Abstract
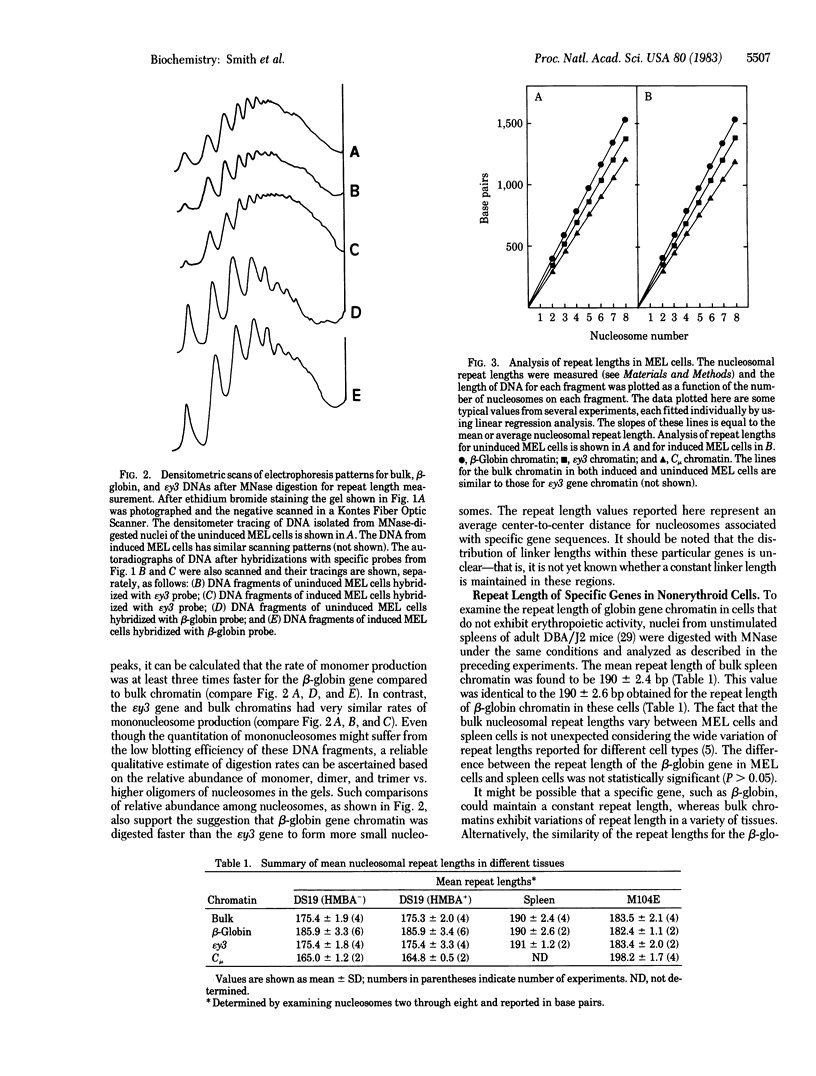
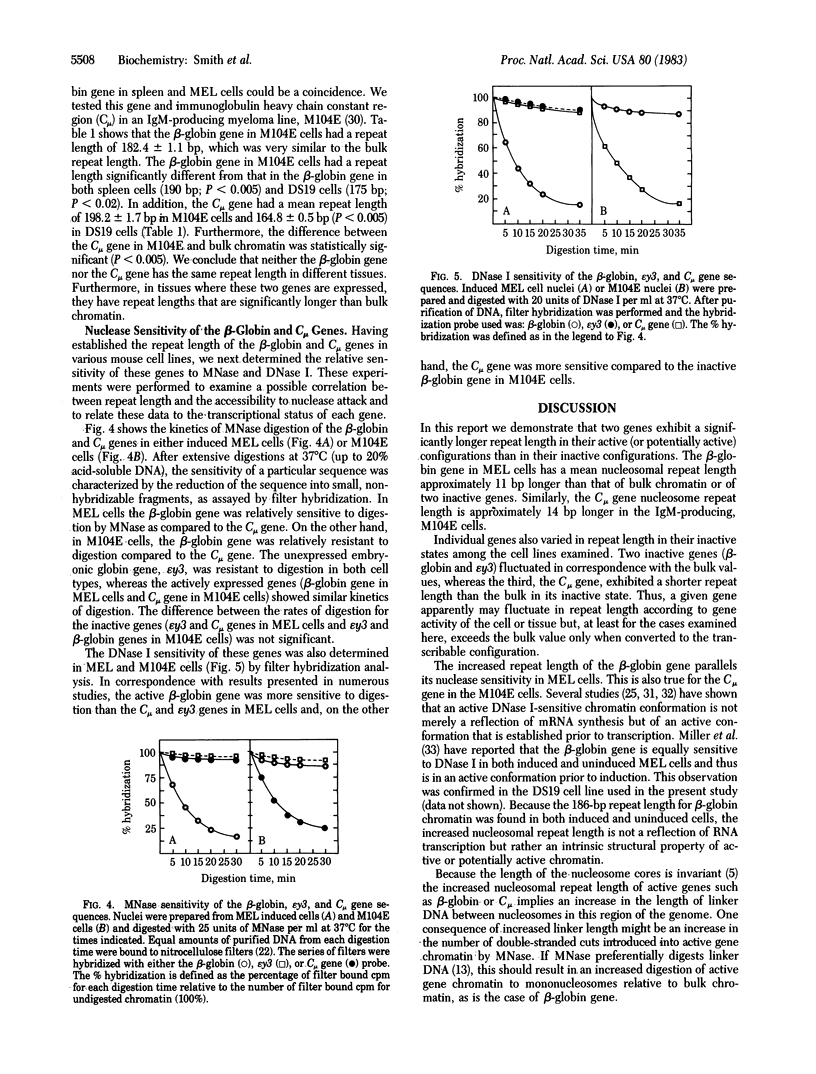
The nucleosomal repeat lengths of bulk chromatin and the chromatin of transcriptionally active and inactive genes were analyzed in two mouse cell lines and adult mouse spleens. The adult beta-globin gene exhibits a nucleosomal repeat length approximately 11 base pairs longer than (i) an inactive embryonic globin gene, epsilon y3; (ii) an immunoglobulin heavy chain gene, Cmu; and (iii) the bulk chromatin in murine erythroleukemia cell line DS19. The repeat length of the Cmu gene was approximately 14 base pairs longer than that of the adult beta-globin or epsilon y3 genes in the IgM-producing cell line M104E. The chromatin of several inactive genes had repeat lengths less than or equal to bulk chromatin. Individual genes were shown to vary in repeat length among the cell types examined. In addition, genes that exhibited an increased nucleosomal spacing were digested to mononucleosomes more rapidly than bulk chromatin or inactive genes with shorter repeats. Increased repeat length was also correlated with an increased sensitivity to DNase I. Thus, increased nucleosomal spacing may be a property of transcriptionally active genes or genes with the potential for transcription.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Annunziato A. T., Seale R. L. Maturation of nucleosomal and nonnucleosomal components of nascent chromatin: differential requirements for concurrent protein synthesis. Biochemistry. 1982 Oct 26;21(22):5431–5438. doi: 10.1021/bi00265a008. [DOI] [PubMed] [Google Scholar]
- Bellard M., Gannon F., Chambon P. Nucleosome structure III: the structure and transcriptional activity of the chromatin containing the ovalbumin and globin genes in chick oviduct nuclei. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):779–791. doi: 10.1101/sqb.1978.042.01.078. [DOI] [PubMed] [Google Scholar]
- Biessmann H., Wadsworth S., Levy B., McCarthy B. J. Correlation of structural changes in chromatin with transcription in the Drosophila heat-shock response. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):829–834. doi: 10.1101/sqb.1978.042.01.083. [DOI] [PubMed] [Google Scholar]
- Caron F., Thomas J. O. Exchange of histone H1 between segments of chromatin. J Mol Biol. 1981 Mar 15;146(4):513–537. doi: 10.1016/0022-2836(81)90045-0. [DOI] [PubMed] [Google Scholar]
- Compton J. L., Bellard M., Chambon P. Biochemical evidence of variability in the DNA repeat length in the chromatin of higher eukaryotes. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4382–4386. doi: 10.1073/pnas.73.12.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. M., Calame K., Early P. W., Livant D. L., Joho R., Weissman I. L., Hood L. An immunoglobulin heavy-chain gene is formed by at least two recombinational events. Nature. 1980 Feb 21;283(5749):733–739. doi: 10.1038/283733a0. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Fantoni A., Bozzoni I., Ullu E., Farace M. G. Construction of a recombinant bacterial plasmid containing DNA sequences for a mouse embryonic globin chain. Nucleic Acids Res. 1979 Aug 10;6(11):3505–3517. doi: 10.1093/nar/6.11.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garel A., Zolan M., Axel R. Genes transcribed at diverse rates have a similar conformation in chromatin. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4867–4871. doi: 10.1073/pnas.74.11.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesfeld J. M., Melton D. A. The length of nucleosome-associated DNA is the same in both transcribed and nontranscribed regions of chromatin. Nature. 1978 May 25;273(5660):317–319. doi: 10.1038/273317a0. [DOI] [PubMed] [Google Scholar]
- Gottesfeld J. M. Organization of 5S genes in chromatin of Xenopus laevis. Nucleic Acids Res. 1980 Feb 25;8(4):905–922. [PMC free article] [PubMed] [Google Scholar]
- Jahn C. L., Hutchison C. A., 3rd, Phillips S. J., Weaver S., Haigwood N. L., Voliva C. F., Edgell M. H. DNA sequence organization of the beta-globin complex in the BALB/c mouse. Cell. 1980 Aug;21(1):159–168. doi: 10.1016/0092-8674(80)90123-3. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- Lohr D., Corden J., Tatchell K., Kovacic R. T., Van Holde K. E. Comparative subunit structure of HeLa, yeast, and chicken erythrocyte chromatin. Proc Natl Acad Sci U S A. 1977 Jan;74(1):79–83. doi: 10.1073/pnas.74.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., Kleid D. G. Nucleotide sequence of the rightward operator of phage lambda. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1184–1188. doi: 10.1073/pnas.72.3.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis D. J., Gorovsky M. A. Structure of rDNA-containing chromatin of Tetrahymena pyriformis analyzed by nuclease digestion. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):773–778. doi: 10.1101/sqb.1978.042.01.077. [DOI] [PubMed] [Google Scholar]
- Miller D. M., Turner P., Nienhuis A. W., Axelrod D. E., Gopalakrishnan T. V. Active conformation of the globin genes in uninduced and induced mouse erythroleukemia cells. Cell. 1978 Jul;14(3):511–521. doi: 10.1016/0092-8674(78)90237-4. [DOI] [PubMed] [Google Scholar]
- Morris A., Cohn K., Scheinman M. M. Right atrial versus left atrial echo zones: a proposed new criterion for determining the atrial site of retrograde preexcitation. J Electrocardiol. 1976;9(4):357–363. doi: 10.1016/s0022-0736(76)80029-5. [DOI] [PubMed] [Google Scholar]
- Noll M. Differences and similarities in chromatin structure of Neurospora crassa and higher eucaryotes. Cell. 1976 Jul;8(3):349–355. doi: 10.1016/0092-8674(76)90146-x. [DOI] [PubMed] [Google Scholar]
- Noll M., Kornberg R. D. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol. 1977 Jan 25;109(3):393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- Nudel U., Salmon J., Fibach E., Terada M., Rifkind R., Marks P. A., Bank A. Accumulation of alpha- and beta-globin messenger RNAs in mouse erythroleukemia cells. Cell. 1977 Oct;12(2):463–469. doi: 10.1016/0092-8674(77)90122-2. [DOI] [PubMed] [Google Scholar]
- Osborne H. B., Bakke A. C., Yu J. Effect of dexamethasone on hexamethylene bisacetamide-induced Friend cell erythrodifferentiation. Cancer Res. 1982 Feb;42(2):513–518. [PubMed] [Google Scholar]
- Reuben R. C., Wife R. L., Breslow R., Rifkind R. A., Marks P. A. A new group of potent inducers of differentiation in murine erythroleukemia cells. Proc Natl Acad Sci U S A. 1976 Mar;73(3):862–866. doi: 10.1073/pnas.73.3.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson S. I., Nelkin B. D., Vogelstein B. The ovalbumin gene is associated with the nuclear matrix of chicken oviduct cells. Cell. 1982 Jan;28(1):99–106. doi: 10.1016/0092-8674(82)90379-8. [DOI] [PubMed] [Google Scholar]
- Rougeon F., Mach B. Cloning and amplification of alpha and beta mouse globin gene sequences synthesised in vitro. Gene. 1977 May;1(3-4):229–239. doi: 10.1016/0378-1119(77)90047-6. [DOI] [PubMed] [Google Scholar]
- Sanders M. M. Fractionation of nucleosomes by salt elution from micrococcal nuclease-digested nuclei. J Cell Biol. 1978 Oct;79(1):97–109. doi: 10.1083/jcb.79.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spadafora C., Bellard M., Compton J. L., Chambon P. The DNA repeat lengths in chromatins from sea urchin sperm and gastrule cells are markedly different. FEBS Lett. 1976 Oct 15;69(1):281–285. doi: 10.1016/0014-5793(76)80704-1. [DOI] [PubMed] [Google Scholar]
- Sperling L., Tardieu A., Weiss M. C. Chromatin repeat length in somatic hybrids. Proc Natl Acad Sci U S A. 1980 May;77(5):2716–2720. doi: 10.1073/pnas.77.5.2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spivak J. L., Marmor J., Dickerman H. W. Studies on splenic erythropoiesis in the mouse. I. Ribosomal ribonucleic acid metabolism. J Lab Clin Med. 1972 Apr;79(4):526–540. [PubMed] [Google Scholar]
- Thomas J. O., Thompson R. J. Variation in chromatin structure in two cell types from the same tissue: a short DNA repeat length in cerebral cortex neurons. Cell. 1977 Apr;10(4):633–640. doi: 10.1016/0092-8674(77)90096-4. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Weintraub H. The nucleosome repeat length increases during erythropoiesis in the chick. Nucleic Acids Res. 1978 Apr;5(4):1179–1188. doi: 10.1093/nar/5.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Wong Y. C., Elgin S. C. The chromatin structure of specific genes: II. Disruption of chromatin structure during gene activity. Cell. 1979 Apr;16(4):807–814. doi: 10.1016/0092-8674(79)90096-5. [DOI] [PubMed] [Google Scholar]




