Abstract
Summary: Database search is the most widely used approach for peptide and protein identification in mass spectrometry-based proteomics studies. Our previous study showed that sample-specific protein databases derived from RNA-Seq data can better approximate the real protein pools in the samples and thus improve protein identification. More importantly, single nucleotide variations, short insertion and deletions and novel junctions identified from RNA-Seq data make protein database more complete and sample-specific. Here, we report an R package customProDB that enables the easy generation of customized databases from RNA-Seq data for proteomics search. This work bridges genomics and proteomics studies and facilitates cross-omics data integration.
Availability and implementation: customProDB and related documents are freely available at http://bioconductor.org/packages/2.13/bioc/html/customProDB.html.
Contact: bing.zhang@vanderbilt.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Sequence database search is the primary method for peptide and protein identification in mass spectrometry-based shotgun proteomics (Nesvizhskii, 2010). The completeness and specificity of the sequence databases directly affect the searching results. We recently showed that sample-specific databases derived from RNA-Seq data can better represent the real protein catalogs in biological samples and thus improve protein identification. In addition, sample-specific databases allow the identification of variant peptides (Wang et al., 2012).
With the advancements of both shotgun proteomics and next-generation sequencing (NGS) technologies, many researchers have started to apply both technologies to the same samples in parallel to gain a multi-dimensional understanding of cellular systems (Chen et al., 2012; Nagaraj et al., 2011). Even for proteomics studies without corresponding RNA-Seq data, it is highly likely to find sequencing data (e.g. whole-genome sequencing, exome sequencing) for similar samples. Here, we report an R package, customProDB, which is dedicated to generate customized database from NGS data, with a focus on RNA-Seq data, for proteomics search.
Based on the assumption that undetected or lowly expressed transcripts are less likely to produce detectable proteins, the package allows users to filter out proteins with undetected or lowly expressed transcripts. It also allows users to incorporate single nucleotide variations, short insertion and deletions and novel junctions identified from RNA-Seq data into the protein database.
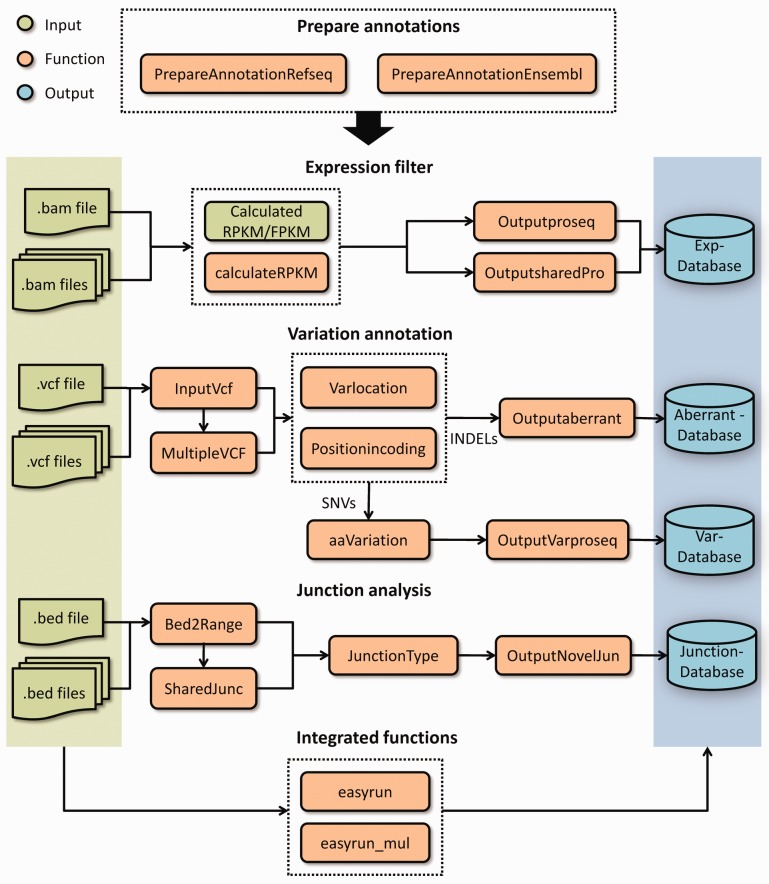
Figure 1 illustrates the overall structure of the package. Methods and functions implemented in the customProDB package are described in detail in a tutorial available as online Supplementary Material (Supplementary File 1). In section 2, we briefly present the main functionalities of the customProDB package.
Fig. 1.
Schematic overview of the customProDB package
2 DESCRIPTION
2.1 Preparing annotation files
For model organisms, customProDB allows users to download annotation data from the University of California, Santa Cruz (UCSC) table browser using rtracklayer (Lawrence et al., 2009) or from ENSEMBL using biomaRt (Durinck et al., 2009) and then process them to generate a standardized data structure. For non-model organisms, users can manually provide the annotation data in the format of UCSC or ENSEMBL.
2.2 Building customized protein databases
2.2.1 Input data
customProDB requires a Binary-sequence Alignment Format (BAM) file and a Variant Call Format (VCF) file as input for each sample of interest. The latter can be generated from a BAM file using single nucleotide polymorphism calling tools such as SAMtools and The Genome Analysis Toolkit (GATK). customProDB also accepts transcript expression estimates when available.
For junction analysis, a Browser Extensible Data (BED) file that contains putative splice junctions is needed. This file can be generated by software such as Tophat (Trapnell et al., 2009) during read alignment.
2.2.2 Expression filter
For a given BAM file, the calculateRPKM function computes the reads per kilobase per million reads sequenced (RPKM) for each transcript based on reads mapped to the exon region. Then the Outputproseq function outputs a FASTA file for proteins with an RPKM value greater than a user-defined cutoff.
For generating a consensus database from n (n > 1) related samples, a function Outputsharedpro is provided to output protein sequences for transcripts with an RPKM value greater than a user-defined cutoff in k (1 < k < n) out of the n samples.
2.2.3 Variation annotation
The InputVcf function generates a GRange (Lawrence et al., 2013) object from a VCF file, which contains variation information from one or multiple samples. The Multiple_VCF function outputs a GRange object with variations presenting in multiple samples.
For a given GRange object, the Varlocation function provides an overview of the genomic locations for all variations. Then protein level variations are identified for both single nucleotide variations and short insertion and deletions.
VCF files derived from whole-genome or exome sequencing data can also be used to generate customized databases.
2.2.4 Junction analysis
Based on an input BED file that contains splice junctions derived from RNA-Seq data, the function JunctionType classifies all junctions into different categories. Then the function OutputNovelJun can be used to generate three-frame translated peptide sequences for all putative novel junctions.
3 APPLICATION
The development of the customProDB package was mainly driven by two demands: (i) to provide a customized protein database from RNA-Seq data for a specific sample, and (ii) to provide a consensus database from a pool of genetically similar samples. Therefore, we provide two integrated functions to help accomplish these tasks in a single step (Fig. 1).
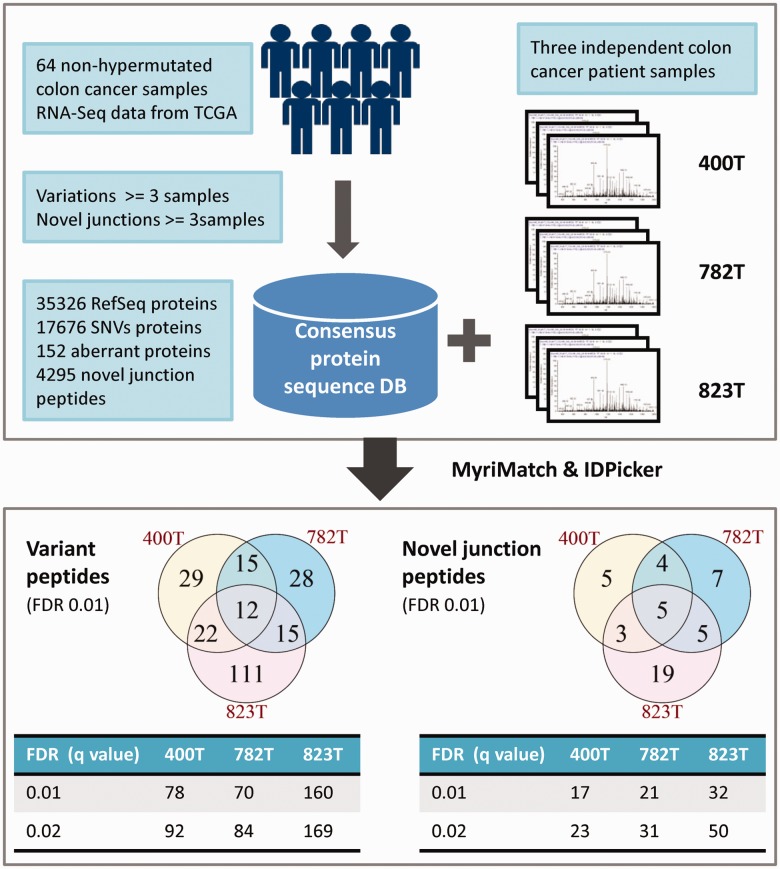
The value of customized databases for individual samples has already been demonstrated (Wang et al., 2012). Here, we provide an example of a consensus database. A consensus database was generated based on RNA-Seq data from 64 colon cancer samples from The Cancer Genome Atlas project (TCGA, 2012). Previously published proteomics data from three colon cancer patients (Li et al., 2011) were searched against the consensus database (Fig. 2). By including variation and novel junction information in the consensus database, we were able to identify variant peptides and novel junction peptides from the proteomics datasets (Supplementary File 2 and 3). We did not gain significant improvements in protein identification by applying the transcript expression threshold, possibly because of the high inter-patient heterogeneity. However, compared with the regular REfSeq database search, more peptide-spectrum matches were identified using the consensus database. This example shows the potential of using a consensus database to capture protein features shared by a cohort of samples.
Fig. 2.
Consensus protein database generation and proteomics search results for three independent colon cancer patients (400T, 782T, 823T)
4 CONCLUSION
The huge amount of genomic and transcriptomic data available from NGS experiments has enhanced and will continue to enhance shotgun proteomics studies. However, it is non-trivial for ordinary proteomics researchers to use such data directly. The customProDB package fills this gap by providing an efficient tool to generate customized protein databases using expression and variation information available from NGS data.
Funding: National Institutes of Health (grant U24 CA159988).
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Chen R, et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148:1293–1307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S, et al. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009;4:1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, et al. rtracklayer: an R package for interfacing with genome browsers. Bioinformatics. 2009;25:1841–1842. doi: 10.1093/bioinformatics/btp328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, et al. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, et al. A bioinformatics workflow for variant peptide detection in shotgun proteomics. Mol. Cell Proteomics. 2011;10:M110.006536. doi: 10.1074/mcp.M110.006536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj N, et al. Deep proteome and transcriptome mapping of a human cancer cell line. Mol. Syst. Biol. 2011;7:548. doi: 10.1038/msb.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI. A survey of computational methods and error rate estimation procedures for peptide and protein identification in shotgun proteomics. J. Proteomics. 2010;73:2092–2123. doi: 10.1016/j.jprot.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, et al. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, et al. Protein identification using customized protein sequence databases derived from RNA-Seq data. J. Proteome Res. 2012;11:1009–1017. doi: 10.1021/pr200766z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.