Abstract
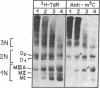
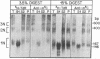
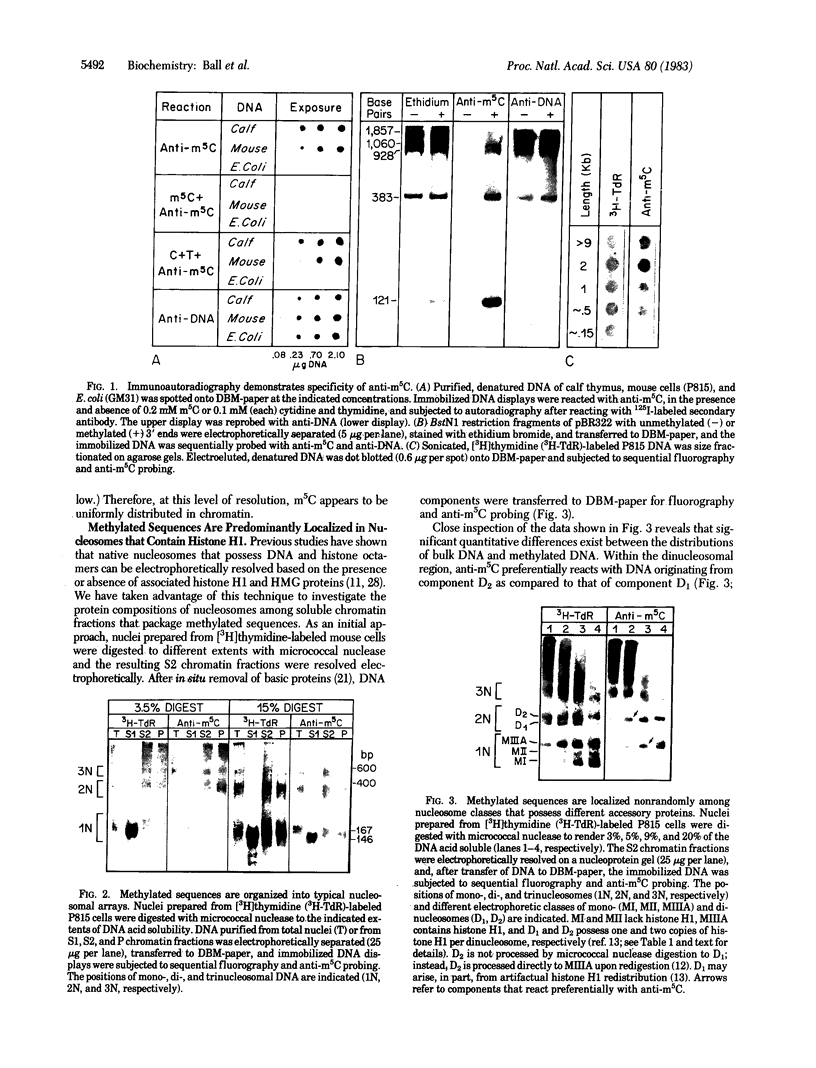
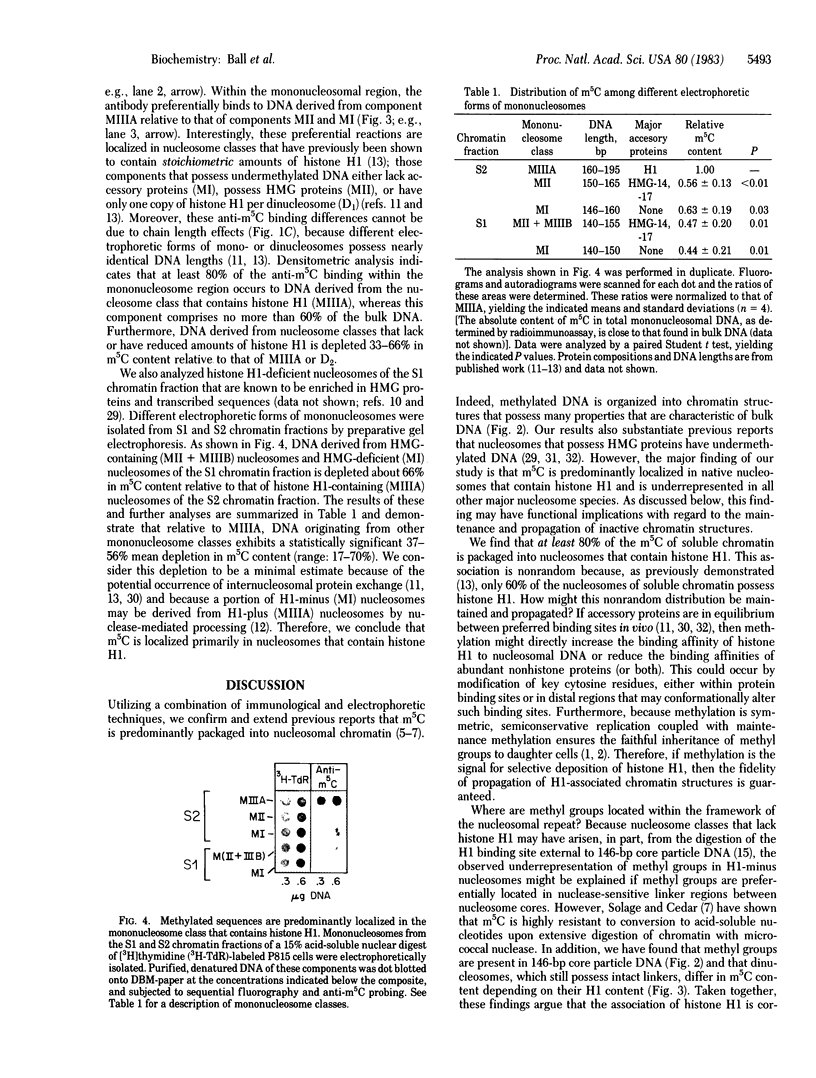
We have developed a procedure for purifying highly specific polyclonal antibodies against 5-methylcytosine. These antibodies were used to probe the distribution of 5-methylcytosine among fractionated nucleosomes of mouse cell chromatin. Our results demonstrate that at least 80% of the 5-methylcytosine is localized in nucleosomes that contain histone H1. Native nucleosomes that lack histone H1 or possess high mobility group proteins package DNA that is 1.6- to 2.3-fold undermethylated. We suggest that the preferential association of methylated sequences with histone H1 has functional significance because DNA methylation has been linked to gene inactivation and histone H1 is known to promote chromatin condensation.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams R. L., Burdon R. H. DNA methylation in eukaryotes. CRC Crit Rev Biochem. 1982;13(4):349–384. doi: 10.3109/10409238209108714. [DOI] [PubMed] [Google Scholar]
- Adams R. L., McKay E. L., Douglas J. T., Burdon R. H. Methylation of nucleosomal and nuclease sensitive DNA. Nucleic Acids Res. 1977 Sep;4(9):3097–3108. doi: 10.1093/nar/4.9.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albright S. C., Nelson P. P., Garrard W. T. Histone molar ratios among different electrophoretic forms of mono- and dinucleosomes. J Biol Chem. 1979 Feb 25;254(4):1065–1073. [PubMed] [Google Scholar]
- Albright S. C., Wiseman J. M., Lange R. A., Garrard W. T. Subunit structures of different electrophoretic forms of nucleosomes. J Biol Chem. 1980 Apr 25;255(8):3673–3684. [PubMed] [Google Scholar]
- Allan J., Cowling G. J., Harborne N., Cattini P., Craigie R., Gould H. Regulation of the higher-order structure of chromatin by histones H1 and H5. J Cell Biol. 1981 Aug;90(2):279–288. doi: 10.1083/jcb.90.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D. L., Thomas J. O. Histones H1 and H5: one or two molecules per nucleosome? Nucleic Acids Res. 1981 Nov 25;9(22):5883–5894. doi: 10.1093/nar/9.22.5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulikas T., Wiseman J. M., Garrard W. T. Points of contact between histone H1 and the histone octamer. Proc Natl Acad Sci U S A. 1980 Jan;77(1):127–131. doi: 10.1073/pnas.77.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette W. N. "Western blotting": electrophoretic transfer of proteins from sodium dodecyl sulfate--polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981 Apr;112(2):195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- Caron F., Thomas J. O. Exchange of histone H1 between segments of chromatin. J Mol Biol. 1981 Mar 15;146(4):513–537. doi: 10.1016/0022-2836(81)90045-0. [DOI] [PubMed] [Google Scholar]
- Davis A. H., Reudelhuber T. L., Garrard W. T. Varigated chromatin structures of mouse ribosomal RNA genes. J Mol Biol. 1983 Jun 15;167(1):133–155. doi: 10.1016/s0022-2836(83)80038-2. [DOI] [PubMed] [Google Scholar]
- ERLANGER B. F., BEISER S. M. ANTIBODIES SPECIFIC FOR RIBONUCLEOSIDES AND RIBONUCLEOTIDES AND THEIR REACTION WITH DNA. Proc Natl Acad Sci U S A. 1964 Jul;52:68–74. doi: 10.1073/pnas.52.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan P. A., Levy-Wilson B. Structure of transcriptionally active and inactive nucleosomes from butyrate-treated and control HeLa cells. Biochemistry. 1981 Jun 23;20(13):3695–3702. doi: 10.1021/bi00516a005. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G., McGhee J. Methylation and gene control. Nature. 1982 Apr 15;296(5858):602–603. doi: 10.1038/296602a0. [DOI] [PubMed] [Google Scholar]
- Fisher E. F., Caruthers M. H. Studies on gene control regions XII. The functional significance of a lac operator constitutive mutation. Nucleic Acids Res. 1979 Sep 25;7(2):401–416. doi: 10.1093/nar/7.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesfeld J. M., Garrard W. T., Bagi G., Wilson R. F., Bonner J. Partial purification of the template-active fraction of chromatin: a preliminary report. Proc Natl Acad Sci U S A. 1974 Jun;71(6):2193–2197. doi: 10.1073/pnas.71.6.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson J. B., Pollock J. M., Jr, Rill R. L. Chromatin fractionation procedure that yields nucleosomes containing near-stoichiometric amounts of high mobility group nonhistone chromosomal proteins. Biochemistry. 1979 Aug 21;18(17):3739–3748. doi: 10.1021/bi00584a015. [DOI] [PubMed] [Google Scholar]
- Kekwick R. A. The serum proteins in multiple myelomatosis. Biochem J. 1940 Sep;34(8-9):1248–1257. doi: 10.1042/bj0341248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D. Quantitative film detection of 3H and 14C in polyacrylamide gels by fluorography. Eur J Biochem. 1975 Aug 15;56(2):335–341. doi: 10.1111/j.1432-1033.1975.tb02238.x. [DOI] [PubMed] [Google Scholar]
- Levinger L., Barsoum J., Varshavsky A. Two-dimensional hybridization mapping of nucleosomes. comparison of DNA and protein patterns. J Mol Biol. 1981 Mar 5;146(3):287–304. doi: 10.1016/0022-2836(81)90389-2. [DOI] [PubMed] [Google Scholar]
- McKeon C., Ohkubo H., Pastan I., de Crombrugghe B. Unusual methylation pattern of the alpha 2 (l) collagen gene. Cell. 1982 May;29(1):203–210. doi: 10.1016/0092-8674(82)90104-0. [DOI] [PubMed] [Google Scholar]
- Naveh-Many T., Cedar H. Active gene sequences are undermethylated. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4246–4250. doi: 10.1073/pnas.78.7.4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson P. P., Albright S. C., Wiseman J. M., Garrard W. T. Reassociation of histone H1 with nucleosomes. J Biol Chem. 1979 Nov 25;254(22):11751–11760. [PubMed] [Google Scholar]
- Razin A., Cedar H. Distribution of 5-methylcytosine in chromatin. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2725–2728. doi: 10.1073/pnas.74.7.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reudelhuber T. L., Ball D. J., Davis A. H., Garrard W. T. Transferring DNA from electrophoretically resolved nucleosomes to diazobenzyloxymethyl cellulose: properties of nucleosomes along mouse satellite DNA. Nucleic Acids Res. 1982 Feb 25;10(4):1311–1325. doi: 10.1093/nar/10.4.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano H., Royer H. D., Sager R. Identification of 5-methylcytosine in DNA fragments immobilized on nitrocellulose paper. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3581–3585. doi: 10.1073/pnas.77.6.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano H., Sager R. Tissue specificity and clustering of methylated cystosines in bovine satellite I DNA. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3584–3588. doi: 10.1073/pnas.79.11.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano H., Spaeth E., Burton W. G. Messenger RNA of the large subunit of ribulose-1,5-bisphosphate carboxylase from Chlamydomonas reinhardi. Isolation and properties. Eur J Biochem. 1979 Jan 2;93(1):173–180. doi: 10.1111/j.1432-1033.1979.tb12808.x. [DOI] [PubMed] [Google Scholar]
- Simpson R. T. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978 Dec 12;17(25):5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- Smith H. O. Nucleotide sequence specificity of restriction endonucleases. Science. 1979 Aug 3;205(4405):455–462. doi: 10.1126/science.377492. [DOI] [PubMed] [Google Scholar]
- Solage A., Cedar H. Organization of 5-methylcytosine in chromosomal DNA. Biochemistry. 1978 Jul 11;17(14):2934–2938. doi: 10.1021/bi00607a036. [DOI] [PubMed] [Google Scholar]
- Stein R., Sciaky-Gallili N., Razin A., Cedar H. Pattern of methylation of two genes coding for housekeeping functions. Proc Natl Acad Sci U S A. 1983 May;80(9):2422–2426. doi: 10.1073/pnas.80.9.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F., Koller T., Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979 Nov;83(2 Pt 1):403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorell J. I., Johansson B. G. Enzymatic iodination of polypeptides with 125I to high specific activity. Biochim Biophys Acta. 1971 Dec 28;251(3):363–369. doi: 10.1016/0005-2795(71)90123-1. [DOI] [PubMed] [Google Scholar]
- Todd R. D., Garrard W. T. Overall pathway of mononucleosome production. J Biol Chem. 1979 Apr 25;254(8):3074–3083. [PubMed] [Google Scholar]
- Urieli-Shoval S., Gruenbaum Y., Sedat J., Razin A. The absence of detectable methylated bases in Drosophila melanogaster DNA. FEBS Lett. 1982 Sep 6;146(1):148–152. doi: 10.1016/0014-5793(82)80723-0. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Beug H., Groudine M., Graf T. Temperature-sensitive changes in the structure of globin chromatin in lines of red cell precursors transformed by ts-AEV. Cell. 1982 Apr;28(4):931–940. doi: 10.1016/0092-8674(82)90072-1. [DOI] [PubMed] [Google Scholar]
- Weisbrod S. T. Properties of active nucleosomes as revealed by HMG 14 and 17 chromatography. Nucleic Acids Res. 1982 Mar 25;10(6):2017–2042. doi: 10.1093/nar/10.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisbrod S., Weintraub H. Isolation of actively transcribed nucleosomes using immobilized HMG 14 and 17 and an analysis of alpha-globin chromatin. Cell. 1981 Feb;23(2):391–400. doi: 10.1016/0092-8674(81)90134-3. [DOI] [PubMed] [Google Scholar]