Abstract
At the cellular level, the process of bone metastasis involves many steps. Circulating cancer cells enter the marrow, proliferate, induce neovascularization, and ultimately expand into a clinically detectable, often symptomatic, metastatic deposit. Although the initial establishment and later expansion of the metastatic deposit in bone require tumor cells to possess invasive capability, the exact proteases responsible for this phenotype are not well known. The objective of our study was to take an unbiased approach to determine which proteases were expressed and functional during the initial interactions between prostate cancer cells and bone marrow stromal (BMS) cells. We found that the combination of human prostate cancer PC3 and BMS cells stimulates the invasive ability of cancer cells through type I collagen. The use of inhibitors for each of the major protease families indicated that 1 or more MMPs was/were responsible for the BMS-induced invasion. Gene profiling and semiquantitative RT-PCR analysis revealed an increased expression of several MMP genes because of PC3/BMS cell interaction. However, only MMP-12 showed an increase in protein expression. Downregulation of MMP-12 expression in PC3 cells by siRNA inhibited the enhanced invasion induced by PC3/BMS cell interaction. In vivo, MMP-12 was found to be primarily expressed by prostate cancer cells growing in bone. Our data suggest that BMS cells induce MMP-12 expression in prostate cancer cells, which results in invasive cells capable of degradation of type I collagen.
Keywords: bone metastasis, matrix metalloproteinases (MMPs), protease inhibitors
Prostate cancer is the second leading cause of cancer-related death and the most common cancer in males, resulting in ~27,000 deaths of men per year in the United States.1 Almost all patients with advanced disease have bone metastasis, and most patients experience painful complications from skeletal involvement.2 Although the biology of tumor growth in bone is beginning to be unraveled, the initial steps of tumor–bone interaction that promote the establishment and expansion of the metastatic deposit are still undefined.
Matrix metalloproteinases (MMPs) are endopeptidases that can cleave virtually any compound of the extracellular matrix (ECM). MMPs are produced by both tumor cells and host cells and have been shown to play a role in regulating cancer cell growth, apoptosis, invasion, metastasis and angiogenesis.3 Although ECM remodeling and invasion are thought to occur by virtue of MMPs’ capacity to destroy underlying cellular foundations, it is now known that MMPs can alter signaling pathways triggered by molecules concealed in the ECM, thereby profoundly influencing the local microenvironment.4 Proliferation, survival and invasiveness of prostate cancer cells depend on their interaction with stromal cells.5,6 This interaction affects cellular behavior through direct cell–cell contact or through diffusible factors using existent intra-cellular signaling pathways.7 In any case, in order to establish a viable metastatic deposit in bone, tumor cells must possess the ability to create space for themselves, and this likely requires proteolytic degradation of ECM.
Previously, we showed that particular MMPs play a role in intraosseous tumor growth and bone degradation. Initially, we studied MMP-9 in the context of bone metastasis because of its known role in basement membrane degradation in primary tumors. We showed an increase in MMP-9 expression by tumor cells when bone was present, and an association between MMP-9 activity and tumor growth and osteolysis in an experimental model of prostate cancer bone metastasis.8,9 We also studied MT1-MMP because of its known crucial role in bone remodeling.10 We showed that MT1-MMP, a tumor-derived membrane-bound MMP, was highly expressed by cancer cells in bone metastases of prostate cancer patients.11 We further showed that MT1-MMP expression profoundly affects the phenotype and growth rate of metastatic bone tumors and that the underlying MT1-MMP-dependent mechanisms included both cleavage of ECM constituents and release of bone-derived, pro-osteolytic cytokines.11
In this study, our aim was to take an unbiased approach to determine which proteases are involved in the initial tumor-stromal interaction. We found that bone marrow stromal (BMS) cells stimulate the invasive ability of human prostate cancer PC3 cells through type I collagen, and that MMP-12 is upregulated during the interaction of BMS and PC3 cells. Moreover, downregulation of MMP-12 expression in PC3 cells attenuated the enhanced invasion induced by PC3/BMS cell interaction. These findings are further supported by our observation that MMP-12 is overexpressed during the interaction of PC3 and the bone microenvironment in vivo. Taken together, our data suggest that MMP-12, also known as macrophage metalloelastase, may be a valid therapeutic target during the early phases of colonization of bone by cancer cells.
Material and methods
Cell culture
The human prostate cancer cell line PC3,12 originally obtained from American Type Culture Collection (Manassas, VA), was used between passages 25 and 32. BMS cells were obtained by scraping the bone marrow cavity of human male fetal femurs (22–24 week gestation), obtained from Advanced Bioscience Resources (Alameda, CA). PC3 and BMS cells were maintained in culture (5% CO2, at 37°C) in RPMI-1640 (Invitrogen, Carlsbad, CA) and supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals, Lawrenceville, GA) and 1% penicillin/streptomycin (Sigma Chemical Co., St Louis, MO).
In vitro invasion assay
Chemoinvasion assays were performed using 24-well Transwell™ inserts (8-μm pore size, Becton Dickinson, Franklin Lakes, NJ), which were precoated with 25-μg type I collagen per filter (83.3 μg/cm2, Trevigen, MD). The Transwell inserts were placed in 24-well plates filled with culture medium supplemented with 5% FBS, used as a chemoattractant. A total of 1 ×105 PC3 and/or BMS cells were seeded in the upper chamber in serum-free medium containing 0.1% bovine serum albumin (BSA, Sigma). When cell types were combined, an equal number of cells (5 ×104 PC3 + 5 ×104 BMS) were used. After 24 hr at 37°C, cells on the upper surface of the filter were removed with a cotton swab, whereas those that transmigrated were stained with Diff-Quik® (Dade Behring, Newark, DE) and counted under 20× magnification. Each experiment was performed in triplicate and repeated at least twice. To test the effect of protease inhibitors, the chemoinvasion assay was performed in the absence or presence of non-toxic doses of inhibitors for serine (aprotinin) (10 μg/ml; Sigma), metallo- (GM-6001) (5 μmol/l; Calbiochem), aspartic (pepstatin A) (1 μmol/l; Sigma), or cysteine (E-64) (10 μmol/l; Sigma) proteinases.
Immunocytochemistry
To detect cytokeratin expression in invasive cells, some of the Transwell inserts used in the chemoinvasion assay were processed for immunocytochemistry. Briefly, cells on the upper surface of the filters were removed with a cotton swab, and the filters were washed with phosphate-buffered saline (PBS) and fixed for 5 min in cold absolute methanol. Endogenous peroxidase activity was quenched with 3% hydrogen peroxide in methanol, and nonspecific immunobinding blocked with Superblock (Scytek Laboratories, Logan, UT) for 10 min at room temperature. Filters were then incubated with an anti-Pan cytokeratin antibody (1:50; Sigma) for 2 hr at room temperature. After rinsing with PBS, Vectastain Elite ABC kit for mouse immunoglobulin G (IgG; Vector Laboratories, Burlingame, CA) was used. Immunoreactive cytokeratins were recognized from the brown color after incubation with diaminobenzidine (Sigma)/H2O2 solution. Slides were lightly counterstained with Mayer hematoxylin before mounting. Cells were counted under 20× magnification, and the percentage of cytokeratin-positive cells was calculated.
WST-1 proliferation assay
PC3 cells were seeded onto 96-well plates at a cell density of 1 × 104 cells/well in serum-free RPMI-1640 containing 0.1% BSA, in the presence or absence of GM-6001 (5 μmol/l). Twenty four hours later, 10 μl of WST-1 reagent (Roche, Indianapolis, IN) were added, followed by incubation for 4 hr at 37°C and absorbance reading at 450 nm. The WST-1 assay was carried out in quadruplicate. The results are shown as mean ± SE.
Analysis of cytokeratin by flow cytometry
BMS and PC3 cells (3 × 105 cells of each cell type) were seeded in 6-well plates coated with type I collagen (83.3 μg/cm2) in serum-free RPMI-1640 containing 0.1% BSA. Twenty four hours later, cells were washed with PBS-BSA, and fixed and permeabilized for 30 min at 4°C by slow addition of cold 100% ethanol (50% final concentration). Cells were then incubated for 30 min with 20 μl of fluorescein isothiocyanate (FITC)-conjugated monoclonal antibody against cytokeratin (CAM 5.2, BD Biosciences, San Jose, CA). As a negative control, cells were incubated with FITC-conjugated isotype nonimmune mouse IgG (BD Biosciences). Cells were then washed with PBS-BSA and analyzed on a FACScan (Becton Dickinson, Mountain View, CA).
Expression analysis using Affymetrix probe arrays
A total of 1 × 106 of BMS and/or PC3 cells (5 × 105 of each cell type when combined) were seeded in 60-mm dishes coated with type I collagen (83.3 μg/cm2), in RPMI medium supplemented with 10% FBS and 1% penicillin/streptomycin. Six hours later, the medium was removed and replaced with serum-free medium for 24 hr. Total RNA was isolated from cells using Trizol® reagent (Invitrogen), and purified using Qiagen RNeasy kit (Qiagen, Valencia, CA), according to the manufacturer’s instructions. RNA was tested for quality to ensure RNA integrity by measuring the 28S:18S ribosomal RNA ratio. Biotin-labeled cRNA transcribed in vitro from cDNA synthesized from each sample was purified and then fragmented and applied to Human Genome U133A Array (Affymetrix, Santa Clara, CA). Following hybridization and staining, arrays were scanned and the gene expression levels normalized and analyzed with Microarray Suite, MicroDB and Data Mining Tool software (Affymetrix) in the Applied Genomics Technology core (Center for Molecular Medicine and Genetics, Wayne State University).
Validation of the microarray analysis by semiquantitative RT-PCR
For RT-PCR, 2 μg of total RNA used for Affymetrix probe arrays, or RNA isolated from human fetal bones injected with PC3 cells (see description below), human fetal bone xenografts or subcutaneous (s.c.) tumors using Trizol reagent, were used to synthesize a first-strand cDNA with an oligo (dT) primer and Super Script II reverse transcriptase (Invitrogen). Primers were based either on published data (MMP-2, MMP-9, GAPDH and MMP-13) or designed using Invitrogen’s OligoPerfect Designer program (MMP-1, MMP-3, MMP-10, MMP-12 and MMP-14, sequences listed in Table I). The real-time PCR analysis was performed with SYBER green PCR core reagents (Stratagene, La Jolla, CA) in a Stratagene Mx4000 cycler, and the data were analyzed using Mx4000 v3.01 software. Relative message levels were calculated with a comparative Ct (threshold cycle method) method,13 by which message levels were normalized to endogenous glyceraldehyde phosphate dehydrogenase (GAPDH) message levels.
TABLE I.
SEQUENCES OF PRIMERS USED IN SEMIQUANTITATIVE REAL TIME-PCR ASSAYS
| Gene | Forward primer | Reverse primer |
|---|---|---|
| MMP-1 | CCTTCTACCCGGAAGTTGAG | TCCGTGTAGCACATTCTGTC |
| MMP-2 | GTCCTGAAGGACACACTAAAG | TTGCCATCCTTCTCAAAGTTGT |
| MMP-3 | GCAGTTTGCTCAGCCTATCC | GAGTGTCGGAGTCCAGCTTC |
| MMP-9 | TGGGCTACGTGACCTATGACAT | GCCCAGCCCACCTCCACTCCTC |
| MMP-10 | CCAGTCTGCTCTGCCTATCC | ACCAACGTCAGGAACTCCAC |
| MMP-12 | ACACATTTCGCCTCTCTGCT | AAGCAGCTTCAATGCCAGAT |
| MMP-13 | GTGGTGTGGGAAGTATCATCA | GCATCTGGAGTAACCGTATTG |
| MMP-14 | CGCTACGCCATCCAGGGTCTC | CGGTCATCATCGGGCAGCACA |
| GAPDH | AAGGTCATCCCTGAGCTGAA | TGACAAAGTGGTCGTTGAGG |
Gelatin zymography
Analysis of gelatinases (MMP-2 and -9) was performed as previously described.8 Briefly, BMS and/or PC3 cells totaling 6 × 105 cells in all cases, and in equal numbers (3 × 105 cells) when combined, were seeded in 6-well plates coated with type I collagen (83.3 μg/cm2) in RPMI media supplemented with 10% FBS and 1% penicillin/streptomycin. Six hours later, the medium was removed and replaced with serum-free medium for 24 hr. Conditioned medium (CM) was used to compare gelatinase production by each cell type, as well as to compare the effect of PC3-BMS cell interaction. Protein concentrations in the CM obtained were determined using the BCA method (Pierce Chemical Company, Rockford, IL). CM derived from PC3/BMS cocultures (total protein concentration 20 μg) were run in parallel with CM obtained separately from each of the cell lines and then combined in equal parts (total protein concentration for each cell line 10 μg, totaling 20 μg). CM were mixed with 4× sample buffer (0.25 mol/l Tris-HCl, pH 6.8, 0.4% sodium dodecyl sulfate, 40% glycerol and bromophenol blue), loaded onto a 10% sodium dodecyl sulfate–polyacrylamide gel containing 1 mg/ml gelatin (Sigma), and run under nonreducing conditions at constant voltage of 125 V. After electrophoresis, the gel was incubated in renaturing buffer (2.5% Triton X-100) at room temperature for 30 min, washed twice with distilled water (10 min each time), and then incubated with the developing buffer (50 mmol/l Tris, pH 8.0, 5 mmol/l CaCl2, 200 mmol/l NaCl and 0.02% Brij-35) at 37°C overnight, stained in 0.5% Coomassie Blue solution for ~2 hr, and then incubated in the destaining buffer (5% acetic acid, 10% methanol in distilled water) until bands of gelatinolytic activity were visualized.
Western blot analysis
Cell lysates (total protein 25 μg) obtained from PC3 and BMS cells cultured separately were used to compare MMP expression in each cell type. Cell lysates obtained from PC3/BMS cocultures (total protein concentration 50 μg) were run in parallel with cell lysates obtained separately from each of the cell lines and then combined in equal parts (total protein concentration for each cell line 25 μg, totaling 50 μg). Cell lysates were resolved under reducing conditions by SDS-polyacrylamide gels and transferred to Hybond C-extra membranes (Amersham Life Science, Arlington Heights, IL), which were then blocked with 5% fat-free milk and immunostained. The antibodies used were mouse anti-human MMP-1 (1 μg/ml), mouse anti-human MMP-3 (1:200), mouse anti-human MMP-13 (1:200), mouse anti-MMP-10 (1:200), rabbit anti-human MMP-12 (1:5,000; Calbiochem, San Diego, CA) and mouse anti-human MMP-14 (LEM-2/15; a generous gift from Dr. A. Arroyo, Hospital de la Princesa, Madrid, Spain). After washing, the membranes were incubated with anti-mouse or anti-rabbit IgG conjugated with horseradish peroxidase (1:2,000 dilution; Pierce). GAPDH (Trevigen, Gaithersburg, MD) was used as loading control. The bands were quantitated by densitometry (UnScan-It, Silk Scientific, Orem, UT).
Small interfering RNA transfection assay
A predesigned small interfering RNA (siRNA; Ambion, Austin, TX) targeting human MMP-12 was used. The sequence of the region targeted by siRNA was 5′-GGACGAATTCTGGACTACA-3′. PC3 cells were transfected with 10–50 nM of MMP-12 siRNA or scrambled (scb) siRNA (Ambion) using Lipofectamine™ 2000 reagent (Invitrogen). Forty eight hours later, Western blot and RT-PCR analysis were performed, and the transfected cells were assessed for invasion.
In vivo models
SCID-human and s.c. xenograft models were used. PC3 cells (2 × 105) were injected into the bone marrow of human male fetal bone fragments (Advanced Bioscience Resources) previously implanted under the skin in 8-week-old male homozygous C.B-17 scid/scid mice (Taconic Farms, Germantown, NY), as described before.14 Previously implanted bone xenografts injected with FBS-free culture medium were used to control for the effects of injection, and tumors obtained by s.c. inoculation of PC3 cells were used as microenvironmental controls. Intraosseous and s.c. PC3 tumors, as well as control bone xenografts, were harvested 14 days after inoculation. Mice were maintained under aseptic conditions, and all procedures were done in compliance with the Animal Investigation Committee of Wayne State University and NIH guidelines.
Immunohistochemistry
Tumors and control bone xenografts were fixed in 10% buffered formalin, decalcified, paraffin-embedded and sectioned. Tissue sections were deparaffinized and rehydrated through graded alcohols. MMP-12 and cytokeratin immunostaining was performed using mouse anti-human MMP-12 antibody (clone 82902, R&D systems, Minneapolis, MN) and mouse anti-pan cytokeratin (Sigma) at a concentration of 8.3 μg/ml and a dilution of 1:50, respectively. The antigens were visualized using the Mouse on Mouse (MOMTM) immunodetection peroxidase kit (Vector Laboratories). The reaction product was developed using diaminobenzidine tetrahydrochloride, and sections were lightly counterstained with Mayer’s hematoxylin. Negative controls consisted of sections where the primary antibody was replaced by isotype-specific non-immune mouse IgG. Digital micrographs were processed using AxioVision (Carl Zeiss AG, Germany) software.
Tartrate-resistant acid phosphatase staining
Histological sections adjacent to those used for immunohistochemistry were stained for tartrate-resistant acid phosphatase (TRAP), an osteoclast-associated enzyme,15 using a commercial leukocyte acid phosphatase kit (Sigma), following the manufacturer’s directions.
Statistical analysis
Data were presented as mean ± SE, and statistically analyzed using Student’s t-test (2 groups) or 1-way analysis of variance (3 or more groups). Whenever statistical significance was obtained with this test, the Tukey–Kramer posttest was used to further determine which specific pair/pairs were statistically different. Values of p < 0.05 were considered statistically different.
Results
The interaction of PC3 and BMS increases the invasive capacity of PC3 cells
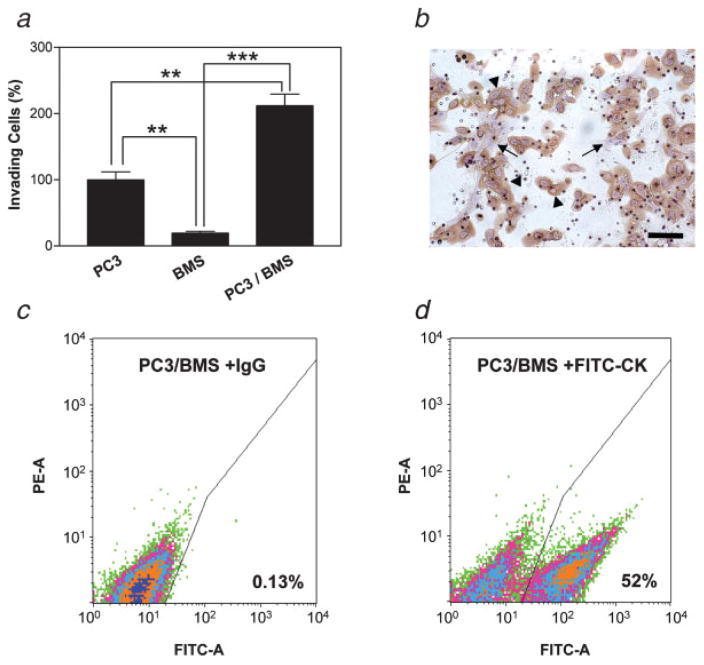
PC3 and BMS cells cultured alone or together were assayed for in vitro invasiveness capacity through collagen I-coated porous filters. The cells that invaded through the filters were stained and counted after 24 hr. As shown in Figure 1a, when cultured alone, a far higher number of PC3 cells traversed the filter than BMS cells. When cultured together, a synergistic and statistically significant increase in the invasiveness was observed (Fig. 1a). Nearly all of the invasive cells were cytokeratin-positive and, therefore, of PC3 nature (Fig. 1b). The differences in the number of cells traversing the filter during coculture is even more significant, given that only half as many PC3 cells were seeded in the coculture as in the single cell culture (see Methods). The enhanced invasiveness induced by PC3/BMS interaction was associated with increased degradation of type I collagen, which covered the pores traversed by the cells. This is supported by an enhanced type I collagenolytic activity found when PC3 and BMS cells were cultured together in the presence of fluorescence-quenched type I collagen with respect to individual cultures of each of the cell types (data not shown).
Figure 1.
Synergistic effect of PC3/BMS cell interaction on chemoinvasion through type I collagen. PC3 and BMS cells were assayed alone (1 × 105 cells each) or in combination (5 × 104 cells each) on top of type I collagen-coated filters in transwells. The number of PC3 cells that traversed the filters toward the chemoattractant when assayed alone was normalized to 100%. Data are expressed as mean ± SE percentage of this control. Each experiment was performed in triplicate. **p < 0.01, ***p < 0.001 (Tukey–Kramer post hoc applied to significant effect of group ANOVA) (a). Immunocytochemistry for cytokeratin was performed to identify epithelial tumor cells (arrowheads) and BMS cells (arrows). Bars, 100 μm (b). Flow cytometry analysis of cytokeratin expression in PC3/BMS cells. Histograms obtained with cells incubated with isotypic IgG (c) and fluorescein-conjugated anti-cytokeratin antibody (d) are depicted. Cytokeratin-positive cells are expressed as percentages. PE-A, phycoerythrin-area; FITC-A, fluorescein isothiocyanate-area (c). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
To exclude the possibility that this enhanced invasiveness induced by PC3/BMS interaction is due to a stimulatory effect of BMS cells on PC3 cell proliferation, we quantitated the number of cells 24 hr after seeding them together or alone onto type I collagen-coated wells, as performed in the invasion assay. We found that the total number of cells was similar in all the cases, with no significant increase 24 hr after seeding. Furthermore, flow cytometry analysis using a monoclonal anti-FITC cytokeratin antibody confirmed that 52% of the cells obtained from the 24-hr coculture were of epithelial origin (Fig. 1c). Taken together, these results demonstrate that PC3/BMS cell interaction does not stimulate cell proliferation during the duration of the invasion assay, excluding the possibility that the enhanced invasiveness shown by PC3/BMS is due to an increase in the cell number of either one or both cell types.
Metalloproteinase inhibitors reduce the enhanced invasion induced by PC3/BMS cell interaction
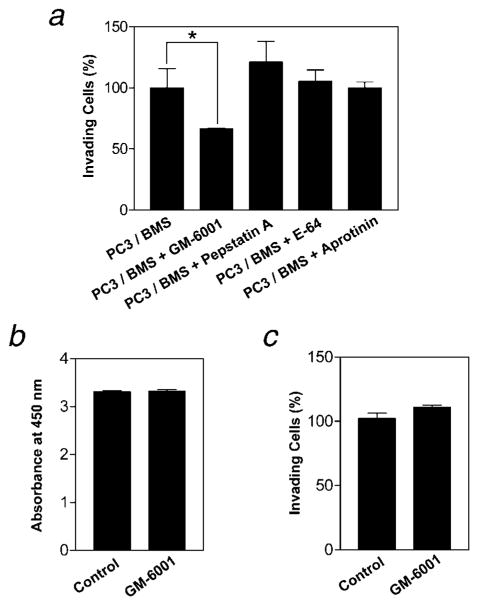
To determine which class of proteases was responsible for the enhanced invasiveness induced by PC3/BMS interaction, several pharmacologic protease inhibitors were used in the in vitro invasion assay. Briefly, cells were seeded in Transwell inserts coated with type I collagen in the presence of nontoxic doses of inhibitors with broad spectrum specificity for serine (aprotinin; 10 μg/ml), aspartic (pepstatin A; 1 μmol/l), metallo-(GM-6001; 5 μmol/l) and cysteine (E-64; 10 μmol/l) proteases, and assayed for invasiveness as described earlier. As shown in Figure 2a, only the treatment with GM-6001, a broad spectrum hydroxamate inhibitor of metalloproteinases (MPs),16,17 reduced the invasion enhanced by PC3/BMS interaction, suggesting that the enzyme responsible for the stimulatory effect belongs to the MP family. Studies with GM-6001 confirmed that the results observed in the invasion assays are not due to an inhibitory effect of the MP inhibitor on PC3 cell proliferation or viability (Fig. 2b). Moreover, GM-6001 did not affect the in vitro invasion of PC3 cells cultured alone at the concentration used (Fig. 2c).
Figure 2.
Metalloproteinases are responsible for the enhanced chemoinvasion due to PC3/BMS cell interaction. PC3 and BMS cells (5 × 104 cells each) were seeded in the upper compartment of trans-wells coated with collagen I (25 μg/filter) in the absence or in the presence of nontoxic doses of inhibitors for serine (aprotinin), matrix metallo- (GM-6001), aspartic (pepstatin A) or cysteine (E-64) proteinases. The number of cells invading was quantified per filter after 24 hr. Results are expressed as mean ± SE percentage of the control (PC3/BMS). *p < 0.05 (Tukey–Kramer post hoc applied to significant effect of group ANOVA) (a). PC3 cell proliferation (b) and invasion (c) in the presence or absence of GM-6001 are shown. Results are expressed as mean ± SE of absorbances and percentage of the control PC3, respectively.
Coculture of PC3 and BMS cells mainly stimulates MMP-12 mRNA and protein expression
To identify which MMPs were stimulated by the coculture interaction of prostate cancer and BMS cells, we performed an Affymetrix Gene chip array on PC3 and BMS cells cultured alone or together for 24 hr on top of type I collagen. The increased gene expression changes were determined by comparing the data array obtained in the PC3/BMS cell combination with those obtained in each cell line when cultured alone, using a 2-fold change as the criterion of analysis. Under this condition, we identified 8 MMP genes to be upregulated: MMP-1, MMP-2, MMP-3, MMP-9, MMP-10, MMP-12, MMP-13 and MMP-14 (Table II). The most pronounced increase was observed for MMP-3 and MMP-12 genes.
TABLE II.
REGULATION OF MMP GENES DUE TO COCULTURE OF PC3 AND BMS (PC3/BMS) CELLS ON TYPE I COLLAGEN
| Gene | Fold increase in mRNA
|
|
|---|---|---|
| PC3/BMS vs. PC3 | PC3/BMS vs. BMS | |
| A. Microarray analysis | ||
| MMP-1 | 2.5 | 2.1 |
| MMP-2 | 445.7 | −2.3 |
| MMP-3 | 119.4 | 36.8 |
| MMP-9 | NC | 2.3 |
| MMP-10 | 2 | 6.5 |
| MMP-12 | 315.2 | 36.8 |
| MMP-13 | 6.1 | 2.6 |
| MMP-14 | 3.2 | 2.1 |
| B. RT-PCR analysis | ||
| MMP-1 | 4.2 | 3.2 |
| MMP-2 | 26467 | 0.2 |
| MMP-3 | 246.9 | 87.3 |
| MMP-9 | 1.1 | 2.7 |
| MMP-10 | 2.3 | 14.2 |
| MMP-12 | 1000 | 269.7 |
| MMP-13 | 9.7 | 4.1 |
| MMP-14 | 2.0 | 0.5 |
Fold increased in mRNA of PC3/BMS relative to PC3 or BMS cells cultured alone, as determined using gene microarray (A) and RT-PCR (B) analyses. NC, no change.
Semiquantitative RT-PCR was used to validate the increase in MMP genes found during gene microarray analysis. As shown in Table II, the results obtained with RT-PCR were mostly in agreement with the microarray data. However, the fold-change obtained using RT-PCR was higher than that obtained using cDNA micro-array analysis, especially for MMP-12.
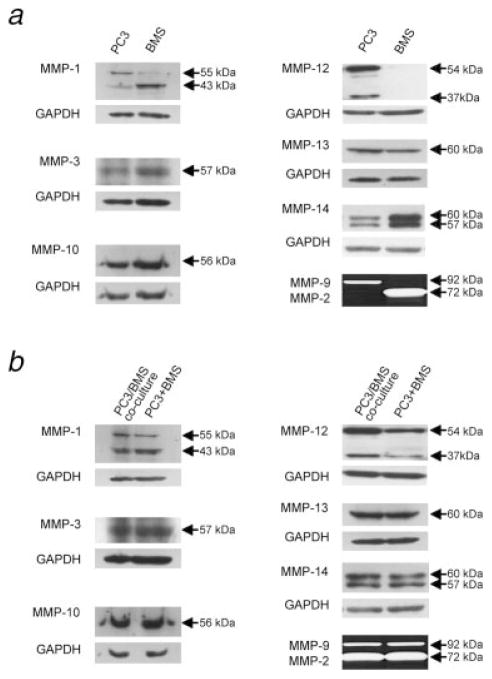
To verify whether the differential expression of MMPs found at the RNA level was also evident at the protein level, Western blot and zymographic analyses were performed. We first used these methods to determine the expression of the MMP under analysis in PC3 and BMS cells independently (Fig. 3a). We found that the 55-kDa latent form of MMP-1 was mostly present in PC3 cells, whereas its 43 kDa species was predominantly expressed by BMS cells. As for MMP-14, both latent (60 kDa) and active (57 kDa) species were found in both cell types, though more strongly expressed by BMS cells. Regarding the gelatinases MMP-2 and -9, zymograms of CM revealed the presence of MMP-9 zymogen only in PC3 cells, while that of MMP-2 proform in BMS cells. MMP-12 was expressed primarily by PC3 cells. All the other MMPs analyzed showed similar expression levels in both PC3 and BMS cells.
Figure 3.
PC3/BMS cell interaction induces MMP-12 expression. PC3 and BMS cells were cultured alone (a) for 24 hr on top of type I collagen-coated dishes, and the expression of various MMPs was examined by immunoblot. The amount of protein in each cell lysate was 25 μg. In another experiment, lysates (50 μg total protein) obtained from PC3 and BMS cells cultured together (PC3/BMS coculture), or separately (PC3+BMS) and combined in equal parts (25 μg total protein each), were compared (b). GADPH loading controls are shown in the lower panel of each Western blot.
To determine the effect of coculture on MMP expression, we prepared 50 μg of total protein from the coculture lysate to be compared with a mixture of 25 μg of protein from the PC3 lysate added to 25 μg of protein from the BMS lysate. To ensure that the proportion of each cell type in the coculture was still the same as the originally seeded 24 hr earlier, cell lysates were subjected to Western blot and then probed with cytokeratin and GAPDH antibodies. While BMS cells were negative for cytokeratin, similar protein levels of cytokeratin and GAPDH were observed in the lysates obtained from coculture of both cell types and from combining equal parts derived from cultures of each cell type alone (data not shown). Therefore, the coculture did not significantly alter the proportion of cells of each type during the course of the experiment. No significant differences in MMP-1, MMP-2, MMP-3, MMP-9, MMP-10, MMP-13 or MMP-14 expression at the protein level were observed when lysates of PC3 and BMS cocultured cells were compared to lysates combined from each cell line cultured alone. On the other hand, a 2-fold increase in both 37 and 54 kDa MMP-12 proteins was shown by densitometry when PC3 and BMS cells were cocultured (Fig. 3b).
MMP12 attenuation in PC3 cells inhibits the enhanced invasion induced by its interaction with BMS cells
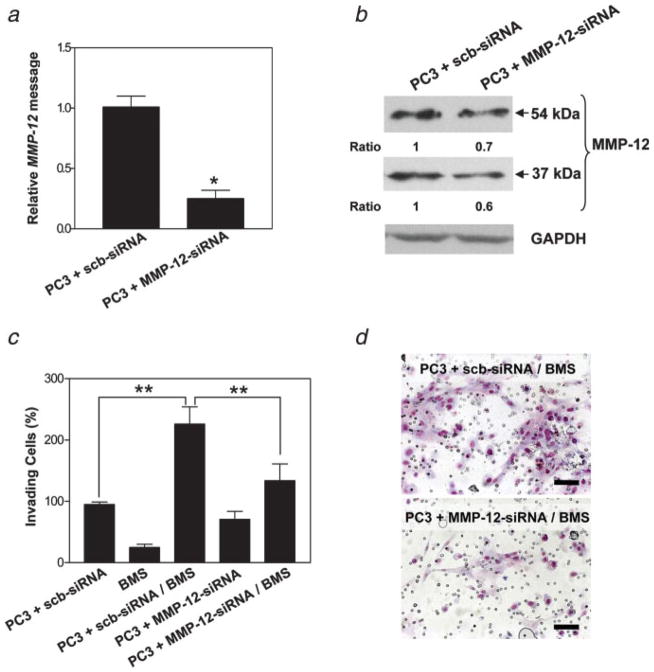
To confirm a functional role for MMP-12 in the enhanced invasive ability induced in PC3/BMS cocultures, we used siRNA directed against human MMP-12. Since this MMP was expressed at the protein level primarily by PC3 cells, we used siRNA to downregulate MMP-12 gene expression in these cells. A dose–response inhibitory effect on MMP-12 expression at the RNA and protein levels could be achieved 48 hr after transient transfection of PC3 cells with different concentrations of siRNA molecules targeting MMP-12 (data not shown). PC3 cells transfected with 50 nM siRNA reduced MMP-12 mRNA and protein expression by ~75% (Fig. 4a) and 30–40% (Fig. 4b), respectively.
Figure 4.
Downregulation of MMP-12 abrogates chemoinvasion induced by PC3/BMS interaction. MMP-12 mRNA in scrambled- and MMP-12-siRNA-transfected PC3 cells, as shown by RT-PCR (a). Western blot bands detected by the antibody against MMP-12 were quantitated by densitometry. Values obtained were normalized using the housekeeping protein GAPDH, and then the ratio between the results for PC3 cells treated with MMP-12-siRNA and scrambled-siRNA (control) was calculated (b). Invasiveness of PC3 cells transiently transfected with either scrambled- or MMP-12-siRNA cultured alone or together with BMS cells (c). Representative micrographs showing invasive capacity of PC3 cells treated with scrambled- or MMP-12-siRNA and combined with BMS cells (d). Bars, 100 μm. Each experiment was performed in triplicate, and results are expressed as mean ±SE percentage of the control (PC3 + scb-siRNA). *p < 0.003 (Student’s t-test) and **p < 0.001 (Tukey–Kramer post hoc applied to significant effect of group ANOVA). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Invasion assays were repeated as mentioned earlier, but using PC3 cells transfected with MMP-12- or scb-siRNA, alone or in combination with BMS cells. As shown in Figures 4c and 4d, downregulation of MMP-12 nearly completely abrogated the enhanced invasion induced by the interaction between scb-siRNA-transfected PC3 and BMS cells. No significant difference in invasion was observed when PC3 cells transfected with either MMP-12- or scb-siRNA were assayed alone (Fig. 4c).
Intraosseous PC3 tumors express MMP-12
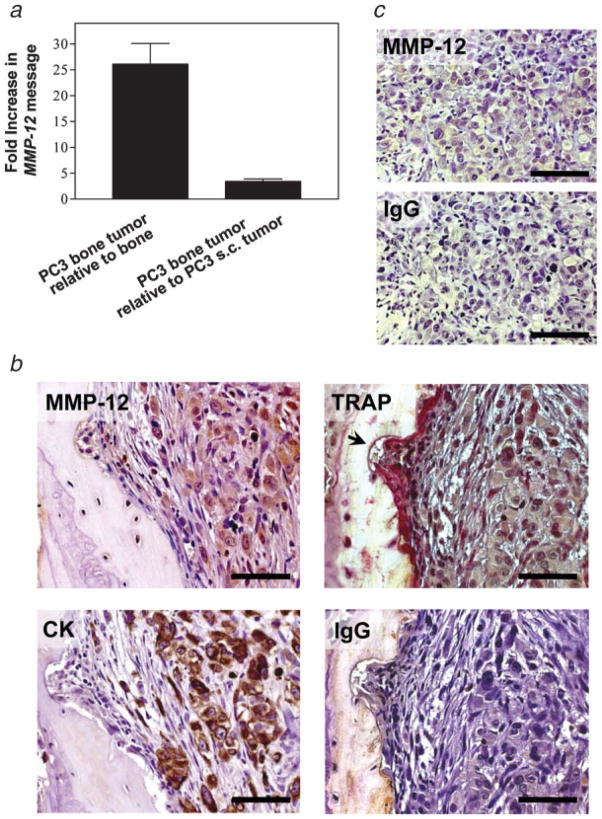
To establish the relevance of our in vitro findings, MMP-12 expression was studied in an experimental model of prostate cancer bone tumor growth. PC3 cells were injected into previously implanted human fetal bone fragments in SCID mice. The subsequent PC3 bone tumors were compared with control human fetal bone xenografts injected with cell culture medium, and with PC3 s.c. tumors. Semiquantitative RT-PCR analysis of cell lysates obtained from the harvested tissues revealed that MMP-12 mRNA level was ~26- and 3-fold higher than in bone xenografts and s.c. PC3 tumors, respectively (Fig. 5a). Immunohistochemi-cal analyses revealed a uniform MMP-12 expression in PC3 bone tumors (Fig. 5b), which was less intense in s.c. PC3 tumors (Fig. 5c). Immunostaining of adjacent sections with an antibody against cytokeratin, which is an epithelial marker, demonstrated that MMP-12 was expressed predominantly by PC3 cells in bone (Fig. 5b). Moreover, staining for TRAP, an osteoclast-associated enzyme, revealed weak MMP-12 expression in TRAP-positive cells associated to resorption bone areas (Fig. 5b). As expected, no MMP-12 expression was evident by immunohistochemistry in the bone xenograft used as a control (data not shown). These results support our in vitro data that showed that MMP-12 protein is primarily expressed by tumor cells, and that this expression is increased when PC3 cells interact with bone microenvironment.
Figure 5.
PC3 cells growing within bone express high levels of MMP-12. RNA message for MMP-12 (RT-PCR) in PC3 bone tumors shown as relative fold increase with respect to control implanted bones and PC3 subcutaneous tumors (a). Immunohistochemical analysis for MMP-12 performed in PC3 bone (b) and subcutaneous (c) tumors. Tartrate-resistant acid phosphatase (TRAP) staining and cytokeratin (CK) immunohistochemistry were also carried out in PC3 bone tumors in adjacent sections. A resorption pit area associated with TRAP-positive cells is shown (arrow). Nonimmune isotype-matched immunoglobulin G (IgG) was used instead of primary antibody as a negative control for immunohistochemistry. Bars, 100 μm.
Discussion
A better understanding of the molecular and cellular interactions occurring between prostate cancer cells and the bone microenvironment is necessary for the development of tailored molecular therapies. The arrival of cancer cells in the marrow disturbs the bone’s normal homeostasis, altering the crosstalk between different bone cells and generating new interactions between the bone microenvironment and cancer cells. A key to understanding how the bone microenvironment and the tumor influence each other is to realize that neither tumor cells nor bone cells have rigid phenotypes with rigid expression patterns; rather, the interaction of bone and cancer cells can influence the pattern of gene expression by each cell type.
We have been interested in the role of proteases in prostate cancer bone metastasis. Proteases are thought to play a crucial role during bone metastasis, either by releasing and/or activating factors stored in the ECM or by degrading organic components of the bone. MMPs, a family of metalloendopeptidases composed of at least 26 members, have been particularly demonstrated to be involved in intraosseous prostate cancer growth and osteolysis. Previously, we found that MMP-9 becomes activated at early stages of bone colonization by prostate cancer cells,8 and that host MMP-9 plays a role in intraosseous tumor growth, osteolysis and angiogenesis.9,18 We also studied MT1-MMP, a membrane-bound protease, which is known to be involved in bone and cartilage metabolism.10 We showed that MT1-MMP is highly expressed in cancer cells in human prostate cancer bone metastasis, and its expression is linked to osteoclastogenesis and intraosseous tumor growth.11 These findings are in accordance with results obtained by others with different in vitro and in vivo models used to study prostate cancer/bone interaction and prostate cancer bone metastasis.19–24 Despite these encouraging preclinical data, there was a general lack of success in human MMP inhibitor clinical trials. There are perhaps several reasons for this, including enrolling patients with high tumor burdens (versus those with primarily skeletal metastasis), the MMP inhibitors were toxic likely due to their broad spectrum of activity, and MMP expression and activity were not evaluated during the course of the trials to determine whether appropriate targets were chosen and then successfully inhibited. If specific protease targets can be delineated and trials carried out more carefully, MMP inhibitors may yet prove to have a role in cancer therapy.
For prostate cancer growth to occur within the bone, many different signals affecting distinct processes like angiogenesis, invasion, ECM degradation, release and activation of growth factors, osteoblast and osteoclast activities, and tumor proliferation and cell survival are necessary. It is not surprising then that different MMPs could be involved in such a plethora of processes. In addition, the activation of many MMPs depends on a complicated proteolytic network. Herein, our focus was to begin to understand how tumor/bone interactions influence that pattern of MMP expression. We therefore started by assessing cancer and BMS cells interaction during an in vitro invasion assay on type I collagen, the most prevalent organic component of the bone ECM. We found that BMS cells increased the invasive capacity of PC3 cells through type I collagen, and that this enhancement is MMP-dependent. Gene and protein expression studies then revealed that MMP-12 was more upregulated than any other MMP. MMP-12 turned out to be mainly derived from PC3 cells, suggesting that the interaction of BMS and PC3 cells led to the increase in MMP-12 expression in the cancer cells. Our finding revealing MMP-12 expression by PC3 cells at the RNA level differs from that reported previously by others.25,26 This discrepancy is probably due to differences in the primers used, as verified in one of the cases in which their sequences were described.26 Furthermore, we confirmed the expression of MMP-12 by PC3 cells at the protein level by Western blot analysis, which strengthens our results.
MMP-12, also known as macrophage metalloelastase,27 degrades a broad spectrum of ECM substrates, such as type IV collagen, fibronectin, laminin, gelatin and vitronectin.28,29 Macrophages from MMP-12 knockout mice have shown a greatly diminished capacity to degrade ECM components.30 Also, MMP-12 is expressed in cells other than macrophages, such as chondrocytes31 and osteoclasts.32 Moreover, this MMP has been found in different cancers, though its role is still controversial. For example, MMP-12 expression has been associated with dedifferentiation and histological aggressiveness in skin cancer,33 with increased invasiveness in squamous cell carcinomas of the vulva and esophagus,34,35 and with metastatic disease in nonsmall cell lung cancer patients.36 On the contrary, over-expression of MMP-12 has been correlated with better prognosis in patients with colorectal and hepatocellular carcinomas,37,38 and with diminished growth of lung metastases.39 These conflicting results might be due to different mechanisms of MMP-12 activity, some that favor tumor progression and others that impede it. For example, MMP-12 has been found to generate angiostatin, a well-recognized endogenous inhibitor of angiogenesis.40,41
MMP-12 siRNA silencing in PC3 cells confirmed a functional role of MMP-12 in BMS-induced invasion through type I collagen. Our findings are in agreement with another study performed with glioma cells, in which MMP-12 was shown to regulate invasiveness through type I collagen.42 However, it is not clear whether MMP-12 contributes to degradation of type I collagen through direct or indirect mechanisms. It is possible that MMP-12 functions by activating type I collagenases. The significance of the in vitro findings was further substantiated by the in vivo results, showing for the first time that prostate tumors express MMP-12 and the expression seems to be enhanced by the bone microenvironment. We are now pursuing studies using PC cells genetically silenced for MMP-12 to define more precisely the role of tumor-derived MMP-12 in intraosseous growth of prostate cancer.
On the basis of our functional and observational studies, performed in vitro and in vivo, we assume that MMP-12 plays an important role in the degradation of the ECM. As suggested by Kerkela et al., macrophage-associated MMP-12 might function during the initial phases of tumor growth, while cancer-associated MMP-12 might promote ECM degradation and tumor growth in the later stages of tumor progression.34 In our experimental models, “later stages” equate to expansion of intraosseous prostate cancer cells into a bone tumor. Also, MMP-12 has been shown to cleave the urokinase-type plasminogen activator receptor and exert a proteolytic activity on other bone matrix proteins such as osteopontin and bone sialoprotein, which have been shown to be involved in the migration and invasion of number of cells including tumor cells,43 and in osteoclast activities,32 respectively.
In summary, we show for the first time that MMP-12 is expressed by human prostate cancer PC3 cells, and its expression is enhanced when prostate cancer PC3 cells interact with bone marrow-derived stromal cells. Moreover, MMP-12 expression was associated with enhanced invasion through type I collagen. Further studies are now under way to confirm whether this stimulatory effect of the bone microenvironment also occurs on other PC cells, which would validate MMP-12 as a molecular therapeutic target for prostate cancer patients with bone metastasis.
Acknowledgments
Grant sponsor: National Institute of Diabetes and Digestive and Kidney Diseases; Grant number: 067687.
Abbreviations
- BMS
bone marrow stromal
- BSA
bovine serum albumin
- CK
cytokeratin
- CM
conditioned medium
- ECM
extracellular matrix
- FBS
fetal bovine serum
- FITC-A
fluorescein isothiocyanate-area
- GAPDH
glyceraldehyde phosphate dehydrogenase
- IgG
immunoglobulin G
- MMP
matrix metalloproteinase
- MP
metalloproteinase
- PBS
phosphate buffer saline
- PE-A
phycoerythrin-area
- s.c
subcutaneous
- scb
scrambled
- siRNA
small interfering RNA
- TRAP
tartrate-resistant acid phosphatase
Footnotes
The last two authors jointly supervised this study and should be considered as senior authors.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Bubendorf L, Schopfer A, Wagner U, Sauter G, Moch H, Willi N, Gasser TC, Mihatsch MJ. Metastatic patterns of prostate cancer: an autopsy study of 1,589 patients. Hum Pathol. 2000;31:578–83. doi: 10.1053/hp.2000.6698. [DOI] [PubMed] [Google Scholar]
- 3.John A, Tuszynski G. The role of matrix metalloproteinases in tumor angiogenesis and tumor metastasis. Pathol Oncol Res. 2001;7:14–23. doi: 10.1007/BF03032599. [DOI] [PubMed] [Google Scholar]
- 4.Sternlicht MD, Werb Z. How matrix metalloproteinases regulate cell behavior. Annu Rev Cell Dev Biol. 2001;17:463–516. doi: 10.1146/annurev.cellbio.17.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olumi AF, Grossfeld GD, Hayward SW, Carroll PR, Tlsty TD, Cunha GR. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Res. 1999;59:5002–11. doi: 10.1186/bcr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lang SH, Stower M, Maitland NJ. In vitro modelling of epithelial and stromal interactions in non-malignant and malignant prostates. Br J Cancer. 2000;82:990–7. doi: 10.1054/bjoc.1999.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sung SY, Chung LW. Prostate tumor-stroma interaction: molecular mechanisms and opportunities for therapeutic targeting. Differentiation. 2002;70:506–21. doi: 10.1046/j.1432-0436.2002.700905.x. [DOI] [PubMed] [Google Scholar]
- 8.Dong Z, Bonfil RD, Chinni S, Deng X, Trindade Filho JC, Bernardo M, Vaishampayan U, Che M, Sloane BF, Sheng S, Fridman R, Cher ML. Matrix metalloproteinase activity and osteoclasts in experimental prostate cancer bone metastasis tissue. Am J Pathol. 2005;166:1173–86. doi: 10.1016/S0002-9440(10)62337-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonfil RD, Sabbota A, Nabha S, Bernardo MM, Dong Z, Meng H, Yamamoto H, Chinni SR, Lim IT, Chang M, Filetti LC, Mobashery S, et al. Inhibition of human prostate cancer growth, osteolysis and angiogenesis in a bone metastasis model by a novel mechanism-based selective gelatinase inhibitor. Int J Cancer. 2006;118:2721–6. doi: 10.1002/ijc.21645. [DOI] [PubMed] [Google Scholar]
- 10.Holmbeck K, Bianco P, Caterina J, Yamada S, Kromer M, Kuznetsov SA, Mankani M, Robey PG, Poole AR, Pidoux I, Ward JM, Birkedal-Hansen H. MT1-MMP-deficient mice develop dwarfism, osteopenia, arthritis, and connective tissue disease due to inadequate collagen turnover. Cell. 1999;99:81–92. doi: 10.1016/s0092-8674(00)80064-1. [DOI] [PubMed] [Google Scholar]
- 11.Bonfil RD, Dong Z, Trindade Filho JC, Sabbota A, Osenkowski P, Nabha S, Yamamoto H, Chinni SR, Zhao H, Mobashery S, Vessella RL, Fridman R, et al. Prostate cancer-associated membrane type 1-matrix metalloproteinase: a pivotal role in bone response and intraosseous tumor growth. Am J Pathol. 2007;170:2100–11. doi: 10.2353/ajpath.2007.060720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaighn ME, Narayan KS, Ohnuki Y, Lechner JF, Jones LW. Establishment and characterization of a human prostatic carcinoma cell line (PC-3) Invest Urol. 1979;17:16–23. [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Nemeth JA, Harb JF, Barroso U, Jr, He Z, Grignon DJ, Cher ML. Severe combined immunodeficient-hu model of human prostate cancer metastasis to human bone. Cancer Res. 1999;59:1987–93. [PubMed] [Google Scholar]
- 15.Minkin C. Bone acid phosphatase: tartrate-resistant acid phosphatase as a marker of osteoclast function. Calcif Tissue Int. 1982;34:285–90. doi: 10.1007/BF02411252. [DOI] [PubMed] [Google Scholar]
- 16.Park HI, Jin Y, Hurst DR, Monroe CA, Lee S, Schwartz MA, Sang QX. The intermediate S1′ pocket of the endometase/matrilysin-2 active site revealed by enzyme inhibition kinetic studies, protein sequence analyses, and homology modeling. J Biol Chem. 2003;278:51646–53. doi: 10.1074/jbc.M310109200. [DOI] [PubMed] [Google Scholar]
- 17.Moss ML, Rasmussen FH. Fluorescent substrates for the proteinases ADAM17, ADAM10, ADAM8, and ADAM12 useful for high-throughput inhibitor screening. Anal Biochem. 2007;366:144–8. doi: 10.1016/j.ab.2007.04.043. [DOI] [PubMed] [Google Scholar]
- 18.Nabha SM, Bonfil RD, Yamamoto HA, Belizi A, Wiesner C, Dong Z, Cher ML. Host matrix metalloproteinase-9 contributes to tumor vascularization without affecting tumor growth in a model of prostate cancer bone metastasis. Clin Exp Metastasis. 2006;23:335–44. doi: 10.1007/s10585-006-9042-x. [DOI] [PubMed] [Google Scholar]
- 19.Duivenvoorden WC, Hirte HW, Singh G. Use of tetracycline as an inhibitor of matrix metalloproteinase activity secreted by human bone-metastasizing cancer cells. Invasion Metastasis. 1997;17:312–22. [PubMed] [Google Scholar]
- 20.Stearns ME, Wang M. Alendronate blocks metalloproteinase secretion and bone collagen I release by PC-3 ML cells in SCID mice. Clin Exp Metastasis. 1998;16:693–702. doi: 10.1023/a:1006524610591. [DOI] [PubMed] [Google Scholar]
- 21.Sanchez-Sweatman OH, Orr FW, Singh G. Human metastatic prostate PC3 cell lines degrade bone using matrix metalloproteinases. Invasion Metastasis. 1998;18:297–305. doi: 10.1159/000024522. [DOI] [PubMed] [Google Scholar]
- 22.Hart CA, Scott LJ, Bagley S, Bryden AA, Clarke NW, Lang SH. Role of proteolytic enzymes in human prostate bone metastasis formation: in vivo and in vitro studies. Br J Cancer. 2002;86:1136–42. doi: 10.1038/sj.bjc.6600207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corey E, Brown LG, Quinn JE, Poot M, Roudier MP, Higano CS, Vessella RL. Zoledronic acid exhibits inhibitory effects on osteoblastic and osteolytic metastases of prostate cancer. Clin Cancer Res. 2003;9:295–306. [PubMed] [Google Scholar]
- 24.Festuccia C, Angelucci A, Gravina GL, Villanova I, Teti A, Albini A, Bologna M. Osteoblast-derived TGF-β1 modulates matrix degrading protease expression and activity in prostate cancer cells. Int J Cancer. 2000;85:407–15. [PubMed] [Google Scholar]
- 25.Singh S, Singh UP, Stiles JK, Grizzle WE, Lillard JW., Jr Expression and functional role of CCR9 in prostate cancer cell migration and invasion. Clin Cancer Res. 2004;10:8743–50. doi: 10.1158/1078-0432.CCR-04-0266. [DOI] [PubMed] [Google Scholar]
- 26.Daja MM, Niu X, Zhao Z, Brown JM, Russell PJ. Characterization of expression of matrix metalloproteinases and tissue inhibitors of metalloproteinases in prostate cancer cell lines. Prostate Cancer Prostatic Dis. 2003;6:15–26. doi: 10.1038/sj.pcan.4500609. [DOI] [PubMed] [Google Scholar]
- 27.Shapiro SD, Kobayashi DK, Pentland AP, Welgus HG. Induction of macrophage metalloproteinases by extracellular matrix. Evidence for enzyme- and substrate-specific responses involving prostaglandin-dependent mechanisms. J Biol Chem. 1993;268:8170–5. [PubMed] [Google Scholar]
- 28.Gronski TJ, Jr, Martin RL, Kobayashi DK, Walsh BC, Holman MC, Huber M, Van Wart HE, Shapiro SD. Hydrolysis of a broad spectrum of extracellular matrix proteins by human macrophage elastase. J Biol Chem. 1997;272:12189–94. doi: 10.1074/jbc.272.18.12189. [DOI] [PubMed] [Google Scholar]
- 29.Chandler S, Cossins J, Lury J, Wells G. Macrophage metalloelastase degrades matrix and myelin proteins and processes a tumour necrosis factor-α fusion protein. Biochem Biophys Res Commun. 1996;228:421–9. doi: 10.1006/bbrc.1996.1677. [DOI] [PubMed] [Google Scholar]
- 30.Shipley JM, Wesselschmidt RL, Kobayashi DK, Ley TJ, Shapiro SD. Metalloelastase is required for macrophage-mediated proteolysis and matrix invasion in mice. Proc Natl Acad Sci USA. 1996;93:3942–6. doi: 10.1073/pnas.93.9.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kerkela E, Bohling T, Herva R, Uria JA, Saarialho-Kere U. Human macrophage metalloelastase (MMP-12) expression is induced in chondrocytes during fetal development and malignant transformation. Bone. 2001;29:487–93. doi: 10.1016/s8756-3282(01)00595-6. [DOI] [PubMed] [Google Scholar]
- 32.Hou P, Troen T, Ovejero MC, Kirkegaard T, Andersen TL, Byrjalsen I, Ferreras M, Sato T, Shapiro SD, Foged NT, Delaisse JM. Matrix metalloproteinase-12 (MMP-12) in osteoclasts: new lesson on the involvement of MMPs in bone resorption. Bone. 2004;34:37–47. doi: 10.1016/j.bone.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 33.Kerkela E, Ala-Aho R, Jeskanen L, Rechardt O, Grenman R, Shapiro SD, Kahari VM, Saarialho-Kere U. Expression of human macrophage metalloelastase (MMP-12) by tumor cells in skin cancer. J Invest Dermatol. 2000;114:1113–19. doi: 10.1046/j.1523-1747.2000.00993.x. [DOI] [PubMed] [Google Scholar]
- 34.Kerkela E, Ala-aho R, Klemi P, Grenman S, Shapiro SD, Kahari VM, Saarialho-Kere U. Metalloelastase (MMP-12) expression by tumour cells in squamous cell carcinoma of the vulva correlates with invasiveness, while that by macrophages predicts better outcome. J Pathol. 2002;198:258–69. doi: 10.1002/path.1198. [DOI] [PubMed] [Google Scholar]
- 35.Ding Y, Shimada Y, Gorrin-Rivas MJ, Itami A, Li Z, Hong T, Maeda M, Komoto I, Kawabe A, Kaganoi J, Imamura M. Clinicopathological significance of human macrophage metalloelastase expression in esophageal squamous cell carcinoma. Oncology. 2002;63:378–84. doi: 10.1159/000066231. [DOI] [PubMed] [Google Scholar]
- 36.Hofmann HS, Hansen G, Richter G, Taege C, Simm A, Silber RE, Burdach S. Matrix metalloproteinase-12 expression correlates with local recurrence and metastatic disease in non-small cell lung cancer patients. Clin Cancer Res. 2005;11:1086–92. [PubMed] [Google Scholar]
- 37.Yang W, Arii S, Gorrin-Rivas MJ, Mori A, Onodera H, Imamura M. Human macrophage metalloelastase gene expression in colorectal carcinoma and its clinicopathologic significance. Cancer. 2001;91:1277–83. [PubMed] [Google Scholar]
- 38.Gorrin Rivas MJ, Arii S, Furutani M, Harada T, Mizumoto M, Nishiyama H, Fujita J, Imamura M. Expression of human macrophage metalloelastase gene in hepatocellular carcinoma: correlation with angiostatin generation and its clinical significance. Hepatology. 1998;28:986–93. doi: 10.1002/hep.510280413. [DOI] [PubMed] [Google Scholar]
- 39.Houghton AM, Grisolano JL, Baumann ML, Kobayashi DK, Hautamaki RD, Nehring LC, Cornelius LA, Shapiro SD. Macrophage elastase (matrix metalloproteinase-12) suppresses growth of lung metastases. Cancer Res. 2006;66:6149–55. doi: 10.1158/0008-5472.CAN-04-0297. [DOI] [PubMed] [Google Scholar]
- 40.Cornelius LA, Nehring LC, Harding E, Bolanowski M, Welgus HG, Kobayashi DK, Pierce RA, Shapiro SD. Matrix metalloproteinases generate angiostatin: effects on neovascularization. J Immunol. 1998;161:6845–52. [PubMed] [Google Scholar]
- 41.Chakraborti S, Mandal M, Das S, Mandal A, Chakraborti T. Regulation of matrix metalloproteinases: an overview. Mol Cell Biochem. 2003;253:269–85. doi: 10.1023/a:1026028303196. [DOI] [PubMed] [Google Scholar]
- 42.Sarkar S, Nuttal RK, Liu S, Edwards DR, Wee Yong V. Tenascin-C stimulates glioma cell invasion through matrix metalloproteinase-12. Cancer Res. 2006;66:11771–80. doi: 10.1158/0008-5472.CAN-05-0470. [DOI] [PubMed] [Google Scholar]
- 43.Koolwijk P, Sidenius N, Peters E, Sier CF, Hanemaaijer R, Blasi F, van Hinsbergh VW. Proteolysis of the urokinase-type plasminogen activator receptor by metalloproteinase-12: implication for angiogenesis in fibrin matrices. Blood. 2001;97:3123–31. doi: 10.1182/blood.v97.10.3123. [DOI] [PubMed] [Google Scholar]