Abstract
As part of large protein complexes, Snf2 family ATPases are responsible for energy supply during chromatin remodeling, but the precise mechanism of action of many of these proteins is largely unknown. They influence many processes in plants, such as the response to environmental stress. This analysis is the first comprehensive study of Snf2 family ATPases in plants. We here present a comparative analysis of 1159 candidate plant Snf2 genes in 33 complete and annotated plant genomes, including two green algae. The number of Snf2 ATPases shows considerable variation across plant genomes (17-63 genes). The DRD1, Rad5/16 and Snf2 subfamily members occur most often. Detailed analysis of the plant-specific DRD1 subfamily in related plant genomes shows the occurrence of a complex series of evolutionary events. Notably tomato carries unexpected gene expansions of DRD1 gene members. Most of these genes are expressed in tomato, although at low levels and with distinct tissue or organ specificity. In contrast, the Snf2 subfamily genes tend to be expressed constitutively in tomato. The results underpin and extend the Snf2 subfamily classification, which could help to determine the various functional roles of Snf2 ATPases and to target environmental stress tolerance and yield in future breeding.
Introduction
In eukaryotes, genomic DNA is organized into chromatin, which is physically restricting the access of regulatory proteins to the genome [1]. The access to the genome can be changed by chromatin modifying activities, altering histone tails or the histone cores covalently; and chromatin remodeling activities, altering DNA–histone interactions non-covalently [1]. Both provide important epigenetic mechanisms to regulate gene expression [2]. The associated ATP-dependent changes in nucleosome organization catalyzed by Snf2-family ATPases accounts for a large part of chromatin remodeling activities [2].
Snf2 ATPases show broad functional diversity and are involved in a variety of genome-wide processes involving DNA, such as transcription, replication, repair and recombination. As ATPase they provide a motor that can translocate and move a complex directionally on double-stranded DNA [2]. In general, Snf2 family ATPases form large complexes with interacting partners [3], although few Snf2 family members can act alone [4,5]. Swapping the ATPase region of two different Snf2 family ATPases in different complexes can also exchange their functionality [6]. The Snf2 ATPases therefore shape the functionality of a complex.
A first analysis of Snf2 family ATPases based on 30 sequences resulted in a classification of eight distinct subfamilies [7]. Snf2 family ATPases are characterized by seven helicase motifs [2,7,8]. The sequence spanning these motifs is called the Snf2 family ATPase region (Figure S1). The conserved ATPase region averages at about 400 amino acids [7] and is supposed to catalyze the translocase activity. A new survey of 1300 Snf2 family ATPases extended the classification to six groups (Snf2-like, Swr1-like, SSO1653-like, Rad54-like, Rad5/16-like and distantly-related Snf2 members) and 24 subfamilies [2]. The division into groups and subfamilies is based on phylogenetic analyses of the Snf2 family ATPase region. In many family members additional (accessory) domains are present, reflecting the sequence-based subfamily classification [3,8]. Not all subfamilies occur in every species or kingdom. An example is the DRD1 (defective in RNA-directed DNA methylation) subfamily occurring only in plant species [9,10].
In plants, functional annotation of Snf2 family members is most advanced in Arabidopsis. The Arabidopsis genome encodes 41 Snf2 family gene loci (http://www.chromdb.org; http://www.snf2.net). Encoded genes are distributed over six groups and 18 subfamilies. The specific function of the majority of the Snf2 proteins in plants is unknown [3], apart from the general contribution to DNA repair and recombination in development [2,11]. Different Snf2 ATPases, including members of the Snf2 and DRD1 subfamilies, have been shown to play a role in plant stress responses. Hence, the exploitation of such genes provides the basis for further functional characterization and could help develop plants that are better able to withstand environmental variation and/or (a)biotic stress. This may result in higher yields in less favorable environments.
We here present the first comprehensive analysis of Snf2 family members within the plant kingdom, to investigate phylogenetic relationships and infer putative specific functions of individual family members. Plant genomes show a high variability of the number of Snf2 genes, ranging from 17 to 63 members. The tomato (S. lycopersicum) genome shows gene expansions of the DRD1 subfamily with distinct expression patterns, suggesting further subfunctionalization of the duplicated members.
Materials and Methods
Genome sequence data, databases and software
Tomato (S. lycopersicum) assembly release 2.40 and iTAG annotation release 2.3 [12] were retrieved from the SGN network (http://www.solgenomics.net).The potato (S. tuberosum group Phureja DM1-3 516R44 (CIP801092)) genome assembly v3 and annotation v3.4 [13] were retrieved from the Potato Genome Sequencing Consortium (http://www.potatogenome.net). Where available, SGN Unigene builds (http://www.solgenomics.net; accessed on 7 October 2011) of other solanaceous species were used. Other green plant genome data were taken from Phytozome [14] (http://www.phytozome.net; version 7). The rice (O. sativa) annotation of Phytozome was enhanced by incorporating the annotation of the Rice Annotation Project Database [15,16]. In addition, protein sequences from ChromDB (http://chromdb.org; accessed on 7 October 2011), UniRef100 (http://www.uniprot.org; accessed on 7 October 2011) and RefSeq [17] (accessed on 7 October 2011) were used. Arabidopsis genome data were obtained from TAIR (http://www.arabidopsis.org/). Snf2 family analysis of Arabidopsis and rice was taken from the general Snf2 family protein resource (http://www.snf2.net/) for reference [8]. Taxonomy information was obtained from the Tree-of-Life project (http://tolweb.org/) and Phytozome.
Phylogenetic Analysis
Data preparation, conversion and filtering were performed with custom Perl scripts, BioPerl [18] and Bio::Phylo [19]. For the Snf2 gene calling in potato, potato protein sequences were determined by aligning all candidate Snf2 ATPase protein sequences against the potato genome using tBlastn [20] (E-value < 10). Hits were clustered into genomic regions with single linkage clustering (distance cut-off of 15kb) using C Clustering Library/Algorithm::Cluster [21]. Final gene models were predicted with Exonerate [22] using the parameters ‘--model protein2genome --showvulgar no --showalignment no --showtargetgff yes’ in the respective regions. Predicted potato gene models, unigenes, cDNAs and transcript sequences were translated using ESTScan2 [23] (additional parameter '-l 200') with the tomato hexamer frequency model obtained from SGN (http://www.solgenomics.net).
Domain detection was performed with HMMER v3.0 [24] and InterproScan [25] using Interpro Database version 35.0 (15 December 2011). Domain profiles were obtained from Pfam [26] and SMART [27]. A domain detection threshold of 1e-3 was used. It was adjusted with Arabidopsis as reference. To create an HMM model of the ATPase region, seed sequences were selected from UniProt, plant section, with the requirement of having the SNF2_N and Helicase_C domains present. Protein sequences smaller than 200 aa or with “putative”, “uncharacterized” or “predicted” in the description were excluded. The ATPase region was selected manually by identifying its conserved motifs Q-N (according to [8]) in the multiple alignment of the seed sequences. The model itself was trained with HMMER v3.0 [24], using hmmbuild with default parameters. A bitscore-based threshold of 200 was used to filter for Snf2 candidates. It was adjusted with Arabidopsis as reference.
Protein alignments were carried out with MAFFT v6.717b [28] using the E-INS-i mode with a maximum of 1000 iterations. Phylogenetic trees were estimated with RAxML v7.7.5 [29,30] using the fast bootstrapping mode and the JTT matrix model (parameters were ‘-x 12345 -p 12345 -f a -m PROTGAMMAJTTF’).
Gene duplications and losses were evaluated with Notung [31]. Intrinsically disordered regions were analyzed with FoldIndex [32] using a score cut-off of -0.2. Phylogenetic trees were visualized with Dendroscope v3 [33] or E.T.E. [34].
Expression data and analysis
Publicly available RNA-seq datasets from tomato (Solanum lycopersicum cv. Heinz 1706; data SRA049915) were retrieved from the SRA database (http://www.ncbi.nlm.nih.gov/sra). Sequence reads were mapped against the tomato reference genome (v. 2.40) with GSNAP [35]. The number of fragments per kb of exon per million fragments mapped (FPKM-values) were estimated for each gene model with cufflinks [36] on the basis of the iTAG 2.3 annotation and in-house enhanced gene models, where applicable. Conversions between SAM and BAM formatted alignments were performed with SAMtools [37]. Genes were categorized in three classes of expression: lowly expressed (FPKM ≤ 5), moderately expressed (5 < FPKM ≤ 200) and highly expressed (FPKM > 200). These categories are similar to a recent analysis of maize RNA-seq data [38], however without the more stringent cut-off proposed. For comparison, the cut-off based on the 95% confidence level was also used for analysis.
RT-PCR analysis
Tomato cultivar Heinz plants were grown in a controlled greenhouse at 23°C in long-day conditions (16 h light/8 h darkness). Seedlings were grown on ½ MS (Murashige & Skoog) agar plates supplemented with 1% sucrose in a growing chamber at 25 °C in long-day conditions. Total RNA was isolated from 10-day-old seedlings, as well as from flowers, leaves and green mature fruits from greenhouse-grown plants using the E.Z.N.A.™ Plant RNA Mini Kit (Omega Bio-Tek, Inc., USA) followed by on column DNase treatment (Qiagen, RNase-free DNase Set). One microgram of RNA was used for cDNA synthesis using the iScriptTM cDNA Synthesis Kit (Bio-Rad Laboratories, Inc., USA) according to the recommendations of the manufacturer. Primers were designed with Primer3Plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi; [39]) and checked for uniqueness in the tomato genome v. 2.40/ ITAG annotation v. 2.3 with the short-sequence BLASTN search of the BLAST 2.2.22+ toolkit (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Primers used are listed in Table S1. All primer pairs were validated by generating positive PCR reactions on genomic DNA. For RT-PCR, 2.5 µl of 10-times diluted cDNA was used. In all cases, actin was used as a reference gene [40]. The conditions used for all RT-PCR were: 95 °C for 4 min, followed by 25 to 35 cycles of 95 °C for 30 s, 60 °C for 30 s, 72 °C for 90 s and final extension at 72 °C for 7 min.
The activity of the primers was tested in a series of PCR reactions on genomic DNA with different concentrations of each primer. The concentration with highest band intensity was determined as the best primer concentration. The specificity of all primer pairs was established in a series of PCR reactions with tomato genomic DNA or cDNA to have only one single band of expected size (data not shown).
Results
Variable numbers of Snf2 family members in plant genomes
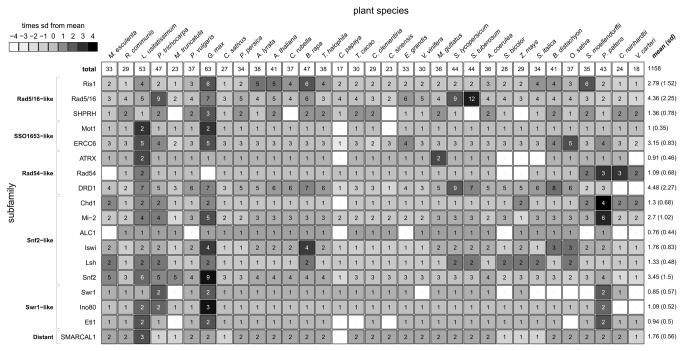
Snf2 family members in the predicted proteomes of 33 plant genomes including two green algae, were identified (Table S2). To prevent the inclusion of peptide fragments in the gene predictions, a cut-off of 200 amino acids (aa) was used, given that the conserved ATPase region has a length of about 400 aa [7]. All protein sequences longer than 200 aa were analyzed for the presence of the SNF2_N and Helicase_C domain. To be considered present, domains required a match in the protein sequence with an E-value smaller than 1e-3. Protein sequences containing at least one SNF2_N domain and one Helicase_C domain were listed as candidate Snf2 ATPase. To improve accuracy, a HMM model spanning the conserved ATPase region was created. The initial result set was filtered with this model and only candidates with a bitscore of at least 200 were used for further analyses. For Arabidopsis, all (41) previously known Snf2 genes (ChromDB; [41]) were identified (Figure 1). In total, 1159 family members were identified (Figure 1).
Figure 1. Distribution of Snf2 family members in plant genomes.
Groupings and subfamilies on the left are named according to the Arabidopsis subfamily classification [3]. Species names on the top are organized on the basis of their phylogenetic relationship according to Phytozome [14]. Snf2 candidate member Cre09.g390000.t1.1 (Chlamydomonas reinhardtii) could not be assigned to any subfamily and was excluded. Subfamily counts are shaded according to the deviation from the subfamily mean in standard deviations (sd). The total count is given on the top right cell. Mean and standard deviations per subfamily are indicated in the last column.
The total number of candidate Snf2 ATPases in plant genomes (Figure S2) shows considerable variation, ranging from 17-63 genes, with an interquartile range of 11, settled between 32 (Q1) and 43 (Q3). The papaya (Carica papaya) genome has only 17 candidate Snf2 family members, whereas in soybean (Glycine max, 63 members) and flax (Linum usitatissimum, 53 members) show an elevated number of family members. We identified 44 candidate Snf2 family members in the tomato genome (Figure 1), whereas the potato genome would carry only 23 candidate members that are also present in the official potato genome annotation. Given that both genomes are closely related in the Solanaceae genus, the surprising difference motivated an identification and re-calling of Snf2 genes in the potato genome. The re-calling identified 21 unannotated candidate Snf2 genes in the potato genome, in addition to the 23 from the first analysis. In other plant annotations, the number of potential Snf2 members was comparable between the genome annotation from Phytozome [14] and the re-calling (data not shown). Hence, all subsequent analyses were carried out with the set of 44 Snf2 family members in potato, the tomato annotation from ITAG and the annotation from Phytozome in all other cases.
Phylogenetic analysis
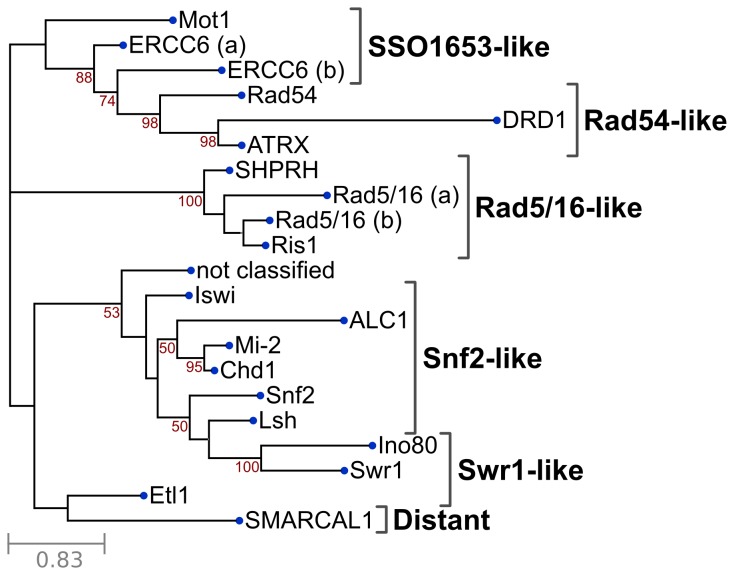
To infer evolutionary and potentially functional relationships of all plant candidate Snf2 genes, a phylogenetic tree was estimated on the basis of the conserved ATPase region of the protein sequence, including 30 aa flanking sequence on both sides to compensate for inaccuracies in domain prediction. To provide a more complete survey with focus on the Solanum genus, also transcriptome and unigene data (Table S2) were included. Each Snf2 subfamily was labeled according to the name of the Arabidopsis Snf2 subfamily in the relevant branch of the estimated tree. The unrooted tree summarizing the evolutionary relationships is presented in Figure 2.
Figure 2. Unrooted phylogenetic tree of all candidate Snf2 genes in plant genomes.
The full tree from which this subset was extracted is presented in Figure S3. The subfamily branches were collapsed to a single node that represents the first split that is part of the subfamily branch. Confidence values (50-100) are indicated at the relevant splits of the branches. The tree is based on 100 bootstrap replicates. The leaf tagged ‘not classified’ indicates candidate Snf2 members that are not part of a known subfamily, including Cre09.g390000.t1.1 (Chlamydomonas reinhardtii) and members of sequence databases.
All 18 subfamilies identified are present in the tree and the overall tree topology of plant Snf2 genes is in agreement with earlier analyses [8], although members of the subfamilies Rad 5/16 and ERCC6 were distributed over two different branches. In green algae, only 3 of the 18 subfamilies are not present (DRD1, ALC1 and Ino80), suggesting a high conservation of Snf2 ATPases in the plant kingdom. The distribution of genes over the various Snf2 subfamilies per plant species is presented in Figure 1. For this estimation, only whole genome data were included. Half of the subfamilies occur in relatively small numbers (mean < 2), whereas 19 of 33 plant species miss one or more of these subfamilies. Four subfamilies (mean ≥ 3) are large: DRD1, Rad 5/16, Snf2 and ERCC6. Largest is the plant-specific DRD1 subfamily (148 members, mean 4.48), followed by the Rad 5/16 subfamily (144 members, mean 4.36) and the Snf2 subfamily (114 members, mean 3.45). Eight Snf2 candidate members originating form ChromDB, RefSeq and UniRef100 and the Snf2 candidate member Cre09.g390000.t1.1 (Chlamydomonas reinhardtii) could not be assigned to any subfamily (not classified). These members were not taken into account. More plant genomes will have to be sequenced to ascertain whether the Snf2 family member distribution reflects any phylogenetic bias in genome sequencing.
Snf2 family members involved in stress responses: DRD1 and Snf2
We focused further analyses on the two subfamilies reported to be connected to stress responses in plants, the DRD1 and Snf2 subfamilies [42–45] and on tomato and potato. Functional annotation of these subfamilies is guided by the functional information available for Arabidopsis genes.
DRD1 subfamily
In Arabidopsis, the DRD1 subfamily has six members. Tomato has eleven members and potato seven. To characterize the phylogenetic relationships between the DRD1 subfamily members of plant species in the Asterid clade (potato, tomato and Mimulus guttatus) and Arabidopsis as model plant at a high resolution, the further analysis was focused on these four plants. According to the species tree (Figure S2), Mimulus is most close to the two solanaceous plants of interest. It has five DRD1 members.
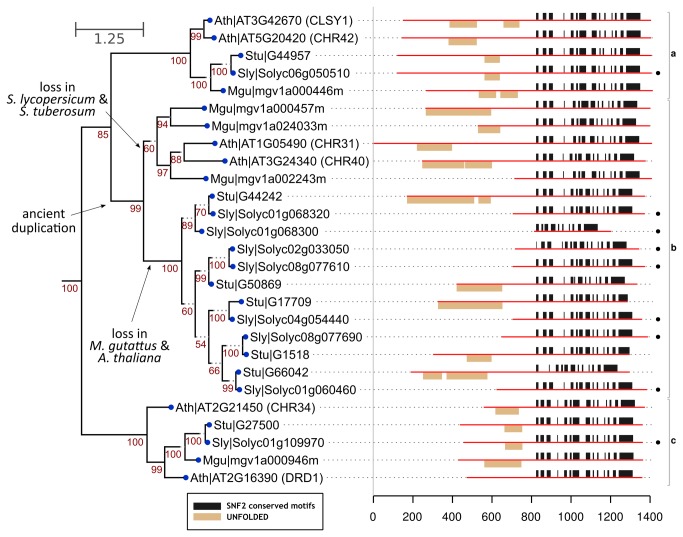
In the unrooted phylogenetic tree based on the data from these four species (Figure 3), the DRD1 members could be grouped in three distinct branches, labeled a, b and c, each containing two Arabidopsis members. AtCHR42 and AtCLSY1 are in branch a, AtCHR31 and AtCHR40 in branch b, whereas AtDRD1 and AtCHR34 are in branch c. In all three branches, DRD1 members from tomato, potato and Mimulus are present. The tree shows that AtCHR42 and AtCSLY1 are in-paralogs [46] with one ortholog in tomato, potato and Mimulus (Figure 3; branch a). Likewise, AtDRD1 and CHR34 are in-paralogs with also one ortholog in tomato, potato and Mimulus (Figure 3, branch c). It is apparent from the tree that branch b is the most complex. In addition to the two members of Arabidopsis in branch b, Mimulus has 3, potato 7 and tomato 9 members. The number of members in branch c is relatively stable in other plant species, ranging from 1 to 3 (mean 1.49, sd 1, tomato and potato excluded). This indicates a relative expansion of DRD1 ATPases in the tomato and potato genomes.
Figure 3. Analysis of the DRD1 subfamily in tomato, potato, Mimulus and Arabidopsis.
The left side shows a detailed view of the DRD1 subfamily branch of an unrooted tree based on 1000 bootstraps of Snf2 data from Arabidopsis thaliana (Ath), Mimulus guttatus (Mgu), Solanum lycopersicum (Sly) and Solanum tuberosum (Stu). Confidence values (50-100) are given at the relevant branches of the tree. Identifiers give the name of the organism in three-letter abbreviations together with gene identifiers. The individual branches identified are indicated by letters in lowercase on the right side. To increase readability, some branch edges have been extended by dotted grey lines. These grey dotted lines are therefore not part of the estimated branch length. The right side shows structural elements (domains and unfolded regions) in the protein sequence of the DRD1 subfamily members in Arabidopsis, Mimulus, tomato and potato. Besides the ATPase region no other domains are present in these genes. A black dot at the right end of the figure indicates the expression of the respective gene in tomato based on the analysis of RNA-seq data.
The potato/tomato members establish a separate sub-branch without members of either Arabidopsis or Mimulus suggesting independent evolution of DRD1 members in tomato and potato. Such evolution requires, the occurrence of a gene duplication in the common ancestor of all four species (labeled ‘ancient duplication’ in Figure 3), followed by independent gene losses in all four species. The high confidence value (99 from 100) for the ancient duplication supports this scenario. Also analysis with Notung [31] supports the mutual gene loss scenario (details not shown). The evolutionary history of solanaceous DRD1 genes suggests specific functions for such genes in tomato and potato.
To infer potential functions of the DRD1 subfamily members, we investigated the presence of additional structural/functional elements in the protein sequences. The DRD1 subfamily members of the four species here investigated had no accessory domains (Figure 3). In many cases, the N-terminal region of DRD1 subfamily members shows a predicted disordered region. In Arabidopsis, this applies to all DRD1 subfamily members, except for the AtDRD1 protein (Figure 3).
Snf2 subfamily
In Arabidopsis, the Snf2 subfamily has four members, while only three were found in tomato, potato and Mimulus. The tree estimated on data from these four species again shows three distinct branches (Figure S4), labeled a, b and c, respectively. The Arabidopsis genes AtCHR12 and AtCHR23 cluster together (Figure S4, branch a), in addition to single genes of the other species. It shows that AtCHR12 and AtCHR23 are in-paralogs with one ortholog in tomato, potato and Mimulus. The two Arabidopsis genes are likely to be the result of a gene duplication event specific to the Arabidopsis genus. The other Arabidopsis genes form one-to-one ortholog relationships with the respective tomato, potato and Mimulus genes (Figure S4). The evolutionary history of the Snf2 subfamily is therefore overall much less eventful than the history of the DRD1 subfamily.
AtCHR12 and AtCHR23 (branch c) carry an unfolded region at the C-terminal end which is not present in any of the other members of the branch (Figure S4). The difference in length of the proteins in this subfamily is remarkable. Whereas branch a consists of relatively short proteins of approx. 1100 amino acids, branch b is characterized by very large proteins, the largest one (AtSYD) carrying 3574 amino acids. AtSYD has a considerably larger C-terminal end compared to all orthologs in its branch and compared to all members in the subfamily. Yet it only shows an unfolded region in the C-terminal end and no other functional or structural domains.
Expression analysis of DRD1 and Snf2 subfamilies
Expression characteristics could also help elucidating the biological function of DRD1 and Snf2 subfamily members. We evaluated the expression profile of these genes in tomato public-domain RNA-seq libraries [12] for flowers, roots, leaves and various stages of fruit of tomato cv Heinz 1706 (Table S3). The FPKM-values of all libraries were calculated and visualized as heat map for the DRD1 and Snf2 subfamilies (Figure S5). All three Snf2 subfamily members of tomato are moderately expressed in the majority of the libraries analyzed. No tissue specificity and/or developmental control are apparent, suggesting a constitutive expression.
In contrast, expression of members of the DRD1 subfamily is more heterogeneous. The highest and most diversely expressed DRD1 subfamily genes are Solyc01g109970 (branch c) and Solyc06g050510 (branch a). Solyc01g109970 is constitutively expressed in all libraries with FPKM values from 5 (leaves) to 37 (fully ripe fruit). Expression of the Solyc06g050510 gene is similar, with the highest FPKM-value of 30 in roots, mature green fruits, immature fruits and 3-cm fruits. The lowest expression shows this gene in breaker and fully ripe fruits (FPKM around 7). The gene Solyc01g068320 shows low specific expression in flower and flower bud tissue. The other 5 members that constitute the solanaceous-specific expansion of branch b in tomato show extremely low expression.
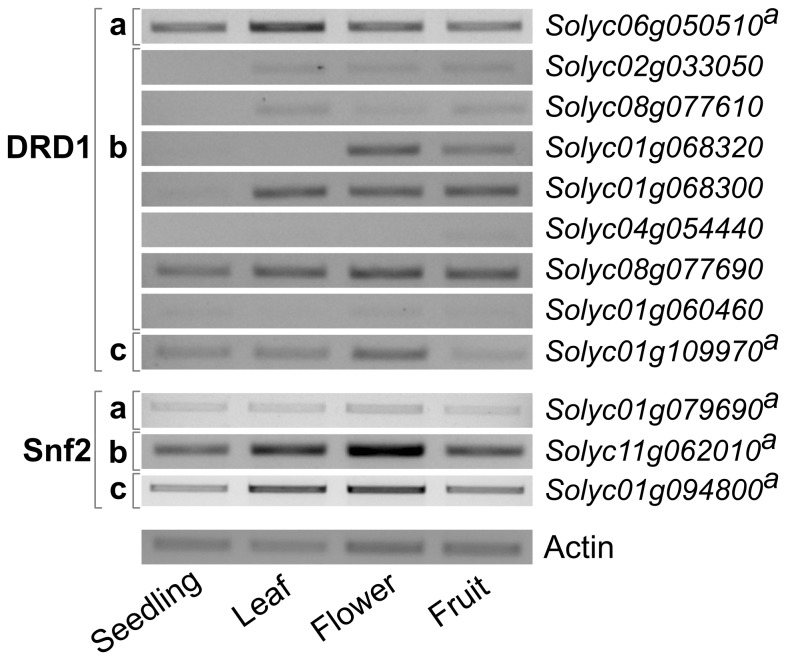
To confirm these expression characteristics, semi-quantitative RT-PCR was performed on leaves, flowers and mature fruits. To be able to extend the analysis to early stages of plant development, 10-day-old in vitro-grown seedlings were included. RT-PCR analysis of the three Snf2 genes confirmed expression in all four tissues analyzed, in concordance with the RNA-seq analysis (Figure 4). It also largely confirmed the RNA-seq results of the DRD1 subfamily genes (Figure 4). Solyc08g077690 is expressed in all tissues examined at the highest level shown by any member in this branch. Expression of Solyc01g068320 is restricted to flower and fruit tissue, the latter at lower levels. For Solyc01g068300 RT-PCR shows a relatively easily detectable product in all tissues except seedlings. Also expression of Solyc02g033050, Solyc01g060460 and Solyc08g077610 is detectable by RT-PCR in all tissues. However, the level of expression is low to very low, approaching the lower limit of reliable detection. Gene Solyc04g054440 is very lowly expressed in possibly only fruits. The highly variable expression patterns of the various DRD1 subfamily genes indicate that the putative function of the encoded DRD1 proteins is likely to be subtle in terms of time or location.
Figure 4. RT-PCR expression analysis of DRD1 and Snf2 subfamilies of tomato Snf2 ATPase genes.
The tissue used is indicated on the x-axis. The individual genes are indicated of the right, the branches identified on the left. The expression of the actin gene (25 cycles) was used as control (lower panel). The number of PCR cycles used for the analysis of the individual gene was adjusted to generate a detectable amount of PCR product. For most of genes, 35 cycles were used. Genes marked with superscript a (a) were amplified with 29 cycles. For the actin gene 25 cycles were used.
Discussion
The Snf2 family of ATPases is a large family of chromatin remodeling enzymes that have versatile roles in a variety of fundamental processes in growth and development. In plants, little is known about the function of individual members of this family, although notably in Arabidopsis functional relationships with gene regulation, DNA recombination, DNA repair and stress tolerance have been reported [42,43,47]. Here, we present the first comprehensive comparative analysis of all Snf2 genes in 33 sequenced and annotated plant genomes, including two green algae. We have identified and analyzed 1159 potential candidate Snf2 family ATPases, of which all but one could be placed in previously established groups and subfamilies and represent genuine plant Snf2 genes. The variation in numbers of Snf2 genes is large, ranging from 17 in papaya to 63 in soybean. This suggests a broad functional diversification of this gene family in the plant kingdom. The high member counts in flax and soybean may originate from recent whole-genome duplications in both species [48,49]
Our results for rice show considerably more differences when compared to another recent study of Snf2 family genes [50], in which 39 putative Snf2 family genes are identified. The overall tree presented [50] does not seem to agree well with the subfamily classification. An example is a branch containing rice genes (Os02g0114000 (Snf2), Os01g0779400 (Ris1), Os05g0150300 (Iswi), Os05g0392400 (DRD1) and Os07g0497000 (Mi-2)) that are distributed in five different subfamilies according to our classification. Possible explanations for the differences are phylogenetic tree modeling based on the complete protein sequence rather than the conserved region, and/or the use of another rice annotation ([15]; http://rapdb.dna.affrc.go.jp).
Surprising sources of error in Snf2 family member identification are the publicly available genome assemblies and annotations. Our example in potato highlights the better performance of gene calling within a protein family opposed to automatic gene calling. Half of the Snf2 family members are absent from the current genome annotation of potato. Assembly and calling of Snf2 genes may be troublesome for the partly automated pipelines in place for overall genome assembly and annotation, despite manual curation effort. Here we show increased sensitivity of candidate Snf2 family gene identification by iterative rounds of homology-based gene prediction. This approach minimizes errors in the predicted coding region that would affect the multiple sequence alignment and phylogenetic reconstruction considerably. For Arabidopsis and rice, the plant species with the richest set of annotation and experimental data, inferred gene models were consistent with the currently available high-quality annotations (not shown). Therefore, the annotation of the potato Snf2 family is likely to have improved markedly with the homology-based prediction routine put in place and is recommended for future analyses. The accuracy of the prediction of the proper coding region is not likely to be improved with the help of (family-) specific gene models or better hexamer models. Such homology-based prediction will not safeguard against errors in assembly.
Not anticipated from the earlier analyses of Snf2 family genes [8] is the relative expansion of the DRD1 (148 genes), Rad5/16 (144 genes) and Snf2 (114 genes) subfamilies in plant genomes. So far, members two of these subfamilies have been associated with environmental stress responses in Arabidopsis, possibly indicating the relative importance of chromatin remodeling in combatting environmental stress in plants. The most abundant subfamily, DRD1, has evolved from apparent non-existence in non-plant species (www.snf2.net) and lower plants, such as Volvox carteri and Chlamydomonas reinhardtii, to the largest and most diverse subfamily in current-day higher plants. It indicates that the DRD1 protein has become an important and possibly diversified asset in the regulation of plant growth and development. Within the expanded DRD1 subfamily, tomato has one of the highest member count of all genomes analyzed, whereas potato, even if higher than average, does not reach this high member count. However, the expansion within this subfamily was not uniform, and while some seem to be unique for Solanaceae (Figure 3, branch b), in other cases, the genome of Arabidopsis carries two genes whereas potato and tomato have only one.
The DRD1 subfamily tree suggests a complex evolutionary history involving a series of independent gene losses, duplication and genomic reshuffling events (recombination, transposition) resulting in a relative expansion of genes in notably tomato. It suggests that the DRD1 subfamily has gained additional functionality in tomato. The results suggest that the relative expansion has been specific for the Solanaceae, although more solanaceous genomes (S. penelli, N. tabacum, S. pimpinellifolium) are required to validate the specificity of this expansion for Solanaceae in general, or for a given species in particular.
It is supposed that the conserved ATPase domain is responsible for the energy release of DRD1 proteins, whereas other parts of the protein specify interaction partners, DNA specificity and/or sub-nuclear localization. The presence of a disordered region that may be characteristic for the expanded branch b. The differences in structure, if any, are so subtle or complex that it is difficult to associate particular sequence determinants with function. The unfolded regions occur regularly at approximately the same position in the N-terminal regions of DRD1 proteins. Such unfolded regions may help or direct protein-protein or protein-nucleic acid interactions [51–53]. Disordered regions in the DRD1 genes may therefore interface the ATPase domain to other proteins or DNA/RNA molecules [54]. This may help to specify interaction partners, whereas the lack of accessory domains indicates that ATPase-mediated remodeling is the main enzymatic function of these DRD1 subfamily members. New interaction partners could determine involvement of DRD1 proteins in new biological processes or conditions.
Given the complex evolution and expression pattern of DRD1 genes in tomato, it is not as straightforward as for the Snf2 subfamily to transfer the function of Arabidopsis genes to the orthologous tomato genes. In Arabidopsis, several genes of this subfamily are important components of RNA-directed DNA methylation (RdDM), the pathway in which specific genomic loci are targeted for methylation by 24 bases small interfering RNAs (siRNA) [44,55]. RdDM operates in many organisms and requires common components such as DNA methyltransferases, histone modifying enzymes and RNAi proteins.
The genes of branch a, CLSY1 and AtCHR42 were found in the Pol-IV polymerase protein complex [56], the RNA polymerase thought to initiate the biogenesis of the targeting siRNAs [57]. In the same complex, ATCHR31 and ATCHR40 (branch b) are also present, suggesting they play a role in the same RdDM pathway [58]. In addition to siRNAs, RdDM is also associated with the accumulation of so-called intergenic noncoding (IGN) transcripts that involves the plant specific RNA-polymerase Pol-V [59]. DRD1 (branch c) was identified in a protein complex critical for the production of Pol-V dependent IGN transcripts [56]. Recently, this gene was also established as an important player in plant immunity. Its knockout mutant showed increased susceptibility to the fungal pathogen Plectosphaerella cucumerina [45]. The second gene of Arabidopsis branch c, At2g21450, was shown to be modulated during early embryogenesis, suggesting a role after fertilization [60]. Related functions affecting small RNA accumulation and cytosine methylation have been shown for RMR1, an Snf2 ortholog in Zea mays (maize), in the context of paramutation [61]. As five out of six Arabidopsis DRD1 genes and RMR1 are implicated in RdDM pathways, a similar function of this subfamily in tomato is likely.
Why tomato would need so much more active DRD1 genes than Arabidopsis? Possibly the continued selection for traits in tomato as agricultural crop has been the driving force for such developments. The functions assigned so far in Arabidopsis point in the direction of protection against biotic and abiotic stresses. The comprehensive analysis here presented shows the evolution and presence of Snf2 genes in plants. Closer evaluation of, e.g. DRD1 subfamily members, could make suitable targets for breeding and plant improvement.
Supporting Information
Schematic layout of Snf2 family ATPases. The conserved Snf2 family ATPase region is part of the protein and consists of two Pfam domains, Snf2_N and Helicase_C, in which seven helicase motifs are present. The average size of the Snf2 family ATPase region is approx. 400aa [1]. In individual proteins, the N-terminal or C-terminal region can be very small [2].
(TIF)
The number of candidate Snf2 genes in annotated plant genomes. The total number of genes estimated for a genome is plotted above the bar in the histogram. Plant species included are organized on the basis of the position in the tree of life (shown at the left). The four species given most attention in this study (Arabidopsis, potato, tomato and Mimulus guttatus) are given in black.
(TIF)
Full phylogenetic tree of all plant Snf2 candidates. The tree is based on the plant data listed in table S2 and calculated with 100 bootstraps due to computational constraints. Branches with a confidence lower than 50 are marked in grey. Members not classified (n.c.) into any subfamily are indicated in light green. To increase readability, the colors of subfamily branches alternate between blue and red.
(TIF)
Analysis of the Snf2 subfamily in tomato, potato, Mimulus and Arabidopsis. The left side shows a detailed view of the DRD1 subfamily branch of an unrooted tree based on 1000 bootstraps of Snf2 data from Arabidopsis thaliana (Ath), Mimulus guttatus (Mgu), Solanum lycopersicum (Sly) and Solanum tuberosum (Stu). Confidence values (50-100) are given at the relevant branches of the tree. Identifiers give the name of the organism in three-letter abbreviations together with gene identifiers. The individual branches identified are indicated by letters in lowercase on the right side. To increase readability, some branch edges have been extended by dotted grey lines. These grey dotted lines are therefore not part of the estimated branch length. The right side shows structural elements in the protein sequence of the Snf2 subfamily members in Arabidopsis, Mimulus, tomato and potato. The individual branches identified are indicated by letters in lowercase. Besides the ATPase region, BROMO (protein-histone interaction), QLQ (protein-protein interaction) and HSA (DNA-binding) domains are present in several members. A black dot at the right end of the figure indicates the expression of the respective gene in tomato based on the analysis of RNA-seq data.
(TIF)
Heat map of the RNA-seq expression data of the tomato DRD1 & Snf2 subfamily genes. The expression is indicated as fragments per kb exon model per million mapped reads-value (FPKM-value). No cut-off was applied. Grey areas correspond to FPKM-values of 0. Gene identifiers are indicated on the x-axis with the corresponding branch name given between brackets. The biological material used to generate the RNA-seq libraries is given on the y-axis. Replicates are indicated by lowercase letters. Details on the RNA-seq libraries used are given in table S3.
(TIF)
Primers used for RT-PCR analysis. The primer sequence of the forward (F) and reversed (R) primer is given for each gene identifier.
(DOC)
Plant data included in the analyses. Sources are the Phytozome annotation (indicated as genome), SGN unigenes (indicated as unigene), de-novo assembled transcriptomes (indicated as transcript) and reference databases (indicated as database). The differences in Snf2 members between the annotation (first value) and the homology-based re-analysis here presented (second value) are indicated for potato (Solanum tuberosum).
(DOC)
RNA-seq libraries included in the analysis. Data are from the short read archive (SRA; http://www.ncbi.nlm.nih.gov/sra). The library and sample IDs refer to the run and sample identifiers in SRA, respectively.
(DOC)
Text file with custom predicted gene models of Solanum tuberosum.
(ZIP)
Text file of the multiple alignment of all plant Snf2 candidates.
(ZIP)
Phylogenetic tree of all plant Snf2 candidates in NEWICK format.
(ZIP)
(DOC)
Acknowledgments
The authors would like to thank Dr. Berend Snel and Dr. Michael Seidl (Utrecht University, the Netherlands) for the help with the interpretation of the phylogenetic analyses. We also would like to thank Dr. Andrew Flaus (NUI Galway, Ireland) for his valuable comments.
Funding Statement
This work was supported by the BioRange program of the Netherlands Bioinformatics Centre (NBIC), which is supported by a BSIK grant through the Netherlands Genomics Initiative (NGI), and by the bioinformatics and biostatistics program of the Centre for BioSystems Genomics (CBSG2012). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kwon CS, Wagner D (2007) Unwinding chromatin for development and growth: a few genes at a time. Trends Genet 23: 403–412. doi: 10.1016/j.tig.2007.05.010. PubMed: 17566593. [DOI] [PubMed] [Google Scholar]
- 2. Flaus A, Owen-Hughes T (2011) Mechanisms for ATP-dependent chromatin remodeling: the means to the end. FEBS J 278: 3579–3595. doi: 10.1111/j.1742-4658.2011.08281.x. PubMed: 21810178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Knizewski L, Ginalski K, Jerzmanowski A (2008) Snf2 proteins in plants: gene silencing and beyond. Trends Plant Sci 13: 557–565. doi: 10.1016/j.tplants.2008.08.004. PubMed: 18786849. [DOI] [PubMed] [Google Scholar]
- 4. Lall S (2011) A bottle opener for TBP. Nat Struct Mol Biol 18: 865. doi: 10.1038/nsmb0811-865. PubMed: 21811312. [DOI] [PubMed] [Google Scholar]
- 5. Hauk G, McKnight JN, Nodelman IM, Bowman GD (2010) The chromodomains of the Chd1 chromatin remodeler regulate DNA access to the ATPase motor. Mol Cell 39: 711–723. doi: 10.1016/j.molcel.2010.08.012. PubMed: 20832723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fan H-Y, Trotter KW, Archer TK, Kingston RE (2005) Swapping function of two chromatin remodeling complexes. Mol Cell 17: 805–815. doi: 10.1016/j.molcel.2005.02.024. PubMed: 15780937. [DOI] [PubMed] [Google Scholar]
- 7. Eisen JA, Sweder KS, Hanawalt PC (1995) Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res 23: 2715–2723. doi: 10.1093/nar/23.14.2715. PubMed: 7651832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Flaus A, Martin DMA, Barton GJ, Owen-Hughes T (2006) Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res 34: 2887–2905. doi: 10.1093/nar/gkl295. PubMed: 16738128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kanno T, Mette MF, Kreil DP, Aufsatz W, Matzke M et al. (2004) Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr Biol 14: 801–805. doi: 10.1016/j.cub.2004.04.037. PubMed: 15120073. [DOI] [PubMed] [Google Scholar]
- 10. Matzke M, Kanno T, Huettel B, Daxinger L, Matzke AJM (2006) RNA-directed DNA methylation and Pol IVb in Arabidopsis. Cold Spring Harb Symp Quant Biol 71: 449–459. doi: 10.1101/sqb.2006.71.028. PubMed: 17381327. [DOI] [PubMed] [Google Scholar]
- 11. Sang Y, Silva-Ortega CO, Wu S, Yamaguchi N, Wu M-F et al. (2012) Mutations in two non-canonical Arabidopsis SWI2/SNF2 chromatin remodeling ATPases cause embryogenesis and stem cell maintenance defects. Plant J: ([MedlinePgn:]) doi: 10.1111/tpj.12009. PubMed: 23062007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sato S, Tabata S, Hirakawa H, Asamizu E, Shirasawa K et al. (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641. doi: 10.1038/nature11119. PubMed: 22660326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xu X, Pan S, Cheng S, Zhang B, Mu D et al. (2011) Genome sequence and analysis of the tuber crop potato. Nature 475: 189–195. doi: 10.1038/nature10158. PubMed: 21743474. [DOI] [PubMed] [Google Scholar]
- 14. Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD et al. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186. doi: 10.1093/nar/gkr944. PubMed: 22110026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rice Annotation Project, Tanaka T, Antonio BA, Kikuchi S, Matsumoto T, Nagamura Y et al. (2008) The Rice Annotation Project Database (RAP-DB): 2008 update. Nucleic Acids Res 36: D1028–D1033. doi: 10.1093/nar/gkm978. PubMed: 18089549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rice Annotation Project, Itoh T, Tanaka T, Barrero RA, Yamasaki C, Fujii Y et al. (2007) Curated genome annotation of Oryza sativa ssp. japonica and comparative genome analysis with Arabidopsis thaliana . Genome Res 17: 175–183. doi: 10.1101/gr.5509507. PubMed: 17210932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pruitt KD, Tatusova T, Maglott DR (2005) NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 33: D501–D504. doi: 10.1093/nar/gki025. PubMed: 15608248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA et al. (2002) The Bioperl toolkit: Perl modules for the life sciences. Genome Res 12: 1611–1618. doi: 10.1101/gr.361602. PubMed: 12368254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vos RA, Caravas J, Hartmann K, Jensen MA, Miller C (2011) BIO::Phylo-phyloinformatic analysis using perl. BMC Bioinformatics 12: 63. doi: 10.1186/1471-2105-12-63. PubMed: 21352572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. doi: 10.1093/nar/25.17.3389. PubMed: 9254694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. De Hoon MJL, Imoto S, Nolan J, Miyano S (2004) Open source clustering software. Bioinformatics 20: 1453–1454. doi: 10.1093/bioinformatics/bth078. PubMed: 14871861. [DOI] [PubMed] [Google Scholar]
- 22. Slater GSC, Birney E (2005) Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6: 31. doi: 10.1186/1471-2105-6-31. PubMed: 15713233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lottaz C, Iseli C, Jongeneel CV, Bucher P (2003) Modeling sequencing errors by combining Hidden Markov models. Bioinformatics 19 Suppl 2: ii103–ii112. doi: 10.1093/bioinformatics/btg1067. PubMed: 14534179. [DOI] [PubMed] [Google Scholar]
- 24. Finn RD, Clements J, Eddy SR (2011) HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39 Suppl 2: W29–W37. doi: 10.1093/nar/gkr367. PubMed: 21593126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zdobnov EM, Apweiler R (2001) InterProScan - an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17: 847–848. doi: 10.1093/bioinformatics/17.9.847. PubMed: 11590104. [DOI] [PubMed] [Google Scholar]
- 26. Finn RD, Mistry J, Tate J, Coggill P, Heger A et al. (2010) The Pfam protein families database. Nucleic Acids Res 38: D211–D222. doi: 10.1093/nar/gkp985. PubMed: 19920124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Letunic I, Doerks T, Bork P (2009) SMART 6: recent updates and new developments. Nucleic Acids Res 37: D229–D232. doi: 10.1093/nar/gkn808. PubMed: 18978020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Katoh K, Kuma K, Toh H, Miyata T (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33: 511–518. doi: 10.1093/nar/gki198. PubMed: 15661851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. doi: 10.1093/bioinformatics/btl446. PubMed: 16928733. [DOI] [PubMed] [Google Scholar]
- 30. Stamatakis A, Hoover P, Rougemont J (2008) A rapid bootstrap algorithm for the RAxML web servers. Syst Biol 57: 758–771. doi: 10.1080/10635150802429642. PubMed: 18853362. [DOI] [PubMed] [Google Scholar]
- 31. Chen K, Durand D, Farach-Colton M (2000) NOTUNG: a program for dating gene duplications and optimizing gene family trees. J Comput Biol 7: 429–447. doi: 10.1089/106652700750050871. PubMed: 11108472. [DOI] [PubMed] [Google Scholar]
- 32. Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O et al. (2005) FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics 21: 3435–3438. doi: 10.1093/bioinformatics/bti537. PubMed: 15955783. [DOI] [PubMed] [Google Scholar]
- 33. Huson DH, Richter DC, Rausch C, Dezulian T, Franz M et al. (2007) Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics 8: 460. doi: 10.1186/1471-2105-8-460. PubMed: 18034891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huerta-Cepas J, Dopazo J, Gabaldón T (2010) ETE: a python Environment for Tree Exploration. BMC Bioinformatics 11: 24. doi: 10.1186/1471-2105-11-24. PubMed: 20070885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu TD, Nacu S (2010) Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26: 873–881. doi: 10.1093/bioinformatics/btq057. PubMed: 20147302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G et al. (2010) Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515. doi: 10.1038/nbt.1621. PubMed: 20436464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J et al. (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. doi: 10.1093/bioinformatics/btp352. PubMed: 19505943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hansey CN, Vaillancourt B, Sekhon RS, de Leon N, Kaeppler SM et al. (2012) Maize (Zea mays L.) genome diversity as revealed by RNA-sequencing. PLOS ONE 7: e33071. doi: 10.1371/journal.pone.0033071. PubMed: 22438891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R et al. (2007) Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res 35: W71–W74. doi: 10.1093/nar/gkm306. PubMed: 17485472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Løvdal T, Lillo C (2009) Reference gene selection for quantitative real-time PCR normalization in tomato subjected to nitrogen, cold, and light stress. Anal Biochem 387: 238–242. doi: 10.1016/j.ab.2009.01.024. PubMed: 19454243. [DOI] [PubMed] [Google Scholar]
- 41. Gendler K, Paulsen T, Napoli C (2008) ChromDB: the chromatin database. Nucleic Acids Res 36: D298–D302. doi: 10.1093/nar/gkm768. PubMed: 17942414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Walley JW, Rowe HC, Xiao Y, Chehab EW, Kliebenstein DJ et al. (2008) The chromatin remodeler SPLAYED regulates specific stress signaling pathways. PLoS Pathog 4: e1000237. doi: 10.1371/journal.ppat.1000237. PubMed: 19079584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mlynárová L, Nap JP, Bisseling T (2007) The SWI/SNF chromatin-remodeling gene AtCHR12 mediates temporary growth arrest in Arabidopsis thaliana upon perceiving environmental stress. Plant J 51: 874–885. doi: 10.1111/j.1365-313X.2007.03185.x. PubMed: 17605754. [DOI] [PubMed] [Google Scholar]
- 44. Huettel B, Kanno T, Daxinger L, Bucher E, van der Winden J et al. (2007) RNA-directed DNA methylation mediated by DRD1 and Pol IVb: a versatile pathway for transcriptional gene silencing in plants. Biochim Biophys Acta 1769: 358–374. doi: 10.1016/j.bbaexp.2007.03.001. PubMed: 17449119. [DOI] [PubMed] [Google Scholar]
- 45. López A, Ramírez V, García-Andrade J, Flors V, Vera P (2011) The RNA silencing enzyme RNA polymerase V is required for plant immunity. PLoS Genet 7: e1002434. doi: 10.1371/journal.pgen.1002434. PubMed: 22242006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Koonin EV (2005) Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet 39: 309–338. doi: 10.1146/annurev.genet.39.073003.114725. PubMed: 16285863. [DOI] [PubMed] [Google Scholar]
- 47. Bezhani S, Winter C, Hershman S, Wagner JD, Kennedy JF et al. (2007) Unique, shared, and redundant roles for the Arabidopsis SWI/SNF chromatin remodeling ATPases BRAHMA and SPLAYED. Plant Cell 19: 403–416. doi: 10.1105/tpc.106.048272. PubMed: 17293567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang Z, Hobson N, Galindo L, Zhu S, Shi D et al. (2012) The genome of flax (Linum usitatissimum) assembled de novo from short shotgun sequence reads. Plant J 72: 461–473. doi: 10.1111/j.1365-313X.2012.05093.x. PubMed: 22757964. [DOI] [PubMed] [Google Scholar]
- 49. Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183. doi: 10.1038/nature08670. PubMed: 20075913. [DOI] [PubMed] [Google Scholar]
- 50. Li X-Y, Wang C, Nie P-P, Lu X-W, Wang M et al. (2011) Characterization and expression analysis of the SNF2 family genes in response to phytohormones and abiotic stresses in rice. Biol Plant 55: 625–633. doi: 10.1007/s10535-011-0160-1. [DOI] [Google Scholar]
- 51. Bolanos-Garcia VM, Wu Q, Ochi T, Chirgadze DY, Sibanda BL et al. (2012) Spatial and temporal organization of multi-protein assemblies: achieving sensitive control in information-rich cell-regulatory systems. Philos. Trans R Soc Ser A 370: 3023–3039. doi: 10.1098/rsta.2011.0268. [DOI] [PubMed] [Google Scholar]
- 52. Uversky VN, Dunker AK (2010) Understanding protein non-folding. Biochim Biophys Acta 1804: 1231–1264. doi: 10.1016/j.bbapap.2010.01.017. PubMed: 20117254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT (2004) Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol 337: 635–645. doi: 10.1016/j.jmb.2004.02.002. PubMed: 15019783. [DOI] [PubMed] [Google Scholar]
- 54. Nguyen Ba AN, Yeh BJ, van Dyk D, Davidson AR, Andrews BJ, et al. (2012) Proteome-wide discovery of evolutionary conserved sequences in disordered regions. Sci Signal 5: rs1 doi: 10.1126/scisignal.2002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mahfouz MM (2010) RNA-directed DNA methylation: mechanisms and functions. Plant Signal Behav 5: 806–816. doi: 10.4161/psb.5.7.11695. PubMed: 20421728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Law JA, Ausin I, Johnson LM, Vashisht AA, Zhu J-K et al. (2010) A protein complex required for polymerase V transcripts and RNA-directed DNA methylation in Arabidopsis. Curr Biol 20: 951–956. doi: 10.1016/j.cub.2010.03.062. PubMed: 20409711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pikaard CS, Haag JR, Ream T, Wierzbicki AT (2008) Roles of RNA polymerase IV in gene silencing. Trends Plant Sci 13: 390–397. doi: 10.1016/j.tplants.2008.04.008. PubMed: 18514566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Law JA, Vashisht AA, Wohlschlegel JA, Jacobsen SE (2011) SHH1, a homeodomain protein required for DNA methylation, as well as RDR2, RDM4, and chromatin remodeling factors, associate with RNA polymerase IV. PLoS Genet 7: e1002195. doi: 10.1371/journal.pgen.1002195. PubMed: 21811420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wierzbicki AT, Haag JR, Pikaard CS (2008) Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135: 635–648. doi: 10.1016/j.cell.2008.09.035. PubMed: 19013275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Xiang D, Venglat P, Tibiche C, Yang H, Risseeuw E et al. (2011) Genome-wide analysis reveals gene expression and metabolic network dynamics during embryo development in Arabidopsis. Plant Physiol 156: 346–356. doi: 10.1104/pp.110.171702. PubMed: 21402797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hale CJ, Stonaker JL, Gross SM, Hollick JB (2007) A novel Snf2 protein maintains trans-generational regulatory states established by paramutation in maize. PLoS Biol 5: e275. doi: 10.1371/journal.pbio.0050275. PubMed: 17941719. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic layout of Snf2 family ATPases. The conserved Snf2 family ATPase region is part of the protein and consists of two Pfam domains, Snf2_N and Helicase_C, in which seven helicase motifs are present. The average size of the Snf2 family ATPase region is approx. 400aa [1]. In individual proteins, the N-terminal or C-terminal region can be very small [2].
(TIF)
The number of candidate Snf2 genes in annotated plant genomes. The total number of genes estimated for a genome is plotted above the bar in the histogram. Plant species included are organized on the basis of the position in the tree of life (shown at the left). The four species given most attention in this study (Arabidopsis, potato, tomato and Mimulus guttatus) are given in black.
(TIF)
Full phylogenetic tree of all plant Snf2 candidates. The tree is based on the plant data listed in table S2 and calculated with 100 bootstraps due to computational constraints. Branches with a confidence lower than 50 are marked in grey. Members not classified (n.c.) into any subfamily are indicated in light green. To increase readability, the colors of subfamily branches alternate between blue and red.
(TIF)
Analysis of the Snf2 subfamily in tomato, potato, Mimulus and Arabidopsis. The left side shows a detailed view of the DRD1 subfamily branch of an unrooted tree based on 1000 bootstraps of Snf2 data from Arabidopsis thaliana (Ath), Mimulus guttatus (Mgu), Solanum lycopersicum (Sly) and Solanum tuberosum (Stu). Confidence values (50-100) are given at the relevant branches of the tree. Identifiers give the name of the organism in three-letter abbreviations together with gene identifiers. The individual branches identified are indicated by letters in lowercase on the right side. To increase readability, some branch edges have been extended by dotted grey lines. These grey dotted lines are therefore not part of the estimated branch length. The right side shows structural elements in the protein sequence of the Snf2 subfamily members in Arabidopsis, Mimulus, tomato and potato. The individual branches identified are indicated by letters in lowercase. Besides the ATPase region, BROMO (protein-histone interaction), QLQ (protein-protein interaction) and HSA (DNA-binding) domains are present in several members. A black dot at the right end of the figure indicates the expression of the respective gene in tomato based on the analysis of RNA-seq data.
(TIF)
Heat map of the RNA-seq expression data of the tomato DRD1 & Snf2 subfamily genes. The expression is indicated as fragments per kb exon model per million mapped reads-value (FPKM-value). No cut-off was applied. Grey areas correspond to FPKM-values of 0. Gene identifiers are indicated on the x-axis with the corresponding branch name given between brackets. The biological material used to generate the RNA-seq libraries is given on the y-axis. Replicates are indicated by lowercase letters. Details on the RNA-seq libraries used are given in table S3.
(TIF)
Primers used for RT-PCR analysis. The primer sequence of the forward (F) and reversed (R) primer is given for each gene identifier.
(DOC)
Plant data included in the analyses. Sources are the Phytozome annotation (indicated as genome), SGN unigenes (indicated as unigene), de-novo assembled transcriptomes (indicated as transcript) and reference databases (indicated as database). The differences in Snf2 members between the annotation (first value) and the homology-based re-analysis here presented (second value) are indicated for potato (Solanum tuberosum).
(DOC)
RNA-seq libraries included in the analysis. Data are from the short read archive (SRA; http://www.ncbi.nlm.nih.gov/sra). The library and sample IDs refer to the run and sample identifiers in SRA, respectively.
(DOC)
Text file with custom predicted gene models of Solanum tuberosum.
(ZIP)
Text file of the multiple alignment of all plant Snf2 candidates.
(ZIP)
Phylogenetic tree of all plant Snf2 candidates in NEWICK format.
(ZIP)
(DOC)