Abstract
Motivation: Novel tools need to be developed to help scientists analyze large amounts of available screening data with the goal to identify entry points for the development of novel chemical probes and drugs. As the largest class of drug targets, G protein-coupled receptors (GPCRs) remain of particular interest and are pursued by numerous academic and industrial research projects.
Results: We report the first GPCR ontology to facilitate integration and aggregation of GPCR-targeting drugs and demonstrate its application to classify and analyze a large subset of the PubChem database. The GPCR ontology, based on previously reported BioAssay Ontology, depicts available pharmacological, biochemical and physiological profiles of GPCRs and their ligands. The novelty of the GPCR ontology lies in the use of diverse experimental datasets linked by a model to formally define these concepts. Using a reasoning system, GPCR ontology offers potential for knowledge-based classification of individuals (such as small molecules) as a function of the data.
Availability: The GPCR ontology is available at http://www.bioassayontology.org/bao_gpcr and the National Center for Biomedical Ontologies Web site.
Contact: sschurer@med.miami.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Most drug targets are receptors expressed on the cell surface. Approximately half of those are G protein-coupled receptors (GPCRs), a class of seven transmembrane domain receptors. GPCRs remain the most pursued drug targets by academic and pharmaceutical groups (Lagerström and Schiöth, 2008). Recently, the National Institutes of Health (NIH) Molecular Libraries Program (MLP) set out to enable large-scale screening capabilities to identify new drug targets and novel chemical probes (Austin et al., 2004). GPCRs were among targets included in high-throughput screening (HTS) campaigns run in the MLP. The resulting large-scale data were deposited in the public repository PubChem. The diversity of these data has highlighted the need for standards to describe HTS datasets to simplify their integration, searching and analysis. Here, we describe an integrated GPCR ontology to aid in aggregating and classifying small molecules based on the results from many screening campaigns into relevant categories including undesirable artifacts or promiscuous molecules and sought-after selective and functionally active compounds as entry points for probe and drug development projects.
The GPCR ontology describes pharmacology, biochemistry and physiology of individual GPCRs inclusive of receptor structure, pharmacologically relevant receptor mutations, tissue distribution, oligomerization and binding affinity of GPCR ligands. Standardized formal descriptions of bioassay targets and screening results facilitate analysis (Schürer et al., 2011). Populating ontology with existing data from public repositories enables capture of GPCR-ligand data, categorization and searching the data by various terms: for example, receptor type, biological activity or chemical properties of ligands.
Publicly available repositories of GPCR-ligand interactions include PubChem (HTS and follow-up screening results from >4000 bioassays from the NIH MLP program of which ∼720 target GPCRs) (Wang et al., 2009), Psychoactive Drug Screening Program affinity values database (PDSP Ki Db, http://pdsp.med.unc.edu/), GLIDA (GPCR-ligand database) (Okuno et al., 2006), IUPHAR (repository of characterization data for GPCRs, ion channels and nuclear hormone receptors from the International Union of Basic and Clinical Pharmacology) (Sharman et al., 2011), GPCR Oligomerization Knowledge Base (GPCR-OKB, computational and experimental information on GPCR oligomerization) (Khelashvili et al., 2010) and GPCRDB (collection of GPCR sequences, ligand binding constants and mutations) (Horn et al., 2003), among others. Although the scientific community perceives these resources as useful, they are not well integrated for the purpose of analyzing HTS data in the context of various characteristics of GPCRs.
The potential of Semantic Web technologies to integrate biological data and manage knowledge has been recognized for some time (Antezana et al., 2009; Pasquier, 2008); this is particularly relevant for areas of rapidly evolving knowledge such as chemical biology and drug discovery where these technologies are now increasingly applied (Wild et al., 2012). Ontologies are formal representations of knowledge and offer distinct advantages (over relational databases) to integrate diverse data from different sources (Martinez-Cruz et al., 2012). As of September 2013, there are 390 ontologies hosted by the Open Biomedical Ontologies initiatives (126 ontologies) (Smith et al., 2007) and the Bioportal (National Center for Biomedical Ontologies, NCBO, 354 ontologies) (Rubin et al., 2006), some of which have become a standard for annotating biomedical data, for example, the Gene Ontology (GO) (Ashburner et al., 2000), which describes cell components, molecular functions and related biological processes. However, currently there is no ontology to describe GPCRs and GPCR-targeting chemical probes and drugs as outlined earlier in the text.
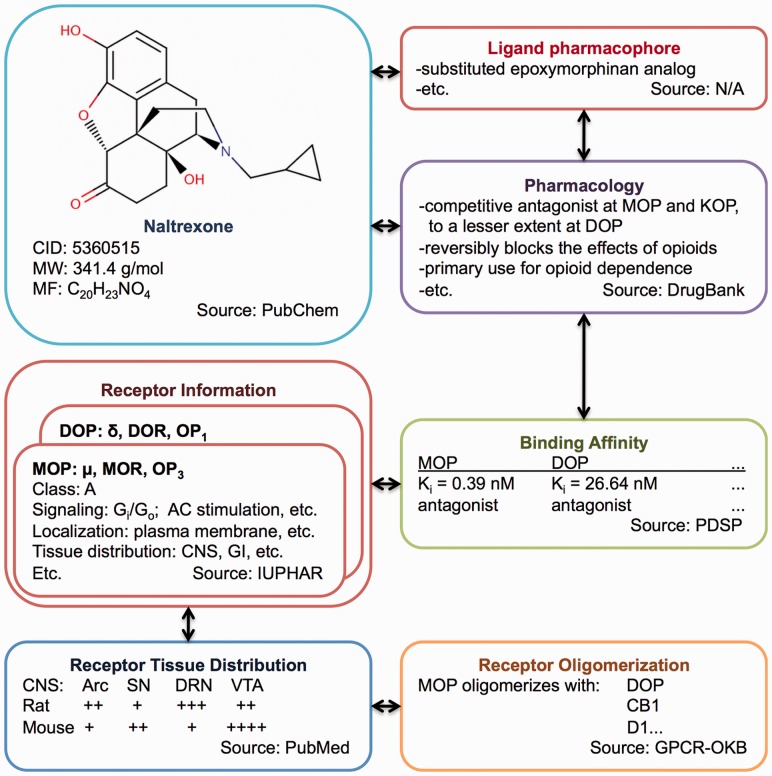
BioAssay Ontology (BAO) (http://bioassayontology.org/) was recently developed to describe and annotate HTS assays and screening results from the MLP (Vempati et al., 2012). The GPCR ontology is intended to work together with BAO and will be integrated as a BAO module. However, it can also be used independently to describe/query structure, function and pharmacology of GPCRs and their ligands. This GPCR ontology formalizes knowledge related to structural and functional characteristics of the receptors and their ligands by making use of description logic and Web Ontology Language (OWL) as recommended by the W3C (Antoniou and Harmelen, 2009). The development, scope and applications for screening data integration and analysis of the first version of an ontology on GPCRs that standardizes and merges several available resources is reported (Fig. 1).
Fig. 1.
GPCR ontology design for integration of GPCR-related information found in public repositories
2 METHODS
2.1 GPCR ontology
The GPCR ontology was developed in OWL 2.0 using the knowledge modeling environment Protégé 4.2. OWLViz and OntoGraph were used for visualization in combination with Pellet as an appropriate description logic reasoning engine. The ontology was primarily constructed at the knowledge level (domain expert-driven) and incorporated information from various public GPCR resources listed earlier in the text (data-driven) (Breitman et al., 2007). The GPCR ontology currently consists of 904 classes, 345 individuals (imported from IUPHAR), 17 object properties (relations) and 19 data properties. To incorporate existing ontologies, ensure interoperability and use ‘consistent and unambiguous formal definitions of the relations expressions’ (Smith et al., 2005), we imported core relations/object properties from the SemanticScience Integrated Ontology (SIO) (Dumontier, 2013). Likewise, relevant biological processes and molecular functions were imported from GO; definitions and gene symbols were added as annotation properties from the Protein Ontology (Natale et al., 2011) where applicable. The ontology will be maintained as a module of BAO, which is currently undergoing a major revision.
2.2 PubChem assay annotation
We iteratively annotated 407 PubChem assays (352 protein target assays, 3 process target assays, 52 summary assays) with >100 descriptor terms from BAO (Vempati et al., 2012). HTS assays that interrogate GPCRs, regulators of G protein signaling (RGS) and G proteins as assay targets were a specific focus of this project. These proteins are part of the GPCR signaling cascade and may be pharmacologically important (Tuteja, 2009). BAO annotations were captured in an Excel spreadsheet, quality checked and loaded into a triple store (Visser et al., 2011). A local mirror of the PubChem database was created to readily compute on the large screening datasets (endpoints) and to readily associate the data with the detailed assay annotations. All assay preprocessing, curation and annotation processed followed previously validated and described approaches (Schürer et al., 2011).
2.3 Calculation of compound promiscuity
To illustrate the utility of the GPCR ontology, compounds were classified based on screening activities against different GPCR target types (namely aminergic, peptidergic and lipidergic). A promiscuity index based on data in PubChem (PCIdx) was calculated for each compound in these categories. A Pipeline Pilot (Accelrys) protocol was developed to query the relational database for the purpose of identifying in how many assays each compound was tested and in how many assays it was found active. This was done for all assays with meta target: molecular target: protein target defined as follows: GPCR, G protein or RGS (352 AIDs total; PubChem Assay IDentifier - AID). To identify ‘active’ compounds, we used previously defined outcome from PubChem. PCIdx was calculated as the ratio of the number of assays in which a compound was ‘active’ and the number of assays in which it was tested. The larger the ratio of ‘active’ assays to assays tested, the higher a compound’s PCIdx. PCIdx was calculated separately for single concentration and concentration-response assays.
2.4 Data aggregation and classification of mechanism of action based on GPCR ontology and BAO classes
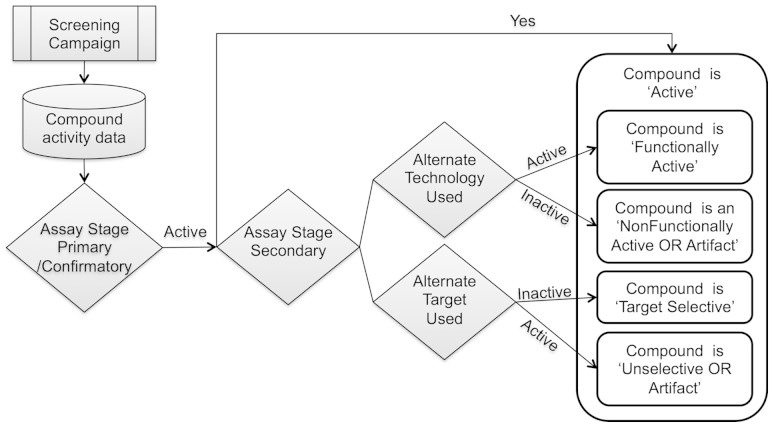
Rule-based classification protocols were developed to illustrate applicability of the GPCR ontology. BAO/GPCR ontology concepts were used to elucidate the compound mechanism of action by filtering and aggregating assay data. The aggregation was based on relationships of assays in a screening campaign, assay stage, assay measurement throughput quality and other relevant categories (Fig. 2). Screening campaign refers to a set of assays performed to identify a compound with a desired efficacy and mechanism of action. Assay stage describes the order of assays in a screening campaign used to identify and filter results. Assay measurement throughput quality categorizes the quality of the results based on tested concentration (single or concentration response) and number of replicates. Detection technology describes the physical method used to record/detect the effect caused by the perturbagen in the assay environment. Molecular target reflects a protein (or nucleic acid) modulated by a perturbagen. Filters can be adjusted based on a desired type of analysis.
Fig. 2.
Flow chart of compound categorization into functionally active, selective, unselective/promiscuous or artifact, based on annotations of assays and the ontology
Analysis was initially performed per single screening campaign with compound categories defined as follows: an ‘active’ compound was one reported efficacious in primary and confirmatory assays annotated in the class assay stage in the ontology and activity outcome ‘active/probe’ in PubChem. These compounds were classified further, namely by activity in alternate confirmatory assays (using alternate assay detection technology) and counterscreen/selectivity assays (using targets other than primary/confirmatory assay). Again, activity here was arbitrarily assigned as a PubChem outcome, but can be modified to include specific cutoff values (e.g. IC50 < 10 µM). An ‘active’ compound tested in an assay with alternate technology and found having activity was termed ‘functionally active’ (confirmed active). A compound lacking activity in this alternate confirmatory assay was classified as ‘non-functionally active OR artifact' (Fig. 2). A ‘target selective’ compound was an ‘active’ compound found to be lacking activity in a secondary assay using an alternate target; an ‘active’ compound that did have activity in an alternate target assay was termed ‘unselective OR artifact’ (also encompassing promiscuous compounds) (Fig. 2).
The ontology was applied to aggregate screening results and classify compounds based on the highest quality of results. Results were obtained and merged by substance identifier (SID) to append all data, producing a file with tags for the different activity categories. Assay measurement throughput quality was used as a measure of confidence for each category. For example compounds were tagged with a selectivity category of ‘+1’ for selective compound and ‘−1’ for unselective compound and an evidence score based on the confidence of the assay results in a screening campaign. Results presented here were based on data with highest confidence.
Next, the compound categories were aggregated across screening campaigns to generate ‘global’ compound category profiles and to identify compounds with relevant activity patterns. A Pipeline Pilot protocol was written to do the following: gather all compound category annotations, merge them across campaigns based on the same SID, compute statistics of the respective categories and their confidence and classify compounds based on these results. Specifically, global selectivity and promiscuity (described earlier in the text) scores were generated and relevant compounds were further categorized by the type of GPCR they targeted (peptidergic, lipidergic or aminergic). Using the scores generated previously, ‘global’ selective and ‘global’ promiscuous compounds were identified. Categorization parameters assured (i) that a ‘global’ selective compound was active at the same target family across various campaigns and (ii) that a ‘global’ promiscuous compound was active across multiple screening campaigns with different targets. Note: a ‘global’ promiscuous compound was defined as one active against multiple GPCR families (sphingosine-1-phosphate, S1P; opioid) not just active at receptors within the same family (muscarinic M1, muscarinic M2). This analysis yields insight into the most potent, selective and promiscuous compounds across the different GPCR families. Similarly, functionally active compounds were characterized across multiple campaigns.
3 RESULTS
3.1 Ontology construction and scope
To create the GPCR application ontology, we first carefully reviewed the currently existing ontologies in the NCBO BioPortal for information on GPCRs. Except for term definitions (National Cancer Institute Thesaurus, http://ncit.nci.nih.gov/) and placeholder for individual GPCRs [Protein Ontology (Natale et al., 2011) and BAO (Vempati et al., 2012)], no granular GPCR-focused ontology exists that encompasses the proposed features of herein described ontology.
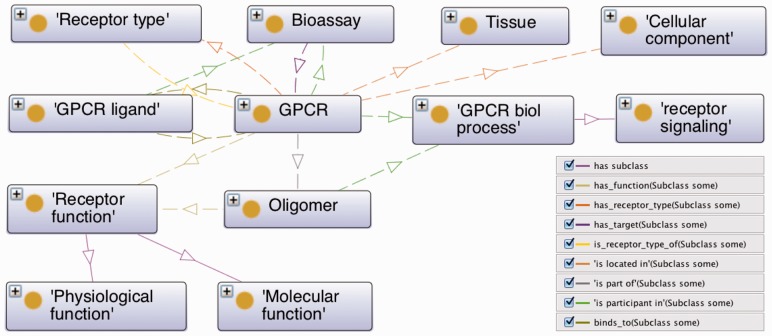
The GPCR ontology provides a framework to link important information about GPCRs and their ligands (Figs 3 and 4, Table 1) with the goal to relate receptors to small molecule and other ligands based on receptor and ligand characteristics (Supplementary Figs S1–S4). The ontology encompasses GPCR categories including receptor type, biological process, receptor signaling, receptor physiological and molecular function, GPCR ligand and others (Fig. 3). These broad categories are then thoroughly detailed by GPCR attributes captured in the ontology as placeholders (example - source), such as: class (Class A - IUPHAR), family (opioid receptor - IUPHAR), species [Homo sapiens (Hs) – IUPHAR], receptor identification: Protein ID [P35372 (Hs) – UniProt], gene ID [4988 (Hs) - Entrez Gene], taxonomy ID (9606 - NCBI Taxonomy), primary sequence (FASTA sequence string - protein sequence/NCBI), cellular component of receptor localization (integral to plasma membrane - GeneOntology/cellular component), tissue distribution (CNS - IUPHAR), physiological function (protein binding - GO/molecular function), receptor structure (4DKL - PDB). With respect to receptor signaling, the signaling pathway (primary versus secondary/IUPHAR), signaling pathway transducer (G alpha i/o family - IUPHAR), signaling pathway effector (adenylate cyclase - GO/biological process) and signal transmission (second messenger signaling - IUPHAR) are captured. With respect to oligomerization, the oligomer component (heteromer or homomer - OKB) and oligomer's physiological relevance (ligand binding - OKB) are included. Furthermore, GPCRs are classified according to the type of ligand they bind, either endogenously or most prevalently (peptidergic - IUPHAR) (Supplementary Fig. S1) (van der Horst et al., 2010).
Fig. 3.
Major classes of the GPCR ontology related to the receptor
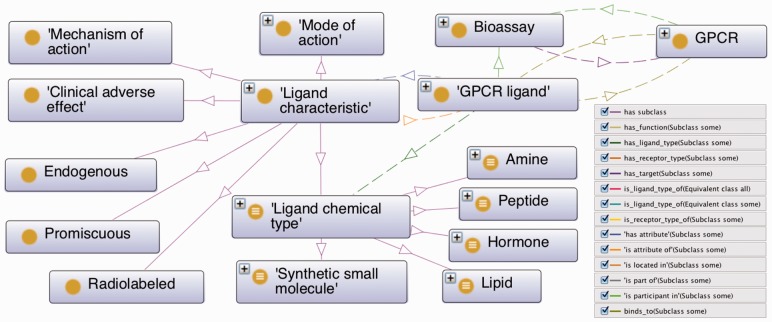
Fig. 4.
Major categories of information contained in the GPCR ontology pertaining to the ligand
Table 1.
Assay attributes captured during annotation
| Attribute class | Attribute | Example | Source DB |
|---|---|---|---|
| Receptor | Receptor identification: Gene symbol | OPRM1 (opioid receptor, mu 1) | EntrezGene |
| Receptor identification: Protein ID | P41597 | UniProt | |
| Receptor identification: Taxonomy ID | 9606 | NCBI | |
| GPCR class | A, B, C | IUPHAR | |
| GPCR name | Mu opioid receptor | IUPHAR | |
| Previous and unofficial names | µ, OP3, MOP, MOR, OPRM | IUPHAR | |
| Receptor type | Peptidergic | — | |
| Signal transmission | Protein Kinase A signaling cascade | Reactome | |
| Signaling pathway | Primary (or major) | IUPHAR | |
| Signaling pathway effector | Adenylate cyclase | IUPHAR | |
| Signaling pathway transducer | G alpha i/o | IUPHAR | |
| Cellular component | Eukaryotic plasma membrane | EntrezGene, GO | |
| Tissue | CNS (in rat, by ICH) | IUPHAR | |
| Physiological function | Addiction | GO | |
| Oligomer components | OPRM1, DRD1 | GPCR-OKB | |
| Oligomer's physiological relevance | Affects ligand binding | GPCR-OKB | |
| Receptor species | Human (Hs), Rat (R) | IUPHAR | |
| Biological Ligand | Ligand Name | Naloxone | DrugBank |
| Ligand function: Mechanism of action | Competitive opioid antagonist | DrugBank | |
| Ligand efficacy | Antagonist | IUPHAR | |
| Ligand chemical type | Amine | — | |
| Ligand endpoint type | IC50 | PubChem | |
| Ligand affinity value | 7.5 | IUPHAR | |
| Ligand affinity unit | nM | UO | |
| Ligand indication | Promiscuous | IUPHAR |
Note: CNS, central nervous system; ICH, immunohistochemistry; DRD1, dopamine receptor D1; IC50, concentration of an inhibitor that produces 50% inhibitory response; nM, nanomolar; UO, unit ontology.
The GPCR concepts (Fig. 3) are modeled in the ontology using several relations, some of which are imported from SIO (Dumontier, 2013). For example, GPCR ‘has_receptor_type’ some ‘receptor type’, ‘is_located_in’ some ‘cellular component’ and ‘is_located_in’ some ‘tissue’. GPCR ‘is_part_of’ some ‘oligomer’, ‘is_participant in’ some ‘biological process’ and ‘has_function’ some ‘receptor function’; GPCR ‘binds_to’ some ‘PCR ligand’, ‘is participant in’ some ‘bioassay’. The class ‘GPCR’ also uses data properties: ‘has gene symbol’, ‘has NCBI taxonomy ID’, ‘has protein sequence’, ‘has species’ (Fig. 3, Supplementary Fig. S1).
GPCR ligands are described by various ligand characteristics including ligand chemical type (aminergic, lipidergic, synthetic small molecule, etc.) and mode of action (agonist, antagonist, etc.). Information captured as placeholders to external resources further includes ligand endpoint (IC50 - IUPHAR/PubChem/BAO), ligand affinity value (0.25 - PubChem), ligand affinity unit (µM - PubChem/BAO), ligand indication (endogenous, promiscuous, radioactive - IUPHAR), ligand type (synthetic small molecule - DrugBank), clinical adverse effects (potential risk of heart attack - Food and Drug Administration Adverse Event Reporting System, http://www.fda.gov/) and ligand’s mechanism of action (pure opiate antagonist - DrugBank) (Fig. 4; Supplementary Fig. S2). We also plan to map terms from the Chemical Entities of Biological Interest ontology (Degtyarenko et al., 2008) to further classify GPCR ligands. Object properties connect ligand to receptors and other classes via axioms such as ‘GPCR ligand’ ‘binds_to’ some GPCR, ‘has_ligand_type’ some ‘ligand chemical type’ and ‘has_attribute’ (SIO) some ‘ligand characteristic’. Data properties include ‘has CAS registry number’, ‘has chemical formula’, ‘has InChI’, ‘has SMILES’, ‘has PubChem ID’, ‘has molecular weight’ and ‘has IUPAC name’ (Fig. 4, Supplementary Fig. S2).
As one example of linking important ligand and receptor properties, the ontology defines what (chemical) types of (endogenous) ligands each receptor type binds. These are associated with corresponding receptor types thus enabling inference (T-box reasoning) of different receptor categories as illustrated for aminergic receptors in Supplementary Figures S3 and S4.
3.2 Data integration
Although BAO describes HTS experiments and their associated results, including ligand binding and functional characteristics (such as allosteric, competitive, irreversible or agonist, antagonist efficacies, etc.), it lacks important details of GPCR binding and functional assays. The GPCR ontology was developed as an extension of BAO, but it can also be used independently to facilitate integration of different GPCR-related data sources. It allows users to interpret GPCR screening results more comprehensively with the potential to infer molecular mechanisms of action. The ontology can be obtained at http://www.bioassayontology.org/bao_gpcr and the NCBO BioPortal and will also be integrated with BAO.
3.3 GPCR bioassay activity data content
To demonstrate identification of GPCR-targeting chemical ligand space via data integration, we performed analysis on this family of receptors. GPCRs activated endogenously or by an exogenous ligand undergo a conformational transformation followed by an exchange of guanosine diphosphate to guanosine triphosphate. GPCR signaling is modulated by various effectors: G proteins, GPCR kinases and arrestins, as well as GTPase accelerating proteins of which RGS are a group. We chose assays with RGS and G protein as assay target for two reasons. First, these targets are important modulators interrelated in the GPCR signaling cascade (Huang and Tesmer, 2011). All are involved in GPCR signaling and interfering in this cascade is one way of effecting a desired pharmacological outcome. Second, there is a large number of PubChem assays targeting these proteins (GPCR: 285 AIDs, G protein: 34 AIDs, RGS: 33 AIDs). RGS are accelerators of signal suppression and have roles in cardiovascular, immune and metabolic functions (Hurst and Hooks, 2009). Drugs targeting RGS could act as potentiators of agonist action, desensitization blockers of exogenous GPCR agonists, specificity enhancers of exogenous agonists or antagonists of effector signaling via an RGS protein (Zhong and Neubig, 2001). For example, inhibitors of RGS8 (or RGS7 or RGS4) could serve as novel analgesics or analgesic potentiators (Zhong and Neubig, 2001). Similarly, a drug affecting the action of RGS complex, RGS-RhoGEF, (a RGS domain coupled to guanine nucleotide exchange factor (GEF) that activates Rho protein), could potentially block the effects of S1P on cancer initiation and tumor vascular maturation (Hurst and Hooks, 2009).
The level of detail captured in assay annotations is exemplified in Table 1 and Supplementary Table S1. The annotated assays related to G protein, RGSs and GPCRs were further classified and counted according to various BAO classes such as protein target, detection technology, measured entity, endpoint and mechanism of action (Supplementary Tables S2–S6). Briefly, protein target refers to the specific protein whose perturbation by interaction with a screened compound is measured in the assay. Measured entity is a molecular entity that is the output of a biological reaction or process and that is detected either directly (by the presence of a tag or probe) or indirectly in a coupled reaction. Endpoint is the final experimental results quantifying or qualifying a given perturbation (e.g. IC50, percent inhibition) (Vempati et al., 2012). Mechanism of action reflects the effect of the compound on the target in an experiment.
3.4 NIH MLPCN probes for the GPCR/RGS receptor family targets
The main goal of the NIH MLP project was ‘to identify chemical probes to study the functions of genes, cells and biochemical pathways’(Austin et al., 2004). As of December 10, 2012, 193 probes were identified from various screening campaigns. Of these, 49 probes targeted GPCR, RGS or G proteins (Supplementary Table S7, http://mli.nih.gov/mli/mlp-probes-2/).
3.5 Data Analysis using GPCR ontology
3.5.1 GPCR ligand types and promiscuity
Using the BAO/GPCR ontology class-based assay annotations, the assays were first categorized by GPCR type (aminergic, lipidergic or peptidergic) followed by computing promiscuity statistics for each compound. The analysis was performed separately for single concentration and concentration-response data. The promiscuity index is listed in Table 2 for the topmost promiscuous compounds in each category (Note: most of these compounds also show activity at non-GPCR targets).
Table 2.
Example of compounds tested in >3 assays for each GPCR category and their promiscuity index
| Single concentration |
Concentration response |
|||||
|---|---|---|---|---|---|---|
| GPCR Type | SID | Number of active| Number of tested | PCIdx | SID | Number of active| Number of tested | PCIdx |
| Peptidergic | 24820674 | 5|7 | 0.714 | 14729216 | 9|10 | 0.900 |
| 47197930 | 5|8 | 0.625 | 14729238 | 5|6 | 0.833 | |
| 47199350 | 5|8 | 0.625 | 26724400 | 5|6 | 0.833 | |
| 49674233 | 5|8 | 0.625 | 26725675 | 4|5 | 0.800 | |
| 49675583 | 5|8 | 0.625 | 24823007 | 4|5 | 0.800 | |
| Lipidergic | 861090 | 3|8 | 0.375 | 855676 | 2|4 | 0.500 |
| 856781 | 3|10 | 0.300 | 851570 | 2|4 | 0.500 | |
| 851570 | 3|11 | 0.273 | — | — | — | |
| Aminergic | 842134 | 2|6 | 0.333 | 842366 | 2|3 | 0.667 |
| 842151 | 2|6 | 0.333 | 842234 | 2|5 | 0.400 | |
| 842453 | 2|6 | 0.333 | 842231 | 2|5 | 0.400 | |
| 842552 | 2|6 | 0.333 | — | — | — | |
| 842383 | 2|7 | 0.286 | — | — | — | |
For example, in the peptidergic GPCR-binding group of compounds, SID 14729216 with PCIdx of 0.9, is a methylbenzylsulfinyl-substituted pyridine that binds to various receptors (S1P2R, NPSR1, S1P4R or M1R) as an antagonist. The functional groups of this compound fit non-specifically into variety of GPCR binding sites. Overall, there were 931 data points for aminergic GPCR type with PCIdx ≥ 0.1, and 9873 and 8981 data points for peptidergic and lipidergic GPCR type with PCIdx ≥ 0.1, respectively. Supplementary Figure S5 depicts complete dataset of compound promiscuity per GPCR type. (A receptor abbreviation list is available in the Supplementary Table S2 legend).
3.5.2 Analysis of individual single screening campaigns
A rule-based aggregation of GPCR ligand activity data was developed and implemented to identify various categories of active compounds within each screening campaign (see flow chart Fig. 2). Overall, there were 73 different screening campaigns that targeted GPCR, G proteins or RGS (Supplementary Table S8); some campaigns were merged during analysis due to assay overlap (e.g. FPR1 and FPR2 screening campaigns included assays that were counter screens for each respective target) and some were omitted due to lack of annotated data. For each of these campaigns, several analyses were performed to classify compounds into the following categories: ‘active’, ‘functionally active’, ‘non-functionally active OR artifact’, ‘target selective’ and ‘unselective OR artifact’ (Fig. 2). Examples of identified compounds for different categories are listed in Table 3.
Table 3.
Examples of compounds identified from individual screening campaigns using the analysis protocol illustrated in Figure 2
| Classification | SID | Target symbol, (species) | SID efficacy | Potency (µM) |
|---|---|---|---|---|
| Functionally Active | 856021 | 5-HT(1A), (Hs) | Agonist | EC50 = 0.018 |
| 22409370 | S1P4R, (Hs) | Antagonist | IC50 = 0.168 | |
| 85285486 | M5R, (Hs) | Allosteric modulator | EC50 = 1.161 | |
| Target Selective (*, probe) | 24428139* | FPR1, (Hs) | Antagonist | Ki = 0.3 |
| 22413249* | NPY-Y2R, (Hs) | Antagonist | IC50 = 3.917 | |
| 4258673* | S1P1R, (Hs) | Agonist | AC50 = 0.207 | |
| Unselective OR Artifact | 17415263 | M1R, (Rn) | Antagonist | AC50 = 3.9811 |
| 85200865 | AVPR1A, (Hs) | Agonist | EC50 = 2.325 | |
| 11532956 | RXFP2, (Hs) | Agonist | AC50 = 0.9742 |
Note: Hs, Homo sapiens; Rn, Rattus norvegicus; Ki, absolute inhibition constant; AC50, potency, concentration at which compound exhibits half maximal efficacy; EC50, half maximal effective concentration.
Interestingly, some of the SIDs identified in our analysis are also confirmed MLP probes (asterisk, probe; Table 3) (Supplementary Table S7). Examples of ‘functionally active’ compounds include the 5-HT(1A) agonist (SID 856021), S1P4R antagonist (SID 22409370) or M5R allosteric ligand (SID 85285486), all with high potencies (Table 3; compound structures in Supplementary Table S9). These compounds were confirmed to be active in secondary assays with alternate technology (e.g. immunoassay versus calcium redistribution assay). Compounds inactive in assays featuring the same target assessed with different technology were most likely artifacts. To address selectivity, compounds were screened in assays featuring various homologous and non-homologous targets. Those inactive in such assays were deemed ‘target selective’ and examples of those include screening campaign probes: FPR1 antagonist (SID 24428139), NPY-Y2R Antagonist (SID 22413249) and S1P1R Agonist (SID 4258673). ‘Unselective OR artifact’ category of compounds includes compounds active in assays with targets other than that of primary interest. Examples of these include compounds screened in the following campaigns: M1R Antagonist (SID 17415263), AVPR1A Agonist (SID 85200865) and RXFP2 Agonist (SID 11532956). Detailed screening information pertaining to a target-selective compound identified within a screening campaign is given in Table 4. SID 24428139 has been screened in multiple assays in the ‘Formyl Peptide Receptor Antagonists’ campaign. It has been tested against FPR1 and FPR2 in five different assays [primary (AID 722), confirmatory (AID 724), counter (AIDs 723 and 725) and alternate confirmatory (AID 863)]. It has been found ‘active’ at FPR1 and ‘inactive’ at FPR2 and was determined by the assay provider to be a probe for this screening campaign. Formyl peptide receptors are GPCRs involved in mediating immune response to infection by acting as potent chemoattractants for neutrophils (Dufton and Perretti, 2010).
Table 4.
Example of a target-selective compound, SID 24428139
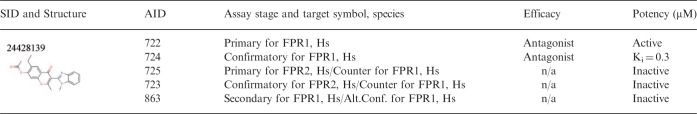 |
Note: Alt.Conf., alternate confirmatory
3.5.3 GPCR ligand analysis across screening campaigns
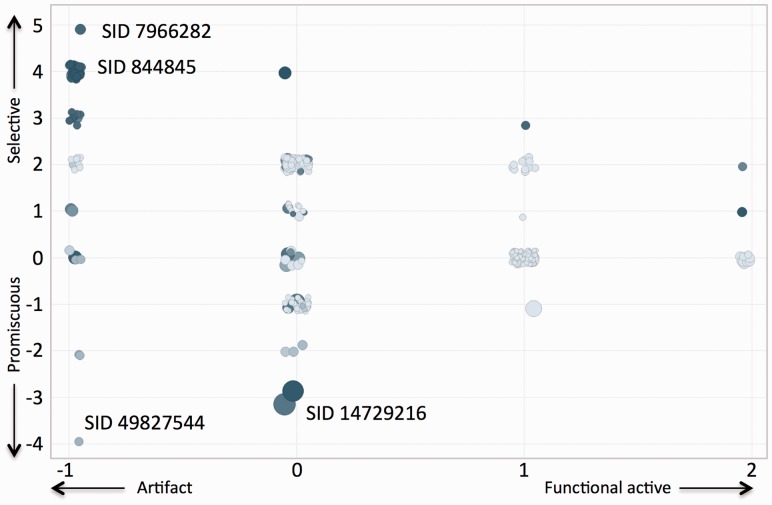
The GPCR ontology was leveraged in combination with BAO to aggregate and categorize compounds screened across multiple campaigns. Activity categories corresponding to individual screening campaign outcomes were aggregated and analyzed by SID as described in Methods to generate ‘global’ compound profiles. The ‘global’ activity map (Fig. 5) illustrates the different cross-campaign profiles for compounds that were identified as selective or promiscuous in at least one campaign.
Fig. 5.
Cross-campaign activity category map of tested compounds. ‘Global’ aggregated scores are shown for the different categories reflecting selectivity and functional activity. Promiscuity is the negative selectivity score, size indicates the aggregate confidence score and color corresponds to the number of screening campaigns (increasing from light to dark). Representative structures and data are in Supplementary Table S12
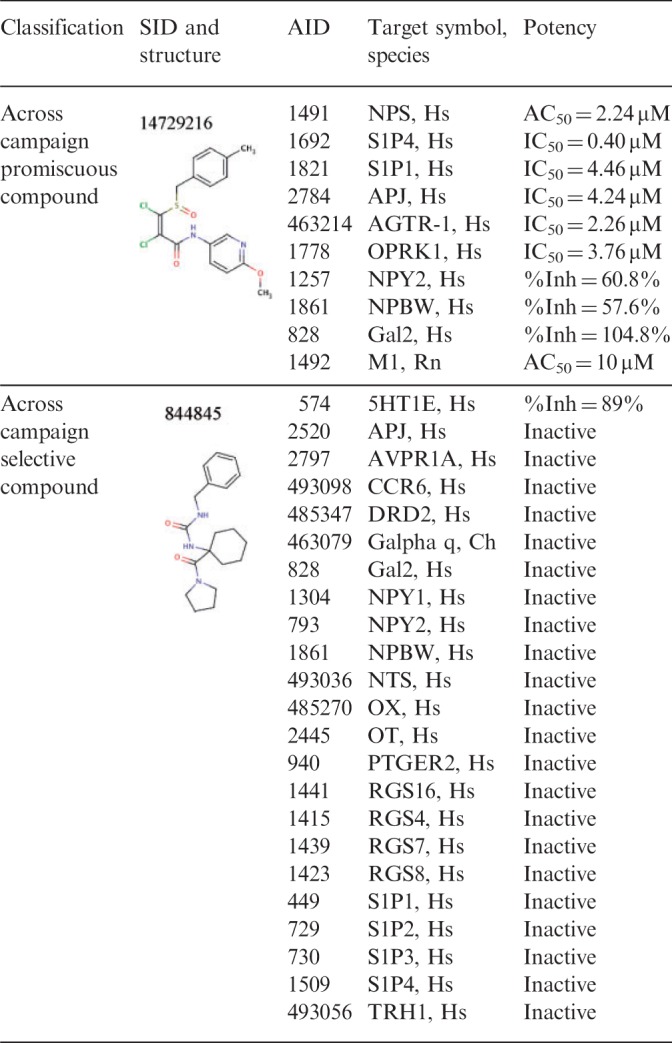
Supplementary Tables S10 and S11 show full details of the most pronounced compounds corresponding to each category. SID 7966282 was identified as the most selective compound (top left in Fig. 5). It was tested in 28 campaigns and 42 assays, rendering it the most selective compound. This compound was active at 5-HT1E receptors and inactive at a number of different GPCRs of all types (aminergic: 5HT1A, DRD2; peptidergic: NTSR, NPY1R, NPY2R, OT, FPR2, NPBWR1, Gal2, APJ; lipidergic: S1P1R, S1P3R, S1P4R, PTGER2, among others). This compound scores in the ‘artifact’ range because it does not have alternate confirmatory assays except for one in which it was found inactive (AID 485270). Likewise, SID 844845 is a high-quality selective compound (top left in Fig. 5). It was tested in 28 campaigns, 43 assays and found active against 1 family of GPCRs: the 5HT1E. It was inactive at peptidergic GPCRs (NTSR, NPY1R, NPY2R, OT, OX, APJ, CCR6 and NPBWR1) as well as lipidergic (S1P3R, S1P4R) and hormonergic (TRH1) GPCRs (Table 5).
Table 5.
Compound activity profile of selected compounds, based on the GPCR ontology
 |
Note: Hs, Homo sapiens; Rn, Rattus norvegicus; Ch, Chinese hamster.
SID 49827544 was identified as the most promiscuous compound (bottom left in Fig. 5). It was tested in 12 campaigns, 20 assays and was found active in 15 assays including alternate target assays, but inactive in alternate technology assays rendering it an artifact; the compound, however, has a low confidence score as inferred from the quality tags of secondary assays (see Methods).
An example of a promiscuous compound with high confidence score is SID 14729216. This compound was tested in 28 campaigns, 48 assays and has activity at numerous GPCR families (large circle, lower left quadrant in Fig. 5). It has inhibitory activity at lipidergic (S1P1R, S1P4R, PTGER2), aminergic (M1) and peptidergic (NPSR, Gal2, NPBWR1, APJ, opioid, AGTR) GPCRs (Table 5 and Supplementary Table S12).
It should be noted again that herein reported results are inclusive of data pertaining only to GPCRs, RGS and G protein targets. Some compounds have reported activity outside of this group of receptors. For example, SID 14729216, the promiscuous compound across GPCR screening campaigns, is also active at other non-GPCR targets [e.g. heat shock protein 70 (Hsp70), E3 Ligase, etc.]. BAOSearch (http://baosearch.ccs.miami.edu/) enables a full view of each compound’s activity.
4 DISCUSSION
GPCRs continue to remain a ‘hot’ target of research as new structural information, signal processing pathways and large-scale compound screening results become validated and available (Manglik et al., 2012; Granier et al., 2012; Thompson et al., 2012; Wojciak et al., 2009; Wu et al., 2012). Acquired data come with challenges of integration and analysis to derive new knowledge. Here, we report GPCR ontology to facilitate integration of various GPCR resources. The GPCR ontology includes relevant classes and relations to characterize and link receptors and compounds that are known binders or show activity on GPCR-related assays. To more effectively disseminate biological data on GPCRs with the goal to drive innovation involving biologists, chemists and bioinformaticians requires a flexible framework to integrate GPCR data from various resources.
To enable identification of novel GPCR probe and drug lead candidates via more efficient data integration and analysis, we first researched and then aggregated available and relevant GPCR pharmacological data sources. GPCR target and ligand features are captured in the ontology to enable more versatile and integrative views on GPCR function. For example, classifying different ligand types based on chemical structure/composition (amine, peptide and lipid) as well as different receptor types (aminergic, peptidergic and lipidergic) can guide compound library design to maximize structure–activity relationship (SAR) studies; we have illustrated how the major receptor categories can be inferred. Ligands are also classified based on their efficacy at a particular GPCR (full agonist, allosteric modulator, etc.) yielding insight into selective agonism issues that are becoming more apparent in GPCR pharmacology (Gesty-Palmer and Luttrell, 2011). In addition, ligands are grouped based on their indication (endogenous, promiscuous and radioactive) and the type of the receptor they interact with. For example, ligand S1P is classified as a full agonist, endogenous, lipid compound targeting the class A, lipidergic, lysophospholipid family of S1P receptors.
To illustrate use of the GPCR ontology the activity of screened compounds was categorized based on assay details (stage, technology, detection, alternate targets, etc.) and receptor information (type, class, etc.). The query to identify how often a compound has been screened against a given type of GPCR-facilitated calculation of receptor type-specific promiscuity. For example, SID 14729216 was tested in 10 assays targeting peptidergic GPCRs and was ‘active’ in 9 of them (PCIdx = 0.9). Categorization of compound data into various activity groups (‘functionally active’, ‘target selective’, ‘non-functionally active’ and ‘unselective’) across a single campaign illustrates how the GPCR ontology can be applied. The process included sorting/tagging the data based on confidence of each data point and using only the highest quality results. Analysis was performed within and across screening campaigns to identify the most interesting and relevant compounds according to several categories. The identification of known MLP probes, in addition to many other compounds that are potential probe or drug development candidates, validates our approach (‘target selective’ category) (Table 3). Compounds not tested in confirmatory assays were filtered out in the analysis pipeline ensuring only high quality likely replicable outcomes were used for the cross campaign categorization. Relaxing the confidence score may thus identify many additional potentially relevant compounds.
Promiscuity calculations and compound categories were performed using a rule-based system, in contrast to reasoning, because we wanted to report statistics and consider confidence of assay results, but also because of the difficulty to reason across large datasets.
The GPCR ontology enables additional levels of analytical complexity, e.g. tissue expression. Linking receptor and tissue expression pattern is another example (among many others) of how the ontology adds value by enabling integration of data sources; this would allow inferring if a compound acts at a receptor in the brain versus another (peripheral) tissue, thus enabling quick identification of potential brain penetrable properties. Analyses as described earlier in the text can be performed for any concept/class in the GPCR ontology.
5 CONCLUSIONS
We present the first GPCR ontology framework to describe functional, structural and physiological aspects of GPCRs and pharmacological and other characteristics of related small molecule acting on the receptors. This GPCR ontology facilitates integration of different resources and provides a new framework to maximize the value of the HTS datasets by bridging the gap between the overflow of HTS data and the bottleneck of integrated analysis. We will make these data available via the BAOSearch (Abeyruwan et al., 2010) software application that facilitates search and exploration of screening data based on various categories from the BAO and chemical structures.
Semantic Web technologies show great promise to link diverse biological data as demonstrated, for example, by the Bio2RDF project (Belleau et al., 2008). Our hope is that this ontology will facilitate incorporating mentioned GPCR resources and in particular the actual screening results into linked data projects with the goal to develop better tools for drug development projects by more effectively ‘repurposing’ already generated datasets.
A future goal is to expand the semantic model to facilitate the integration of screening data with biological pathways, disease networks and structural biology with the goal to analyze screening results in the context of molecular mechanisms of action. This approach offers the potential to generate new knowledge by inference (using a reasoning engine). The development of additional ontologies modules is currently underway.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge resources of the Center for Computational Science at the University of Miami. They thank Hande Küçük, Saminda Abeyruwan and Ubbo Visser for discussions related to the ontology and Caty Chung for publishing the ontology.
Funding: NIH (grant number NIH RC2 HG005668 and RC2 HG005668-02S1).
Conflict of Interest: None declared.
REFERENCES
- Abeyruwan S, et al. Semantic Web Challenge, 9th International Semantic Web Conference (ISWC) Shanghai, China: 2010. BAOSearch: a semantic web application for biological screening and drug discovery research. http://challenge.semanticweb.org/submissions/swc2010_submission_20.pdf (15 October 2013, date last accessed) [Google Scholar]
- Antezana E, et al. Biological knowledge management: the emerging role of the Semantic Web technologies. Brief. Bioinformatics. 2009;10:392–407. doi: 10.1093/bib/bbp024. [DOI] [PubMed] [Google Scholar]
- Antoniou G, Harmelen FV. Web Ontology Language: OWL. In: Staab S, Studer R, editors. Handbook on Ontologies. Berlin Heidelberg: Springer-Verlag; 2009. pp. 91–110. [Google Scholar]
- Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin CP, et al. NIH molecular libraries initiative. Science. 2004;306:1138–1139. doi: 10.1126/science.1105511. [DOI] [PubMed] [Google Scholar]
- Belleau F, et al. Bio2RDF: towards a mashup to build bioinformatics knowledge systems. J. Biomed Inform. 2008;41:706–716. doi: 10.1016/j.jbi.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Breitman K, et al. Semantic Web: Concepts, Technologies and Applications. London: Springer London; 2007. Methods for Ontology Development; pp. 155–173. [Google Scholar]
- Degtyarenko K, et al. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res. 2008;36:D344–D350. doi: 10.1093/nar/gkm791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufton N, Perretti M. Therapeutic anti-inflammatory potential of formyl-peptide receptor agonists. Pharmacol. Ther. 2010;127:175–188. doi: 10.1016/j.pharmthera.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Dumontier M. SemanticScience Integrated Ontology. 2013 doi: 10.1186/2041-1480-5-14. http://semanticscience.org (15 October 2013, date last accessed) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesty-Palmer D, Luttrell LM. Refining efficacy: exploiting functional selectivity for drug discovery. Adv. Pharmacol. 2011;62:79–107. doi: 10.1016/B978-0-12-385952-5.00009-9. [DOI] [PubMed] [Google Scholar]
- Granier S, et al. Structure of the δ-opioid receptor bound to naltrindole. Nature. 2012;485:400–404. doi: 10.1038/nature11111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn F, et al. GPCRDB information system for G protein-coupled receptors. Nucleic Acids Res. 2003;31:294–297. doi: 10.1093/nar/gkg103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Horst E, et al. A novel chemogenomics analysis of G protein-coupled receptors (GPCRs) and their ligands: a potential strategy for receptor de-orphanization. BMC Bioinformatics. 2010;11:316. doi: 10.1186/1471-2105-11-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CC, Tesmer JG. Recognition in the face of diversity: interactions of heterotrimeric G proteins and G protein-coupled receptor (GPCR) kinases with activated GPCRs. J. Biol. Chem. 2011;286:7715–7721. doi: 10.1074/jbc.R109.051847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst JH, Hooks SB. Regulator of G-protein signaling (RGS) proteins in cancer biology. Biochem. Pharmacol. 2009;78:1289–1297. doi: 10.1016/j.bcp.2009.06.028. [DOI] [PubMed] [Google Scholar]
- Khelashvili G, et al. GPCR-OKB: the G protein coupled receptor oligomer knowledge base. Bioinformatics. 2010;26:1804–1805. doi: 10.1093/bioinformatics/btq264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagerström MC, Schiöth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nature. Rev. Drug Discov. 2008;7:339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- Manglik A, et al. Crystal structure of the µ-opioid receptor bound to a morphinan antagonist. Nature. 2012;485:321–326. doi: 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Cruz C, et al. Ontologies versus relational databases: are they so different? A comparison. Artif. Intell. Rev. 2012;38:271–290. [Google Scholar]
- Natale DA, et al. The Protein Ontology: a structured representation of protein forms and complexes. Nucleic Acids Res. 2011;39:D539–D545. doi: 10.1093/nar/gkq907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno Y, et al. GLIDA: GPCR-ligand database for chemical genomic drug discovery. Nucleic Acids Res. 2006;34:D673–D677. doi: 10.1093/nar/gkj028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquier C. Biological data integration using Semantic Web technologies. Biochimie. 2008;90:584–594. doi: 10.1016/j.biochi.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Rubin, et al. National Center for Biomedical Ontology: advancing biomedicine through structured organization of scientific knowledge. Omics. 2006;10:185–198. doi: 10.1089/omi.2006.10.185. [DOI] [PubMed] [Google Scholar]
- Schürer, et al. BioAssay ontology annotations facilitate cross-analysis of diverse high-throughput screening data sets. J. Biomol. Screening. 2011;16:415–426. doi: 10.1177/1087057111400191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharman JL, et al. IUPHAR-DB: new receptors and tools for easy searching and visualization of pharmacological data. Nucleic Acids Res. 2011;39:D534–D538. doi: 10.1093/nar/gkq1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B, et al. Relations in biomedical ontologies. Genome Biol. 2005;6:R46. doi: 10.1186/gb-2005-6-5-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat. Biotechnol. 2007;25:1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AA, et al. Structure of the nociceptin/orphanin FQ receptor in complex with a peptide mimetic. Nature. 2012;485:395–399. doi: 10.1038/nature11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja N. Signaling through G protein coupled receptors. Plant Signal. Behav. 2009;4:942–947. doi: 10.4161/psb.4.10.9530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vempati UD, et al. Formalization, annotation and analysis of diverse drug and probe screening assay datasets using the BioAssay Ontology (BAO) PLoS One. 2012;7:e49198. doi: 10.1371/journal.pone.0049198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser U, et al. BioAssay Ontology (BAO): a semantic description of bioassays and high-throughput screening results. BMC Bioinformatics. 2011;12:257. doi: 10.1186/1471-2105-12-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, et al. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009;37:W623–W633. doi: 10.1093/nar/gkp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild D, et al. Systems chemical biology and the Semantic Web: what they mean for the future of drug discovery research. Drug Discov. Today. 2012;17:469–474. doi: 10.1016/j.drudis.2011.12.019. [DOI] [PubMed] [Google Scholar]
- Wojciak JM, et al. The crystal structure of sphingosine-1-phosphate in complex with a Fab fragment reveals metal bridging of an antibody and its antigen. Proc. Natl Acad. Sci. USA. 2009;106:17717–17722. doi: 10.1073/pnas.0906153106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, et al. Structure of the human κ-opioid receptor in complex with JDTic. Nature. 2012;485:327–332. doi: 10.1038/nature10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, Neubig RR. Regulator of G protein signaling proteins: novel multifunctional drug targets. J. Pharmacol. Exp. Ther. 2001;297:837–845. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.