Abstract
On the basis of analysis of 17,151 records on zebrafish identified from Zebrafish Information Network: the zebrafish model organism database and Web of Science, the research performance on this model organism has been evaluated. The earliest research work on zebrafish as reflected in the databases goes back to 1951. After a rather slow growth till the 1980s, research on zebrafish gained momentum in the 1990s. Analysis shows a rapid and consistent increase in the publication output with 226 publications in the year 1996, to 1929 publications in the year 2012. The prominent areas of zebrafish research, journals, and leading authors as reflected from the research output have been identified. USA is the most productive country with 8196 articles. The most frequently used keywords were also determined to gain insights about the research trends and some of the commonly used keywords other than zebrafish and Danio rerio are development, retina, and gene expression.
Introduction
Zebrafish (Danio rerio) is a freshwater fish that inhabits rivers in India, Pakistan, and other places in Asia. It is a small teleost fish that has long been a favorite in home aquariums due to its hardy nature and has emerged as one of the leading models for studying development. As a vertebrate organism, the zebrafish presents many organs and cell types similar to that of mammals. It has attracted researchers from various fields, such as neuroscience, hematopoiesis, or cardiovascular research. It has been described as “the canonical vertebrate”, due to the similarities between zebrafish and mammalian biology.1 In the last few years, the use of zebrafish in scientific research is growing very rapidly. Initially, it was popular as a model of vertebrate development because zebrafish embryos are transparent and also develop rapidly. Presently, the research using zebrafish is expanding into other areas such as pharmacology, clinical research as a disease model, and in drug discovery.2 Biomedical researches depend on the use of animal models to understand the pathogenesis of human disease at a cellular and molecular level and to provide systems for developing and testing new therapies.3 In addition zebrafish proved to be more advantageous over previous model organisms in many ways. Early developmental processes are less accessible in the mouse because they occur in utero. Practically, space requirements are much higher, and maintenance and breeding are prohibitively expensive. Other established systems such as Drosophila and Caenorhabditis elegans can serve as powerful model systems for many biological processes and are amenable to large-scale screens; however, they cannot be utilized to address the development and function of vertebrate-specific features such as the kidney, multichambered heart, multilineage hematopoiesis, notochord, and neural crest cells.4
Scientometric analysis of research areas enable studying the growth of a research field and helps in identifying prominent authors, institutes, etc. Such studies have been carried out in many areas5–7; however, an analytical study on zebrafish research output has not been carried out so far. A scientometric study of this important organism will enable us to understand the trends in zebrafish research as we use this model organism to explore new research frontiers.
Objective
This study was carried out with the aim to
map significant publication patterns in zebrafish research;
list research performance on multiple perspectives, such as author, institution, country, journal, and keywords;
and present a supplementary evaluation of research development.
Materials and Methods
Data for such scientometric studies are generally gathered from major abstracting and citation databases, including Medline, Scopus, Web of Science, and so on. These general databases are well structured and include several fields for carrying out analysis. However, as these are general databases, keyword or subject searches can include unrelated or marginally related records. A preliminary search based only on Web of Science (Science Citation Index-Expanded and Conference Proceedings Citation Index- Science) was conducted by using the term “Zebrafish” OR “Danio rerio” OR “Zebra fish” on the search interface under the “Topic” field and the search was limited to those publications that contained the selected search terms. A total of 23,598 records were retrieved for the period from 1945 to December 2012 and a random check revealed that many records were irrelevant.
For ensuring accuracy and comprehensiveness of the data, the Zebrafish Information Network (ZFIN) database was also used. The ZFIN is a web-based community resource that serves as a centralized location for the curation and integration of zebrafish genetic, genomic, and developmental data. ZFIN provides an integrated representation of mutants, genes, genetic markers, mapping panels, publications, and community contact data. ZFIN participates in collaboration with the National Center for Biotechnology Information (NCBI), the Sanger Institute, and SWISS-PROT by exchanging manually curated data.8 The ZFIN website (http://zfin.org/) also has a bibliography of the research work and articles on zebrafish.
All the 17,222 bibliographic records available in the ZFIN database for the period 1945–2012 were downloaded. The ZFIN database, although a curated database, has a limitation in the number of bibliographic elements or fields that it offers. The database includes only the basic fields such as the author, year, title, journal, volume, and page, which are insufficient for detailed analysis.
Therefore, a combination of both the databases (ZFIN and Web of Science) was used to have an accurate as well as comprehensive dataset as far as possible.
We matched each and every one of the 17,222 records in the ZFIN database with the Web of Science data (23,598 records) using Excel. It was found that 11,972 records in ZFIN matched exactly with as many Web of Science records. The ZFIN database indexes dissertations, theses, films, and videos, which do not appear in the Web of Science database and as the present study focuses only on research articles, these nonjournal bibliographic records, about 1200 in number appearing in the ZFIN database, were not included for analysis.
There were 11,626 unmatched records in the Web of Science database and as it was impossible to manually check each and every record, we considered only those that had “Danio rerio” OR “Zebrafish” OR “Zebra fish” in the title of the article. This yielded 4837 records, which we randomly checked and found that most of them were present in the remaining ZFIN records. We then manually compared each of the remaining 6789 records of Web of Science that did not have “Danio rerio” OR “Zebrafish” OR “Zebra fish” in the title and found that 451 records matched with those in the ZFIN database. These did not match automatically in Excel earlier due to typographical variations in records between the two datasets.
The method used resulted in 17,260 records. Of these, 109 records were corrections as per the document type field in Web of Science and so these were removed. This left us with 17,151 records that have been taken up for analysis on various aspects except finding the most productive institutions. For identifying the productive institutions in Zebrafish research, we relied only on the Web of Science data as it was found that there are artifacts owing to variations in rendering institution names in the downloaded data, which could have been corrected only by checking each author affiliation manually in the 17,151 records. Considering the large number of records and institutions that we are dealing with, we went by the WoS listing of institutes by using the “Organization-Enhanced” refining option in the database.
Results
Document types and languages
Thirteen document types were found and the most frequent document type was articles (12,625), which accounted for 73.6% of the total publication. The remaining were meeting abstracts, reviews, proceeding articles, editorial materials, book chapters, news items, notes, letters, biographical items, book reviews, database reviews, and reprints.
As for the publication language of the articles, 17,095 or 99.7% of the 17,151 articles were in English. Other publication languages included French (21), Chinese (13), German (11), Japanese (4), Russian (3), Italian (1), Portuguese (1), Korean (1), and Romanian (1).
Publication outputs
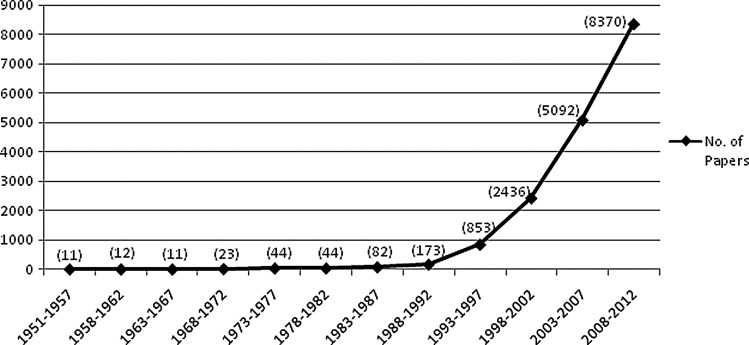
From the available data, it is seen that the first article on zebrafish goes back to 1951. As research literature on zebrafish was very low in the early years, we aggregated the periods into 5-year blocks to assess the growth of research on zebrafish (Fig. 1). Research output indicators suggest a significant increase in the zebrafish research since 1996 with 226 publications in the year 1996 to 1929 publications in the year 2012. There was over 390% growth in research articles from 1993 to 1997 (853 articles) over the previous block of period 1988–1992 (173 articles).
FIG. 1.
Annual growth trend of articles from the year 1951 to 2012 in zebrafish research.
The importance of zebrafish was recognized very early and as noted in the editorial of the first issue of Zebrafish, “the work of George Streisinger and his colleagues in the late 1970s planted the seed from which today's large vibrant community of researchers using zebrafish and other aquarium fish models has grown.”9 The article by Kimmel et al. (1995) on stages of embryonic development of zebrafish is also a pathbreaking work.10 The article has received over 3500 citations so far.
Subject categories and major journals
Research output on zebrafish spanned across 101 subject categories. The four most common subject categories were developmental biology (4662 articles, 27.010% of the total), biochemistry/molecular biology (3058; 17.717%), cell biology (2423; 14.038%), and neurosciences/neurology (1872; 10.846%) followed by genetics/heredity, zoology, science technology other topics, toxicology, life sciences/biomedicine other topics, and anatomy/morphology. The 25 most active subject areas of zebrafish research are presented in Table 1.
Table 1.
Top 25 Subjects in the Area of Zebrafish Research
| Rank | Research areas | Publications | Percenta |
|---|---|---|---|
| 1 | Developmental Biology | 4662 | 27.010 |
| 2 | Biochemistry/Molecular Biology | 3058 | 17.717 |
| 3 | Cell Biology | 2423 | 14.038 |
| 4 | Neurosciences/Neurology | 1872 | 10.846 |
| 5 | Genetics/Heredity | 1681 | 9.739 |
| 6 | Zoology | 1134 | 6.570 |
| 7 | Science & Technology Other Topics | 1114 | 6.454 |
| 8 | Toxicology | 995 | 5.765 |
| 9 | Life Sciences/Biomedicine Other Topics | 699 | 4.050 |
| 10 | Anatomy/Morphology | 680 | 3.940 |
| 11 | Biotechnology/Applied Microbiology | 551 | 3.192 |
| 12 | Environmental Sciences/Ecology | 549 | 3.181 |
| 13 | Marine/Freshwater Biology | 513 | 2.972 |
| 14 | Physiology | 505 | 2.926 |
| 15 | Hematology | 503 | 2.914 |
| 16 | Endocrinology/Metabolism | 479 | 2.775 |
| 17 | Pharmacology/Pharmacy | 429 | 2.486 |
| 18 | Biophysics | 388 | 2.248 |
| 19 | Immunology | 355 | 2.057 |
| 20 | Ophthalmology | 326 | 1.889 |
| 21 | Evolutionary Biology | 318 | 1.842 |
| 22 | Fisheries | 268 | 1.553 |
| 23 | Research/Experimental Medicine | 266 | 1.541 |
| 24 | Chemistry | 251 | 1.460 |
| 25 | Veterinary Sciences | 245 | 1.419 |
Total percent is more than 100 because an article can be assigned to more than one research area.
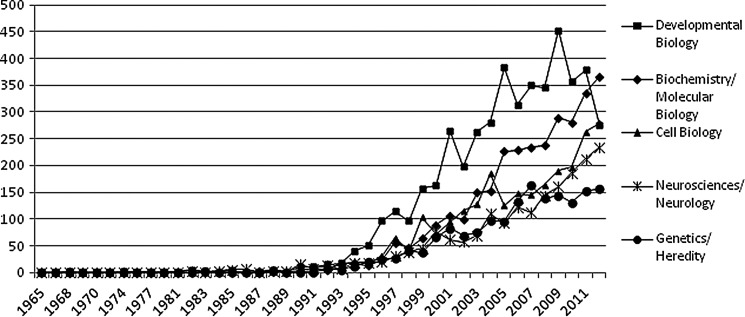
The annual growth rate of articles in five most active subjects was also determined (Fig. 2). The plot reflects growth in the five areas and expectedly, developmental biology is the focus of about 27% of the articles followed by biochemistry/molecular biology. It is also seen that despite developmental biology being the leading area of research, there has been a dip in this area in the recent years, but the areas of biochemistry/molecular biology, cell biology, and neurosciences/neurology in zebrafish research is rapidly growing in the recent years.
FIG. 2.
Annual growth rates of articles in top five active subjects.
Zebrafish research appeared in more than 1500 journals. The top 25 journals that published most zebrafish studies have been presented (Table 2) along with their impact factor and the number of articles published by them. The top three journals that report Zebrafish research—Developmental Biology, Development, and Developmental Dynamics—clearly reflect that the focus of zebrafish research has been on developmental aspects. There are five journals that are of more recent origin, which include PLoS One, Zebrafish, Gene Expression Pattern, Developmental Cell, and BMC Developmental Biology. Most of the journals falling in the top 25 category belong to the United States (17 journals) followed by Netherlands (4), England (3), and Germany (1).
Table 2.
Top 25 Journals in the Area of Zebrafish Research
| Rank | Source titles | Published since | Records | Percent | Impact factor (2012) |
|---|---|---|---|---|---|
| 1 | Developmental Biology | 1959 | 1383 | 8.013 | 3.868 |
| 2 | Development | 1953 | 830 | 4.809 | 6.208 |
| 3 | Developmental Dynamics | 1901 | 553 | 3.204 | 2.590 |
| 4 | PLOS One | 2006 | 430 | 2.491 | 3.730 |
| 5 | Mechanisms of Development | 1972 | 429 | 2.486 | 2.383 |
| 6 | Proceedings of the National Academy of Sciences of the United States of America | 1941 | 327 | 1.895 | 9.737 |
| 7 | FASEB Journal | 1987 | 293 | 1.698 | 5.704 |
| 8 | Blood | 1946 | 285 | 1.651 | 9.060 |
| 9 | Investigative Ophthalmology & Visual Science | 1962 | 219 | 1.269 | 3.441 |
| 10 | Journal of Neuroscience | 1981 | 216 | 1.251 | 6.908 |
| 11 | Zebrafish | 2004 | 215 | 1.246 | 2.883 |
| 12 | Aquatic Toxicology | 1981 | 177 | 1.025 | 3.730 |
| 13 | Journal of Biological Chemistry | 1905 | 169 | 0.979 | 4.651 |
| 14 | Current Biology | 1991 | 164 | 0.950 | 9.494 |
| 15 | Journal of Comparative Neurology | 1891 | 158 | 0.915 | 3.661 |
| 16 | Gene Expression Patterns | 2001 | 156 | 0.904 | 1.640 |
| 17 | Biochemical and Biophysical Research Communications | 1959 | 156 | 0.904 | 2.406 |
| 18 | Gene | 1976 | 131 | 0.759 | 2.196 |
| 19 | Developmental Cell | 2001 | 124 | 0.718 | 12.861 |
| 20 | Nature | 1869 | 123 | 0.713 | 38.597 |
| 21 | Integrative and Comparative Biology | 1961 | 122 | 0.707 | 3.023 |
| 22 | Development Genes and Evolution | 1894 | 111 | 0.643 | 1.695 |
| 23 | Molecular Biology of the Cell | 1992 | 111 | 0.643 | 4.604 |
| 24 | Comparative Biochemistry and Physiology A-Molecular & Integrative Physiology | 1960 | 110 | 0.637 | 2.167 |
| 25 | BMC Developmental Biology | 2001 | 101 | 0.585 | 2.728 |
Highly cited articles
Garfield referred to articles that have received an unusually large number of citations as citation classics.11 A number of citations received, including self-citations, have been studied. The article by Kimmel CB, Ballard WW, Kimmel SR, et al. entitled “Stages of embryonic-development of the zebrafish” published in Developmental Dynamics has received the largest number of citations (3544). We found that 14,867 (86.68%) articles have received less than 50 citations and 1422 articles have received 50 or more citations but less than 100 citations. Six hundred and thirty articles have received 100 or more citations but below 200 citations and 208 articles have citations in the range of 200–499. Table 3 gives the list of articles with 500 and more citations, of which 17 articles have been cited more than 600 times so far.
Table 3.
Articles Receiving ≥500 Citations
| S. No. | Article details | Total no. of citations |
|---|---|---|
| 1 | Kimmel CB, Ballard WW, Kimmel SR, et al. Stages of embryonic-development of the zebrafish. Developmental Dynamics 203, 3, 253–310, 1995 | 3544 |
| 2 | Force A, Lynch M, Pickett FB, et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 4, 1531–1545, 1999 | 1677 |
| 3 | Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nature Genetics 26, 2, 216–220, 2000 | 1442 |
| 4 | Amores A, Force A, Yan YL, et al. Zebrafish hox clusters and vertebrate genome evolution. Science 282, 5394, 1711–1714, 1998 | 987 |
| 5 | Burns JC, Friedmann T, Driever W, et al. Vesicular stomatitis-virus G glycoprotein pseudotyped retroviral vectors—concentration to very high-titer and efficient gene-transfer into mammalian and nonmammalian cells. Proceedings of the National Academy of Sciences of the United States of America 90, 17, 8033–8037, 1993 | 917 |
| 6 | Haffter P, Granato M, Brand M, et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development 123 Special, SI, 1–36, 1996 | 900 |
| 7 | Krauss S, Concordet JP, Ingham PW. A functionally conserved homology of the drosophila segment polarity gene-HH is expressed in tissues with polarizing activity in zebrafish embryos. Cell 75, 7, 1431–1444, 1993 | 831 |
| 8 | Pasquinelli AE, Reinhart BJ, Slack F, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408, 6808, 86–89, 2000 | 821 |
| 9 | Donovan A, Brownlie A, Zhou Y, et al. Positional cloning of zebrafish ferroportin1 identifies a conserved vertebrate iron exporter. Nature 403, 6771, 776–781, 2000 | 772 |
| 10 | Driever W, SolnicaKrezel L, Schier AF, et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development 123 Special, SI, 37–46, 1996 | 692 |
| 11 | Esko JD, Selleck SB. Order out of chaos: Assembly of ligand binding sites in heparan sulfate. Annual Review of Biochemistry 71, 435–471, 2002 | 675 |
| 12 | Roelink H, Augsburger A, Heemskerk J, et al. Floor plate and motor-neuron induction by VHH-1, a vertebrate homolog of hedgehog expressed by the notochord. Cell 76, 4, 761–775, 1994 | 646 |
| 13 | Wienholds E, Kloosterman WP, Miska E, et al. MicroRNA expression in zebrafish embryonic development. Science 309, 5732, 310–311, 2005 | 635 |
| 14 | Thisse C, Thisse B, Schilling TF, et al. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development 119, 4, 1203–1215, 1993 | 632 |
| 15 | Streisinger G, Walker C, Dower N, et al. Production of Clones of Homozygous Diploid Zebra Fish (Brachydanio-Rerio). Nature 291, 5813, 293–296, 1981 | 629 |
| 16 | Ivics Z, Hackett PB, Plasterk RH, et al. Molecular reconstruction of Sleeping beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell 91, 4, 501–510, 1997 | 618 |
| 17 | Giraldez AJ, Mishima Y, Rihel J, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312, 5770, 75–79, 2006 | 613 |
| 18 | Giraldez AJ, Cinalli RM, Glasner ME, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science 308, 5723, 833–838, 2005 | 588 |
| 19 | Pruitt KD, Maglott DR, RefSeq, LocusLink. NCBI gene-centered resources. Nucleic Acids Research 29, 1, 137–140, 2001 | 581 |
| 20 | Heisenberg CP, Tada M, Rauch GJ, et al. Silberblick/Wnt11 mediates convergent extension movements during zebrafish gastrulation. Nature 405, 6782, 76–81, 2000 | 579 |
| 21 | Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Developmental Biology 248, 2, 307–318, 2002 | 558 |
| 22 | Oxtoby E, Jowett T. Cloning of the zebrafish krox-20 gene (krx-20) and its expression during hindbrain development. Nucleic Acids Research 21, 5, 1087–1095, 1993 | 533 |
| 23 | Schultemerker S, Ho RK, Herrmann BG, et al. The protein product of the zebrafish homolog of the mouse t-gene is expressed in nuclei of the germ ring and the notochord of the early embryo, Development 116, 4, 1021, 1992 | 522 |
| 24 | Postlethwait JH, Yan YL, Gates MA, et al. Vertebrate genome evolution and the zebrafish gene map. Nature Genetics 18, 4, 345–349, 1998 | 500 |
Prominent authors
The most productive authors in zebrafish research were Zon LI with 278 publications followed by Stainier DYR with 162 articles, Lin S (129), Hammerschmidt M (120), and Thisse B with 116 articles. The 25 most productive authors and number of articles are presented in Table 4. Although we have taken care to identify them as unique authors, the problem owing to identical names for different persons cannot be ruled out.
Table 4.
The 25 Most Prominent Authors
| Rank | Authors | Records |
|---|---|---|
| 1 | Zon LI | 278 |
| 2 | Stainier DYR | 162 |
| 3 | Lin S | 129 |
| 4 | Hammerschmidt M | 120 |
| 5 | Thisse B | 116 |
| 6 | Driever W | 117 |
| 7 | Thisse C | 115 |
| 8 | Kimmel CB | 114 |
| 9 | Postlethwait JH | 110 |
| 10 | Kawakami K | 109 |
| 11 | Dowling JE | 105 |
| 12 | Westerfield M | 105 |
| 13 | Brand M | 103 |
| 14 | Korzh V | 103 |
| 15 | Schier AF | 102 |
| 16 | Strahle, U | 98 |
| 17 | Wilson SW | 94 |
| 18 | Look AT | 94 |
| 19 | Gong ZY | 92 |
| 20 | Johnson SL | 92 |
| 21 | Weinstein BM | 92 |
| 22 | Okamoto H | 91 |
| 23 | Solnica-Krezel L | 91 |
| 24 | Fishman MC | 90 |
| 25 | Eisen JS | 88 |
Major countries and institutes of zebrafish research
Research articles on zebrafish have originated from 103 countries. The top 25 most productive countries are presented in Table 5. Among these top 25 productive countries, 18 countries were of developed economy. Five countries (People's Republic of China, Singapore, Brazil, Israel, and India) are of developing economy and two (Taiwan and South Korea) of advanced economy. As per the World Bank, the countries with a large stock of physical capital, in which most people undertake highly specialized activities, are classified as developed economies. Countries with low or middle levels of gross national product per capita as well as five high-income developing economies—Hong Kong (China), Israel, Kuwait, Singapore, and the United Arab Emirates are classified as developing economy.12
Table 5.
Top 25 Productive Countries
| Rank | Countries/Territories | Records | No. of institutes doing zebrafish research | Records/Institute | Percenta |
|---|---|---|---|---|---|
| 1 | USA | 8196 | 877 | 9.35 | 47.625 |
| 2 | Germany | 1871 | 359 | 5.21 | 10.869 |
| 3 | England | 1393 | 180 | 7.74 | 8.094 |
| 4 | Japan | 1284 | 228 | 5.63 | 7.451 |
| 5 | People's Republic of China | 1151 | 255 | 4.51 | 6.703 |
| 6 | Canada | 902 | 113 | 7.98 | 5.243 |
| 7 | France | 877 | 219 | 4.00 | 5.098 |
| 8 | Netherlands | 501 | 77 | 6.51 | 2.914 |
| 9 | Spain | 449 | 138 | 3.25 | 2.601 |
| 10 | Taiwan | 431 | 84 | 5.13 | 2.509 |
| 11 | Singapore | 421 | 34 | 12.38 | 2.445 |
| 12 | Italy | 419 | 167 | 2.51 | 2.433 |
| 13 | Australia | 322 | 74 | 4.35 | 1.866 |
| 14 | South Korea | 275 | 99 | 2.78 | 1.593 |
| 15 | Switzerland | 251 | 54 | 4.65 | 1.454 |
| 16 | Sweden | 238 | 34 | 7.00 | 1.379 |
| 17 | Belgium | 215 | 47 | 4.57 | 1.246 |
| 18 | Norway | 208 | 43 | 4.84 | 1.211 |
| 19 | Israel | 180 | 30 | 6.00 | 1.043 |
| 20 | Scotland | 176 | 17 | 10.35 | 1.020 |
| 21 | India | 170 | 50 | 3.40 | 0.991 |
| 22 | Brazil | 151 | 51 | 2.96 | 0.875 |
| 23 | Austria | 131 | 27 | 4.85 | 0.765 |
| 24 | Chile | 103 | 16 | 6.44 | 0.597 |
| 25 | Portugal | 103 | 39 | 2.64 | 0.597 |
Total percent is more than 100 because many articles have authors from different countries.
In terms of the number of articles, USA is the most productive country with 8196 records followed by Germany (1871), England (1393), Japan (1284), and People's Republic of China (1151). India ranks 21st with 170 publications. We also looked at the number of institutions engaged in zebrafish research in each country and found that Singapore is the most productive with an average of 12 publications produced per institution followed by Scotland with about 10 articles per institution.
With regard to the top 25 productive institutions in zebrafish research, the Harvard University topped with 1106 publications followed by the University of California with 1015, the University of Washington with 705, the Max Planck Society with 693, and the University of London with 541 publications (Table 6).
Table 6.
Prominent Institutions
| Rank | Organizations | Recordsa | % |
|---|---|---|---|
| 1 | Harvard University, USA | 1106 | 5.424 |
| 2 | University of California System, USA | 1015 | 4.977 |
| 3 | University of Washington, USA | 705 | 3.457 |
| 4 | Max Planck Society, Germany | 693 | 3.398 |
| 5 | University of London, UK | 541 | 2.653 |
| 6 | University of Oregon, USA | 473 | 2.32 |
| 7 | National Institutes of Health, USA | 468 | 2.295 |
| 8 | Chinese Academy of Sciences, China | 430 | 2.109 |
| 9 | National University of Singapore, Singapore | 358 | 1.756 |
| 10 | University of Tokyo, Japan | 337 | 1.653 |
| 11 | University of California San Francisco, USA | 334 | 1.638 |
| 12 | University of Pennsylvania, USA | 296 | 1.452 |
| 13 | University of Michigan, USA | 294 | 1.442 |
| 14 | University College London, UK | 284 | 1.393 |
| 15 | University of Utah, USA | 281 | 1.378 |
| 16 | University of Utrecht, Netherlands | 252 | 1.236 |
| 17 | University of California Los Angeles, USA | 233 | 1.143 |
| 18 | Helmholtz Association, Germany | 227 | 1.113 |
| 19 | Academia Sinica Taiwan, Taiwan | 223 | 1.094 |
| 20 | Kings College London, UK | 220 | 1.079 |
| 21 | University of Wisconsin System, USA | 218 | 1.069 |
| 22 | Vanderbilt University, USA | 216 | 1.059 |
| 23 | Dana Farber Cancer Center, USA | 211 | 1.035 |
| 24 | Riken, Japan | 203 | 0.995 |
| 25 | Massachusetts Institute of Technology MIT, | 194 | 0.951 |
Indicative table based on Web of Science.
Keyword analysis
The keyword analysis was also performed to obtain some information about zebrafish research trends. The keywords of a research article reflect the focus of research. A total of 17,872 unique keywords were obtained with a total of 52,152 occurrences. However 12,557 out of these 17,872 keywords appeared only once and 17,158 keywords appeared less than 10 times. The large number of single occurrence author keywords probably indicates a lack of continuity in research and a wide disparity in research foci. Furthermore, these keywords might not be standard or widely accepted by researchers as has been the case of a similar study on the global climatic change.13
Fifty most frequently used keywords are presented in Table 7 in 5-year intervals from 1991 to 2012 except for the period 2011–2012. The count and ranks of the keywords have varied during the different periods. The most commonly used keywords were zebrafish, Danio rerio, development, retina, gene expression, and fish. However, the keywords zebrafish and Danio rerio are used in less than 10% of the articles although it is expected that either of these two keywords should appear in all the articles. This highlights the inconsistencies in the author assigned keywords in research articles and also the shortcomings in indexing even by prominent citation databases. Despite the limitations in the keywords, the analysis here reflected the general trend.
Table 7.
Temporal Evolution of Most Frequently Used Keywords
| |
1991–1995 |
1996–2000 |
2001–2005 |
2006–2010 |
2011–2012 |
|
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Keywords | Cnt | % | Rank | Cnt | % | Rank | Cnt | % | Rank | Cnt | % | Rank | Cnt | % | Rank | Total Cnt | Rank |
| Zebrafish | 88 | 8.95 | 1 | 506 | 9.65 | 1 | 1200 | 9.09 | 1 | 1971 | 9.09 | 1 | 1134 | 10.28 | 1 | 4910 | 1 |
| Danio rerio | 12 | 1.22 | 4 | 106 | 2.02 | 2 | 202 | 1.53 | 2 | 203 | 0.94 | 2 | 94 | 0.85 | 2 | 617 | 2 |
| Development | 3 | 0.31 | 12 | 49 | 0.93 | 3 | 139 | 1.05 | 3 | 173 | 0.80 | 3 | 86 | 0.78 | 3 | 450 | 3 |
| Retina | 6 | 0.61 | 9 | 41 | 0.78 | 4 | 79 | 0.60 | 4 | 98 | 0.45 | 5 | 34 | 0.31 | 7 | 258 | 4 |
| Gene expression | 6 | 0.61 | 9 | 18 | 0.34 | 12 | 30 | 0.23 | 21 | 102 | 0.47 | 4 | 48 | 0.44 | 5 | 205 | 5 |
| Fish | 3 | 0.31 | 12 | 31 | 0.59 | 5 | 55 | 0.42 | 7 | 86 | 0.40 | 6 | 28 | 0.25 | 10 | 203 | 6 |
| Evolution | 3 | 0.31 | 12 | 25 | 0.48 | 8 | 61 | 0.46 | 6 | 80 | 0.37 | 7 | 26 | 0.24 | 11 | 195 | 7 |
| Neural crest | 5 | 0.51 | 10 | 31 | 0.59 | 5 | 52 | 0.39 | 9 | 64 | 0.30 | 9 | 17 | 0.15 | 17 | 169 | 8 |
| Hindbrain | 3 | 0.31 | 12 | 27 | 0.52 | 6 | 71 | 0.54 | 5 | 44 | 0.20 | 20 | 14 | 0.13 | 20 | 159 | 9 |
| Embryo | 1 | 0.10 | 14 | 16 | 0.31 | 14 | 52 | 0.39 | 9 | 63 | 0.29 | 10 | 20 | 0.18 | 14 | 152 | 10 |
| Teleost | 6 | 0.61 | 9 | 17 | 0.32 | 13 | 39 | 0.30 | 13 | 60 | 0.28 | 11 | 29 | 0.26 | 9 | 151 | 11 |
| Apoptosis | 1 | 0.10 | 14 | 11 | 0.21 | 19 | 35 | 0.27 | 17 | 72 | 0.33 | 8 | 30 | 0.27 | 8 | 149 | 12 |
| Gastrulation | 21 | 2.14 | 2 | 20 | 0.38 | 11 | 37 | 0.28 | 15 | 53 | 0.24 | 15 | 16 | 0.15 | 18 | 147 | 13 |
| Angiogenesis | 0 | 0.00 | 0 | 3 | 0.06 | 27 | 21 | 0.16 | 28 | 63 | 0.29 | 10 | 56 | 0.51 | 4 | 143 | 14 |
| Neurogenesis | 4 | 0.41 | 11 | 20 | 0.38 | 11 | 36 | 0.27 | 16 | 56 | 0.26 | 14 | 25 | 0.23 | 12 | 141 | 15 |
| Brain | 3 | 0.31 | 12 | 13 | 0.25 | 17 | 41 | 0.31 | 12 | 52 | 0.24 | 16 | 26 | 0.24 | 11 | 135 | 16 |
| Morpholino | 0 | 0.00 | 0 | 2 | 0.04 | 28 | 53 | 0.40 | 8 | 57 | 0.26 | 13 | 21 | 0.19 | 13 | 133 | 17 |
| Embryogenesis | 12 | 1.22 | 4 | 26 | 0.50 | 7 | 36 | 0.27 | 16 | 33 | 0.15 | 28 | 19 | 0.17 | 15 | 126 | 18 |
| Spinal cord | 11 | 1.12 | 5 | 22 | 0.42 | 9 | 31 | 0.23 | 20 | 40 | 0.18 | 23 | 15 | 0.14 | 19 | 119 | 19 |
| Regeneration | 3 | 0.31 | 12 | 15 | 0.29 | 15 | 21 | 0.16 | 28 | 58 | 0.27 | 12 | 19 | 0.17 | 15 | 116 | 20 |
| Heart | 2 | 0.20 | 13 | 14 | 0.27 | 16 | 32 | 0.24 | 19 | 50 | 0.23 | 17 | 16 | 0.15 | 18 | 114 | 21 |
| WNT | 3 | 0.31 | 12 | 10 | 0.19 | 20 | 46 | 0.35 | 10 | 36 | 0.17 | 25 | 18 | 0.16 | 16 | 113 | 22 |
| Notch | 0 | 0.00 | 0 | 5 | 0.10 | 25 | 38 | 0.29 | 14 | 46 | 0.21 | 19 | 18 | 0.16 | 16 | 107 | 23 |
| Retinoic acid | 9 | 0.92 | 6 | 9 | 0.17 | 21 | 24 | 0.18 | 26 | 38 | 0.18 | 24 | 20 | 0.18 | 14 | 100 | 24 |
| Gene duplication | 1 | 0.10 | 14 | 7 | 0.13 | 23 | 43 | 0.33 | 11 | 35 | 0.16 | 26 | 13 | 0.12 | 21 | 98 | 25 |
| Transgenic | 1 | 0.10 | 14 | 8 | 0.15 | 22 | 22 | 0.17 | 27 | 48 | 0.22 | 18 | 19 | 0.17 | 15 | 97 | 26 |
| FGF | 0 | 0.00 | 0 | 3 | 0.06 | 27 | 30 | 0.23 | 21 | 41 | 0.19 | 22 | 18 | 0.16 | 16 | 93 | 27 |
| Notochord | 7 | 0.71 | 8 | 21 | 0.40 | 10 | 27 | 0.20 | 24 | 28 | 0.13 | 33 | 10 | 0.09 | 24 | 93 | 28 |
| In situ hybridization | 3 | 0.31 | 12 | 21 | 0.40 | 10 | 22 | 0.17 | 27 | 35 | 0.16 | 26 | 12 | 0.11 | 22 | 93 | 29 |
| Toxicity | 1 | 0.10 | 14 | 7 | 0.13 | 23 | 15 | 0.11 | 34 | 41 | 0.19 | 21 | 28 | 0.25 | 10 | 92 | 30 |
| BMP | 0 | 0.00 | 0 | 11 | 0.21 | 19 | 36 | 0.27 | 16 | 27 | 0.12 | 34 | 15 | 0.14 | 19 | 89 | 31 |
| Nodal | 1 | 0.10 | 14 | 15 | 0.29 | 15 | 43 | 0.33 | 11 | 25 | 0.12 | 36 | 5 | 0.05 | 29 | 89 | 32 |
| Hematopoiesis | 1 | 0.10 | 14 | 16 | 0.31 | 14 | 22 | 0.17 | 27 | 33 | 0.15 | 28 | 15 | 0.14 | 19 | 87 | 33 |
| Behavior | 0 | 0.00 | 0 | 5 | 0.10 | 25 | 19 | 0.14 | 30 | 35 | 0.16 | 26 | 37 | 0.34 | 6 | 85 | 34 |
| Muscle | 1 | 0.10 | 14 | 7 | 0.13 | 23 | 20 | 0.15 | 29 | 29 | 0.13 | 32 | 17 | 0.15 | 17 | 85 | 35 |
| Hedgehog | 3 | 0.31 | 12 | 6 | 0.11 | 24 | 29 | 0.22 | 22 | 34 | 0.16 | 27 | 12 | 0.11 | 22 | 84 | 36 |
| Mouse | 1 | 0.10 | 14 | 7 | 0.13 | 23 | 33 | 0.25 | 18 | 29 | 0.13 | 32 | 14 | 0.13 | 20 | 84 | 37 |
| Morphogenesis | 3 | 0.31 | 12 | 11 | 0.21 | 19 | 19 | 0.14 | 30 | 35 | 0.16 | 26 | 11 | 0.10 | 23 | 79 | 38 |
| Ovary | 0 | 0.00 | 0 | 4 | 0.08 | 26 | 32 | 0.24 | 19 | 25 | 0.12 | 36 | 16 | 0.15 | 18 | 77 | 39 |
| Eye | 1 | 0.10 | 14 | 12 | 0.23 | 18 | 21 | 0.16 | 28 | 35 | 0.16 | 26 | 7 | 0.06 | 27 | 76 | 40 |
| Somitogenesis | 2 | 0.20 | 13 | 20 | 0.38 | 11 | 21 | 0.16 | 28 | 25 | 0.12 | 36 | 6 | 0.05 | 28 | 74 | 41 |
| Lateral line | 0 | 0.00 | 0 | 9 | 0.17 | 21 | 19 | 0.14 | 30 | 35 | 0.16 | 26 | 11 | 0.10 | 23 | 74 | 42 |
| Sonic hedgehog | 1 | 0.10 | 14 | 25 | 0.48 | 8 | 19 | 0.14 | 30 | 21 | 0.10 | 40 | 7 | 0.06 | 27 | 73 | 43 |
| Somite | 2 | 0.20 | 13 | 25 | 0.48 | 8 | 32 | 0.24 | 19 | 36 | 0.17 | 25 | 4 | 0.04 | 30 | 73 | 44 |
| Axon guidance | 1 | 0.10 | 14 | 12 | 0.23 | 18 | 28 | 0.21 | 23 | 24 | 0.11 | 37 | 7 | 0.06 | 27 | 72 | 45 |
| Microarray | 0 | 0.00 | 0 | 0 | 0.00 | 0 | 6 | 0.05 | 43 | 52 | 0.24 | 16 | 18 | 0.16 | 16 | 71 | 46 |
| Zebrafish embryo | 3 | 0.31 | 12 | 5 | 0.10 | 25 | 20 | 0.15 | 29 | 32 | 0.15 | 29 | 30 | 0.27 | 8 | 71 | 47 |
| Forebrain | 3 | 0.31 | 12 | 21 | 0.40 | 10 | 27 | 0.20 | 24 | 15 | 0.07 | 46 | 4 | 0.04 | 30 | 70 | 48 |
| Xenopus | 2 | 0.20 | 13 | 13 | 0.25 | 17 | 18 | 0.14 | 31 | 27 | 0.12 | 34 | 8 | 0.07 | 26 | 68 | 49 |
| Medaka | 1 | 0.10 | 14 | 2 | 0.04 | 28 | 18 | 0.14 | 31 | 35 | 0.16 | 26 | 12 | 0.11 | 22 | 68 | 50 |
Conclusion
In the last few years, the use of zebrafish in scientific research is growing very rapidly. Our analysis suggested that there has been a steady growth in zebrafish research output. The importance of the subject is also evidenced by the launch of the journal, Zebrafish, which in a short span of time has gone on to become an important journal with an impact factor 2.883. From a rather slow beginning, zebrafish research picked up momentum in the 1990s. Our study reveals that more than fifty percent of the highly cited articles (500+ citations) have appeared in the 1990s with Kimmel CB et al. (1995) being the iconic article that has been cited over 3500 times. Developmental biology of zebrafish has been the most studied aspect and continues to contribute the bulk of the literature. Whereas interest in developmental biology was evident since the beginning, more intensive research in this area commenced in the 1990s, after publication of the article by Kimmel CB on the stages of embryonic development in zebrafish. Developmental biology research on zebrafish has been the key area of research for all period, but in recent years, it is seen that there is a drop in the number of articles published in this area, whereas the areas of biochemistry and molecular biology, cell biology, neurosciences/neurology and genetics, and heredity are on the ascent. It is expected that in the coming years, biochemistry and molecular biology and neurosciences with regard to zebrafish will emerge as leading areas of research.
Acknowledgment
The authors thank the anonymous reviewers whose insightful comments enabled us to improve several sections of the article.
Disclosure Statement
No competing financial interests exist.
References
- 1.Rubinstein AL. Zebrafish: From disease modeling to drug discovery. Curr Opin Drug Discov Devel. 2003;6:218–223. [PubMed] [Google Scholar]
- 2.Chakraborty C. Hsu CH. Wen ZH. Lin CS. Agoramoorthy G. Zebrafish: a complete animal model for in vivo drug discovery and development. Curr Drug Metab. 2009;10:116–124. doi: 10.2174/138920009787522197. [DOI] [PubMed] [Google Scholar]
- 3.Lieschke GJ. Currie PD. Animal models of human disease: zebrafish swim into view. Nat Rev Genet. 2007;8:353–367. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- 4.Dooley K. Zon LI. Zebrafish: a model system for the study of human disease. Curr Opin Genet Dev. 2000;10:252–256. doi: 10.1016/s0959-437x(00)00074-5. [DOI] [PubMed] [Google Scholar]
- 5.Vitzthum K. Scutaru C. Musial-Bright L. Quarcoo D. Welte T. Groneberg-Kloft MSB. Scientometric analysis and combined density equalizing mapping of environmental tobacco smoke (ETS) research. PLoS One. 2010;5:1–10. doi: 10.1371/journal.pone.0011254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta BM. Bala A. A scientometric analysis of Indian research output in medicine during 1999–2008. J Nat Sci Biol Med. 2011;2:87–100. doi: 10.4103/0976-9668.82313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta BM. Heredity blood disorders (HBD): a scientometric analysis of publications output from India during 2002–2011. J Blood Disord Transf. 2012;3:126. [Google Scholar]
- 8.Sprague J. Clements D. Conlin T. Edwards P, et al. The Zebrafish Information Network (ZFIN): the zebrafish model organism database. Nucleic Acids Res. 2003;31:241–243. doi: 10.1093/nar/gkg027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collodi P. A unique journal for a unique experimental model. Zebrafish. 2004;1:1. doi: 10.1089/154585404774101608. [DOI] [PubMed] [Google Scholar]
- 10.Kimmel CB. Ballard WW. Kimmel SR. Ullmann B. Schilling TF. Stages of Embryonic-Development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 11.Garfield E. Introducing citation classics: the human side of scientific reports. Curr Contents. 1997;3:6–7. [Google Scholar]
- 12.Glossary of World Bank. www.worldbank.org/depweb/english/beyond/global/glossary.html www.worldbank.org/depweb/english/beyond/global/glossary.html
- 13.Li J. Wang MH. Ho YS. Trends in research on global climate change: a science citation index expanded-based analysis. Global Planet Change. 2011;77:13–20. [Google Scholar]