Abstract
Membrane lipid composition is an important correlate of the rate of aging of animals and, therefore, the determination of their longevity. In the present work, the use of high-throughput technologies allowed us to determine the plasma lipidomic profile of 11 mammalian species ranging in maximum longevity from 3.5 to 120 years. The non-targeted approach revealed a specie-specific lipidomic profile that accurately predicts the animal longevity. The regression analysis between lipid species and longevity demonstrated that the longer the longevity of a species, the lower is its plasma long-chain free fatty acid (LC-FFA) concentrations, peroxidizability index, and lipid peroxidation-derived products content. The inverse association between longevity and LC-FFA persisted after correction for body mass and phylogenetic interdependence. These results indicate that the lipidomic signature is an optimized feature associated with animal longevity, emerging LC-FFA as a potential biomarker of longevity.
The involvement of oxidative stress in determining the animal longevity (maximum lifespan potential or maximum longevity, MLSP) is intrinsically related to its key role in the origin and evolution of life1. Oxidative stress is not only a natural consequence of aerobic life, but it has also been a determinant factor which demanded structural and functional adaptations to the cells and organisms that, in turn, determined their longevities2. Available evidences suggest that aerobic life, and long-lived species, evolved by reducing the relative abundance of those structural components that are highly susceptible to oxidative damage, thus conferring to the cellular constituents a higher structural stability and lower susceptibility to oxidative damage2. In this context, a recent study3 with a phylogenomic approach to identify the genetic targets of natural selection for increased longevity in mammals shows that genes involved in fatty acid biosynthesis (elongases, desaturases and lipoxidation repair) have collectively undergone increased selective pressure in long-lived species, while most of the candidate genes experimentally associated with aging (e.g., DNA repair, uncoupling proteins, antioxidants, PolG, Sod, Foxo) have played no detectable role in the evolution of longevity in mammals. Hence, it can be inferred that cellular membrane has been an optimized feature during evolution. Indeed, membrane fatty acid composition differs between animal species and the susceptibility to oxidative damage of membrane lipids is inversely related to the maximum longevity4,5,6. Such a correlation has been documented in vertebrates and also in a more restricted number of investigations of invertebrate species2,6. Reinforcing these findings, two recent works specifically focused to humans (as exceptionally long-lived species) also demonstrated that erythrocyte membranes7 and plasma lipids8 of longevous individuals are more resistant to oxidative stress.
Despite the fact that systems biology-based approaches allow a global characterization (at a molecular level) of complex global biological systems, up to date no lipidomic studies investigating differences in plasma or tissue lipid molecules across animal species have been reported. To overcome these limitations in knowledge, here we have determined the plasma lipidomic profile by using high-throughput lipidome profiling technologies of 11 mammalian species, including humans, ranging in MLSP from 3.5 to 120 years. Plasma was also elected to identify lipid species that could arise as human longevity potential biomarkers. This has allowed us to define a robust lipidomic signature that accurately predicts the animal longevity.
Results
Specific plasma lipidomic profiles for mammalian species
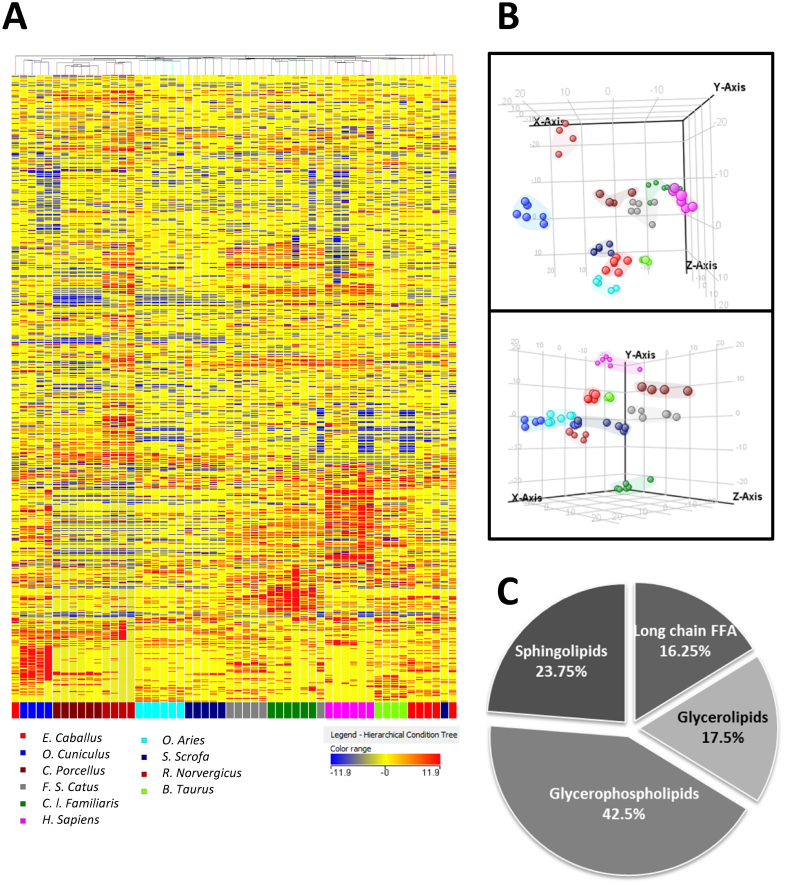
Using a non-targeted approach we detected about 14,000 different ionic species in plasma (considering both positive and negative ionizations). After selecting only those which were present in at least 75% of the samples of the same group, clustering analysis were applied. As the hierarchical clustering (Fig. 1A) analysis and multivariate statistics (specifically partial least square discrimination analyses (PLS-DA)) shows, animal species is the factor which explains better the variability of the samples suggesting a specific lipidome signature for each species. The statistical analysis (ANOVA, multiple testing correction Benjamini-Hodgberg, p < 0.05) revealed 1,034 differential lipids between species, 682 of them correlated significantly with MLSP (p < 0.05) (data not shown). Among them, 80 were identified belonging to long-chain free fatty acids (LC-FFA) (16.25%), glycerolipids (17.5%), sphingolipids (23.75%) and glycerophopholipids (42.5%) families (Fig. 1, Supplementary Table S1). Interestingly, all the LC-FFA identified correlated negatively to MLSP. This fact, jointly with the importance of circulating LC-FFA in human diseases, led us to further analyse the correlation between them and MLSP.
Figure 1. Specific plasma lipidomic profiles for mammalian species.
(A). Heat map representation of hierarchical clustering of molecular features found in each plasma sample. Each line of this graphic represents an accurate mass ordered by retention time, coloured by its abundance intensity normalized to internal standard and baselining to median/mean across the samples. The scale from −11.9 (blue) to 11.9 (red) represents this normalized abundance in arbitrary units. (B). Tridimensional PLS-DA graphs (up: representation of positive ionization molecules; down: representation of negative ionization molecules) demonstrating a differentiation effect of animal species on plasma lipidomic profiles. Overall accuracy = 0.78. (C). Distribution by families of statistical differential lipid molecules that correlate with longevity. Among them, 100% long-chain free fatty acids, 92.8% glycerolipids, 79.4% glycerophospholipids and 86.9% sphingolipids presents an inverse correlation with maximum longevity. N × group: 4–6 animals.
Relationship between MLSP and long-chain free fatty acids
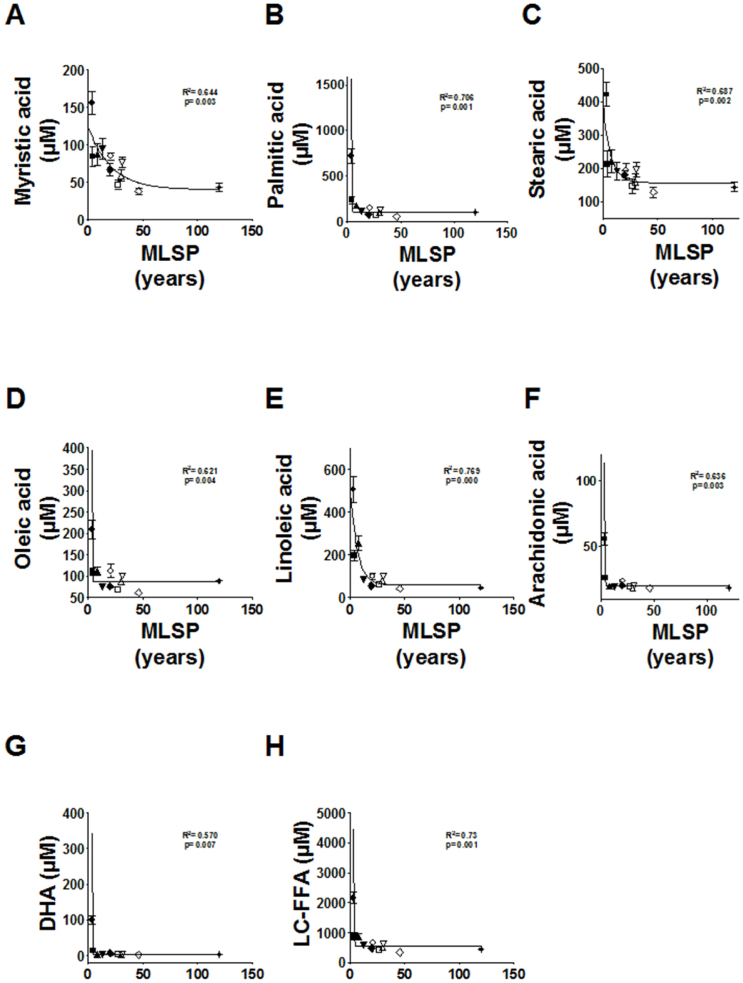
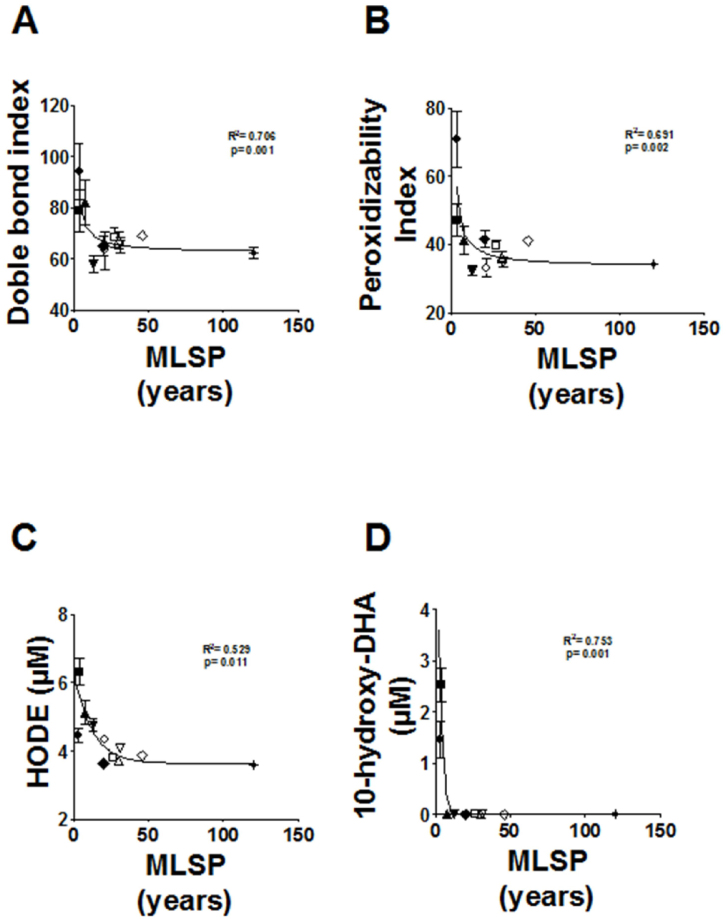
Regression curve analysis shows that there is a negative correlation between animal longevity and plasma FFAs. Indeed the longer the longevity of a species, the lower is its plasma LC-FFA concentration. The plasma LC-FFA composition and derived parameters in the 11 animal species studied in this investigation is shown in Supplementary Table S2. This was due to the negative correlation with MLSP for all the LC-FFAs [myristic acid (C14:0): R2 = 0.644, p < 0.003; palmitic acid (C16:0): R2 = 0.706, p < 0.001; stearic acid (C18:0): R2 = 0.687, p < 0.002; oleic acid (C18:1n − 9): R2 = 0.621, p < 0.004; linoleic acid (C18:2n − 6): R2 = 0.769, p < 0.0001; arachidonic acid (C20:4n − 6): R2 = 0.636, p < 0.003; and docosahexaenoic acid (DHA) (C22:6n − 3): R2 = 0.570, p < 0.007] (Fig. 2) being also significant the correlation between total LC-FFAs and the MLSP (R2 = 0.73, p < 0.001). In contrast, no correlations were observed for medium-chain FFAs (capric acid, C10:0; and lauric acid, C12:0) nor for the average chain length (ACL) (data not shown). The density of double bonds in FFAs (double bond index (DBI)) and susceptibility to peroxidation (peroxidizability index, PI, which takes into account that the sensitivity to peroxidation increases as an exponential function of the number of double bonds per fatty acid) (Fig. 3) showed significant negative correlations with MLSP (R2 = 0.706, p < 0.001; and R2 = 0.691, p < 0.002, respectively). In addition, due to the importance of oxidative stress in animal longevity2 we focused in several lipid peroxidation products, by using a targeted lipidomic approach. Among them, we specifically detected and quantified the hydroperoxides 9- and 13-hydroxy-octadecadienoic acid (HODE) and the 10-hydroxy-DHA. In agreement with the lower DBI and PI of long-lived animals, this was also reflected in the endogenous content of lipid peroxidation products from very LC-FFAs, since the in vivo lipid peroxidation of plasma samples was also negatively correlated with the MLSP of the animal species (HODE: R2 = 0.529, p < 0.011; 10-hydroxy-DHA: R2 = 0.753, p < 0.001) (Fig. 3).
Figure 2. Relationship between maximum longevity (MLSP) and long-chain free fatty acid (LC-FFA) concentrations.
From A to G, for specific FFA; H, for total concentration of plasma LC-FFA. Curve estimation (inverse mode) equation: Y = b0 + (b1/MLSP) (Myristic acid: b0 = 51.397, b1 = 95.311; Palmitic acid: b0 = 27.754, b1 = 1664.309; Stearic acid: b0 = 137.742, b1 = 699.445; Oleic acid: b0 = 70.313, b1 = 336.26; Linoleic acid: b0 = 25.156, b1 = 1284.553; Arachidonic acid: b0 = 15.920, b1 = 91.108; Docosahexaenoic acid (DHA): b0 = −7.023, b1 = 230.665; LC-FFA: b0 = 319.938, b1 = 4609.232). N × group: 9–15 animals. Values are means ± SEM.
Figure 3. Relationship between maximum longevity (MLSP) and double bond index (DBI) (A), peroxidizability index (PI) (B) and specific lipid peroxidation-derived products of plasma in mammalian species (C–D).
Curve estimation (inverse mode) equation: Y = b0 + (b1/MLSP): Doble bond index: b0 = 62.235, b1 = 93.398; Peroxidizability index: b0 = 32.687, b1 = 95.311; hydroxyoctadecadienoic acid (HODE): b0 = 3.78, b1 = 6.338; 10-hydroxy-docosahexaenoic acid (10-hydroxy-DHA): b0 = −0.326, b1 = 7.827. N × group: 9–15 animals. Values are means ± SEM.
Corrections for body mass and phylogeny
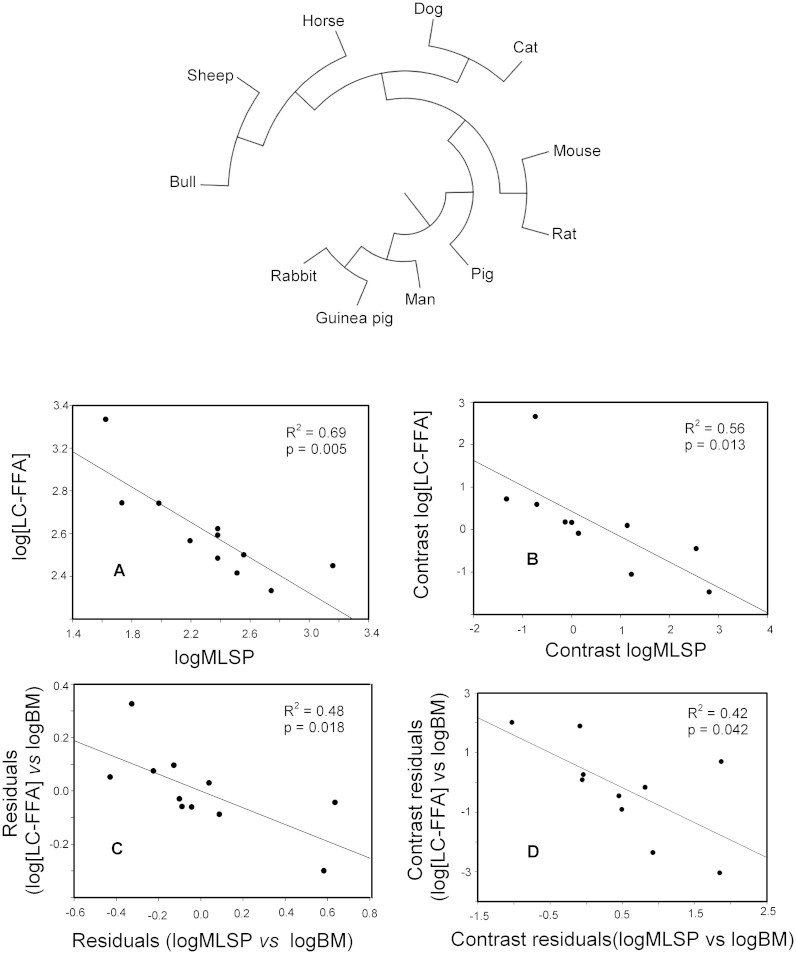
Since species are part of a hierarchically structured phylogeny, it may happen that data from the eleven species used in the current study may not necessarily be statistically independent from one another. Thus, we addressed whether the above described correlations remained significant after correcting for the phylogenetic inertia. To this end, for each analysed variable, its corresponding phylogenetically independent contrast was computed according to Felsenstein's method9. The negative association between MLSP and the total concentration of LC-FFA remained significant after phylogenetic corrections (Fig. 4). When the relationships between the longevity contrast and the contrast of specific FFA were assessed, statistically significant negative correlations were observed for all the analysed FFA, with the exception of docosahexaenoic acid, which exhibited a marginally significant correlation with MLSP (see Supplementary Table S3). On the other hand, when the examined variables were the peroxidizability index and longevity, their contrasts showed indeed a negative covariance, but the correlation failed to be significant (p = 0.127).
Figure 4. Long-chain free fatty acid (LC-FFA) concentrations are negatively related to longevity even after correction for phylogenetic inertia and body size.
The observed relationship between log[LC-FFA] and logMLSP (A), remained statistically significant even after correction for phylogenetic inertia (B). Partial correlation of log[LC-FFA] and logMLSP after correction for body mass was also significant (C), as well as the correlation between their respective phylogenetically independent contrasts (D). The phylogenetic tree used to calculate the contrasts analysed in (B) and (D), is shown in the upper part of the figure.
Because of the well-known relationship between longevity and body mass, we next addressed the potential confounding effect of body mass. To this end, residual analyses were carried out. Neither the relationship between MLSP and LC-FFA (Fig. 4) nor that between MLSP and PI (Table S4), could be accounted for the body mass variable.
Discussion
Metabolism is adapted and optimized to achieve balance and economy, and one of the attractors involved in biomolecular evolution is the minimizing of unwanted side reactions10. However, uncontrolled and potentially deleterious endogenous reactions occur, even under physiological conditions. Available evidences support an important role for membrane unsaturation in the determination of longevity4,5,6. It is now well known that the PI of membrane fatty acids decreases as longevity increases when different mammalian species are compared2,6,10 in different tissues including heart, liver, and skeletal muscle. A low degree of fatty acid unsaturation also occurs in extraordinarily long-lived animals (vertebrates and invertebrates) like birds12,13,11, naked mole-rats14, the short-beaker echidna15, queen honey bees16, long-lived bivalves17, or C. elegans18 and correlates with natural variation in longevity even within a species as it occurs in the case of different mice strains19. The results obtained in the present study also support that fact, extending the relevance of lipid metabolism in determining longevity to plasma LC-FFAs in a specific way for long-lived mammalian species. Furthermore, lipid peroxidation is a kind of oxidative damage quantitatively, functionally and pathologically important inside and outside cells20. In this line, a low degree of FFA unsaturation in plasma (proposed and determined for first time in the present work) should be advantageous, in oxidative stress terms, by decreasing their sensitivity to lipid peroxidation. This would also protect other molecules against lipoxidation-derived damage20. In agreement with this idea, we found a strongly decreased total content of FFAs, as well as the number of fatty acid double bonds (expressed as DBI) and PI in plasma from long-lived animals. The decrease in PI confers to the plasma, similarly to cell membranes4,5,6, superior resistance to lipid peroxidation and lowers potential lipoxidation-dependent damage to macromolecules.
Comparative studies among species have been revealed as a powerful source of information that may assist our understanding of the aging process2. However, this kind of studies face two problems that need to be properly addressed before any conclusion can be drawn. First, body mass is a pervasive trait that influences all levels of organismal biology21. Therefore, the problem is that simply correlating MLSP with plasma LC-FFA concentrations, does not allow us to conclude any underlying causality. In other words, we do not know whether these differences in plasma lipid concentrations contribute to the differences in MLSP, or alternatively, whether these differences in plasma lipid composition arise only because both variable (lipid composition and longevity) are related to body mass. To overcome this limitation, we have carried out statistical analyses that aimed to separate the alternative causes that might generate an interspecific association between MLSP and specific plasma lipid concentrations. In this way, we have reached the conclusion that body mass does not seem to be behind the negative correlation between LC-FFA and MLSP. Similarly, our results suggest that the observed negative relation between MLSP and the PI is not mediated by body mass.
The second problem that needs attention when carrying out interspecific comparative studies is the potential lack of independence of the data due to a shared phylogenetic history of the species involved in the analyses. Because independence of the individual data included in any analysis is a prerequisite of most statistical procedures, including correlation analysis, the use of raw data may lead to overestimate the significativeness of the correlation between the analysed variables. This problem in comparative biology has been appreciated for a long time. A whole series of sophisticated statistical approaches have been developed that aim to transform raw interspecific data into what are generally termed phylogenetically independent contrast22. Herein we have used Felsenstein's method that transforms the original variables into contrasts, using the known phylogeny of the involved species9. The fact that the contrasts of logMLSP were significantly correlated with the contrasts of different LC-FFA concentrations, supports the view of a genuine relationship between longevity and plasma concentration of certain lipids, which is in line with the suggestion that the lipidomic signature is an optimized feature associated with longevity.
On the other hand, although we have observed statistically significant relationships between longevity and indexes such as PI and DBI, which remained significant after correcting for body mass (see Table S4), we could not accept a significant correlation between these variables after correcting for phylogenetic inertia (p-values, 0.12 and 0.09, respectively). Nevertheless, it should be pointed out that this negative result does not necessarily mean that a true relationship between these variables should be ruled out. For instance, if we accept that 27% of the PI variance can be truly accounted for by the MLSP (R2 = 0.27), then analyzing only 11 mammalian species (n = 10, for phylogenetic independent contrasts) entails a beta-type error of 0.52. In other words, it may well be that we are accepting the null hypothesis when it is actually false, just because of a lack of statistical power. As pointed out by Felsenstein, we should realize that different traits may exhibit different degree of phylogenetic inertia, depending on how fast they respond to natural selection7. Thus, for those correlated characters exhibiting a high degree of inertia, correlation analyses would require a large number of species in order to detect the underlying relationship between these inert characters.
LC-FFAs are essential for physiological homeostasis, providing, for example, a major portion of mammalian energy needs23. We now recognize that FFAs are the vehicle by which triacylglycerol stored in adipose tissue is transported to its sites of utilization. FFA turnover is rapid, with a plasma half-life around 2–4 min23. The only significant site of FFA liberation into plasma is adipose tissue23. Therefore, results of the present study seem to suggest an evolutionary adaptation in the sense that: i) adipose tissue regulates the fatty acid type stored and released to plasma in a species-specific fashion maintaining a lower degree of unsaturation in long-lived species, and ii) adipose tissue regulates LC-FFA concentration in a species-specific fashion as expression of the different metabolic needs.
LC-FFAs are not only essential nutrient components, but they also function as signalling molecules in numerous physiological processes25,26,24. Among them, it is particularly relevant their direct effect mediated by specific FFA receptors (FFARs) on insulin secretion25,26,24. Additionally, the insulin signalling pathway is an evolutionary conserved mechanism involved in the determination of animal longevity28,29,30,27. Indeed a diminished insulin signalling has been described in long-lived mutants and strains of worms, flies and mice, as well as from the pro-longevity effect of nutritional interventions such as caloric restriction when applied to different animal species28,29,30,27. Therefore, it is plausible to postulate that long-lived mammals (including humans) exhibit a low LC-FFA concentration to maintain an attenuated activity of the insulin signalling pathway. In this line, the lower levels of LC-FFA observed in humans could be an evolutionary adaptive response that could explain the pathological states such as insulin resistance linked to an increased LC-FFA plasma concentration23 leading to a shortened MLSP.
In conclusion, these results indicate that the lipidomic signature is an optimized feature associated with animal longevity, emerging LC-FFAs as a potential biomarker of longevity.
Methods
Animals
All animals, namely, mouse (Mus musculus), rat (Rattus norvergicus), guinea pig (Cavia porcellus), rabbit (Oryctolagus cuniculus), sheep (Ovis aries), dog (Cannis familiaris), pig (Sus scrofa), bull (Bos taurus), cat (Felis catus), horse (Equus caballus), and man (Homo sapiens) whose MLSP vary from 3.5 to 120 years, were male adult specimens with an age at 15–30% of their MLSP (equivalent to an age ranging from 0.5 to 40 years). The recorded values of MLSP (in yrs) were: mouse, 3.5; rat, 4.5; guinea pig, 8; rabbit, 13; sheep, 20; dog, 20; pig, 27; bull, 30; cat, 20; horse, 46; and man, 120. The animal care protocols were approved by the University of Lleida Animal Experimentation Ethics Committee. Human protocols were approved by the Committee for Ethics in clinical research of the Hospital Universitari Arnau de Vilanova (in accordance with the Declaration of Helsinki). All subjects were fully informed of the aims and scope of the research and signed an informed consent. Blood samples were obtained after fasting (8–12 h) by venipunction, centrifuge to separate plasma fraction, and immediately frozen in liquid nitrogen, and transferred before 4 h to a −80°C freezer for storage, to be used later for lipidomic analyses.
Lipidomic analyses
Lipidomic analysis was based on previously described method31. In order to precipitate the protein samples 600 μl of cold acetone were added to 200 μl of sample (25 μl of plasma and 175 μl of phosphate buffer). After the addition, the samples were vigorously agitated for 10 s and softly agitated during 30 min at 4°C. Then, the samples were centrifuged 10 min at 1400 rpm and the supernatants were evaporated using a Speed Vac (Thermo Fisher Scientific, Barcelona, Spain). Lipid class representative internal standards were added, as described previously32 (Supplementary Table S5). Plasma lipids were extracted by adding methanol: chloroform: 0.7% KCl (1:2:1, v/v/v). After the samples were agitated and centrifugated 10 min at 1400 rpm the chloroformic part was separated and evaporated using a Speed Vac (Thermo Fisher Scientific, Barcelona, Spain). Finally, the samples were resuspended in chloroform:methanol (1:3, v/v) and injected onto the chromatographic system. Lipid extracts were subjected to mass-spectrometry using an Agilent LC 1200 series coupled to the Q-TOF MS/MS 6520 (Agilent Technologies, Barcelona, Spain). LC method was modified by Sartain et al33. Briefly, 2 μl of sample was applied onto a reverse phase column (LXBridge C18 column, 2.1 × 150 mm, 3.5 μm, Waters Corp.; Milford, MA; USA) equipped with a reverse phase guard column (XBridge C18, 2.1 × 10 mm, 3.5 μm, USA) heated to 55°C. The flow rate was 250 μl/min with solvent A composed of 5 mM ammonium acetate in methanol-water (90:10, v/v) and solvent B composed of 5 mM ammonium acetate in 1-propanol-hexane-water (79:20:1, v/v/v). The gradient consisted of 100% solvent A for 2 min, from 0% to 100% in 30 minutes, held at 100% B for 3 min, return to 0% B in 20 minutes, and re-equilibrated at 0% solvent B for 4 min. Data were collected in positive and negative electrospray ionization mode TOF operated in full-scan mode at 100 to 3000 m/z in an extended dynamic range (2 GHz), using N2 as nebulizer gas (5 L/min, 350°C). The capillary voltage was 3500 V with a scan rate of 1 scan/s. The MassHunter Data Analysis Software (Agilent Technologies, Barcelona, Spain) was used to collect the results and the MassHunter Qualitative Analysis Software (Agilent Technologies, Barcelona, Spain) to obtain the molecular features of the samples, representing different, co-migrating ionic species of a given molecular entity using the Molecular Feature Extractor (MFE) algorithm (Agilent Technologies, Barcelona, Spain)34. Briefly, the MFE algorithm uses the accuracy of the mass measurements to group related ions (basing on charge-state envelope, isotopic distribution and/or the presence of different adducts and dimers/trimers) assigning multiple species (ions) to a single compound referred to as a feature. Finally, the MassHunter Mass Profiler Professional Software (Agilent Technologies, Barcelona, Spain) was used to perform a non-targeted lipidomic analysis over the extracted features. Only common features (found in at least 75% of the samples of the same condition) were taken into account to correct for individual bias. Multivariate statistics (Hirerchical clustering and PLS-DA analyses) were done using this software. The masses representing significant differences by Student T-test (fold change ≥ 2, p < 0.05 with Benjamini-Hochberg Multiple Testing Correction) were searched against the LIPID MAPS35 database (exact mass ppm < 20). The identities obtained were then compared to retention time of the authentic standards added.
Targeted lipidomic analysis was performed using the same chromatographic and spectrometric method as in the untargetted approach. We looked for the following free fatty acids: capric acid, lauric acid, myristic acid, palmitic acid, palmitoleic acid, stearic acid, oleic acid, linoleic acid, linolenic acid, arachidonic acid, docosahexaenoic acid (DHA). The m/z values used for quantification were: m/z 227.2017 [M-H]− for myristic acid, m/z 255.233 [M-H]− for palmitic acid, m/z 283.2643 [M-H]− for stearic acid, m/z 245.2486 [M-H]− for oleic acid, m/z 279.233 [M-H]− for linoleic acid, m/z 303.233 [M-H]− for arachidonic acid and m/z 327.233 [M-H]− for DHA. We detected and quantified the 9-, 13-hydroxyoctadecadienoic acid (HODE) and 10-,17-hydroxy-DHA as lipid peroxidation-derived markers (the m/z value used for quantification was: m/z 295.2279 [M-H]− for HODE and m/z 343.2279 [M-H]− for 10-,17-hydroxy-DHA). Quantification was performed by external standardization using internal standards for correction.
Phylogenetic analyses
The amino acid sequences of the mtDNA-encoded cytochrome c oxidase subunits (COX 1, COX 2 and COX 3) were concatenated for each species. Afterwards, a multiple sequence alignment was performed using ClustalW. The phylogeny was reconstructed using this alignment and a method of maximum likelihood implemented by MEGA536. The obtained tree was used to compute Felsenstein's phylogenetically independent contrasts9 of the variables under study, using the Contrast program from the PHYLIP package.
Calculations and statistics
To determine FFA-derived parameters, FFA concentrations were converted to mol%. The average chain length was calculated as ACL = [(Σ %Total16 × 16) + ... + (Σ %Totaln × n)]/100 (n = carbon atom number); DBI = (Σ %Monoenoic × 1) + (Σ %Dienoic × 2) + (Σ %Trienoic × 3) + (Σ %Tetraenoic × 4) + (Σ %Pentaenoic × 5) + (Σ %Hexaenoic × 6); PI = [(0.025 × Σ mol% monoenoic) + (1 × Σ mol% dienoic) + (2 × Σ mol% trienoic) + (4 × Σmol % tetraenoic) + (6 × Σ mol% pentaenoic) + (8 × Σ mol% hexaenoic)].
Correlation, partial correlation and residuals analyses were carried out with SPSS. Regression equations were obtained by nonlinear regression analyses with the curve estimation statistic by SPSS/PC software for Windows (SPSS, Chicago, IL). These regressions were determined and tested for significance using the mean values for each species. The 0.05 level was selected as the point of minimal statistical significance. Values in tables and figures are expressed as mean ± SEM.
Author Contributions
G.B. and R.P. designed the experiments. M.J., M.P.-O. and R.P. analysed the data. M.J., A.N., R.C. and V.A. performed the experiments. J.C.A. performed the phylogenetic analyses. R.P. supervised the design and data interpretation. The manuscript was written by M.J., J.C.A. and R.P. and edited by R.P. All authors discussed the results and commented on the manuscript.
Supplementary Material
Table S1-S5
Acknowledgments
We acknowledge funding from the Spanish Ministry of Science and Innovation (CGL2010-18124) to J.C.A.; from the Spanish Ministry of Health (PI11-01532) to M.P.O.; and from the Spanish Ministry of Science and Innovation (BFU2009-11879/BFI), the Generalitat of Catalonia (2009SGR735), and the La MaratoTV3 Foundation to R.P. R.C. was supported by a Generalitat of Catalonia Predoctoral Fellowship. We thank David Argiles for excellent technical assistance. We thank Dr. Montserrat Rué for her support in statistical analyses.
References
- Dowling D. K. & Simmons L. W. Reactive oxygen species as universal constraints in life-history evolution. Proc. Biol. Sci. 276, 1737–1745 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamplona R. & Barja G. An evolutionary comparative scan for longevity-related oxidative stress resistance mechanisms in homeotherms. Biogerontology 12, 409–435 (2011). [DOI] [PubMed] [Google Scholar]
- Jobson R. W., Nabholz B. & Galtier N. An evolutionary genome scan for longevity-related natural selection in mammals. Mol. Biol. Evol. 27, 840–847 (2010). [DOI] [PubMed] [Google Scholar]
- Pamplona R., Barja G. & Portero-Otin M. Membrane fatty acid unsaturation, protection against oxidative stress, and maximum life span: a homeoviscous-longevity adaptation? Ann. N. Y. Acad. Sci. 959, 475–490 (2002). [DOI] [PubMed] [Google Scholar]
- Pamplona R. Membrane phospholipids, lipoxidative damage and molecular integrity: a causal role in aging and longevity. Biochim. Biophys. Acta 1777, 1249–1262 (2008). [DOI] [PubMed] [Google Scholar]
- Hulbert A. J., Pamplona R., Buffenstein R. & Buttemer W. A. Life and death: metabolic rate, membrane composition, and life span of animals. Physiol. Rev. 87, 1175–1213 (2007). [DOI] [PubMed] [Google Scholar]
- Puca A. A. et al. Fatty acid profile of erythrocyte membranes as possible biomarker of longevity. Rejuvenation Res. 11, 63–72 (2008). [DOI] [PubMed] [Google Scholar]
- Gonzalez-Covarrubias V. et al. Lipidomics of familial longevity. Aging Cell. 12, 426–434 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. Am. Nat. 125, 1–15 (1985). [DOI] [PubMed] [Google Scholar]
- Pamplona R. & Barja G. Highly resistant macromolecular components and low rate of generation of endogenous damage: two key traits of longevity. Ageing Res. Rev. 6, 189–210 (2007). [DOI] [PubMed] [Google Scholar]
- Pamplona R. et al. Low fatty acid unsaturation protects against lipid peroxidation in liver mitochondria from long-lived species: the pigeon and human case. Mech. Ageing Dev. 86, 53–66 (1996). [DOI] [PubMed] [Google Scholar]
- Pamplona R. et al. Protein and lipid oxidative damage and complex I content are lower in the brain of parakeets and canaries than in mice. Relation to aging rate. AGE 27, 267–280 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttemer W. A., Battam H. & Hulbert A. J. Fowl play and the price of petrel: long-living Procellariiformes have peroxidation-resistant membrane composition compared with short-living Galliformes. Biol. Lett. 4, 351–354 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell T. W., Buffenstein R. & Hulbert A. J. Membrane phospholipid composition may contribute to exceptional longevity of the naked mole-rat (Heterocephalus glaber): a comparative study using shotgun lipidomics. Exp. Gerontol. 42, 1053–1062 (2007). [DOI] [PubMed] [Google Scholar]
- Hulbert A. J., Beard L. A. & Grigg G. C. The exceptional longevity of an egg-laying mammal, the short-beaked echidna (Tachyglossus aculeatus) is associated with peroxidation-resistant membrane composition. Exp. Gerontol. 43, 729–733 (2008). [DOI] [PubMed] [Google Scholar]
- Haddad L. S., Kelbert L. & Hulbert A. J. Extended longevity of queen honey bees compared to workers is associated with peroxidation-resistant membranes. Exp. Gerontol. 42, 601–609 (2007). [DOI] [PubMed] [Google Scholar]
- Munro D. & Blier P. U. The extreme longevity of Arctica islandica is associated with increased peroxidation resistance in mitochondrial membranes. Aging Cell. 11, 845–855 (2012). [DOI] [PubMed] [Google Scholar]
- Shmookler Reis R. J. et al. Modulation of lipid biosynthesis contributes to stress resistance and longevity of C. elegans mutants. Aging (Albany NY) 3, 125–147 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulbert A. J., Faulks S. C., Harper J. M., Miller R. A. & Buffenstein R. Extended longevity of wild-derived mice is associated with peroxidation-resistant membranes. Mech. Ageing Dev. 127, 653–657 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamplona R. Advanced lipoxidation end-products. Chem. Biol. Interact. 192, 14–20 (2011). [DOI] [PubMed] [Google Scholar]
- Speakman J. R. Correlations between physiology and lifespan–two widely ignored problems with comparative studies. Aging Cell. 4, 167–175 (2005). [DOI] [PubMed] [Google Scholar]
- Harvey P. & Pagel M. in The comparative method in evolutionary biology (Oxford University Press, 1998). [Google Scholar]
- Karpe F., Dickmann J. R. & Frayn K. N. Fatty acids, obesity, and insulin resistance: time for a reevaluation. Diabetes 60, 2441–2449 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T., Kimura I., Inoue D., Ichimura A. & Hirasawa A. Free Fatty Acid Receptors and Their Role in Regulation of Energy Metabolism. Rev. Physiol. Biochem. Pharmacol. (2013). [DOI] [PubMed] [Google Scholar]
- Graciano M. F., Valle M. M., Kowluru A., Curi R. & Carpinelli A. R. Regulation of insulin secretion and reactive oxygen species production by free fatty acids in pancreatic islets. Islets 3, 213–223 (2011). [DOI] [PubMed] [Google Scholar]
- Ichimura A., Hirasawa A., Hara T. & Tsujimoto G. Free fatty acid receptors act as nutrient sensors to regulate energy homeostasis. Prostaglandins Other Lipid Mediat. 89, 82–88 (2009). [DOI] [PubMed] [Google Scholar]
- Guarente L. & Kenyon C. Genetic pathways that regulate ageing in model organisms. Nature 408, 255–262 (2000). [DOI] [PubMed] [Google Scholar]
- Kenyon C. A conserved regulatory system for aging. Cell 105, 165–168 (2001). [DOI] [PubMed] [Google Scholar]
- Kenyon C. J. The genetics of ageing. Nature 464, 504–512 (2010). [DOI] [PubMed] [Google Scholar]
- Fontana L., Partridge L. & Longo V. D. Extending healthy life span–from yeast to humans. Science 328, 321–326 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo H. et al. Lipidome analysis in multiple sclerosis reveals protein lipoxidative damage as a potential pathogenic mechanism. J. Neurochem. 123, 622–634 (2012). [DOI] [PubMed] [Google Scholar]
- Laaksonen R. et al. A systems biology strategy reveals biological pathways and plasma biomarker candidates for potentially toxic statin-induced changes in muscle. PLoS One 1, e97 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartain M. J., Dick D. L., Rithner C. D., Crick D. C. & Belisle J. T. Lipidomic analyses of Mycobacterium tuberculosis based on accurate mass measurements and the novel “Mtb LipidDB”. J. Lipid Res. 52, 861–872 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jove M. et al. Multicompartmental LC-Q-ToF-based metabonomics as an exploratory tool to identify novel pathways affected by polyphenol rich diets in mice. J. Proteome Res. 10, 3501–3512 (2011). [DOI] [PubMed] [Google Scholar]
- The LIPID MAPS Lipidomics Gateway, http://www.lipidmaps.org/.
- Tamura K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1-S5