]
- Flytzanis C. N., Britten R. J., Davidson E. H. Ontogenic activation of a fusion gene introduced into sea urchin eggs. Proc Natl Acad Sci U S A. 1987 Jan;84(1):151–155. doi: 10.1073/pnas.84.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks R. R., Anderson R., Moore J. G., Hough-Evans B. R., Britten R. J., Davidson E. H. Competitive titration in living sea urchin embryos of regulatory factors required for expression of the CyIIIa actin gene. Development. 1990 Sep;110(1):31–40. doi: 10.1242/dev.110.1.31. [DOI] [PubMed] [Google Scholar]
- Frasch M., Chen X., Lufkin T. Evolutionary-conserved enhancers direct region-specific expression of the murine Hoxa-1 and Hoxa-2 loci in both mice and Drosophila. Development. 1995 Apr;121(4):957–974. doi: 10.1242/dev.121.4.957. [DOI] [PubMed] [Google Scholar]
- Gan L., Klein W. H. A positive cis-regulatory element with a bicoid target site lies within the sea urchin Spec2a enhancer. Dev Biol. 1993 May;157(1):119–132. doi: 10.1006/dbio.1993.1117. [DOI] [PubMed] [Google Scholar]
- Gan L., Zhang W., Klein W. H. Repetitive DNA sequences linked to the sea urchin spec genes contain transcriptional enhancer-like elements. Dev Biol. 1990 May;139(1):186–196. doi: 10.1016/0012-1606(90)90287-s. [DOI] [PubMed] [Google Scholar]
- Gray S., Szymanski P., Levine M. Short-range repression permits multiple enhancers to function autonomously within a complex promoter. Genes Dev. 1994 Aug 1;8(15):1829–1838. doi: 10.1101/gad.8.15.1829. [DOI] [PubMed] [Google Scholar]
- Gérard M., Duboule D., Zákány J. Structure and activity of regulatory elements involved in the activation of the Hoxd-11 gene during late gastrulation. EMBO J. 1993 Sep;12(9):3539–3550. doi: 10.1002/j.1460-2075.1993.tb06028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Skarmeta J. L., Rodríguez I., Martínez C., Culí J., Ferrés-Marcó D., Beamonte D., Modolell J. Cis-regulation of achaete and scute: shared enhancer-like elements drive their coexpression in proneural clusters of the imaginal discs. Genes Dev. 1995 Aug 1;9(15):1869–1882. doi: 10.1101/gad.9.15.1869. [DOI] [PubMed] [Google Scholar]
- Haenlin M., Kunisch M., Kramatschek B., Campos-Ortega J. A. Genomic regions regulating early embryonic expression of the Drosophila neurogenic gene Delta. Mech Dev. 1994 Jul;47(1):99–110. doi: 10.1016/0925-4773(94)90099-x. [DOI] [PubMed] [Google Scholar]
- Harding K., Hoey T., Warrior R., Levine M. Autoregulatory and gap gene response elements of the even-skipped promoter of Drosophila. EMBO J. 1989 Apr;8(4):1205–1212. doi: 10.1002/j.1460-2075.1989.tb03493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch M., Schröder C., Seifert E., Jäckle H. cis-acting control elements for Krüppel expression in the Drosophila embryo. EMBO J. 1990 Aug;9(8):2587–2595. doi: 10.1002/j.1460-2075.1990.tb07440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hough-Evans B. R., Franks R. R., Cameron R. A., Britten R. J., Davidson E. H. Correct cell-type-specific expression of a fusion gene injected into sea urchin eggs. Dev Biol. 1987 Jun;121(2):576–579. doi: 10.1016/0012-1606(87)90193-x. [DOI] [PubMed] [Google Scholar]
- Hough-Evans B. R., Franks R. R., Zeller R. W., Britten R. J., Davidson E. H. Negative spatial regulation of the lineage specific CyIIIa actin gene in the sea urchin embryo. Development. 1990 Sep;110(1):41–50. doi: 10.1242/dev.110.1.41. [DOI] [PubMed] [Google Scholar]
- Howard K. R., Struhl G. Decoding positional information: regulation of the pair-rule gene hairy. Development. 1990 Dec;110(4):1223–1231. doi: 10.1242/dev.110.4.1223. [DOI] [PubMed] [Google Scholar]
- Huang J. D., Schwyter D. H., Shirokawa J. M., Courey A. J. The interplay between multiple enhancer and silencer elements defines the pattern of decapentaplegic expression. Genes Dev. 1993 Apr;7(4):694–704. doi: 10.1101/gad.7.4.694. [DOI] [PubMed] [Google Scholar]
- Hursh D. A., Padgett R. W., Gelbart W. M. Cross regulation of decapentaplegic and Ultrabithorax transcription in the embryonic visceral mesoderm of Drosophila. Development. 1993 Apr;117(4):1211–1222. doi: 10.1242/dev.117.4.1211. [DOI] [PubMed] [Google Scholar]
- Ip Y. T., Levine M., Bier E. Neurogenic expression of snail is controlled by separable CNS and PNS promoter elements. Development. 1994 Jan;120(1):199–207. doi: 10.1242/dev.120.1.199. [DOI] [PubMed] [Google Scholar]
- Ip Y. T., Park R. E., Kosman D., Bier E., Levine M. The dorsal gradient morphogen regulates stripes of rhomboid expression in the presumptive neuroectoderm of the Drosophila embryo. Genes Dev. 1992 Sep;6(9):1728–1739. doi: 10.1101/gad.6.9.1728. [DOI] [PubMed] [Google Scholar]
- Jackson P. D., Hoffmann F. M. Embryonic expression patterns of the Drosophila decapentaplegic gene: separate regulatory elements control blastoderm expression and lateral ectodermal expression. Dev Dyn. 1994 Jan;199(1):28–44. doi: 10.1002/aja.1001990104. [DOI] [PubMed] [Google Scholar]
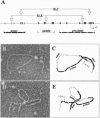
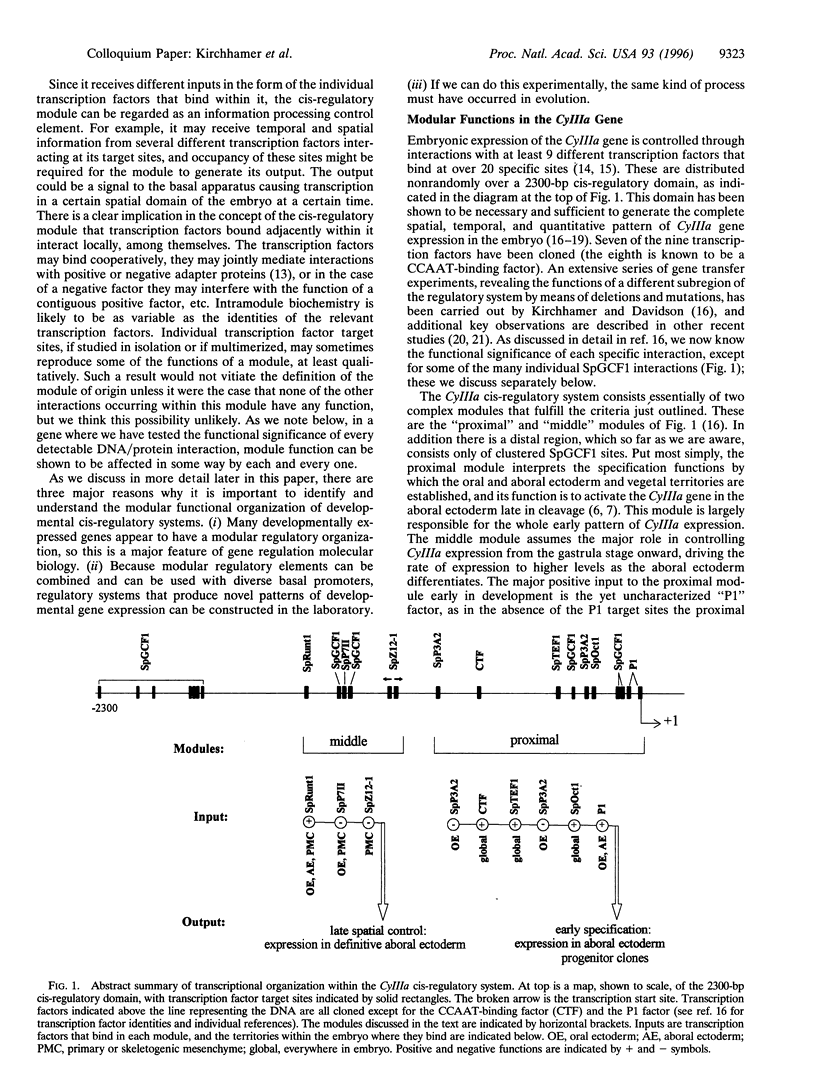
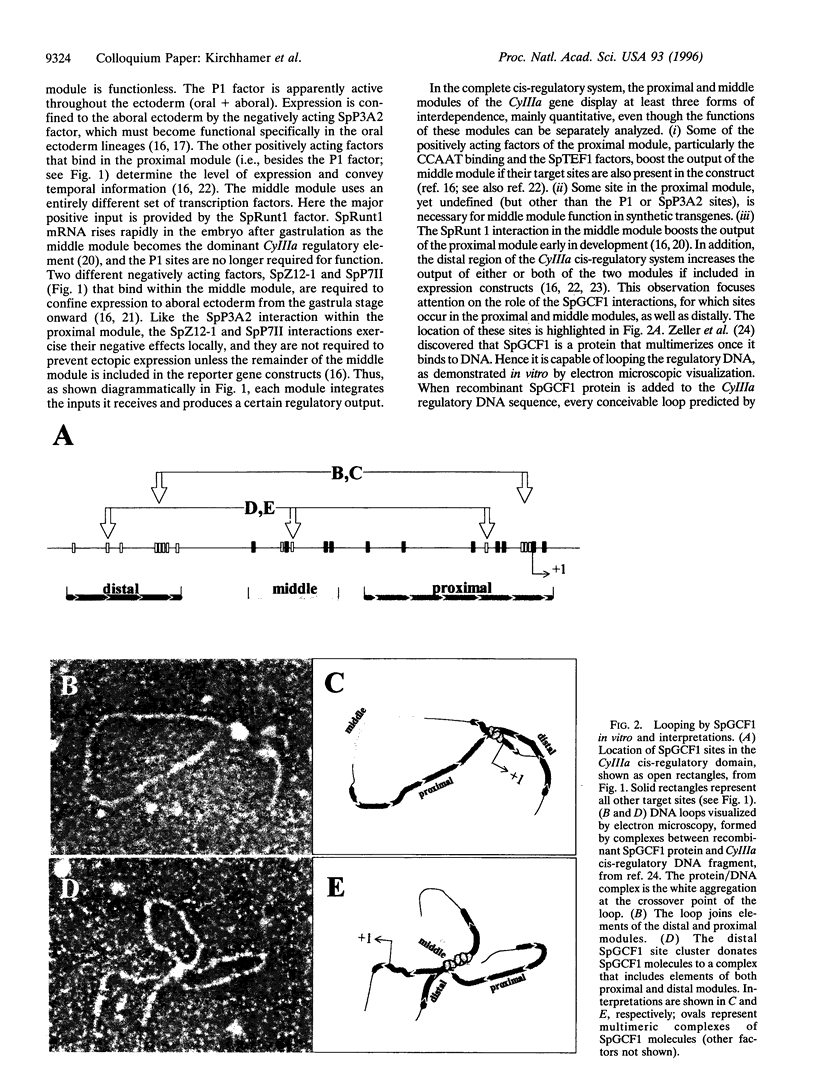
- Kirchhamer C. V., Davidson E. H. Spatial and temporal information processing in the sea urchin embryo: modular and intramodular organization of the CyIIIa gene cis-regulatory system. Development. 1996 Jan;122(1):333–348. doi: 10.1242/dev.122.1.333. [DOI] [PubMed] [Google Scholar]
- Komachi K., Redd M. J., Johnson A. D. The WD repeats of Tup1 interact with the homeo domain protein alpha 2. Genes Dev. 1994 Dec 1;8(23):2857–2867. doi: 10.1101/gad.8.23.2857. [DOI] [PubMed] [Google Scholar]
- Langeland J. A., Carroll S. B. Conservation of regulatory elements controlling hairy pair-rule stripe formation. Development. 1993 Feb;117(2):585–596. doi: 10.1242/dev.117.2.585. [DOI] [PubMed] [Google Scholar]
- Lee J. J., Calzone F. J., Davidson E. H. Modulation of sea urchin actin mRNA prevalence during embryogenesis: nuclear synthesis and decay rate measurements of transcripts from five different genes. Dev Biol. 1992 Feb;149(2):415–431. doi: 10.1016/0012-1606(92)90296-s. [DOI] [PubMed] [Google Scholar]
- Livant D. L., Cutting A. E., Britten R. J., Davidson E. H. An in vivo titration of regulatory factors required for expression of a fusion gene in transgenic sea urchin embryos. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7607–7611. doi: 10.1073/pnas.85.20.7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan S. K., Garabedian M. J., Wensink P. C. DNA regions that regulate the ovarian transcriptional specificity of Drosophila yolk protein genes. Genes Dev. 1989 Sep;3(9):1453–1461. doi: 10.1101/gad.3.9.1453. [DOI] [PubMed] [Google Scholar]
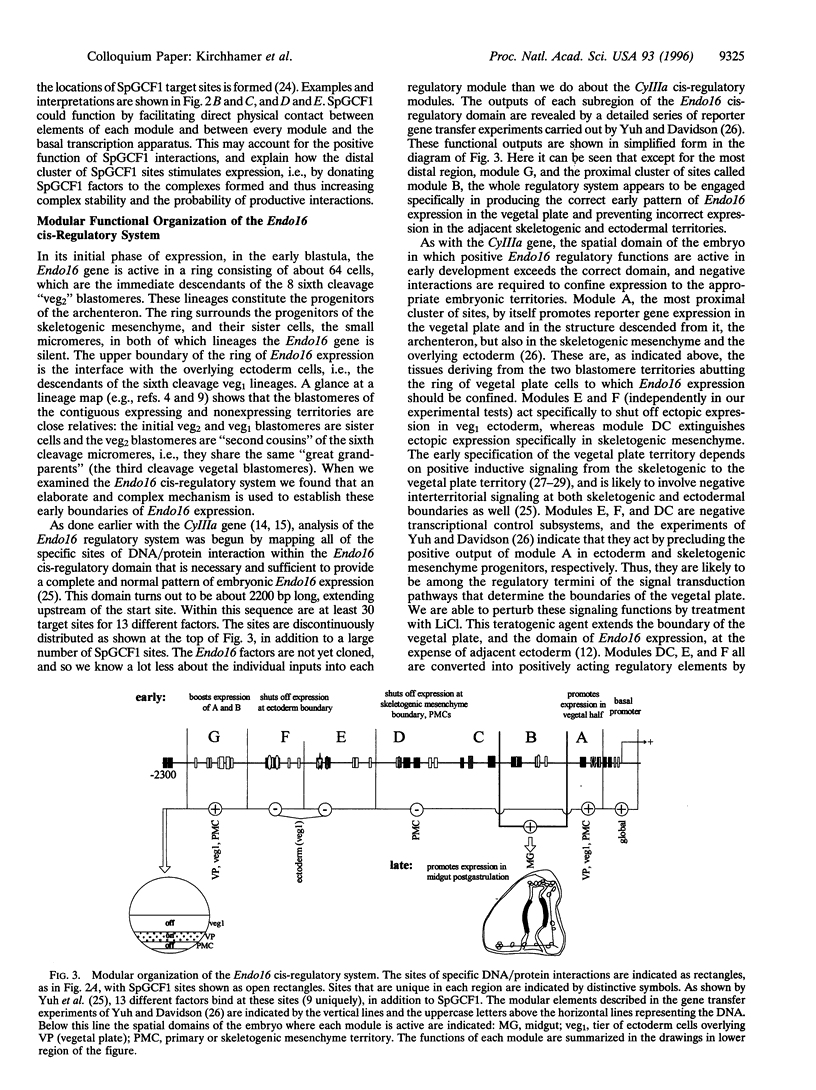
- Makabe K. W., Kirchhamer C. V., Britten R. J., Davidson E. H. Cis-regulatory control of the SM50 gene, an early marker of skeletogenic lineage specification in the sea urchin embryo. Development. 1995 Jul;121(7):1957–1970. doi: 10.1242/dev.121.7.1957. [DOI] [PubMed] [Google Scholar]
- Marshall H., Studer M., Pöpperl H., Aparicio S., Kuroiwa A., Brenner S., Krumlauf R. A conserved retinoic acid response element required for early expression of the homeobox gene Hoxb-1. Nature. 1994 Aug 18;370(6490):567–571. doi: 10.1038/370567a0. [DOI] [PubMed] [Google Scholar]
- Masucci J. D., Hoffmann F. M. Identification of two regions from the Drosophila decapentaplegic gene required for embryonic midgut development and larval viability. Dev Biol. 1993 Sep;159(1):276–287. doi: 10.1006/dbio.1993.1240. [DOI] [PubMed] [Google Scholar]
- Morrison A., Chaudhuri C., Ariza-McNaughton L., Muchamore I., Kuroiwa A., Krumlauf R. Comparative analysis of chicken Hoxb-4 regulation in transgenic mice. Mech Dev. 1995 Sep;53(1):47–59. doi: 10.1016/0925-4773(95)00423-8. [DOI] [PubMed] [Google Scholar]
- Nemer M., Bai G., Stuebing E. W. Highly identical cassettes of gene regulatory elements, genomically repetitive and present in RNA. Proc Natl Acad Sci U S A. 1993 Nov 15;90(22):10851–10855. doi: 10.1073/pnas.90.22.10851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemer M., Stuebing E. W., Bai G., Parker H. R. Spatial regulation of SpMTA metallothionein gene expression in sea urchin embryos by a regulatory cassette in intron 1. Mech Dev. 1995 Apr;50(2-3):131–137. doi: 10.1016/0925-4773(94)00330-p. [DOI] [PubMed] [Google Scholar]
- Nocente-McGrath C., Brenner C. A., Ernst S. G. Endo16, a lineage-specific protein of the sea urchin embryo, is first expressed just prior to gastrulation. Dev Biol. 1989 Nov;136(1):264–272. doi: 10.1016/0012-1606(89)90147-4. [DOI] [PubMed] [Google Scholar]
- Püschel A. W., Balling R., Gruss P. Position-specific activity of the Hox1.1 promoter in transgenic mice. Development. 1990 Mar;108(3):435–442. doi: 10.1242/dev.108.3.435. [DOI] [PubMed] [Google Scholar]
- Qian S., Capovilla M., Pirrotta V. The bx region enhancer, a distant cis-control element of the Drosophila Ubx gene and its regulation by hunchback and other segmentation genes. EMBO J. 1991 Jun;10(6):1415–1425. doi: 10.1002/j.1460-2075.1991.tb07662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransick A., Davidson E. H. A complete second gut induced by transplanted micromeres in the sea urchin embryo. Science. 1993 Feb 19;259(5098):1134–1138. doi: 10.1126/science.8438164. [DOI] [PubMed] [Google Scholar]
- Ransick A., Davidson E. H. Micromeres are required for normal vegetal plate specification in sea urchin embryos. Development. 1995 Oct;121(10):3215–3222. doi: 10.1242/dev.121.10.3215. [DOI] [PubMed] [Google Scholar]
- Ransick A., Ernst S., Britten R. J., Davidson E. H. Whole mount in situ hybridization shows Endo 16 to be a marker for the vegetal plate territory in sea urchin embryos. Mech Dev. 1993 Aug;42(3):117–124. doi: 10.1016/0925-4773(93)90001-e. [DOI] [PubMed] [Google Scholar]
- Riddihough G., Ish-Horowicz D. Individual stripe regulatory elements in the Drosophila hairy promoter respond to maternal, gap, and pair-rule genes. Genes Dev. 1991 May;5(5):840–854. doi: 10.1101/gad.5.5.840. [DOI] [PubMed] [Google Scholar]
- Soltysik-Española M., Klinzing D. C., Pfarr K., Burke R. D., Ernst S. G. Endo16, a large multidomain protein found on the surface and ECM of endodermal cells during sea urchin gastrulation, binds calcium. Dev Biol. 1994 Sep;165(1):73–85. doi: 10.1006/dbio.1994.1235. [DOI] [PubMed] [Google Scholar]
- Studer M., Pöpperl H., Marshall H., Kuroiwa A., Krumlauf R. Role of a conserved retinoic acid response element in rhombomere restriction of Hoxb-1. Science. 1994 Sep 16;265(5179):1728–1732. doi: 10.1126/science.7916164. [DOI] [PubMed] [Google Scholar]
- Thézé N., Calzone F. J., Thiebaud P., Hill R. L., Britten R. J., Davidson E. H. Sequences of the CyIIIa actin gene regulatory domain bound specifically by sea urchin embryo nuclear proteins. Mol Reprod Dev. 1990 Feb;25(2):110–122. doi: 10.1002/mrd.1080250203. [DOI] [PubMed] [Google Scholar]
- Tjian R., Maniatis T. Transcriptional activation: a complex puzzle with few easy pieces. Cell. 1994 Apr 8;77(1):5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- Topol J., Dearolf C. R., Prakash K., Parker C. S. Synthetic oligonucleotides recreate Drosophila fushi tarazu zebra-stripe expression. Genes Dev. 1991 May;5(5):855–867. doi: 10.1101/gad.5.5.855. [DOI] [PubMed] [Google Scholar]
- Vogels R., Charité J., de Graaff W., Deschamps J. Proximal cis-acting elements cooperate to set Hoxb-7 (Hox-2.3) expression boundaries in transgenic mice. Development. 1993 May;118(1):71–82. doi: 10.1242/dev.118.1.71. [DOI] [PubMed] [Google Scholar]
- Wang D. G., Kirchhamer C. V., Britten R. J., Davidson E. H. SpZ12-1, a negative regulator required for spatial control of the territory-specific CyIIIa gene in the sea urchin embryo. Development. 1995 Apr;121(4):1111–1122. doi: 10.1242/dev.121.4.1111. [DOI] [PubMed] [Google Scholar]
- Whiting J., Marshall H., Cook M., Krumlauf R., Rigby P. W., Stott D., Allemann R. K. Multiple spatially specific enhancers are required to reconstruct the pattern of Hox-2.6 gene expression. Genes Dev. 1991 Nov;5(11):2048–2059. doi: 10.1101/gad.5.11.2048. [DOI] [PubMed] [Google Scholar]
- Yuh C. H., Davidson E. H. Modular cis-regulatory organization of Endo16, a gut-specific gene of the sea urchin embryo. Development. 1996 Apr;122(4):1069–1082. doi: 10.1242/dev.122.4.1069. [DOI] [PubMed] [Google Scholar]
- Yuh C. H., Ransick A., Martinez P., Britten R. J., Davidson E. H. Complexity and organization of DNA-protein interactions in the 5'-regulatory region of an endoderm-specific marker gene in the sea urchin embryo. Mech Dev. 1994 Aug;47(2):165–186. doi: 10.1016/0925-4773(94)90088-4. [DOI] [PubMed] [Google Scholar]
- Zeller R. W., Griffith J. D., Moore J. G., Kirchhamer C. V., Britten R. J., Davidson E. H. A multimerizing transcription factor of sea urchin embryos capable of looping DNA. Proc Natl Acad Sci U S A. 1995 Mar 28;92(7):2989–2993. doi: 10.1073/pnas.92.7.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C. C., Müller J., Hoch M., Jäckle H., Bienz M. Target sequences for hunchback in a control region conferring Ultrabithorax expression boundaries. Development. 1991 Dec;113(4):1171–1179. doi: 10.1242/dev.113.4.1171. [DOI] [PubMed] [Google Scholar]