Abstract
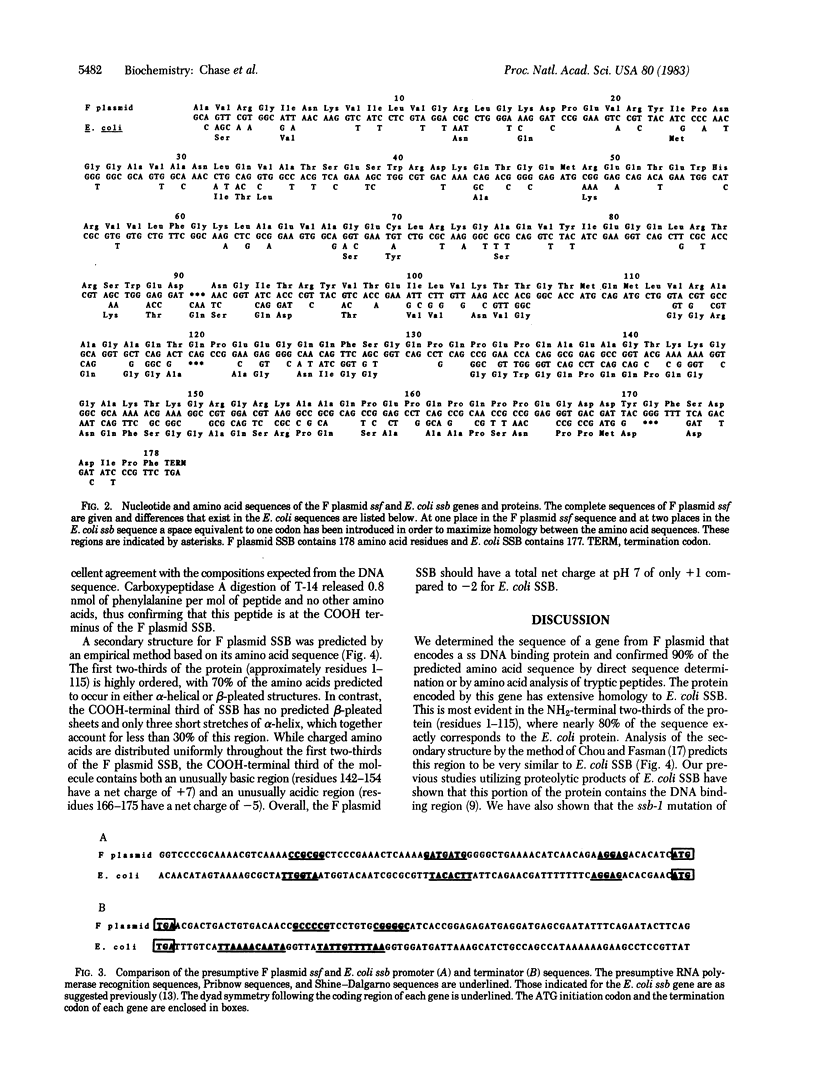
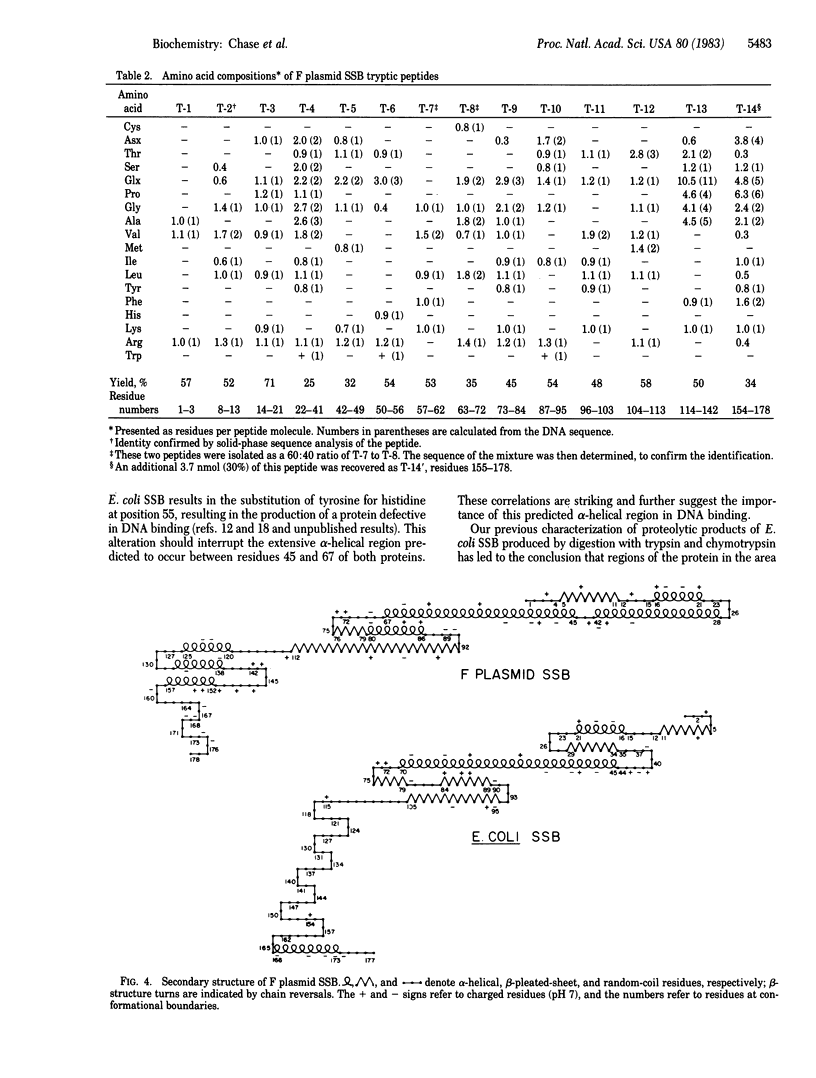
We have determined the sequence of the gene encoding a single-stranded DNA (ss DNA) binding protein (SSB) from the Escherichia coli F sex factor and the amino acid sequence of the protein it encodes. The protein has extensive homology with E. coli SSB, particularly within its NH2-terminal region, where 87 of the first 115 amino acid residues are identical to those of the E. coli protein. We have previously shown that this portion of E. coli SSB contains the DNA binding region. The sequences diverge extensively in their COOH-terminal regions, although small areas of homology exist in several places. Six of the last seven amino acid residues of the two proteins are identical, which may have implications in terms of the direct interactions of these proteins with other proteins required for DNA replication, recombination, and repair. The coding region of the F plasmid ssf gene is 537 base pairs. The protein encoded by the gene contains 178 amino acids (one more than E. coli SSB) and has a calculated molecular weight of 19,505. Other than the presumptive Shine-Dalgarno sequence, the promoter and terminator regions of both genes are not similar. The most significant feature in this regard may be the lack of a region of dyad symmetry within the presumptive promoter of the F plasmid ssf gene as is found in the region of the presumptive E. coli ssb promoter. In this report the predicted secondary structures of both the F plasmid and E. coli SSB proteins are compared and the evolutionary significance of their sequence and structural similarities to the functional domains of the proteins are discussed.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chase J. W., Murphy J. B., Whittier R. F., Lorensen E., Sninsky J. J. Amplification of ssb-1 mutant single-stranded DNA-binding protein in Escherichia coli. J Mol Biol. 1983 Feb 25;164(2):193–211. doi: 10.1016/0022-2836(83)90075-x. [DOI] [PubMed] [Google Scholar]
- Chase J. W., Whittier R. F., Auerbach J., Sancar A., Rupp W. D. Amplification of single-strand DNA binding protein in Escherichia coli. Nucleic Acids Res. 1980 Jul 25;8(14):3215–3227. doi: 10.1093/nar/8.14.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Coleman J. E., Oakley J. L. Physical chemical studies of the structure and function of DNA binding (helix-destabilizing) proteins. CRC Crit Rev Biochem. 1980 Jan;7(3):247–289. doi: 10.3109/10409238009105463. [DOI] [PubMed] [Google Scholar]
- Glassberg J., Meyer R. R., Kornberg A. Mutant single-strand binding protein of Escherichia coli: genetic and physiological characterization. J Bacteriol. 1979 Oct;140(1):14–19. doi: 10.1128/jb.140.1.14-19.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub E. I., Low K. B. Indirect stimulation of genetic recombination. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1401–1405. doi: 10.1073/pnas.80.5.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C. J., Walker G. C. Expression of the E. coli uvrA gene is inducible. Nature. 1981 Feb 26;289(5800):808–810. doi: 10.1038/289808a0. [DOI] [PubMed] [Google Scholar]
- Kolodkin A. L., Capage M. A., Golub E. I., Low K. B. F sex factor of Escherichia coli K-12 codes for a single-stranded DNA binding protein. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4422–4426. doi: 10.1073/pnas.80.14.4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman H. B., Witkin E. M. DNA degradation, UV sensitivity and SOS-mediated mutagenesis in strains of Escherichia coli deficient in single-strand DNA binding protein: effects of mutations and treatments that alter levels of Exonuclease V or recA protein. Mol Gen Genet. 1983;190(1):92–100. doi: 10.1007/BF00330329. [DOI] [PubMed] [Google Scholar]
- Lovett M. A., Helinski D. R. Method for the isolation of the replication region of a bacterial replicon: construction of a mini-F'kn plasmid. J Bacteriol. 1976 Aug;127(2):982–987. doi: 10.1128/jb.127.2.982-987.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Meyer R. R., Glassberg J., Kornberg A. An Escherichia coli mutant defective in single-strand binding protein is defective in DNA replication. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1702–1705. doi: 10.1073/pnas.76.4.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Salles B., Paoletti C., Villani G. Lack of single-strand DNA-binding protein amplification under conditions of SOS induction in E. coli. Mol Gen Genet. 1983;189(1):175–177. doi: 10.1007/BF00326074. [DOI] [PubMed] [Google Scholar]
- Sancar A., Williams K. R., Chase J. W., Rupp W. D. Sequences of the ssb gene and protein. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4274–4278. doi: 10.1073/pnas.78.7.4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmis K., Cabello F., Cohen S. N. Cloning, isolation, and characterization of replication regions of complex plasmid genomes. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2242–2246. doi: 10.1073/pnas.72.6.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittier R. F., Chase J. W. A filter assay specific to Eco helix-destablizing Protein I in crude extracts. Anal Biochem. 1980 Jul 15;106(1):99–108. doi: 10.1016/0003-2697(80)90123-2. [DOI] [PubMed] [Google Scholar]
- Whittier R. F., Chase J. W. DNA repair properties of Escherichia coli tif-1, recAo281 and lexA1 strains deficient in single-strand DNA binding protein. Mol Gen Genet. 1983;190(1):101–111. doi: 10.1007/BF00330330. [DOI] [PubMed] [Google Scholar]
- Williams K. R., LoPresti M. B., Setoguchi M., Konigsberg W. H. Amino acid sequence of the T4 DNA helix-destabilizing protein. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4614–4617. doi: 10.1073/pnas.77.8.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K. R., Spicer E. K., LoPresti M. B., Guggenheimer R. A., Chase J. W. Limited proteolysis studies on the Escherichia coli single-stranded DNA binding protein. Evidence for a functionally homologous domain in both the Escherichia coli and T4 DNA binding proteins. J Biol Chem. 1983 Mar 10;258(5):3346–3355. [PubMed] [Google Scholar]



