Escape of aberrant proteins from protein quality control leads to accumulation of toxic protein species. Sti1 interacts with Hsp70 to mediate spatial PQC of amyloid-like proteins by regulating their distribution in different intracellular protein-handling depots. Sti1 suppresses proteotoxicity by targeting amyloid-like proteins to perinuclear foci.
Abstract
Conformational diseases are associated with the conversion of normal proteins into aggregation-prone toxic conformers with structures similar to that of β-amyloid. Spatial distribution of amyloid-like proteins into intracellular quality control centers can be beneficial, but cellular mechanisms for protective aggregation remain unclear. We used a high-copy suppressor screen in yeast to identify roles for the Hsp70 system in spatial organization of toxic polyglutamine-expanded Huntingtin (Huntingtin with 103Q glutamine stretch [Htt103Q]) into benign assemblies. Under toxic conditions, Htt103Q accumulates in unassembled states and speckled cytosolic foci. Subtle modulation of Sti1 activity reciprocally affects Htt toxicity and the packaging of Htt103Q into foci. Loss of Sti1 exacerbates Htt toxicity and hinders foci formation, whereas elevation of Sti1 suppresses Htt toxicity while organizing small Htt103Q foci into larger assemblies. Sti1 also suppresses cytotoxicity of the glutamine-rich yeast prion [RNQ+] while reorganizing speckled Rnq1–monomeric red fluorescent protein into distinct foci. Sti1-inducible foci are perinuclear and contain proteins that are bound by the amyloid indicator dye thioflavin-T. Sti1 is an Hsp70 cochaperone that regulates the spatial organization of amyloid-like proteins in the cytosol and thereby buffers proteotoxicity caused by amyloid-like proteins.
INTRODUCTION
Protein conformational diseases represent a collection of disorders in which structurally diverse proteins are converted into a common cytotoxic conformational state that has features of β-amyloid (Carrell and Lomas, 1997). A large number of normal proteins are converted to an amyloid-like state that is rich in β-structure, detergent insoluble, and binds indicator dyes such as thioflavin-T (Chiti and Dobson, 2006). β-Amyloid is deposited in extracellular plaques in the brains of Alzheimer's patients (Chiti and Dobson, 2006; Haass and Selkoe, 2007; Treusch et al., 2009), and there are also numerous instances in which protein conformational disease is associated with intracellular accumulation of amyloid-like assemblies (Carrell and Lomas, 1997). Flux of proteins through the amyloid assembly pathway is associated with disease, but whether the amyloid-like aggregates themselves or small oligomers of converted proteins are toxic is debatable (Bucciantini et al., 2004; Cohen et al., 2006; Wolfe and Cyr, 2011). There is mounting evidence that molecular chaperones facilitate the packaging of toxic protein species into protein-handling depots to suppress proteotoxicity (Muchowski and Wacker, 2005; Douglas et al., 2008). Yet the cellular mechanisms for molecular chaperone function in protective protein aggregation remain unclear.
Hsp70 functions with specialized cochaperones to suppress protein aggregation, refold clients, and promote protein degradation (Cyr et al., 1994, 2002; Meacham et al., 2001; Tyedmers et al., 2010a). Hsp70 binds extended regions of polypeptide chains in an ATP-dependent reaction cycle that is regulated by Hsp40 (Liberek et al., 1991; Cyr et al., 1992; Langer et al., 1992). Hsp40 serves as a substrate selector and helps stabilize Hsp70:polypeptide complexes by converting Hsp70-ATP to Hsp70-ADP. Substrate release from Hsp70 is regulated by nucleotide exchange factors and tetratricopeptide repeat (TPR) cochaperones (Scheufler et al., 2000; Steel et al., 2004; Shaner et al., 2005). TPR cochaperones contain a TPR repeat, which serves as an Hsp70/Hsp90 interaction domain that recognizes a conserved C-terminal EEVD motif (Scheufler et al., 2000), and specialized functional domains that dictate the fate of chaperone-bound clients. Sti1 is a TRP-repeat cochaperone whose mammalian homologue is HOP, which contains three separate TPR domains that serve to bring Hsp70 and Hsp90 together in complexes and act sequentially on clients (Hernandez et al., 2002). Sti1 also functions with Hsp70 and Hsp40 to modulate the biogenesis and function of prions in yeast and humans (Jones et al., 2004; Reidy and Masison, 2010; Roffe et al., 2010). CyP40 contains a cis-trans-prolyl-isomerase domain that facilitates conformational maturation of late-stage protein-folding intermediates (Chen et al., 1998); CHIP is a ubiquitin ligase that regulates the heat stress response (Dai et al., 2003; Qian et al., 2006) and targets misfolded proteins for proteasomal degradation (Jiang et al., 2001; Meacham et al., 2001). Thus TPR-repeat cochaperones of Hsp70/Hsp90 act downstream of Hsp40s and participate in multiple aspects of protein homeostasis.
In protein aggregates, hydrophobic surfaces that would be recognized by components of the Hsp70 system are buried, so they escape recognition by protein quality control machines. Thus aggregates are inefficiently cleared from cells and cause cytotoxicity by damaging cell membranes, altering the conformation of other proteins, and sequestering essential cellular proteins (Haass and Selkoe, 2007; Last et al., 2011; Olzscha et al., 2011). Aberrant proteins that are not initially cleared from the cytosol can be segregated into distinct cytosolic foci, observed in both yeast and mammalian systems, which appear to store oligomers and aggregates until conditions favor refolding or degradation (Johnston et al., 1998; Kaganovich et al., 2008; Wang et al., 2009; Tyedmers et al., 2010b; Specht et al., 2011; Malinovska et al., 2012; Weisberg et al., 2012; Shiber et al., 2013; Summers et al., 2013). Remarkably, detergent-soluble and amyloid-like protein conformers are segregated into different protein-handling depots, and this process is influenced by components in the Hsp70 system (Douglas et al., 2008, 2009; Malinovska et al., 2012; Summers et al., 2013). Amyloid-like and detergent-insoluble protein assemblies accumulate in perinuclear aggresomes (Johnston et al., 1998; Wang et al., 2009) and insoluble protein deposits (IPODs), which are located in the cell periphery (Kaganovich et al., 2008; Tyedmers et al., 2010b). Protein oligomers and aggregates that are soluble in detergents accumulate in the spatially distinct juxtanuclear quality control (JUNQ; Kaganovich et al., 2008; Weisberg et al., 2012) and peripheral compartments (Specht et al., 2011). Ubiquitinated proteins that evade the proteasome and terminally misfolded proteins that escape recognition by protein quality control (PQC) E3 ligases are swept into the JUNQ (Kaganovich et al., 2008). Proteins in the JUNQ can be solubilized and degraded by the proteasome in a pathway that involves the PQC E3 ligase Ubr1 (Summers et al., 2013). The peripheral compartment contains Hsp42 and holds proteins in a folding-competent state (Specht et al., 2011). The IPOD may function in disaggregation of detergent-insoluble protein aggregates (Tyedmers et al., 2010b), whereas the peripheral compartment and JUNQ aid in refolding (Specht et al., 2011) and degradation (Kaganovich et al., 2008) of detergent-soluble protein assemblies, respectively. The aggresome resembles an inclusion body and may represent a long-term storage site for aberrant proteins (Johnston et al., 1998; Wang et al., 2009). Thus cells have the capacity to sequester toxic protein species, but the mechanism for spatial segregation of protein handling centers is a mystery.
Although flux through protein aggregation pathways is associated with cell death, chaperone-dependent sequestration of amyloid-like proteins into protein-handling centers correlates with suppression of proteotoxicity (Cohen et al., 2006; Douglas et al., 2008; Summers et al., 2009; Treusch et al., 2009). In yeast models, overexpression of the yeast prion protein Rnq1 is toxic, but only if preexisting [RNQ+] prions are present to seed its conversion to an amyloid-like conformation (Sondheimer and Lindquist, 2000; Derkatch et al., 2001; Douglas et al., 2008). The Hsp70, Ssa1, cooperates with the Hsp40, Sis1, to increase the efficiency by which seeded forms of Rnq1 assemble into amyloid-like [RNQ+] prions and thereby suppress proteotoxicity (Douglas et al., 2008). Of interest, a fragment of Huntingtin (Htt) similar to those found in inclusions from Huntington's disease brains (Htt with 103Q glutamine stretch [Htt103Q]), is toxic when expressed ectopically in yeast, but only if [RNQ+] prions are present (Meriin et al., 2002). In yeast, the aggregation state of Htt103Q and its toxicity are inversely correlated with very tight, detergent-insoluble aggregates being benign and loose, detergent-soluble aggregates being toxic (Duennwald et al., 2006). Aggregation and toxicity of Htt103Q are influenced by its subcellular environment and also by the sequences of amino acids that flank its polyglutamine (polyQ) stretch (Dehay and Bertolotti, 2006; Duennwald et al., 2006; Douglas et al., 2009). Forms of Htt103Q that contain the polyproline region that follows the polyQ domain in full-length Htt are benign due to their sequestration into the aggresome in a process that requires the ringed molecular chaperone p97 (Wang et al., 2009). Forms of Htt103Q that lack the polyproline region are toxic, as they accumulate in loose aggregates that are distributed throughout the cytosol in speckled foci and tight aggregates that accumulate in the IPOD (Kaganovich et al., 2008; Douglas et al., 2009). Thus studies on proteotoxicity of Htt103Q and Rnq1 in yeast provide an excellent model system in which to uncover mechanisms for Hsp70 function in protein detoxification through protective protein aggregation.
Here we uncover a role for the cochaperone Sti1 in orchestrating spatial organization of Htt103Q and Rnq1 into benign amyloid-like assemblies. We use the toxic form of Htt103Q lacking the proline-rich region and screen for proteins that suppress proteotoxicity by facilitating its aggregation. Sti1 cooperates with the Hsp70 system to control the spatial organization of amyloid-like protein assemblies, thereby suppressing proteotoxicity.
RESULTS
The Hsp70/Hsp90 cochaperone Sti1 suppresses Htt103Q toxicity
To identify cellular factors that defend against proteotoxicity, we carried out a high-copy screen in Saccharomyces cerevisiae to discover proteins that suppressed Htt103Q toxicity. A construct encoding the first 17 amino acids of the Htt gene followed by 103 glutamines and a green fluorescent protein (GFP) moiety (Htt103Q) was integrated into a W303α strain under control of the galactose promoter. Htt103Q expression caused a growth defect in a glutamine length–dependent manner (Meriin et al., 2002; Duennwald et al., 2006), and differential sensitivity to Htt103Q was exhibited by individual strains (Figure 1A). The yeast Hsp70/Hsp90 cochaperone Sti1 (Nicolet and Craig, 1989) was identified as a high-copy suppressor of the severe Htt103Q-dependent growth defect (see Materials and Methods for details of the suppressor screen). Expression of the TPR family cochaperones Cns1 (Dolinski et al., 1998) and Sgt2 (Kordes et al., 1998) did not suppress Htt103Q toxicity (Supplemental Figure S1A), indicating that this activity is not a characteristic of TPR proteins in general. Of importance, deletion of STI1 caused Htt103Q to become toxic in a strain in which it was normally benign (Figure 1A, right). Parallel growth assays using wild-type and ΔSti1 Htt25Q strains demonstrate that deletion of STI1 does not reduce growth on galactose (Figure 1A, left). So, loss of Sti1 function reduces the capacity of cells to tolerate expression of Htt103Q.
FIGURE 1:
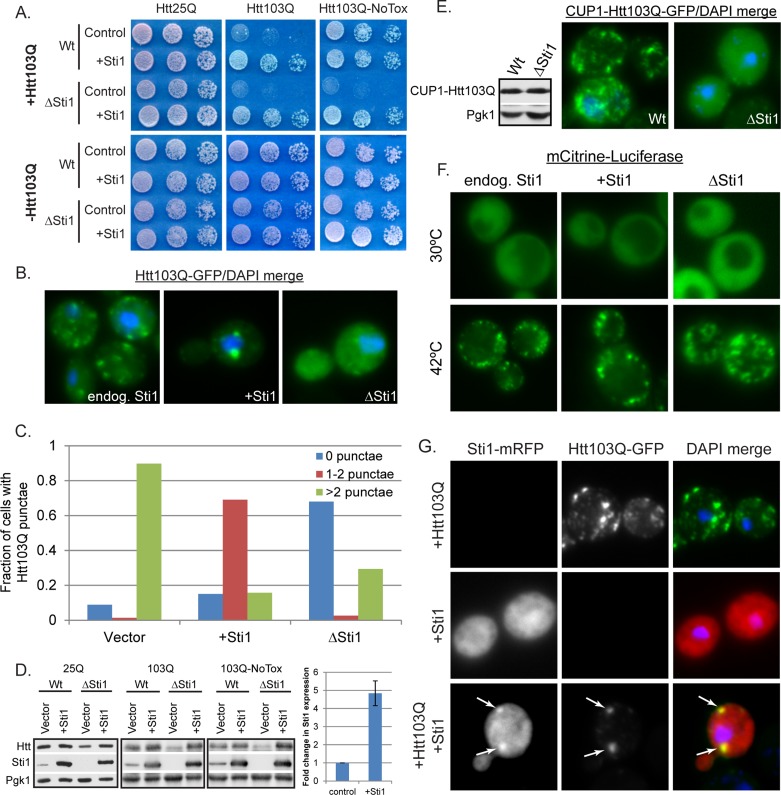
Sti1 suppresses Htt103Q toxicity and reorganizes Htt103Q-GFP foci. (A) Effect of Sti1 on Htt103Q growth defect as monitored by cell growth assay plated in fivefold dilutions. Htt103Q and Htt103Q-NoTox are two integration strains with similar Htt expression that differ in severity of the growth defect. (B, C) Effect of Sti1 on organization of Htt103Q-GFP foci. Cells expressing Htt103Q-GFP for 4 h from the GAL1 promoter were fixed as indicated in Materials and Methods. Quantitation indicates 150–300 individual cells counted from different experiments using ImageJ. (D) Expression levels of Htt as detected by Western blot analysis of lysates taken from cultures grown from strains in A. Graph depicts quantitation of Sti1 overexpression compared with endogenous Sti1 (n = 6). (E) Expression level and localization of Htt103Q-GFP as in B and D. Here Htt103Q was expressed from a CUP1 promoter induced with 500 μM CuSO4 for 4 h. (F) Influence of Sti1 on organization of mCitrine-luciferase during heat shock. Cells were imaged live during growth at 30°C and then again after a shift to 42°C for 20 min. (G) Colocalization of Htt103Q-GFP and Sti1-mRFP. White arrows indicate colocalization. Nuclei were visualized using DAPI staining. Lysates for Western blot analysis came from cells grown under the same conditions used for fluorescence microscopy.
Elevation of Sti1 caused the redistribution of Htt103Q from a mixture of diffuse material and foci that are dispersed throughout the cytosol into several distinct large foci that were often found in close proximity to the nucleus (Figure 1, B and C). Conversely, deletion of STI1 greatly reduced Htt103Q punctae formation, with the majority of Htt dispersed in a diffuse cytosolic pattern (Figure 1, B and C). Sti1 overexpression had no effect upon Htt103Q levels, yet deletion reduced expression (Figure 1D), likely due to delayed induction from the galactose promoter (Floer et al., 2008). Deletion of STI1 clearly sensitizes the cell to Htt103Q because even though there was reduced Htt103Q expression, growth defects caused by Htt103Q were exacerbated.
Sti1 may suppress Htt103Q toxicity by organizing it into foci; however, fewer foci may be observed in the ΔSti1 strain because there is less Htt103Q being expressed. To rule out this possibility, Htt103Q-GFP localization was examined when expressed to equivalent levels via copper-inducible promoter in wild-type and ΔSti1 strains (Figure 1E). Again Sti1 was required for efficient organization of Htt103Q into foci. We also examined the influence of Sti1 expression upon temporal aspects of Htt103Q foci accumulation. Over a 4-h time course, Sti1 accelerated Htt103Q foci formation while reducing number of foci per cell (Supplemental Figure S1B). Cycloheximide chase experiments demonstrate that once formed, the Sti1-dependent foci containing Htt103Q were stable for at least 5 h (Supplemental Figure S1C). These data provide a correlation between Sti1 action in suppression of Htt toxicity and packaging of Htt103Q into perinuclear foci.
Does Sti1 act only on Htt103Q, or can it also modulate the organization of other cytosolic protein aggregates? To answer this question, we used mCitrine-tagged luciferase, an Hsp70 client that aggregates during heat stress (Specht et al., 2011). At 30°C, mCitrine-luciferase is diffuse throughout the cytosol and was not affected by Sti1 overexpression (Figure 1F, top). During thermal stress at 42°C for 20 min, luciferase accumulated in numerous aggregates throughout the cytosol (Figure 1F). Neither elevation nor deletion of STI1 changed the appearance of the luciferase aggregates (Figure 1F, bottom). Thus, since Sti1 does not suppress aggregation of heat-damaged luciferase or organize aggregated luciferase into foci, it may act specifically on amyloid-like forms of Htt103Q.
The TPR cochaperone CHIP inhibits Hsp70 function in protein folding (Jiang et al., 2001), so it is possible that Sti1 affects Htt103Q assembly into foci by similarly interfering with Hsp70. This is unlikely because Sti1 overexpression does not cause mCitrine-luciferase to aggregate (Figure 1F). In addition, Sti1 is implicated as an independent activator of Ssa1 ATPase activity and is able to stimulate Ydj1-dependent refolding of luciferase by Hsp70 (Wegele et al., 2003, 2006). Nevertheless, we examined in the effect of purified Sti1 on the ability of Ssa1 and Sis1 to refold denatured luciferase. When Sti1 was added at a fourfold molar excess over Ssa1 and Sis1 it had little detectable effect on luciferase refolding (Supplemental Figure S1, D and E). In contrast, the TPR-repeat protein CHIP, which has antichaperone functions, inhibited Ssa1- and Sis1-dependent refolding of luciferase. Thus, when paired with Ydj1, Sti1 can stimulate luciferase refolding, but under the conditions used here it does not stimulate the refolding activity of Sis1 and Ssa1. In contrast to CHIP, Sti1 is a positive regulator of Hsp70 function and appears capable of enhancing interactions between Ssa1 and Htt103Q (Wegele et al., 2003) and may thereby affect spatial quality control.
If Sti1 has a direct effect on Htt103Q, it should be present with Htt103Q foci. When expressed alone, Sti1 tagged with a monomeric red fluorescent protein (mRFP) exhibited a diffuse localization pattern in the cytosol (Figure 1G). When coexpressed with Htt103Q, it redistributed the polyQ protein into distinct punctae, and Sti1-mRFP was indeed concentrated in foci with Htt103Q (Figure 1G, arrows). These data demonstrate that Sti1 promotes Htt103Q foci formation and this is beneficial to the cell, so there is an inverse relationship between Htt103Q aggregation and toxicity that is modulated by Sti1.
Sti1 interacts with high–molecular weight forms of Htt103Q
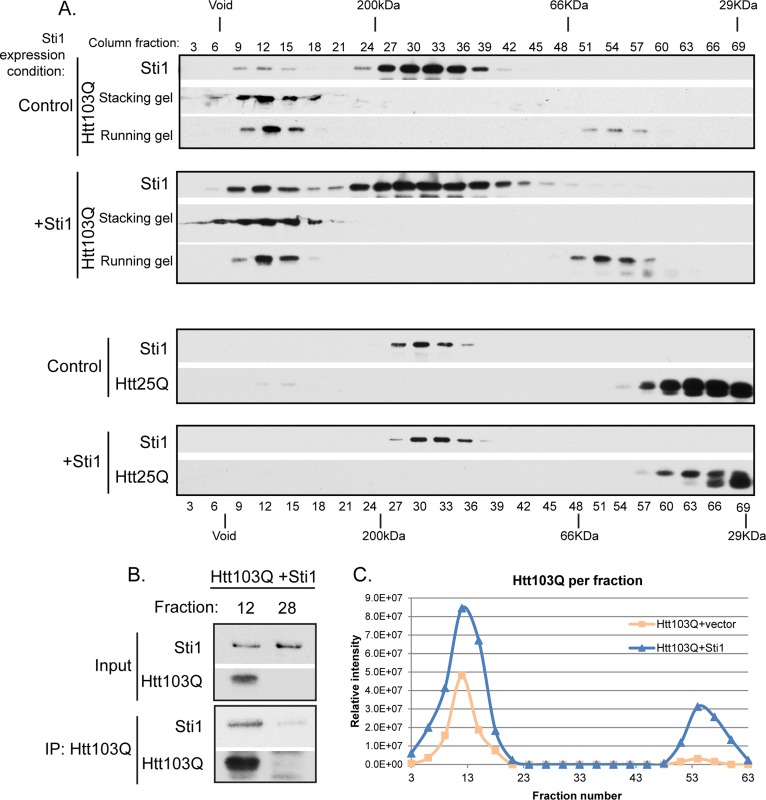
To understand the mechanism for Sti1 action, we asked whether Sti1 interacts with low–molecular weight Htt103Q or high–molecular weight oligomeric assemblies via gel filtration chromatography. Cell lysates prepared by homogenizing spheroplasts in the absence of detergent were immediately applied to a Sephacryl S-200 column. Proteins in every third fraction were resolved on an SDS–PAGE gel and probed by Western blot for levels of Htt103Q and Sti1 (Figure 2, A–C). A portion of high–molecular weight, SDS-resistant Htt103Q is retained at the stacking front of the gel (Douglas et al., 2009), so both the stacking and running gel sections are shown and quantitated. Htt expression was similar in the presence of endogenous and overexpressed Sti1 (Figure 1C). Under all conditions, a pool of Sti1 migrated as a low–molecular weight form (Figure 2A); Sti1 is proposed to function as a dimer (Prodromou et al., 1999) as well as a monomer and is also bound to Hsp70/Hsp90 (Li et al., 2011). The separating power of gel filtration chromatography is insufficient to discern between these different low–molecular weight species of Sti1. Yet the mobility of endogenous low–molecular weight Sti1 is shifted to a high–molecular weight fraction in the presence of Htt103Q, whereas low–molecular weight Htt103Q did not comigrate with low–molecular weight Sti1 (Figure 2A, top/control). Of importance, endogenous Sti1 did not migrate in a high–molecular weight fraction when non–aggregation-prone Htt25Q was expressed (Figure 2A, bottom). On overexpression of Sti1, the high–molecular weight pools of Sti1 and Htt103Q both increased proportionally (Figure 2, A, top/+Sti1, and C), yet this was not observed when Sti1 levels were elevated in the presence of Htt25Q (Figure 2A, bottom/+Sti1). Sti1 was also present in immune complexes when high–molecular weight Htt103Q was precipitated from column fraction 12 (Figure 2B). This interaction was specific because there was no Sti1 signal detected in the absence of Htt103Q (Figure 2B). While reorganizing foci and suppressing toxicity, Sti1 appears to interact with high–molecular weight forms of Htt103Q.
FIGURE 2:
Sti1 cofractionates with high–molecular weight forms of Htt103Q. (A) Migration of indicated forms of Htt with Sti1 on Sephacryl S200 gel filtration column at endogenous (control) or overexpressed Sti1 levels as detected by Western blot. (B) Detection of Sti1 in immune precipitates with FLAG-Htt103Q-GFP. Immunoprecipitation was performed with anti-FLAG conjugated beads from aliquots of fraction 12 containing both Htt103Q and Sti1 or from fraction 28 containing only Sti1 as a background control. The immunoprecipitation was carried out using the column sample with elevated levels of Sti1, and precipitates were analyzed by Western blot. (C) Quantitation of total Htt103Q in column fractions that was detected in both the stacking and running gel plotted vs. the fraction number. The total signal in the gel and the quantitation shown in the graph reflect each other. Nondenaturing lysates were prepared as described in Materials and Methods, and equivalent amounts of total protein per sample were applied to the S200 column.
Under conditions in which Sti1 increases pools of high–molecular weight Htt103Q, an increase in the pool size of low–molecular weight Htt103Q also occurs (Figure 2A). We believe that this happens because the higher–molecular weight pools of Htt103Q are elevated, and assembled Htt103Q is more stable than unassembled forms. Amyloid-like assemblies are dynamic, so a pool of Htt103Q monomers should be in equilibrium with the higher–molecular weight species. Thus the corresponding increase in low–molecular weight as well as high–molecular weight forms of Htt103Q is not unexpected. We speculate that since more low–molecular weight Htt103Q is detected when Sti1 is overexpressesed and growth defects are suppressed, the conformer of low–molecular weight Htt103Q detected is nontoxic. Sti1 appears to increase accumulation of a benign high–molecular weight form of Htt103Q and may also hinder conversion of monomeric Htt103Q to an aberrant toxic conformer.
Sti1 directs amyloidogenic proteins to a perinuclear location
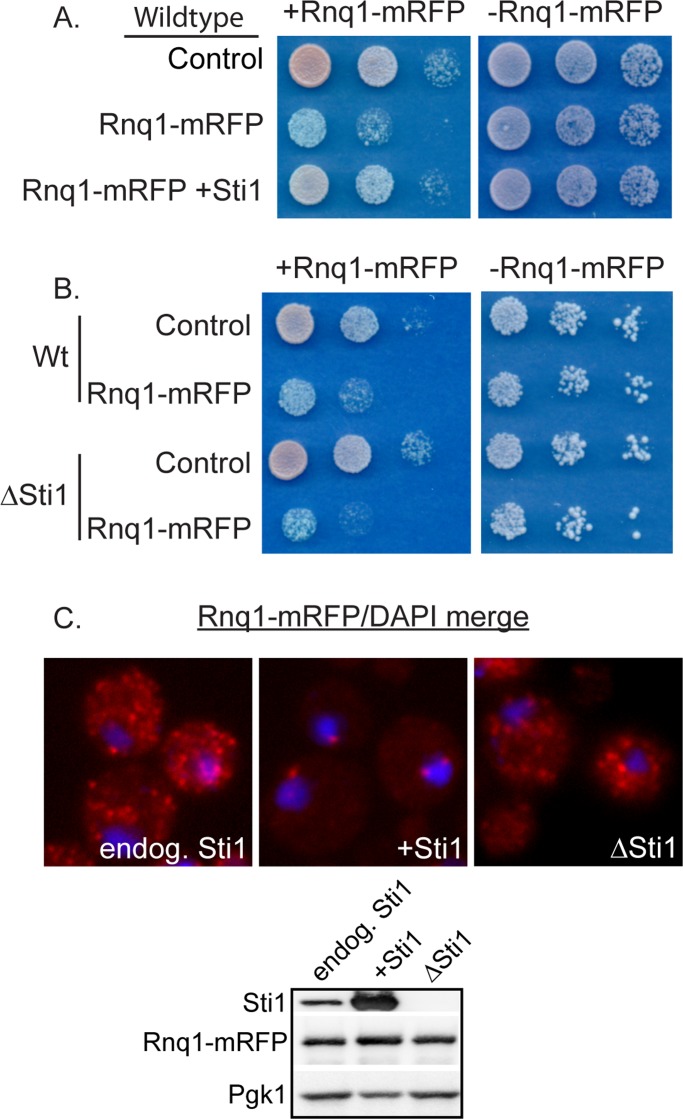
Htt103Q is toxic to yeast upon undergoing a conformational change driven by interaction with the [RNQ+] prion (Derkatch et al., 2001; Meriin et al., 2002). [RNQ+] prion biogenesis and toxicity are modulated by Hsp70 and Hsp40 (Sondheimer et al., 2001; Aron et al., 2007; Douglas et al., 2008; Tipton et al., 2008), and weak variants of the prion [PSI+] are eliminated from yeast by elevation of Sti1 activity (Kryndushkin et al., 2002). Thus we evaluated the effect of Sti1 on [RNQ+] prion toxicity and subcellular organization. As with Htt103Q, an increase in Sti1 suppressed the toxicity associated with Rnq1 overexpression (Figure 3A). Under the conditions used, Rnq1 expression is highly toxic, and so deletion of STI1 led to only modest increase in growth defects caused by Rnq1 overexpression (Figure 3B). At endogenous levels of Sti1, Rnq1-mRFP foci were speckled throughout the cytosol, but upon overexpression of Sti1, Rnq1-mRFP became localized to one or two distinct juxtanuclear foci (Figure 3C). In contrast to what is observed with Htt103Q-GFP, speckled Rnq1-mRFP foci were still detected in a ΔSti1 strain, indicating that this strain remains in the [RNQ+] prion state (Figure 3C, ΔSti1). Sti1 is not required for maintenance of [RNQ+] prions, and Sti1 overexpression does not eliminate [RNQ+] prions. Instead, suppression of Rnq1 and Htt103Q toxicity by Sti1 is associated with organization of multiple punctae containing these proteins into one or two distinct perinuclear foci.
FIGURE 3:
Sti1 suppresses Rnq1 toxicity and reorganizes Rnq1-mRFP foci. (A, B) Effect of Sti1 on the growth defect caused by Rnq1-mRFP overexpression as monitored by cell growth assay plated in fivefold dilutions. Toxicity assay was plated on synthetic media containing 2% glucose and 500 μM CuSO4. (C) Effect of Sti1 on organization of Rnq1-mRFP foci. Expression levels of Rnq1-mRFP were detected by Western blot analysis under conditions identical to those used for fluorescence microscopy and described in Materials and Methods.
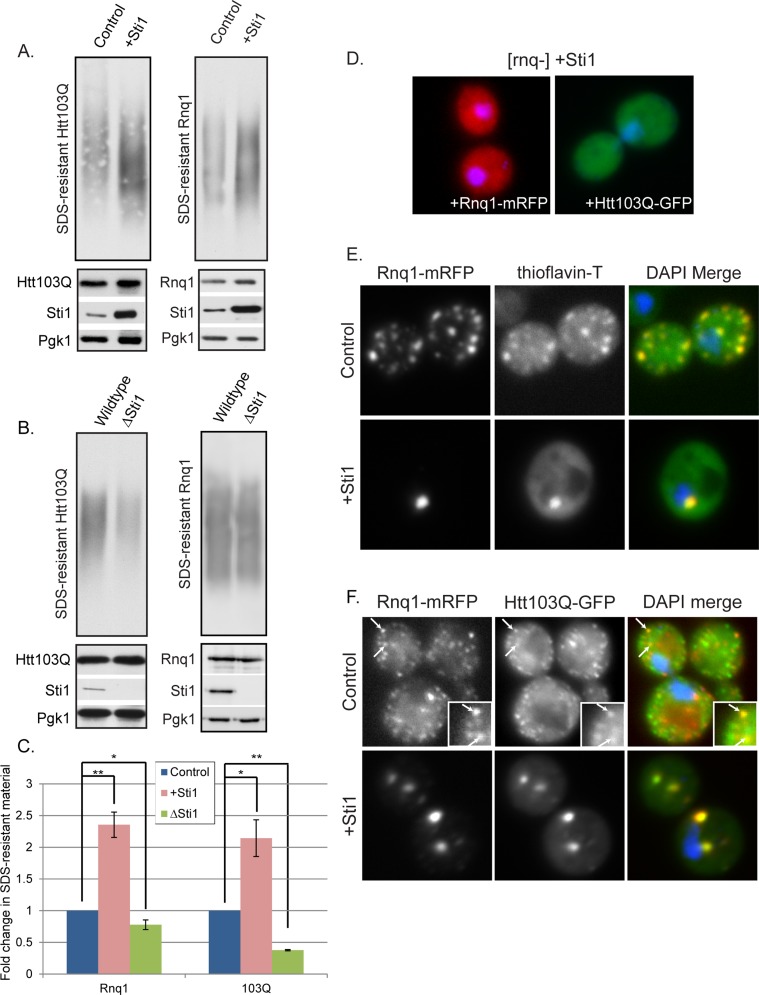
We next sought to determine what conformational state of Rnq1 and Htt103Q is favored under conditions in which Sti1 suppresses their proteotoxicty. Sti1 increased accumulation of oligomeric SDS-resistant forms of Rnq1 and Htt103Q by more than twofold (Figure 4, A–C). Deletion of STI1 reduced Htt103Q SDS-resistant material by >50%, but there was little effect on Rnq1 (Figure 4, A–C). Sti1 appears to act on the oligomeric assemblies of Htt103Q and Rnq1 that only form in [RNQ+] prion strains because its overexpression has no effect on localization of Htt103Q or Rnq1 in [rnq-] strains (Figure 4D).
FIGURE 4:
Sti1 modulates assembly of amyloidogenic substrates. (A–C) The effect of Sti1 on SDS-resistant Htt103Q-GFP or Rnq1-mRFP. SDS-resistant material was measured by SDD-AGE. (A, B) Representative images of quantitation shown in C (values represent means ± SE, n ≥ 3, *p < 0.05, **p < 0.005). (B) Inability of Sti1 to reorganize Htt103Q-GFP or Rnq1-mRFP to foci in an [rnq-] background. (C) Staining of Rnq1-mRFP foci with the amyloid indicator dye thioflavin-T, as well as with DAPI to indicate nuclei. (D) Colocalization of Htt103-GFP and Rnq1-mRFP during Sti1 overexpression. White arrows indicate colocalization. Insets represent the same punctae indicated with white arrows in the main images.
Conversion to an SDS-resistant oligomer is a characteristic of amyloid, so Sti1 appears to target amyloid-like forms of proteins to perinuclear foci. To gain experimental support for this conclusion, we asked whether Sti1-inducible foci stain positive for the amyloid-specific dye thioflavin-T (Chiti and Dobson, 2006). Rnq1-mRFP was used as a tool instead of Htt103Q-GFP because, upon binding amyloid, thioflavin-T emission is enhanced to a wavelength in the excitation range of GFP (LeVine, 1993). [RNQ+] prions have features of amyloid-like protein species, as they bind thioflavin-T, whereas the dye does not bind to amorphous aggregates of Rnq1 that accumulate in an [rnq-] strain (Douglas et al., 2008). Speckled Rnq1-mRFP foci in both the cell periphery and perinuclear locations were positive for thioflavin-T staining. Perinuclear Rnq1-mRFP foci whose formation is induced by Sti1 are also thioflavin-T positive (Figure 4E).
To ascertain whether Htt103Q is also present in foci with amyloid-like [RNQ+] prions, we asked whether they colocalize with each other in the presence and absence of Sti1 overexpression (Figure 4F). When Htt103Q-GFP and Rnq1-mRFP were coexpressed, a mixed population of speckled foci was detected, with ∼20% of foci containing both proteins (Figure 4F, inset and arrows). Sti1 elevation resulted in redistribution of most of the Htt103Q and Rnq1 to one or two larger perinuclear foci that contain both proteins (Figure 4F).
Owing to the colocalization of Htt103Q and Rnq1 in foci that contain amyloid-like proteins, we asked whether Sti1 is necessary for these proteins to interact. To answer this question, we took advantage of the fact that addition of a nuclear localization signal (NLS) to Rnq1 directs [RNQ+] prions to the nucleus and Htt103Q follows (Douglas et al., 2009). Thus, if Sti1 is required for interaction between Htt103Q and Rnq1, STI1 deletion of would prevent this relocalization. However, Rnq1-NLS still redistributed Htt103Q to the nucleus in both the presence and absence of Sti1 (Supplemental Figure S2A). Thus Sti1 promotes accumulation of Htt103Q and foci that contain amyloid-like species of Rnq1 but does not appear to be necessary for Rnq1 and Htt to interact. Instead, Sti1 may control the spatial organization of amyloid-like protein-handling centers.
Sti1-inducible foci colocalize with markers for cytosolic protein-handling depots
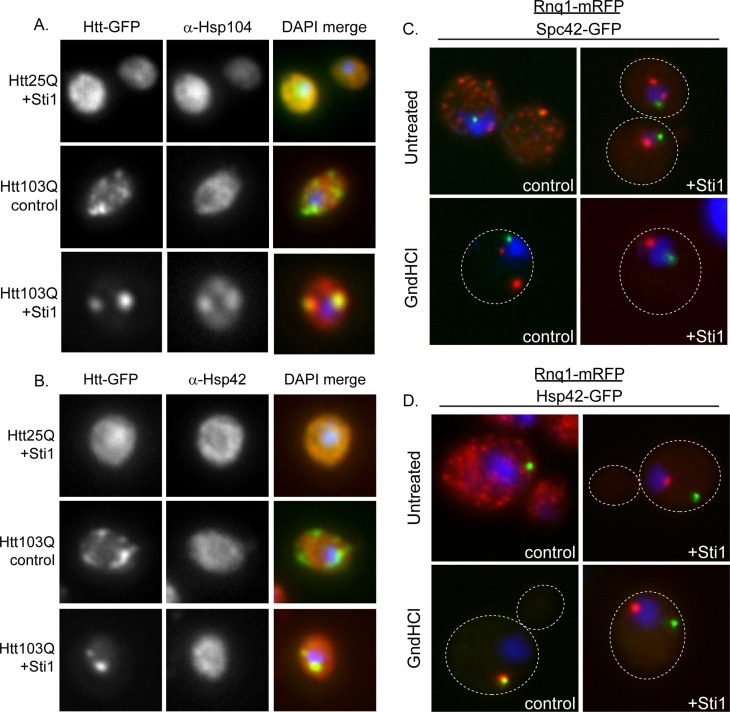
Sti1-inducible foci (StiF) are perinuclear and contain amyloid-like material, yet it is unclear whether they have features of protein-handling depots such as the IPOD and JUNQ. To characterize the StiF in more detail, we determined whether these foci contain PQC machinery that would help protect cells from proteotoxic stress. Under normal conditions, Hsp104 and Hsp42 were not detected in the speckled foci that contain Htt103Q-GFP (Figure 5, A and B), but Hsp104 and Hsp42 were present in StiF. Sti1 expression caused Hsp104 to become concentrated in Stif because its signal intensity in the cytosol versus the Stif has a 1:1.5 ratio. Although Hsp42 did colocalize with Htt103Q, the signal for Hsp42 in foci and cytosol had a 1:1 ratio, so it is difficult to judge whether the colocalization observed is due to overlap in staining or concentration of Hsp42 to the StiF.
FIGURE 5:
Sti1-inducible foci colocalize with markers for cytosolic protein-handling depots. (A, B) Localization of endogenous (A) Hsp104 or (B) Hsp42 as monitored by immunofluorescence using Htt103Q as a StiF marker. (C, D) Effect of Hsp104 inhibition on Rnq1-mRFP localization at the StiF. Fluorescence microscopy was carried out in strains with (C) Spc42 or (D) Hsp42 tagged at the endogenous locus with GFP. Rnq1-mRFP was induced with 50 μM CuSO4, and Hsp104 was inhibited with 3 mM GndHCl for 2 h before cells were fixed. Nuclei were visualized using DAPI staining. Dotted lines indicate the outline of the cell.
Because the StiF is an Hsp104-positive inclusion, we next determined whether StiF formation is affected by modulation of Hsp104 activity. Here we treated cells containing endogenous or overexpressed Sti1 with guanidinium hydrochloride (GndHCl) for 4 h to inhibit Hsp104 (Ferreira et al., 2001; Jung and Masison, 2001) and monitored the effect this had on organization of Rnq1-mRFP into foci. To determine the nature of the foci observed, we used strains containing a GFP-tagged version of Hsp42 or Spc42, a spindle pole body component that marks the aggresome (Johnston et al., 1998). Hsp104 inhibition condensed most of the Rnq1-mRFP material to one peripherally localized puncta, which colocalized with Hsp42 but not with Spc42 (Figure 5, C and D). Foci formed during Sti1 overexpression in the untreated samples did not colocalize with Spc42 (Figure 5C). Furthermore, Htt103Q-GFP containing the proline-rich region is targeted in a p97-dependent manner to aggresomes (Wang et al., 2009), and deletion of STI1 has no effect on aggregation of Htt103Q-Pro (Supplemental Figure S2B). Therefore Sti1 is not promoting the accumulation of Htt103Q in the yeast aggresome.
Strikingly, Sti1 appears to prevent accumulation of Rnq1-mRFP in the IPOD and by promoting its accumulation in StiF. Rnq1 is considered to be an IPOD marker (Kaganovich et al., 2008; Tyedmers et al., 2010b), and the single Rnq1-mRFP and Hsp42-GFP–containing peripheral foci detected when Hsp104 is inhibited for 2 h appear to be the IPOD (Figure 5, C and D). Elevation of Sti1 in the presence of GndHCl prevents Rnq1-mRFP from accumulating in peripheral foci with Hsp42-GFP and instead promotes its accumulation in the StiF.
These data demonstrate that organization of Rnq1 in speckled foci is highly sensitive to modulation of Hsp104 and Hsp70 system activity. Impairment of Hsp104 leads Rnq1-mRFP to accumulate in a single cytosolic puncta instead of speckled foci. The effects of Sti1 on Rnq1-mRFP foci resemble those of Hsp104, except that Sti1 promotes accumulation of amyloid-like proteins in StiF rather than a peripheral puncta. The StiF is a perinuclear quality control depot that contains Sti1, Hsp104, and amyloid-like proteins. It appears to be distinct from the aggresome, as it does not form at the spindle pole body (Wang et al., 2009) and may serve as a detoxification center for amyloid-like proteins.
Sti1 reorganizes complexes that contain Htt103Q and Hsp70
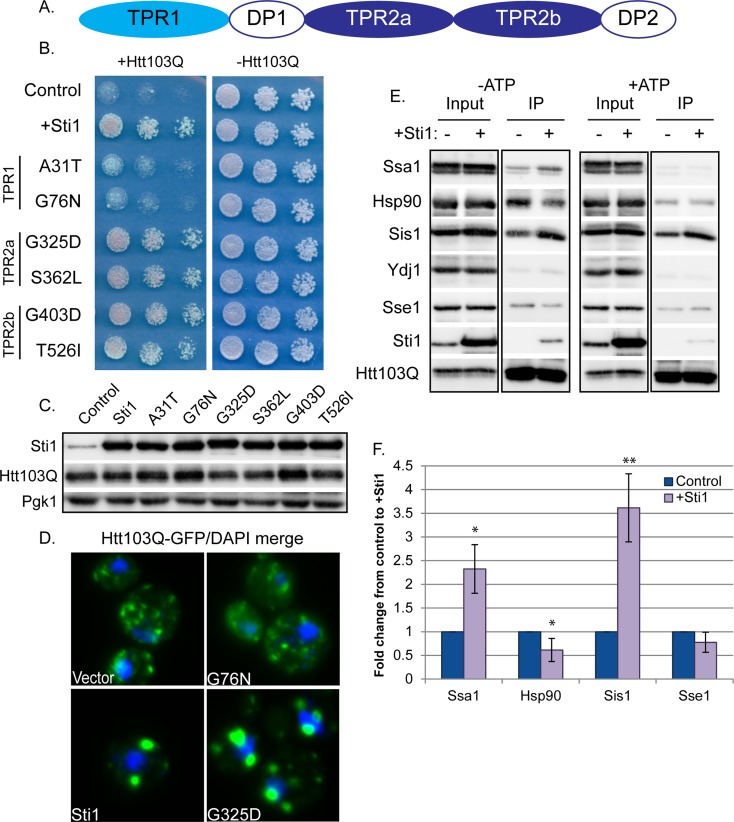
Sti1 associates with oligomeric forms of Htt103Q (Figure 2) and colocalizes with amyloid-like proteins in the StiF (Figure 4), which might enable it to suppress Htt toxicity by acting alone or in concert with Hsp70/Hsp90. The cochaperone function of Sti1 is conferred by three TPR repeats, which are important for interaction with Hsp70 and Hsp90 (Figure 6A). TPR1 is implicated in interaction with Hsp70, and TPR2a with Hsp90, whereas both chaperones bind with lower affinity to the TPR2b region (Hernandez et al., 2002; Song and Masison, 2005; Flom et al., 2007). Point mutations in TPR1 and Tpr2a the hinder Sti1’s ability to regulate Hsp70 and Hsp90 function, respectively (Song and Masison, 2005; Flom et al., 2007). Such mutants were used to evaluate whether Sti1 interacts with Hsp70 or Hsp90 to modulate spatial quality control of Htt103Q toxicity (Figure 6B). Although mutations in TPR2a and TPR2b had no effect (Figure 6, B–D), mutations in TPR1 hindered the ability of Sti1 to reorganize Htt103Q to the StiF (Figure 6D). TPR1 mutants also lost their ability to suppress growth defects caused by Htt103Q (Figure 6B). Thus StiF formation and suppression of Htt103Q toxicity require functional domains of Sti1 that enable it to interact with the Hsp70 system.
FIGURE 6:
Sti1 reorganizes complexes that contain Htt103Q and Hsp70. (A) Diagram of Sti1’s domain structure. (B) Effect of mutating different Sti1 TPR domains on ability to suppress Htt103Q growth defect as monitored by cell growth assay plated in fivefold dilutions. (C) Sti1 and Htt103Q expression as monitored by Western blot analysis of strains shown in B. (D) Effect of select Sti1 point mutations on Sti1’s ability to reorganize Htt103Q. (E) Effect of elevated Sti1 on the multichaperone:substrate complex as monitored by coimmunoprecipitation of Htt103Q and Western blot of indicated chaperones. Coimmunoprecipitations were carried out in the absence or presence of ATP as indicated above each group of images. (F) Quantitation of chaperones that coimmunoprecipitated with Htt103Q in E. (values represent means ± SE, n ≥ 3, *p < 0.05, **p < 0.005). Experiments were carried out in the wild-type Htt103Q integration strain.
To determine the effect of Sti1 on interactions between Hsp70 and Hsp90 with Htt103Q, we evaluated complex formation between these proteins by coimmunoprecipitation (Figure 6, E and F). Htt103Q was immunoprecipitated from yeast lysates prepared under nondenaturing conditions, and the presence of Ssa1, Hsp90, Ydj1, Sis1, and Sse1 in precipitates was determined by Western blot. Entrance of Sti1 into complexes altered relative quantities of different molecular chaperones that associated with Htt103Q (Figure 6, E and F). The influence of Sti1 on Hsp70/Hsp90 interactions with Htt103Q was sensitive to changes in ATP levels, so its effects are specific for the nucleotide bound state of the chaperones (Figure 6E). Of interest, Sti1 increased levels of Htt103Q that associated with the type II Hsp40, Sis1, by around fourfold and Ssa1 by more than twofold (Figure 6, E and F). In contrast, little of the type I Hsp40, Ydj1, was present, and Sti1 reduced the amount of coprecipitated Hsp90 and Sse1. These collective data indicate that spatial quality control of amyloid-like assemblies involves Sti1-dependent reorganization of complexes containing Hsp70 and Htt103Q, with the greatest effect being a dramatic increase in the presence of Sis1.
Sti1 and Sis1 cooperate to modulate amyloid toxicity
Sis1 interacts with oligomeric forms of Rnq1 to facilitate the propagation and affect the spatial organization of [RNQ+] prions (Sondheimer et al., 2001; Aron et al., 2007; Douglas et al., 2009). Thus Sti1 and Sis1 may cooperate to suppress growth defects caused by the presence of amyloid-like protein in the cytosol. We therefore investigated whether Sti1 is required for Sis1 to suppress Rnq1 and Htt103Q toxicity (Figure 7, A and B). Elevation of Sis1 diminished Rnq1 toxicity in a wild-type strain, but its protective effect was severely diminished in a ΔSti1 strain, even though deletion of STI1 had no effect on Sis1 expression (Figure 7A). Similarly, elevated Sis1 also suppressed the growth defect caused by Htt103Q, and this protective effect was reduced in a ΔSti1 strain (Figure 7B).
FIGURE 7:
Sti1 and Sis1 cooperate to regulate Rnq1 and Htt103Q toxicity. (A, B) Inability of Sis1 to suppress growth defect of (A) Rnq1-mRFP (p423-CUP1 plasmid) or (B) Htt103Q (toxic integration strain) in a Sti1 deletion strain as monitored by cell growth assay plated in fivefold dilutions. Expression levels of Sti1, Sis1, and Htt103Q or Rnq1 were measured by Western blot analysis. (C) Effect of elevated Sti1 on subcellular localization of Sis1 during Rnq1-mRFP expression. Experiments were carried out in a strain in which Sis1 was tagged at the endogenous locus with GFP.
Further support for functional interaction between Sti1 and Sis1 in buffering the toxicity of amyloid-like proteins comes from the demonstration that Sti1 drives the redistribution of Sis1 to the StiF (Figure 7C). Under normal conditions, Rnq1-mRFP was found in speckled foci and Sis1-GFP was distributed between the cytosol and nucleus (Figure 7C; Douglas et al., 2009). On elevation of Sti1, the localization pattern of Sis1 changed dramatically, and a large pool of it was concentrated in the StiF with Rnq1-mRFP (Figure 7C). Data presented in Figures 5–7 suggest that Sti1 helps suppress proteotoxicity via stabilization of complexes between Hsp70, Sis1, and amyloid-like protein assemblies to promote their accumulation in the perinuclear StiF.
DISCUSSION
The Hsp70/90 cochaperone, Sti1, suppresses proteotoxicity by organizing amyloid-like assemblies into the StiF. The StiF is a juxtanuclear compartment containing thioflavin-T–positive assemblies of Rnq1 and templated Htt103Q. Amyloid-like protein accumulation in the StiF correlates with suppression of proteotoxicity caused by Rnq1 and Htt103Q. Partitioning of aberrant proteins between the StiF and other protein-handling centers is controlled by interactions between Sti1 and the Hsp70 chaperone system. Loss of Sti1 exacerbates Htt proteotoxicity, whereas increases in Sti1 levels suppress it by increasing protein accumulation in the StiF. Sti1 is identified as a component of the Hsp70/Hsp90 system that acts in spatial PQC to promote protective protein aggregation.
Sti1 aids in organization of myeloid-like forms of Rnq1 and Htt103Q into one or two perinuclear foci (Figure 8). Studies with marker proteins show that the StiF exhibits similarities and differences with respect to the IPOD, JUNQ, and yeast aggresome (Kaganovich et al., 2008; Wang et al., 2009; Weisberg et al., 2012). Therefore whether the StiF represents a perinuclear IPOD, a JUNQ that contains amyloid-like protein, or a novel protein-handling depot is not clear. The aggresome and the IPOD contain detergent-insoluble and amyloid-like protein species (Kaganovich et al., 2008; Wang et al., 2009), whereas the peripheral compartment and JUNQ contain detergent-soluble protein aggregates (Kaganovich et al., 2008; Specht et al., 2011). Therefore Stif could be hybrid protein deposition sites formed as a result of cooperation between Hsp70 and Sti1 in spatial quality control.
FIGURE 8:
Model for chaperone-dependent steps in Sti1-inducible foci formation (StiF). The StiF is perinucear but is not localized at the spindle pole body (SPB).
Sti1 is a stress-inducible protein, and StiF are most visible when Sti1 levels are elevated fivefold to sixfold above normal. Sti1 is induced around threefold in response to heat stress (Supplemental Figure S4), so the effect of overexpressed Sti1 on spatial quality control of amyloid-like aggregates occurs in a concentration range relative to that of endogenous Sti1. Spatial organization of Htt103Q is also affected by deletion of STI1, and so endogenous Sti1, as well as overexpressed Sti1, affects spatial organization of foci that contain amyloid-like proteins. There is also potential for Sti1 to aid in cell stress protection by affecting the spatial organization of amyloid-like aggregates that form in response to proteotoxic growth conditions.
Why is accumulation of Htt103Q or [RNQ+] prions in the StiF cytoprotective? Onset of [RNQ+] prion or Htt103Q toxicity is detected within 4 h of expression (Douglas et al., 2008, 2009), and at this time they appear in the cytosol as speckled punctae and diffuse assemblies. Elevation of Sti1 accelerates the sequestration of amyloid-like proteins into the StiF and increases levels of detergent-insoluble oligomers of [RNQ+] and Htt103Q more than twofold. This may occur via diversion of amyloidogenic proteins away from the IPOD to limit shearing by Hsp104, which could prevent formation of toxic oligomers (Chernoff et al., 1995; Tyedmers et al., 2010b). Alternatively, Hsp70 functions via a molecular switch mechanism to activate Hsp104 (Lee et al., 2013). Thus elevation of Sti1 could hinder Hsp70’s ability to activate Hsp104 and thereby increase pools of benign amyloid-like protein aggregates.
[RNQ+] prions sequester Spc42 away from its normal function in the spindle pole body, leading to cell death (Treusch and Lindquist, 2012), so increasing accumulation of [RNQ+] prions to the StiF would be protective. Accumulation of amyloid-like protein species in the JUNQ is cytotoxic, as it inhibits normal PQC function (Kaganovich et al., 2008; Weisberg et al., 2012; Shiber et al., 2013; Summers et al., 2013), and thus diverting these proteins to the StiF would also ensure normal function of the ubiquitin proteasome system. The fate of proteins that enter the foci induced by Sti1 is not entirely clear, yet these assemblies are highly stable, so Stif may serve as a long-term storage vault for otherwise toxic proteins. Thus the StiF may function as an IPOD-like perinuclear deposition site for amyloid-like proteins.
How does modulation of Sti1 alter spatial partitioning of amyloid-like aggregates? Sti1 promotes association of Ssa1 and Sis1 with Htt103Q while reducing the presence of Hsp90. Sti1’s cooperation with Sis1 in spatial distribution of amyloid-like proteins is consistent with observations that Sis1 affects the subcellular localization of prions and misfolded proteins (Douglas et al., 2009; Malinovska et al., 2012). During heat stress, Sis1 is proposed to interact with the aggregate sorting factor Btn2, targeting misfolded proteins to a juxtanuclear location (Malinovska et al., 2012). Sis1 function is also implicated in targeting of [RNQ+] prions to the nucleus (Douglas et al., 2008, 2009) in a process that may involve Cur1 (Malinovska et al., 2012). Levels of Btn2 and Cur1 during heat stress are believed to regulate the relative cytosolic concentrations of Sis1, thereby dictating whether misfolded protein accumulates with Sis1 at a juxtanuclear site or with Hsp42 at a peripheral cytosolic site (Malinovska et al., 2012). Thus there are protein-sorting factors that interact with Sis1 and affect targeting of protein aggregates to different protein-handling centers. Cur1, Btn2, and Hsp42 were deleted from strains expressing Htt103Q but were dispensable for Sti1-dependent accumulation of Htt103Q in the StiF and suppression of Htt toxicity (Supplemental Figure S3). Nevertheless, the existence of such spatial PQC factors suggests the possibility that Sti1 and Sis1 function in an alternate spatial quality control process for amyloid-like proteins.
How might Sti1 reorganize the subcellular foci that contain the amyloid-like oligomers with which it interacts? Hsp104 functionally interacts with Ssa1, Sis1, and Sti1 in maintenance of the yeast prion [PSI+] (Kryndushkin et al., 2002; Reidy and Masison, 2010), as evidenced by the observation that deletion of STI1 inhibits the ability of overexpressed Hsp104 to eliminate [PSI+] (Reidy and Masison, 2010). In addition, Sti1 can directly interact with Hsp104, likely through its TPR1 domain (Abbas-Terki et al., 2001). Of interest, both the elevation of Sti1 and inhibition of Hsp104 for a short period with GndHCl prevent accumulation of speckled Rnq1-mRFP foci. Yet GndHCl promotes formation of foci in a peripheral location and Sti1 in a perinuclear location, with Sti1 being dominant over GndHCl. Inhibition of Hsp104 eliminates [RNQ+] prions from cells (Sondheimer and Lindquist, 2000), but Sti1 overexpression does not cure yeast of [RNQ+] prions (Supplemental Figure S4A, left). However, when Hsp104 is inhibited, Sti1 elevation accelerates the rate of [RNQ+] prion curing caused by inhibition of Hsp104 (Supplemental Figure S4). Thus Sti1 does not appear to be an inhibitor of Hsp104. Instead, it appears to interact with Sis1 and possibly other spatial PQC factors to direct amyloid-like proteins to the StiF, and this sequesters pools of Rnq1 and Htt103Q away from the machinery that would normally break them into smaller speckled foci.
The cochaperones CHIP and BAG3 target Hsp70 clients for degradation via the ubiquitin proteasome or autophagy, respectively (Cyr et al., 1994; Meacham et al., 2001; Gamerdinger et al., 2011). Sti1 is now reported to enable Hsp70 to function in spatial PQC by targeting amyloid-like proteins to the StiF. Sti1 is stress inducible, and, in humans, CHIP and Sti1 differentially bind to Hsp70 and Hsp90 based on changes in phosphorylation (Muller et al., 2013). Thus, under normal and stress conditions, subtle changes in Sti1 levels or phosphorylation status could affect targeting of Hsp70 clients to an alternate fate. Sti1 is differentially expressed in various brain regions of both healthy humans and neurodegenerative disease patients (Hawrylycz et al., 2012; Martins-de-Souza et al., 2012). Thus differences in Sti1 activity in different tissues or cell types could affect the efficiency of mammalian spatial PQC, influencing the onset of protein conformational disorders. In the future, it will be interesting to determine whether differences in the cellular set point of spatial quality control factors in healthy and diseased tissues correlate with neuronal subpopulations in which neurodegenerative diseases originate.
MATERIALS AND METHODS
Strains and plasmids
Yeast strains and plasmids are listed in Supplemental Tables S1 and S2 (Douglas et al., 2008, 2009). All strains harbored Rnq1 in its [RNQ+] prion form unless otherwise indicated. The generation of isogenic [rnq-] strains was accomplished via sequential passage of cells on plates containing 3 mM guanidinium-HCl. Strains were transformed with plasmids and cultured in synthetic media as described previously (Douglas et al., 2008). Constructs containing a copper-inducible promoter (CUP1) were induced at a low constitutive level from basal levels of copper in media unless it is specified that media were supplemented with additional CuSO4. This is the case for all Rnq1-mRFP microscopy unless otherwise indicated. For Sti1-overexpression experiments, Sti1 was always expressed from its endogenous promoter on a high-copy plasmid. Each of the Sti1 point mutants in Figure 6 was created using site-directed mutagenesis of the parent yEPlac-Sti1 plasmid and verified by sequencing.
Htt25Q and Htt103Q integration strains were generated using the pRS305 plasmid backbone. This vector carried a construct containing an amino-terminal FLAG tag, the first 17 amino acids of Htt, a 25- or 103-polyQ stretch, and a carboxy-terminal GFP tag. The Htt constructs were integrated under control of a galactose-inducible promoter (GAL1). Htt103Q-NoTox was an integration strain that lacked a growth defect and thus was used to demonstrate that deletion of STI1 elevates Htt103Q toxicity. The toxic Htt103Q integration strain was used in every other instance in the article where Htt103Q was expressed, except in Figure 1D. Deletion strains for Sti1, Hsp42, Cur1, and Btn2 were generated in the toxic Htt103Q integration strain and the Htt25Q integration strain by inserting a G418 resistance cassette at the genomic locus. This cassette was amplified from genomic DNA isolated from the BY4742 deletion library (YKO Collection; Open Biosystems, Huntsville, AL). Each deletion was verified by PCR from genomic DNA. Sti1 and Hsp42 deletions were further verified by Western blot, but Cur1 and Btn2 were not, as no antibody for these proteins was available. A deletion strain for Sti1 was also generated in the same manner in the W303 strain.
Expression of Htt103Q
Htt cultures were grown in synthetic media containing 2% glucose until cultures reached a density of 4 OD600 or greater. They were then diluted back in 2% raffinose, allowed to recover to mid–log phase growth, and finally induced for indicated times with 2% galactose. Except during time courses, Htt25Q and Htt103Q were induced for 4 h. The only exception to these expression conditions was when we used the Htt103Q-Pro construct. This construct lacked the N terminal FLAG tag and contained a proline-rich region following the polyQ stretch. Expression of Htt103Q-Pro was induced with 2% galactose for 24 h from a low-copy plasmid and galactose-inducible promoter.
High-copy screen for toxicity suppressors
An integration strain carrying a galactose-inducible Htt103Q with NLS was transformed with a high-copy yeast expression library in which the backbone plasmid carried a 5- to 10-kb fragment of DNA (Carlson and Botstein, 1982). Cells were plated on galactose to induce expression of the Htt103Q-NLS. Transformants that grew under these toxic conditions were verified by 5-fluoroorotic acid (5-FOA) screening to be plasmid-dependent suppressors. Suppressor plasmids were rescued from transformants that grew upon galactose initially but not after passage on 5-FOA. Each full-length screen hit also was found to suppress toxicity of the Htt103Q integration strain used in this study. The screen hit containing Sti1 came up twice in the screen. Other genes within the same DNA fragments of these two hits were YOR029W and CIN5. Although we subcloned each gene with the 5′ and 3′ untranslated regions (from this and the other screen hits), we also asked various sources for readily available constructs to expedite the verification process. Sti1 was one of these genes.
Toxicity assays
Yeast samples were normalized to the same OD600 and then fivefold serially diluted in a sterile 96-well plate. Each dilution was plated in 10-μl spots on appropriate synthetic dropout media. Most images of yeast grown on glucose were scanned at 48 h and galactose at 72 h. Lysates for Western blots from toxicity assays were prepared using cultures picked from control growth plates after they were scanned. Rnq1 toxicity was assessed on plates containing 2% glucose with 500 μM CuSO4, and Htt toxicity plates contained 2% galactose.
SDS–PAGE and Western blotting
Lysates were prepared from pellets of yeast by either mechanical disruption via glass bead breaking (for semidenaturing detergent agarose gel electrophoresis [SDD–AGE] and coimmunoprecipitation) as described previously (Douglas et al., 2008, 2009) or an alkali lysis method as described previously (Summers et al., 2013; for samples that were directly run on SDS–PAGE gels). For mechanical disruption, the lysis buffer used was 0.1% Triton X-100, 75 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and Sigma-Aldrich (St. Louis, MO) protease inhibitor cocktail. Standard separation techniques were used to analyze lysates via SDS–PAGE. Protein was transferred to nitrocellulose for 75–90 min at 110 V and probed using the indicated antibodies. Antibodies used in this study are listed in Supplemental Table S3 (Lee et al., 2002). The Sti1 antibody was raised in rabbit against purified full-length Sti1 protein. The antisera were not purified or concentrated any further, as the Western blot signal was strong and clean.
Fluorescence microscopy
Fluorescence microscopy was carried out as described previously (Douglas et al., 2009). Briefly, cultures were fixed with 4% formaldehyde and 0.1 M KPO4, pH 7.4, and stored in 0.1 M KPO4, pH 7.4, with 1.2 M sorbitol. When 4′,6-diamidino-2-phenylindole (DAPI) stained, cells were permeabilized with 0.1% Triton in phosphate-buffered saline (PBS), washed twice with PBS, incubated with 0.5 μg/ml DAPI for 10 min, washed twice again with PBS, and resuspended in the sorbitol solution. An Olympus IX81 motorized inverted fluorescence microscope (Olympus, Tokyo, Japan) paired with MetaMorph software (Molecular Devices, Sunnyvale, CA) was used to collect images. Micrographs were merged and processed using ImageJ (National Institutes of Health, Bethesda, MD).
Immunofluorescence and thioflavin-T staining
Cells were collected and fixed as described. After fixation, spheroplasts were generated using 5 mg/ml Zymolyase as to be described for size exclusion chromatography. For thioflavin-T staining, spheroplasts were treated with 0.1% Triton in PBS for 10 min and then washed with sorbitol solution. After this permeabilization, cells were incubated with 0.001% thioflavin-T for 10 min, washed, and resuspended in sorbitol solution. For immunofluorescence, spheroplasts were applied to Teflon-coated glass slides (Electron Microscopy Sciences, Hatfield, PA) with poly-l-lysine–coated wells. Then wells were then blocked using buffer containing 0.1 M KPO4, pH 7.5, 0.5% bovine serum albumin, and 0.02% Tween-20. Primary and secondary antibodies were applied with washes after each. Antibody incubation and washes all were carried out with the same blocking buffer.
SDD–AGE
SDD–AGE was carried out as described previously (Douglas et al., 2008). Briefly, samples were prepared via bead breaking as described and normalized by a protein determination assay (DC protein assay kit; Bio-Rad, Hercules, CA). Equivalent amounts of total protein were loaded on a 1.4% agarose gel containing 0.1% SDS, run at 90–100 V for approximately 2 h, and then transferred to polyvinylidene fluoride at 12 V for 15 h.
Size exclusion chromatography
Nondenaturing extracts from the Htt103Q strain were prepared via spheroplasting as follows. Approximately 150 OD of mid–log phase cells were resuspended in 2 ml of Z buffer containing 50 mM Tris-Cl, pH 7.5, 10 mM MgCl2, 1 M sorbitol, and 30 mM dithiothreitol (DTT) and then incubated at room temperature for 15 min. Sample was then centrifuged and resuspended in 1 ml of Z buffer with 5 mg of Zymolyase added (30 mM DTT) and incubated at 30°C in shaker at 50 rpm for 45 min. After Zymolyase treatment, spheroplasts were centrifuged and carefully washed twice with 1 ml of cold Z buffer (1 mM DTT) and then thoroughly resuspend with 800 μl of lysis buffer (listed previously, without Triton) using a cold glass disposable pipet. Samples were precleared to remove cell debris and normalized so that the same amount of protein was loaded to the column each time. A 500-μl loop was loaded into a Sephacryl S200 column, and 500-μl fractions were collected. Western blots show every third fraction starting slightly before the void volume as determined by thyroglobulin.
Coimmunoprecipitation
The Htt103Q strain carried either an empty vector or the YepLac-Sti1 construct and was induced and lysed via bead breaking as described. For experiments in which ATP was present, the lysis buffer contained 75 mM Tris, pH 7.4, 150 mM NaCl, 0.1% Triton, 10 mM MgCl, and 4 mM ATP. Lysates were incubated with anti–FLAG M2 affinity gel (Sigma-Aldrich). Htt103Q that was captured was then eluted by addition of 0.2 mg/ml FLAG peptide (Sigma-Aldrich) and incubation at 37°C for 20 min. After incubation, samples were spun at 4000 rpm for 5 min, and supernatant was harvested, combined with sample buffer, and loaded on an SDS–PAGE gel along with an input sample. In the case of coimmunoprecipitation from column fractions, two 500-μl fractions were combined and a small amount reserved for an input sample, and the remaining sample was used for the immunoprecipitation.
Luciferase refolding assay
Refolding of denatured luciferase by Ssa1 and Sis1 was performed as described previously (Lee et al., 2002). Briefly, firefly luciferase (13.3 mg/ml; Promega, Madison, WI) was diluted 42-fold into 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 7.4, 50 M KCl, 5 mM MgCl2, 6 M GndHCl, and 5 mM DTT and denatured by incubation at 25°C for 1 h. Aliquots (1 μl) were removed and added to 124 μl of refolding buffer that was composed of 25 mM HEPES, pH 7.4, 50 mM KCl, 5 mM MgCl2, and 1 mM ATP and contained the indicated concentration of Ssa1, Sis1, Sti1, and/or CHIP. Reaction mixtures were incubated at 30°C, and 1-μl aliquots were removed and mixed with 60 μl of luciferase assay buffer (Promega) at the indicated time points; luminescence was collected for 10 s with a TD20/20 luminometer (Turner Designs, Sunnyvale, CA).
Analysis of Rnq1 to determine Hsp104 shearing activity
The W303α yeast strain harbored either an empty vector (control) or Sti1 construct expressed from its endogenous promoter and p315-Cup1-Rnq1-GFP. Cultures were maintained at log phase in synthetic media containing glucose. After addition of 3 mM guanidinium HCl, cultures were monitored at OD600 for doubling time. At the indicated generations, a portion of the culture was pelleted and frozen. After all time points were collected, pellets were thawed on ice, lysed via bead breaking, and normalized. An aliquot was separated to use as the total fraction, and then the remaining lysate was subjected to 100,000 × g spin in an ultracentrifuge at 4°C. An aliquot of the supernatant was collected, and then the pellet was resuspended in the beginning volume with the same lysis buffer. A 20-μl portion of each sample (total, supernatant, and pellet) was run on a 9% SDS–PAGE gel and transferred to nitrocellulose, then probed using anti-GFP antibody to monitor Rnq1 solubility. A control culture was grown for the same amount of time without GndHCl and analyzed in the same way.
Supplementary Material
Acknowledgments
We thank P. Brennwald, J. Buchner, B. Bukau, V. Kushnirov, J. Johnson, D. Picard, and S. Lindquist for reagents. We acknowledge Pete Douglas for helpful discussions and Daniel Summers for critical reading of the manuscript. The Cyr laboratory is supported by the National Institutes of Health (R01GM56981 and R01GM067785), and K.W. is supported by F31NS074777.
Abbreviations used:
- GFP
green fluorescent protein
- GndHCl
guanidinium hydrochloride
- Htt103Q
Huntingtin with 103Q glutamine stretch
- IPOD
insoluble protein deposit
- JUNQ
juxtanuclear quality control compartment
- mRFP
monomeric red fluorescent protein
- PolyQ
polyglutamine
- PQC
protein quality control
- StiF
Sti1-inducible foci
- TPR
tetratricopeptide repeat
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-06-0315) on October 9, 2013.
REFERENCES
- Abbas-Terki T, Donze O, Briand PA, Picard D. Hsp104 interacts with Hsp90 cochaperones in respiring yeast. Mol Cell Biol. 2001;21:7569–7575. doi: 10.1128/MCB.21.22.7569-7575.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aron R, Higurashi T, Sahi C, Craig EA. J-protein co-chaperone Sis1 required for generation of [RNQ+] seeds necessary for prion propagation. EMBO J. 2007;26:3794–3803. doi: 10.1038/sj.emboj.7601811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucciantini M, Calloni G, Chiti F, Formigli L, Nosi D, Dobson CM, Stefani M. Prefibrillar amyloid protein aggregates share common features of cytotoxicity. J Biol Chem. 2004;279:31374–31382. doi: 10.1074/jbc.M400348200. [DOI] [PubMed] [Google Scholar]
- Carlson M, Botstein D. Two differentially regulated mRNAs with different 5’ ends encode secreted with intracellular forms of yeast invertase. Cell. 1982;28:145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Carrell RW, Lomas DA. Conformational disease. Lancet. 1997;350:134–138. doi: 10.1016/S0140-6736(97)02073-4. [DOI] [PubMed] [Google Scholar]
- Chen S, Sullivan WP, Toft DO, Smith DF. Differential interactions of p23 and the TPR-containing proteins Hop, Cyp40, FKBP52 and FKBP51 with Hsp90 mutants. Cell Stress Chaperones. 1998;3:118–129. doi: 10.1379/1466-1268(1998)003<0118:diopat>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313:1604–1610. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- Cyr DM, Hohfeld J, Patterson C. Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem Sci. 2002;27:368–375. doi: 10.1016/s0968-0004(02)02125-4. [DOI] [PubMed] [Google Scholar]
- Cyr DM, Langer T, Douglas MG. DnaJ-like proteins: molecular chaperones and specific regulators of Hsp70. Trends Biochem Sci. 1994;19:176–181. doi: 10.1016/0968-0004(94)90281-x. [DOI] [PubMed] [Google Scholar]
- Cyr DM, Lu X, Douglas MG. Regulation of Hsp70 function by a eukaryotic DnaJ homolog. J Biol Chem. 1992;267:20927–20931. [PubMed] [Google Scholar]
- Dai Q, et al. CHIP activates HSF1 and confers protection against apoptosis and cellular stress. EMBO J. 2003;22:5446–5458. doi: 10.1093/emboj/cdg529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehay B, Bertolotti A. Critical role of the proline-rich region in Huntingtin for aggregation and cytotoxicity in yeast. J Biol Chem. 2006;281:35608–35615. doi: 10.1074/jbc.M605558200. [DOI] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN(+)] Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- Dolinski KJ, Cardenas ME, Heitman J. CNS1 encodes an essential p60/Sti1 homolog in Saccharomyces cerevisiae that suppresses cyclophilin 40 mutations and interacts with Hsp90. Mol Cell Biol. 1998;18:7344–7352. doi: 10.1128/mcb.18.12.7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas PM, Summers DW, Ren HY, Cyr DM. Reciprocal efficiency of RNQ1 and polyglutamine detoxification in the cytosol and nucleus. Mol Biol Cell. 2009;20:4162–4173. doi: 10.1091/mbc.E09-02-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas PM, Treusch S, Ren HY, Halfmann R, Duennwald ML, Lindquist S, Cyr DM. Chaperone-dependent amyloid assembly protects cells from prion toxicity. Proc Natl Acad Sci USA. 2008;105:7206–7211. doi: 10.1073/pnas.0802593105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duennwald ML, Jagadish S, Muchowski PJ, Lindquist S. Flanking sequences profoundly alter polyglutamine toxicity in yeast. Proc Natl Acad Sci USA. 2006;103:11045–11050. doi: 10.1073/pnas.0604547103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira PC, Ness F, Edwards SR, Cox BS, Tuite MF. The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol Microbiol. 2001;40:1357–1369. doi: 10.1046/j.1365-2958.2001.02478.x. [DOI] [PubMed] [Google Scholar]
- Floer M, Bryant GO, Ptashne M. HSP90/70 chaperones are required for rapid nucleosome removal upon induction of the GAL genes of yeast. Proc Natl Acad Sci USA. 2008;105:2975–2980. doi: 10.1073/pnas.0800053105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flom G, Behal RH, Rosen L, Cole DG, Johnson JL. Definition of the minimal fragments of Sti1 required for dimerization, interaction with Hsp70 and Hsp90 and in vivo functions. Biochem J. 2007;404:159–167. doi: 10.1042/BJ20070084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamerdinger M, Kaya AM, Wolfrum U, Clement AM, Behl C. BAG3 mediates chaperone-based aggresome-targeting and selective autophagy of misfolded proteins. EMBO Rep. 2011;12:149–156. doi: 10.1038/embor.2010.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer's amyloid beta-peptide. Nat Rev Mol Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- Hawrylycz MJ, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez MP, Sullivan WP, Toft DO. The assembly and intermolecular properties of the hsp70-Hop-hsp90 molecular chaperone complex. J Biol Chem. 2002;277:38294–38304. doi: 10.1074/jbc.M206566200. [DOI] [PubMed] [Google Scholar]
- Jiang J, Ballinger CA, Wu Y, Dai Q, Cyr DM, Hohfeld J, Patterson C. CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J Biol Chem. 2001;276:42938–42944. doi: 10.1074/jbc.M101968200. [DOI] [PubMed] [Google Scholar]
- Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G, Song Y, Chung S, Masison DC. Propagation of Saccharomyces cerevisiae [PSI+] prion is impaired by factors that regulate Hsp70 substrate binding. Mol Cell Biol. 2004;24:3928–3937. doi: 10.1128/MCB.24.9.3928-3937.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G, Masison DC. Guanidine hydrochloride inhibits Hsp104 activity in vivo: a possible explanation for its effect in curing yeast prions. Curr Microbiol. 2001;43:7–10. doi: 10.1007/s002840010251. [DOI] [PubMed] [Google Scholar]
- Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordes E, Savelyeva L, Schwab M, Rommelaere J, Jauniaux JC, Cziepluch C. Isolation and characterization of human SGT and identification of homologues in Saccharomyces cerevisiae and Caenorhabditis elegans. Genomics. 1998;52:90–94. doi: 10.1006/geno.1998.5385. [DOI] [PubMed] [Google Scholar]
- Kryndushkin DS, Smirnov VN, Ter-Avanesyan MD, Kushnirov VV. Increased expression of Hsp40 chaperones, transcriptional factors, and ribosomal protein Rpp0 can cure yeast prions. J Biol Chem. 2002;277:23702–23708. doi: 10.1074/jbc.M111547200. [DOI] [PubMed] [Google Scholar]
- Langer T, Lu C, Echols H, Flanagan J, Hayer MK, Hartl FU. Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature. 1992;356:683–689. doi: 10.1038/356683a0. [DOI] [PubMed] [Google Scholar]
- Last NB, Rhoades E, Miranker AD. Islet amyloid polypeptide demonstrates a persistent capacity to disrupt membrane integrity. Proc Natl Acad Sci USA. 2011;108:9460–9465. doi: 10.1073/pnas.1102356108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kim JH, Biter AB, Sielaff B, Lee S, Tsai FT. Heat shock protein (Hsp) 70 is an activator of the Hsp104 motor. Proc Natl Acad Sci USA. 2013;21:8513–8518. doi: 10.1073/pnas.1217988110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Fan CY, Younger JM, Ren H, Cyr DM. Identification of essential residues in the type II Hsp40 Sis1 that function in polypeptide binding. J Biol Chem. 2002;277:21675–21682. doi: 10.1074/jbc.M111075200. [DOI] [PubMed] [Google Scholar]
- LeVine H., 3rd Thioflavine T interaction with synthetic Alzheimer's disease beta-amyloid peptides: detection of amyloid aggregation in solution. Protein Sci. 1993;2:404–410. doi: 10.1002/pro.5560020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Richter K, Buchner J. Mixed Hsp90-cochaperone complexes are important for the progression of the reaction cycle. Nat Struct Mol Biol. 2011;18:61–66. doi: 10.1038/nsmb.1965. [DOI] [PubMed] [Google Scholar]
- Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci USA. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinovska L, Kroschwald S, Munder MC, Richter D, Alberti S. Molecular chaperones and stress-inducible protein sorting factors coordinate the spatio-temporal distribution of protein aggregates. Mol Biol Cell. 2012;23:3041–3056. doi: 10.1091/mbc.E12-03-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins-de-Souza D, Guest PC, Mann DM, Roeber S, Rahmoune H, Bauder C, Kretzschmar H, Volk B, Baborie A, Bahn S. Proteomic analysis identifies dysfunction in cellular transport, energy, and protein metabolism in different brain regions of atypical frontotemporal lobar degeneration. J Proteome Res. 2012;11:2533–2543. doi: 10.1021/pr2012279. [DOI] [PubMed] [Google Scholar]
- Meacham GC, Patterson C, Zhang W, Younger JM, Cyr DM. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat Cell Biol. 2001;3:100–105. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- Meriin AB, Zhang X, He X, Newnam GP, Chernoff YO, Sherman MY. Huntington toxicity in yeast model depends on polyglutamine aggregation mediated by a prion-like protein Rnq1. J Cell Biol. 2002;157:997–1004. doi: 10.1083/jcb.200112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchowski PJ, Wacker JL. Modulation of neurodegeneration by molecular chaperones. Nat Rev Neurosci. 2005;6:11–22. doi: 10.1038/nrn1587. [DOI] [PubMed] [Google Scholar]
- Muller P, Ruckova E, Halada P, Coates PJ, Hrstka R, Lane DP, Vojtesek B. C-terminal phosphorylation of Hsp70 and Hsp90 regulates alternate binding to co-chaperones CHIP and HOP to determine cellular protein folding/degradation balances. Oncogene. 2013;32:3101–3110. doi: 10.1038/onc.2012.314. [DOI] [PubMed] [Google Scholar]
- Nicolet CM, Craig EA. Isolation and characterization of STI1, a stress-inducible gene from Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:3638–3646. doi: 10.1128/mcb.9.9.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzscha H, Schermann SM, Woerner AC, Pinkert S, Hecht MH, Tartaglia GG, Vendruscolo M, Hayer-Hartl M, Hartl FU, Vabulas RM. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell. 2011;144:67–78. doi: 10.1016/j.cell.2010.11.050. [DOI] [PubMed] [Google Scholar]
- Prodromou C, Siligardi G, O'Brien R, Woolfson DN, Regan L, Panaretou B, Ladbury JE, Piper PW, Pearl LH. Regulation of Hsp90 ATPase activity by tetratricopeptide repeat (TPR)-domain co-chaperones. EMBO J. 1999;18:754–762. doi: 10.1093/emboj/18.3.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian SB, McDonough H, Boellmann F, Cyr DM, Patterson C. CHIP-mediated stress recovery by sequential ubiquitination of substrates and Hsp70. Nature. 2006;440:551–555. doi: 10.1038/nature04600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidy M, Masison DC. Sti1 regulation of Hsp70 and Hsp90 is critical for curing of Saccharomyces cerevisiae [PSI+] prions by Hsp104. Mol Cell Biol. 2010;30:3542–3552. doi: 10.1128/MCB.01292-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roffe M, et al. Prion protein interaction with stress-inducible protein 1 enhances neuronal protein synthesis via mTOR. Proc Natl Acad Sci USA. 2010;107:13147–13152. doi: 10.1073/pnas.1000784107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl FU, Moarefi I. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- Shaner L, Wegele H, Buchner J, Morano KA. The yeast Hsp110 Sse1 functionally interacts with the Hsp70 chaperones Ssa and Ssb. J Biol Chem. 2005;280:41262–41269. doi: 10.1074/jbc.M503614200. [DOI] [PubMed] [Google Scholar]
- Shiber A, Breuer W, Brandeis M, Ravid T. Ubiquitin conjugation triggers misfolded protein sequestration into quality-control foci when Hsp70 chaperone levels are limiting. Mol Biol Cell. 2013;24:2076–2087. doi: 10.1091/mbc.E13-01-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondheimer N, Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Mol Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- Sondheimer N, Lopez N, Craig EA, Lindquist S. The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J. 2001;20:2435–2442. doi: 10.1093/emboj/20.10.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Masison DC. Independent regulation of Hsp70 and Hsp90 chaperones by Hsp70/Hsp90-organizing protein Sti1 (Hop1) J Biol Chem. 2005;280:34178–34185. doi: 10.1074/jbc.M505420200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specht S, Miller SB, Mogk A, Bukau B. Hsp42 is required for sequestration of protein aggregates into deposition sites in Saccharomyces cerevisiae. J Cell Biol. 2011;195:617–629. doi: 10.1083/jcb.201106037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel GJ, Fullerton DM, Tyson JR, Stirling CJ. Coordinated activation of Hsp70 chaperones. Science. 2004;303:98–101. doi: 10.1126/science.1092287. [DOI] [PubMed] [Google Scholar]
- Summers DW, Douglas PM, Ren HY, Cyr DM. The type I Hsp40 Ydj1 utilizes a farnesyl moiety and zinc finger-like region to suppress prion toxicity. J Biol Chem. 2009;284:3628–3639. doi: 10.1074/jbc.M807369200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers DW, Wolfe KJ, Ren HY, Cyr DM. The Type II Hsp40 Sis1 cooperates with Hsp70 and the E3 ligase Ubr1 to promote degradation of terminally misfolded cytosolic protein. PLoS One. 2013;8:e52099. doi: 10.1371/journal.pone.0052099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipton KA, Verges KJ, Weissman JS. In vivo monitoring of the prion replication cycle reveals a critical role for Sis1 in delivering substrates to Hsp104. Mol Cell. 2008;32:584–591. doi: 10.1016/j.molcel.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treusch S, Cyr DM, Lindquist S. Amyloid deposits: protection against toxic protein species. Cell Cycle. 2009;8:1668–1674. doi: 10.4161/cc.8.11.8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treusch S, Lindquist S. An intrinsically disordered yeast prion arrests the cell cycle by sequestering a spindle pole body component. J Cell Biol. 2012;197:369–379. doi: 10.1083/jcb.201108146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyedmers J, Mogk A, Bukau B. Cellular strategies for controlling protein aggregation. Nat Rev Mol Cell Biol. 2010a;11:777–788. doi: 10.1038/nrm2993. [DOI] [PubMed] [Google Scholar]
- Tyedmers J, Treusch S, Dong J, McCaffery JM, Bevis B, Lindquist S. Prion induction involves an ancient system for the sequestration of aggregated proteins and heritable changes in prion fragmentation. Proc Natl Acad Sci USA. 2010b;107:8633–8638. doi: 10.1073/pnas.1003895107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Meriin AB, Zaarur N, Romanova NV, Chernoff YO, Costello CE, Sherman MY. Abnormal proteins can form aggresome in yeast: aggresome-targeting signals and components of the machinery. FASEB J. 2009;23:451–463. doi: 10.1096/fj.08-117614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegele H, Haslbeck M, Reinstein J, Buchner J. Sti1 is a novel activator of the Ssa proteins. J Biol Chem. 2003;278:25970–25976. doi: 10.1074/jbc.M301548200. [DOI] [PubMed] [Google Scholar]
- Wegele H, Wandinger SK, Schmid AB, Reinstein J, Buchner J. Substrate transfer from the chaperone Hsp70 to Hsp90. J Mol Biol. 2006;356:802–811. doi: 10.1016/j.jmb.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Weisberg SJ, Lyakhovetsky R, Werdiger AC, Gitler AD, Soen Y, Kaganovich D. Compartmentalization of superoxide dismutase 1 (SOD1G93A) aggregates determines their toxicity. Proc Natl Acad Sci USA. 2012;109:15811–15816. doi: 10.1073/pnas.1205829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KJ, Cyr DM. Amyloid in neurodegenerative diseases: friend or foe. Semin Cell Dev Biol. 2011;22:476–481. doi: 10.1016/j.semcdb.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.