Abstract
The elaborate spatial organization of cells enhances, restricts, and regulates protein–protein interactions. However, the biological significance of this organization has been difficult to study without ways of directly perturbing it. We highlight synthetic biology tools for engineering novel cellular organization, describing how they have been, and can be, used to advance cell biology.
INTRODUCTION
Synthetic biology seeks to use cellular modularity to predictably build living systems that do useful things. Historically, many of the fruits of the field have been genetic circuits—elements that can remember, report on stimuli, and perform simple computational tasks. Other engineered products include synthetic genomes, as well as proteins endowed by design with novel functions.
Synthetic biology's engineering principles can also be applied to interrogate and perturb natural cellular function. In fact, nearly every article published in Molecular Biology of the Cell already makes use of engineered proteins through fluorescent or affinity tags that long ago entered into the standard cell biological toolkit. Although these methods have permitted the observation of cellular organization at a fine scale, our tools for perturbing that organization are somewhat blunt. Drugs, mutagenesis, and tunable chemical and physical environments may have imprecise or incomplete effects. The growing field of synthetic biology, however, offers new approaches for modifying cellular organization at a molecular level.
In this Perspective, we first present prevailing views on the importance of mechanisms for generating cellular order. Next we highlight examples of how the engineering of molecular interactions has enabled researchers to understand principles of cellular organization. Finally, we discuss how new developments in synthetic biology will advance our knowledge of cellular function, which, in turn, will enable us to design more complex living systems.
WHY ORGANIZE?
Specifying molecular localization provides a powerful way of selecting for and against biological interactions resulting in tunable mechanisms for regulation (Figure 1). Thus, confining free diffusion, either by reducing its dimensionality or by corralling it in a physical compartment, not only can protect proteins from off-target effects, but also can enhance desirable interactions. For example, the reduction of diffusion dimensionality can reduce the search time for finding an on-target interaction. The classic case study is DNA-binding proteins, which appear to find their targets two orders of magnitude faster than would be predicted by free diffusion (Riggs et al., 1970; Elf et al., 2007). This effect is also seen in the confinement of membrane components to two-dimensional spaces (Saffman and Delbrück, 1975).
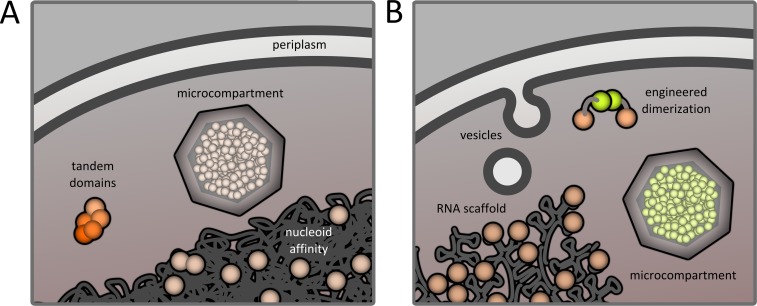
FIGURE 1:
Natural and synthetic cellular organization. (A) In a bacterial cell, protein–protein interactions may be facilitated by tandem domains or shared affinity for a certain intracellular region or surface (such as the nucleoid). Furthermore, proteins may be encapsulated in compartments comprising either membranes (i.e., the periplasm) or proteins (microcompartments). (B) Engineered localization tools can be as simple as fusion to dimerizing domains. Scaffolds, whether protein or RNA based, can also organize interactions. Finally, cargoes can be corralled in membranous vesicles and proteinaceous compartments.
The establishment of a physical compartment offers several additional benefits over colocalization, namely 1) creating unique microenvironments, 2) protecting contents from side reactions, and 3) isolating toxicity from the rest of the cell. These microenvironments, such as specialized pH or redox states, can specifically enable cargo function within the compartment. Classically, the proton gradient of mitochondria is essential for function of the electron transport chain, and in the lysozome, acid hydrolases are pH regulated, such that they are active only within their compartment. This protects cellular contents from off-target degradation. The isolation works both ways, however, as compartmentalization insulates contents from inefficient side reactions. For example, one model suggests that the shell of the carboxysome, a proteinaceous carbon-fixing compartment in cyanobacteria, is selectively impermeable to O2, a high-affinity, but unproductive, substrate for ribulose-1,5-bisphosphate carboxylase oxygenase (Kinney et al., 2012). These cases highlight the incentive to build synthetic compartments to optimize redesigned biological functions.
STRATEGIES FOR ENGINEERING SUBCELLULAR ORGANIZATION
How can cell biologists probe, perturb, and build upon the spatial organization of the cell? For directing interactions between proteins, many tools are entering into common use. For example, there are a variety of dimerization tags that can be used to induce colocalization, such as leucine zippers, LIM domains, and glutathione S-transferase. The most popular family is arguably that of the inducible heterodimerizing or homodimerizing FKBP domains (Rivera et al., 1996; Rollins et al., 2000), which create interactions that can be modulated by the addition of small molecules. For more complicated signaling needs, scaffolding proteins can also be engineered to control and rewire signaling cascades (reviewed in Good et al., 2011).
Engineering protein interactions on other types of scaffolds and surfaces, such as membranes or nucleic acids, are less explored. RNA scaffolds, originally designed as a modular and scalable way to engineer reactions, could present a way to study diffusion on a surface (Delebecque et al., 2012). Diverse classes of biomolecules can be localized to RNA scaffolds, and their spacing, orientation, and stoichiometry can all be controlled. DNA origami offers similar opportunities for specific molecular localization in vitro; this technology was recently used to observe the motile behavior of dynein and kinesin (Derr et al., 2012). RNA scaffolds may permit experiments like these to be carried out inside cells.
The tools of synthetic biology have also been effectively used to study how compartments interact with one another. Kornmann et al. (2009) constructed a synthetic tether between mitochondria and the endoplasmic reticulum (ER), enabling its use as a “crutch” in a screen for proteins responsible for ER–mitochondrial contacts. Flux through the Golgi apparatus has been investigated with the expression of procollagen, which forms an aggregate too large to fit into transport vesicles (Bonfanti et al., 1998). Recently FKBP repeats were used to create inducible “staples” in the lumen of the Golgi, enabling researchers to study whether cisternae move through the stack (Lavieu et al., 2013; Rizzo et al., 2013).
We can now also consider creating unique environments with engineered compartments. For example, vesicles can be formed in bacteria expressing eukaryotic calveolin (Walser et al., 2012). Because this approach causes vesicles to be pinched from the cell membrane, the contents and chemical environment inside the vesicles are identical to the periplasm. However, targeting other proteins such as transporters to these vesicles may facilitate modulation of the internal microenvironment. Protein-based microcompartments such as carboxysomes could prove to be an even more tractable solution, as they can be heterologously expressed, with cargoes targeted to their lumens by fusion to their endogenous cargoes (Bonacci et al., 2012) or, for certain compartments, short peptide-targeting sequences (Fan et al., 2010).
Approaching extreme forms of synthetic intracellular complexity, we can engineer endosymbiotic relationships that may mimic early evolutionary events such as creation of the chloroplast. Recently cyanobacteria have been shown to survive and divide for limited periods inside human cells and those of growing zebrafish (Agapakis et al., 2011). To create a truly mutualistic relationship, the bacteria must be able to export significant quantities of beneficial products. We engineered cyanobacteria to export sucrose (Ducat et al., 2012), but not in a manner compatible with intracellular life. Briefly, an Escherichia coli sucrose symporter was cloned into Synechococcus elongatus. Because the cyanobacteria basify their media, the symporter runs in the opposite direction in these cells, pumping sucrose out instead of in. Sucrose is naturally produced by cyanobacteria to balance osmotic pressure; therefore adding salt to the symporter strain causes accumulation of >10 mM sucrose in the media. However, the cyanobacterial proliferative capability is likely to be limiting in providing a significant benefit to the host.
Sucrose is only one of the many commodities being produced by engineered cells; so are alkanes (Howard et al., 2013) and a precursor to the antimalarial drug artemisinin (Paddon et al., 2013). One of the benefits of synthetic biology is that these products or proteins need not be natural; for example, E. coli has been engineered to respond to Pseudomonas aeruginosa quorum-sensing molecules with the production of a chimeric bacteriocidal agent that specifically inhibits the growth of the pathogens (Gupta et al., 2013). In the future, the introduction of alternative biosynthetic pathways will enable the engineering of stable endosymbiosis.
NEW TOOLS FOR UNANSWERED QUESTIONS
Much of cell biology relies on three major techniques to modify proteins: 1) removing them with a knockout or knockdown, 2) locking them into a given state, as with phospho-mimetic and phospho-null mutants, and 3) labeling them with synthetic fusions to reporters like green fluorescent protein. Synthetic biology aims to expand the biological toolkit to control localization and compartmentalization, allowing us to address new, unanswered questions. For example, what is the role of diffusion in biochemical reactions, and what are the thermodynamic and kinetic benefits of localization at different scales? Whereas theoretical models may inform our understanding of interactions between proteins in solution, the complex viscoelastic properties of the cytoplasm (Weber et al., 2010) are difficult to specifically recreate in a test tube, as are affinities for scaffolds and confinement within compartments. Thus, engineering localization and compartmentalization may offer a way to study interactions in their native chemical context, offering the additional benefit of allowing understanding of the interplay of confinement to scaffolds and insoluble phases in facilitating intermolecular interactions.
Furthermore, as synthetic biology advances toward its engineering goals, the artificial test cases created uncover basic biological questions. For example, although engineering a true symbiotic relationship is left to the future, the finding that Gram-negative cyanobacteria do not kill human and zebrafish cells after injection, as E. coli does, raises questions about how and why specificity in cellular response to invasion evolved. Expanding our knowledge of cellular function will, in turn, enable us to engineer living systems in ways we cannot yet imagine.
Acknowledgments
We thank Tyler Ford, Ethan Garner, and Tim Mitchison for helpful discussions. Funding was provided by the Jane Coffin Childs Fund (to J.K.P.), the Defense Advanced Research Projects Agency, and the National Institutes of Health (to P.A.S.).
Abbreviations used:
- FKBP
FK506 binding protein
- LIM
Lin11, IsI-1, Mec-3
Footnotes
REFERENCES
- Agapakis CM, Niederholtmeyer H, Noche RR, Lieberman TD, Megason SG, Way JC, Silver PA. Towards a synthetic chloroplast. PLoS One. 2011;6:e18877. doi: 10.1371/journal.pone.0018877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonacci W, Teng PK, Afonso B, Niederholtmeyer H, Grob P, Silver PA, Savage DF. Modularity of a carbon-fixing protein organelle. Proc Natl Acad Sci USA. 2012;109:478–483. doi: 10.1073/pnas.1108557109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfanti L, Mironov AA, Jr, Martínez-Menárguez JA, Martella O, Fusella A, Baldassarre M, Buccione R, Geuze HJ, Mironov AA, Luini A. Procollagen traverses the Golgi stack without leaving the lumen of cisternae: evidence for cisternal maturation. Cell. 1998;95:993–1003. doi: 10.1016/s0092-8674(00)81723-7. [DOI] [PubMed] [Google Scholar]
- Delebecque CJ, Silver PA, Lindner AB. Designing and using RNA scaffolds to assemble proteins in vivo. Nat Protoc. 2012;7:1797–1807. doi: 10.1038/nprot.2012.102. [DOI] [PubMed] [Google Scholar]
- Derr ND, Goodman BS, Jungmann R, Leschziner AE, Shih WM, Reck-Peterson SL. Tug-of-war in motor protein ensembles revealed with a programmable DNA origami scaffold. Science. 2012;338:662–665. doi: 10.1126/science.1226734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducat DC, Avelar-Rivas JA, Way JC, Silver PA. Rerouting carbon flux to enhance photosynthetic productivity. Appl Environ Microbiol. 2012;78:2660–2668. doi: 10.1128/AEM.07901-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elf J, Li GW, Xie XS. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 2007;316:1191–1194. doi: 10.1126/science.1141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Cheng S, Liu Y, Escobar CM, Crowley CS, Jefferson RE, Yeates TO, Bobik TA. Short N-terminal sequences package proteins into bacterial microcompartments. Proc Natl Acad Sci USA. 2010;107:7509–7514. doi: 10.1073/pnas.0913199107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good MC, Zalatan JG, Lim WA. Scaffold proteins: hubs for controlling the flow of cellular information. Science. 2011;332:680–686. doi: 10.1126/science.1198701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Bram EE, Weiss R. Genetically programmable pathogen sense and destroy. ACS Synth Biol. 2013 doi: 10.1021/sb4000417. DOI: 10.1021/sb400417. [DOI] [PubMed] [Google Scholar]
- Howard TP, et al. Synthesis of customized petroleum-replica fuel molecules by targeted modification of free fatty acid pools in Escherichia coli. Proc Natl Acad Sci USA. 2013;110:7636–7641. doi: 10.1073/pnas.1215966110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney JN, Salmeen A, Cai F, Kerfeld CA. Elucidating essential role of conserved carboxysomal protein CcmN reveals common feature of bacterial microcompartment assembly. J Biol Chem. 2012;287:17729–17736. doi: 10.1074/jbc.M112.355305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, Walter P. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325:477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavieu G, Zheng H, Rothman JE. Stapled Golgi cisternae remain in place as cargo passes through the stack. eLife. 2013;2:e00558. doi: 10.7554/eLife.00558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddon CJ, et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 2013;496:528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- Riggs AD, Bourgeois S, Cohn M. The lac represser-operator interaction: III. Kinetic studies. J Mol Biol. 1970;53:401–417. doi: 10.1016/0022-2836(70)90074-4. [DOI] [PubMed] [Google Scholar]
- Rivera VM, et al. A humanized system for pharmacologic control of gene expression. Nat Med. 1996;2:1028–1032. doi: 10.1038/nm0996-1028. [DOI] [PubMed] [Google Scholar]
- Rizzo R, Parashuraman S, Mirabelli P, Puri C, Lucocq J, Luini A. The dynamics of engineered resident proteins in the mammalian Golgi complex relies on cisternal maturation. J Cell Biol. 2013;201:1027–1036. doi: 10.1083/jcb.201211147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins CT, et al. A ligand-reversible dimerization system for controlling protein–protein interactions. Proc Natl Acad Sci USA. 2000;97:7096–7101. doi: 10.1073/pnas.100101997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffman PG, Delbrück M. Brownian motion in biological membranes. Proc Natl Acad Sci USA. 1975;72:3111–3113. doi: 10.1073/pnas.72.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walser PJ, et al. Constitutive formation of caveolae in a bacterium. Cell. 2012;150:752–763. doi: 10.1016/j.cell.2012.06.042. [DOI] [PubMed] [Google Scholar]
- Weber SC, Spakowitz AJ, Theriot JA. Bacterial chromosomal loci move subdiffusively through a viscoelastic cytoplasm. Phys Rev Lett. 2010;104:238102. doi: 10.1103/PhysRevLett.104.238102. [DOI] [PMC free article] [PubMed] [Google Scholar]