Cell stress caused by repression of 54 individual ribosomal genes in Saccharomyces cerevisiae is analyzed. Cell cycle progression and cell morphology responses are specific to the protein whose synthesis is repressed but bud site selection is not. Proteins that generate G2/M and G1 phenotypes map to separate areas of the ribosomal particle.
Abstract
The biogenesis of ribosomes is coordinated with cell growth and proliferation. Distortion of the coordinated synthesis of ribosomal components affects not only ribosome formation, but also cell fate. However, the connection between ribosome biogenesis and cell fate is not well understood. To establish a model system for inquiries into these processes, we systematically analyzed cell cycle progression, cell morphology, and bud site selection after repression of 54 individual ribosomal protein (r-protein) genes in Saccharomyces cerevisiae. We found that repression of nine 60S r-protein genes results in arrest in the G2/M phase, whereas repression of nine other 60S and 22 40S r-protein genes causes arrest in the G1 phase. Furthermore, bud morphology changes after repression of some r-protein genes. For example, very elongated buds form after repression of seven 60S r-protein genes. These genes overlap with, but are not identical to, those causing the G2/M cell cycle phenotype. Finally, repression of most r-protein genes results in changed sites of bud formation. Strikingly, the r-proteins whose repression generates similar effects on cell cycle progression cluster in the ribosome physical structure, suggesting that different topological areas of the precursor and/or mature ribosome are mechanistically connected to separate aspects of the cell cycle.
INTRODUCTION
It has been known for >50 years that ribosomes make up as much as 50% of cell mass and that their synthesis is regulated in response to the cell's environment. Originally demonstrated for bacteria (Schaechter et al., 1958; Maaløe and Kjeldgaard, 1966), this phenomenon has also been observed in Saccharomyces cerevisiae (referred to as “yeast” in the following; Kief and Warner, 1981) and other eukaryotes, including mammalian cells (Derenzini et al., 1998, 2000; Fujita, 1999; Montanaro et al., 2012). The mechanisms coupling growth, cell proliferation, and ribosome biogenesis have remained a subject of intense study for decades. In eukaryotes, ribosome formation is regulated by cues from the cell environment via phosphorylation cascades in the target of rapamycin and ras pathways (Mayer and Grummt, 2006; Lempiainen et al., 2009). In addition, cell cycle progression responds to translation capacity and ribosome biogenesis (Thomas et al., 1979; Zaragoza et al., 1998; Jorgensen et al., 2004). However, our understanding of the molecular mechanisms for coordinating the complex networks controlling cell cycle and ribosome formation remains incomplete.
Normal ribosome biogenesis depends on a balanced supply of the ribosomal components (four types of rRNA and 80 ribosomal proteins [r-proteins] in yeast). It also requires ∼200 ribosomal biogenesis factors, proteins, and small RNAs that convert the primary rRNA precursor transcript into mature rRNAs, assist assembly of the ribosomal components and shaping of the ribosomal particles, monitor the quality of the ribosomal precursor particles, and ensure their transport to the cytoplasm (Henras et al., 2008; Woolford and Baserga, 2013). These ancillary ribosome assembly factors are not components of the mature translation-competent ribosomes, even though many become temporarily incorporated into ribosomal precursor particles.
Disturbing the balanced synthesis of ribosomal components and assembly factors leads to changes in rRNA processing and abolition of the formation of one or both ribosomal subunits (Ferreira-Cerca et al., 2005; Farrar et al., 2008; Pöll et al., 2009; Woolford and Baserga, 2013). This is often termed “nucleolar stress” and can broadly affect cellular functions. However, this phenomenon is not well understood. The purpose of the work reported here is to establish a systematic phenomenological framework defining the relationships between nucleolar stress and cell cycle progression and cell morphology in the simple eukaryote yeast.
RESULTS
Strategy
Fifty-seven of the 80 r-proteins in yeast are encoded by two chromosomal genes whose protein products in some cases are identical and in many other cases differ by only one or several amino acids. To determine the effects of repression of the synthesis of individual r-proteins, we used strains in which normal control of the expression of a single r-protein was abolished by deleting the chromosomal gene(s) for the protein and replacing it or them with a single plasmid-borne gene for that protein under control of a galactose-inducible promoter. Alternatively, one of the chromosomal genes for a given r-protein was deleted and the other brought under control of a galactose-inducible promoter inserted into the chromosome (see Materials and Methods for details). Most of the strains were from the collections of the Milkereit, Tschochner, and Woolford labs and were previously used to determine the effect of r-protein gene repression on the processing of rRNA (Ferreira-Cerca et al., 2005, 2007; Pöll et al., 2009; Jakovljevic et al., 2012; Gamalinda et al., 2013). Each strain will be referred to as Pgal-RPSxx or Pgal-RPLxx, where RPSxx and RPLxx indicate the specific r-protein gene controlled by the galactose promoter. The corresponding proteins are termed Sxx and Lxx, respectively. Strain BY4741, in which the r-protein genes have not been manipulated, was used a control. Traditional nomenclature for yeast r-proteins is used. For comparison with bacterial and human ribosomal protein nomenclature, see Jenner et al. (2012).
The essentiality of the 54 different r-proteins considered here was tested by streaking strains, each with a given r-protein gene under galactose control, on media with galactose or glucose as carbon sources. All grew on galactose but not on glucose, that is, each of the 54 proteins we analyzed is essential for growth; 28 are from the 60S ribosomal subunit, and 26 are from the 40S subunit. Subsequently, the 54 strains were grown in liquid medium with galactose as carbon source (“unrepressed condition”) and then shifted to glucose medium (“repressed condition”). Sixteen hours after the shift, we determined cell DNA content, cell and bud morphology, cell dimensions, and the number of buds on mother cells. Northern analysis of mRNA abundance for eight r-protein genes representing all the phenotypes to be described confirmed repression of transcription in glucose medium (Supplemental Figure S1). The concentration of ribosomal proteins after the repression of the galactose promoter was not followed because the ribosomal protein genes in the strains used were not tagged. However, the Milkereit and Woolford groups previously documented the distortion of ribosome biogenesis (Ferreira-Cerca et al., 2005, 2007; Pöll et al., 2009; Jakovljevic et al., 2012; Gamalinda et al., 2013). Furthermore, it is not clear that signals for changes in the parameters measured here are derived from a decrease in total ribosomal protein. They may come from the availability of newly synthesized proteins and the consequences for ribosome assembly.
Flow cytometric analysis of DNA content and cell size
We used flow cytometry of cells fixed in ethanol and stained with propidium iodide to measure the relative amount of DNA in each cell. Note that mother cells with associated buds are registered as “single cells” by the flow cytometer. Because the cultures were nonsynchronous, we observed two peaks representing 1N and 2N DNA equivalents (Figure 1). These 1N and 2N peaks correspond to cells in the G1 and G2/M phase, respectively, whereas the “valley” between the peaks contains cells in S phase. The distribution of the number of cells in the peaks was determined using Summit Software (Beckman Coulter).
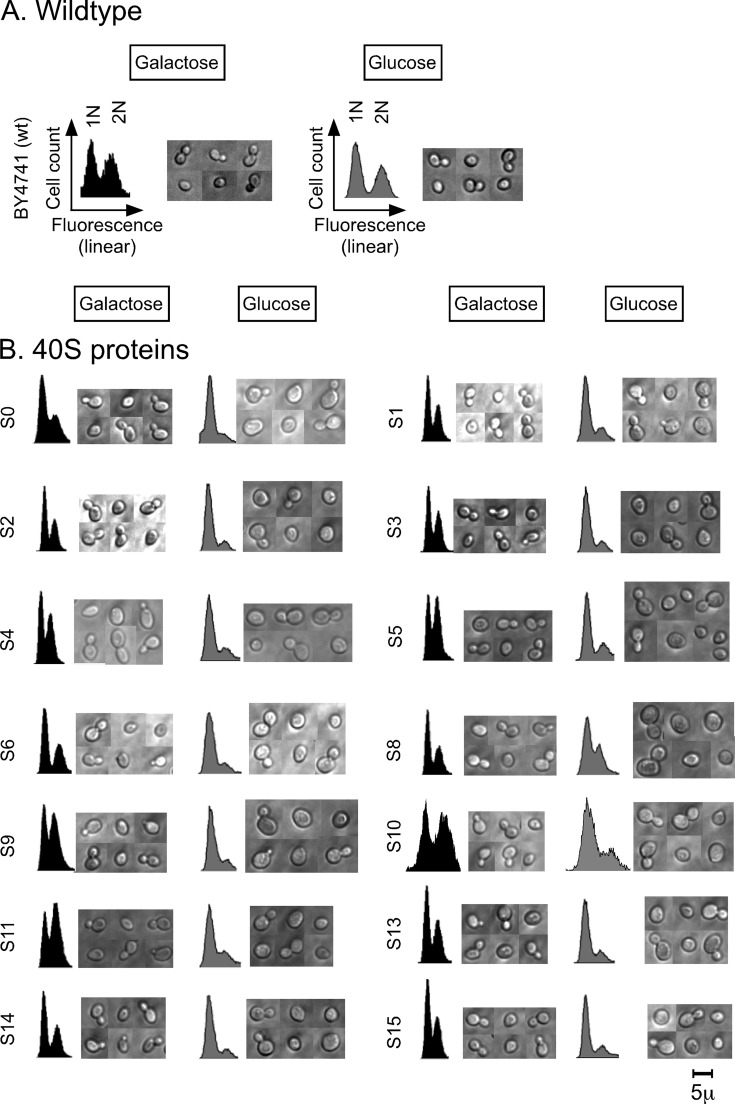
FIGURE 1:
Flow cytometry and microscopy of cells after cessation of the synthesis of individual r-proteins. Synthesis of a single r-protein in each of 54 strains was repressed by shifting each strain from galactose to glucose medium (see Supplemental Table S1 for genotypes). Cells were collected 16 h after the shift and prepared for flow cytometry by fixing and staining of DNA or for microscopy of live cells. (A–C) Examples of raw data. (A) BY4741 (all chromosomal r-protein genes were intact). (B) Strains for repression of individual protein genes specific to the 40S ribosomal subunit. (C) Strains for repression of individual r-protein genes specific to the 60S ribosomal subunit. (D–G) Quantification of data from experiments exemplified in A–C. (D) Fraction of cells in the 1N peak before and after cessation of the synthesis of the indicated 40S proteins. The light gray and open bars indicate the fraction of cells in the 1N peaks in galactose and glucose medium, respectively. The black bars indicate the ratio between the fraction of cells in the 1N peak in glucose and the fraction of cells in the 1N peak in galactose medium, that is, the change caused by the medium shift. (E) Fraction of cells with bud (at least 100 cells counted) before and after cessation of the synthesis of the indicated 40S proteins. The light gray and open bars indicate data for galactose and glucose medium, respectively. The black bars indicate the ratio between the fraction of budded cells in glucose and the fraction of budded cells in galactose medium. (F) Fraction of cells in the 1N and 3N peaks after cessation of the synthesis of the indicated 60S proteins. The light gray and open bars indicate the fraction of cells in the 1N peaks in galactose and glucose medium, respectively. The black bars indicate the ratio between the fraction of cells in the 1N peak in glucose and the fraction of cells in the 1N peak in galactose medium. The dark gray bars show the fraction of cells in the 3N peak or trail (for L17) in glucose medium. (G) Budding status (at least 100 cells counted) before and after cessation of the synthesis of the indicated 60S proteins. The light gray bars indicate fraction of budded cells in galactose medium, the open bars indicate the fraction of cells with one bud in glucose medium, and the black bars indicate the ratio between the fraction of single-budded cells in glucose and the fraction of budded cells in galactose. The dark gray bars indicate the fraction of cells with two or more buds in glucose. All strains are identified with the traditional yeast name of the r-protein whose synthesis was repressed. For correspondence to r-protein nomenclature for bacteria and humans, see Jenner et al. (2012). Assignment of cell cycle phenotypes: G1, 1N(glu) ≥0.7 and 1N(glu)/1N(gal) ≥ 1.4 (these values are marked by the dotted lines in D and G); (G1), 1N(glu) ≥ 0.6 and 1N(glu)/1N(gal) ≥ 1.3; G2/M, 3N or trail ≥ 0.1.
In nearly all strains growing in galactose medium, the cells distributed with about half (40–60%) in each of the 1N and 2N peaks (Figure 1, A–D and F). After the shift of the BY4741 control strain to glucose medium, the relative distribution of cells between the 1N and 2N peaks was essentially unchanged (Figure 1, A and D). In contrast, transfer to glucose of 22 of the 26 strains with 40S proteins under Pgal control resulted in redistribution from the 2N peak to the 1N peak (Figure 1, B and D). This indicates that repression of the synthesis of most 40S proteins causes G1 cell cycle–arrest or significantly prolonged G1 phase. Even though the distribution of cells between the flow cytometry peaks is not a discrete parameter, we have assigned a “G1” phenotype to strains in which the fraction of cells in the 1N peak is at least 70% after the shift and the ratio of 1N fraction in glucose to the 1N fraction in galactose exceeds 1.3–1.4 (Figure 1D). The 1N/2N ratio changed little for S20, but we have nevertheless assigned this strain as G1 in glucose because it has a high preponderance of 1N cells already in galactose and may be somewhat abnormal even under nonrepressing conditions. Accordingly, we conclude that a G1 phenotype follows after repression of the synthesis of S0–S6, S9–S11, S13–S15, S17, S19–S22, S26, S27, S29, and S30 (Figure 1, B and D). Repression of the remaining four 40S proteins (S8, S23, S24, and S31) did not elicit a significant change in the flow cytometry profile; these strains were designated as N for “no change” (Figure 1, B and D).
The flow cytometry response after repression of 60S protein synthesis was more complex than that observed for the 40S proteins. In particular, after repression of the synthesis of L4, L7, L18, and L40, we observed an unexpected third peak, containing 20–30% of the cells (Figure 1, C and F). The position of this peak is consistent with cells, or mother cells with associated buds, containing more than 2N DNA equivalents. Accordingly, we named the peak 3N, although we do not know whether cells in this peak contain exactly 3N equivalents of DNA. Repression of the synthesis of L3, L28, L35, or L37 resulted in a weaker 3N peak or, in the case of L17, a somewhat more heterogeneous “trail” of cells or cell clusters with >2N DNA content accounting for 10–15% of the cells (Figure 1, C and F). Strains forming the 3N peak or >2N trail in glucose medium were assigned as G2/M (Figure 1F). These strains will be analyzed in more detail later.
Repression of the synthesis of nine large ribosomal subunit proteins (L1, L3, L9, L16, L19, L21, L25, L30, and L43) shifted cells from the 2N to the 1N peak (Figure 1, C and F), similar to the results obtained with the majority of the 40S proteins, again indicative of G1 arrest or delay. Note that the phenotype of the Pgal-RPL3 strain was mixed and scored as both G2/M and a weak G1. Similar experiments previously reported by the de la Cruz lab also suggested that repression of RPL3 promotes G1 arrest or delay (Rosado et al., 2007). The remaining 60S proteins (L2, L5, L8, L10, L13, L20, L23, L27, L32–L34) did not cause any significant changes in the flow cytometry profile and were designated as N (Figure 2, C and F).
In summary, our flow cytometric analysis showed that repression of synthesis of 60S proteins yields two different protein-specific effects on DNA content, suggesting that certain 60S r-proteins are required for progression through the G2/M phase, whereas others are required for passage through the G1 phase. However, repression of the synthesis of 11 60S proteins had no effect on the distribution of cells between the cell cycle stages. In contrast, most 40S r-proteins are required for progression through G1.
Bright-field microscopy of cells
As with the flow cytometry experiments, light microscopy analysis of cells after repression of 40S and 60S r-protein synthesis revealed a complex response spectrum. We saw three morphological phenotypes after repression of the synthesis of r-proteins from the 40S subunit (Figure 1B). 1) For most strains, we observed a larger cell cross section after repression. This was supported by increased mean forward light scatter in the flow cytometer, which correlates with cell volume (Koch and Ehrenfeld, 1968; Lamouille and Derynck, 2007; Supplemental Figure S2). Thus our data suggest that repression of the synthesis of most 40S r-proteins results in increased cell size. 2) Repression of S20 synthesis generated irregularly shaped cells: cells were not just extremely large, but also unevenly shaped with elongated buds, appearing as if there were surface projections. 3) Repression of S17 synthesis generated no change—neither a defect in cell morphology nor an increase in cell size.
The morphological changes observed by bright-field microscopy after repression of 60S protein synthesis were more diverse and included phenotypes not observed for the 40S proteins (Figure 1C). We classified the morphological effects after the medium shift of 60S strains into six categories. 1) Mother cells with dibuds or tribuds were observed after depletion of L2, L4, L7, L18, and L40. Furthermore, the mother cells were exceptionally large after repression of L40 synthesis. 2) Mother cells with elongated buds were observed after repressed synthesis of L2, L4, L7–L9, L18, and L19. In some strains, we found small ellipsoid single cells, which may have budded off from the mother cells with long buds. 3) Repressing the expression of RPL17 led to a unique cell morphology defect with chains of three or more cells. 4) A bulge at the distal end of the mother cell emerged after repression of L34, L35, and L37 synthesis. 5) Increased cell dimensions were observed after repressing most of the 60S r-protein genes (see also Supplemental Figure S2). 6) No morphological or cell size defects were observed after repression of the synthesis of L31–L33. Note that there is some overlap between categories; that is, two or more of these characteristics can apply to a given strain. In summary, the microscopic observations show that both cell cycle progression and bud morphology control were disturbed in a protein-specific pattern by repressed synthesis of individual r-proteins.
We also counted the number of budded cells after the shift to glucose medium in each strain. The fraction of budded cells in the control strain, BY4741, remained virtually constant at ∼50% (Figure 1E). In galactose medium, the fraction of budding cells in the strains with r-protein genes under Pgal control was also 40–60%. However, after the shift to glucose medium, the fraction of budding cells in most of the gal-dependent cells decreased by twofold to fourfold (Figure 1, E and G). This was expected for the strains that were classified as G1 according to the flow cytometry, but the fraction of budding cells also decreased even in strains that showed no shift between the 1N and 2N peaks (i.e., the “no-change” group) with the exception of L23. This suggests that the control of budding and DNA synthesis was uncoupled in these strains.
Further analysis of the flow cytometry profiles for r-proteins L4, L17, L18, and L40
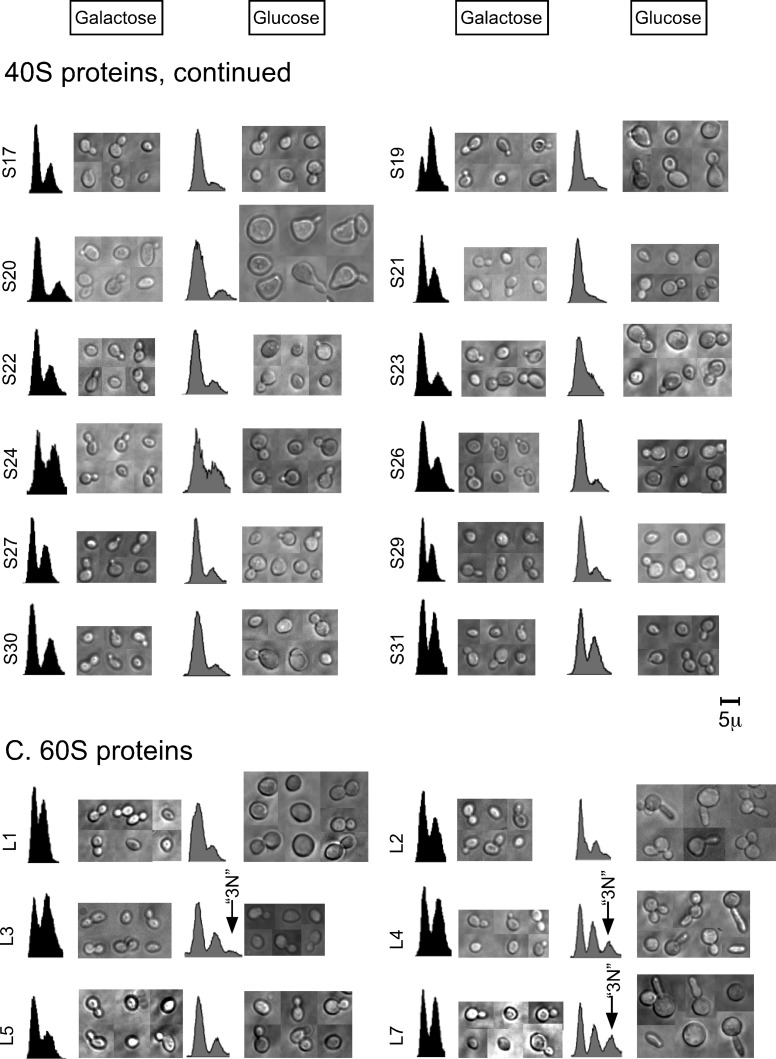
As already mentioned, repressed synthesis of L4, L7, L17, L18, and L40 resulted in a flow cytometry profile in which 20–30% of the cells were located in a peak or a trail containing cells with >2N equivalents of DNA (Figure 1, C and F). Repression of the synthesis of these proteins also generated mother cells with two or more buds or with chains of cells (Figure 1C and Supplemental Figure S3). These results suggest that the >2N portion of the flow cytometry profile is populated by multibudded or chained cells. To test this hypothesis, we analyzed cells repressed for L4, L17, L18, or L40 synthesis using a cell sorter according to their fluorescence intensity.
Microscopy of the cell pools originating from each peak showed that the 3N peak in the L4, L18, and L40 strains contained cells with two or three buds (Figure 2A), as we had hypothesized. Also as expected, the 1N peak contained single cells; the 2N peaks contained mostly single budded cells, but also some with two buds, suggesting that elongated buds can be formed before substantial excess DNA synthesis (i.e., >2N) has commenced. We do not know how much, if any, DNA has been transported into the buds. Mother cell and each of the two buds may have contained ∼1N equivalent of DNA, so that the mother cell–bud complex contained, in total, approximately 3N equivalents of DNA. However, it is also possible that all of the DNA was retained in the mother cell or that the buds contained incomplete genomes. Overall we interpret these results to mean that one or more late steps in the cell cycle is delayed or arrested but that a significant fraction of the mother cells can bypass this step and begin a new round of cell cycle, leading to the second bud. As also hypothesized, the >2N trail observed after repression of L17 synthesis contained chains of cells, which also is consistent with inhibition of completion of late steps in the cell cycle (Figure 2A).
FIGURE 2:
Analysis of cells from different parts of the flow-cytometry spectrum. Cells were collected and fixed 16 h after the shift from galactose to glucose. The cell suspensions were then sorted according to the indicated DNA content. (A) Cells from the 1N, 2N, and 3N or trail were inspected by manual microscopy. (B) Imaging (multidimensional) flow cytometry of cells from the Pgal-RPL17. The database was queried according to the fluorescence intensity, and images from the 1N, 2N, and trail are shown. Bf, bright field; PI, propidium iodide; Bf/PI, merge. Strains are identified by the traditional yeast name of the protein whose synthesis was repressed.
The bud morphology observed in the sorted cells matched what we observed for live cells; that is, elongated buds were seen after depletion of L4 and L18, whereas buds after L40 depletion were mostly of normal morphology, as also observed with the live cells (Figure 1C). The 1N peak of the L4- and L18-depleted strains contained both normal-looking cells and oval-shaped single cells possibly generated by cytokinesis of the elongated buds.
To further dissect the flow cytometry profile, we shifted cultures of the Pgal-RPL17 cells to glucose and subjected them to multispectral imaging flow cytometry. This procedure generates low-resolution images of cells as they pass through the flow cytometer, and the images are stored in a database along with the data for propidium fluorescence intensity for each imaged cell. Images of randomly chosen cells from the different peaks in the flow cytometer profile were printed (Figure 2B). The results confirmed the microscopy of the sorted cell populations described previously. That is, the 1N peak contained single cells with one nucleus (Figure 2B), the region between the 1N and 2N peaks contained a mother cell with a single nucleus and a small bud (not shown), the 2N peak contained mother cells with larger buds and two nuclei, and the 2N “trail” comprised cell chains.
Collectively the results shown in Figures 1C and 2 suggest that repression of r-proteins L4, L17, L18, and L40 activates a checkpoint, affecting passage through G2/M and/or cytokinesis. By inference, we suggest that the remaining proteins in the 3N group (L3, L7, L28, L35, and L37) also activate such a checkpoint.
Budding pattern and distribution of nuclei
To further characterize the cells after repression of specific r-protein genes, we investigated budding patterns and 4′,6-diamidino-2-phenylindole (DAPI; DNA) staining after inhibiting the synthesis of S1, S5, S19, S20, L2, L5, L17, L18, and L25. These proteins were chosen because they represent a variety of phenotypes in the bright-field microscopy and flow cytometry experiments. Budding patterns were examined by staining with calcofluor, which binds to the “bud scars,” that is, to the chitin left on the mother cell at the site(s) where buds have been pinched off. In haploid wild-type strains, the bud scars on the mother cells are “axial,” that is, adjacent to one another, whereas budding in diploid wild-type strains is “bipolar,” with some scars at opposite poles (Slaughter et al., 2009). Previous studies showed that many different deletions in diploid strains, including haploinsufficiency for several r-protein genes, distort the normal control of budding site selection (Ni and Snyder, 2001).
The BY4741 control strain, in which all chromosomal r-protein genes are intact, generated the expected haploid cell axial budding pattern (i.e., bud scars from successive divisions were adjacent on the cell surface) in both galactose medium and 16 h after a shift to glucose medium (Figure 3). Furthermore, the DAPI staining showed nuclei in all large but not small buds (Figure 3). The same calcofluor and DAPI patterns were seen in both galactose and glucose medium in the S1 strain (Figure 3), indicating that repression of S1 synthesis does not affect bud site selection.
FIGURE 3:
Nuclear content of, and bud scar distribution, on cells before and 16 h after the repression of the indicated r-protein genes. In each group of images the bright-field image is to the left and the confocal image is to the right. DAPI staining appears in turquoise staining to the left, and calcofluor appears to the right in gold stain.
All of the other strains we examined also showed normal wild-type budding patterns when grown in galactose medium. However, after the shift to glucose medium, the budding pattern changed to a bipolar (S19, L5, L18, L35) or random (S20) arrangement or a combination of both (S5, L2, L17; Figure 3). Bud scars were seen at mother–daughter cell interfaces after repression of L2, L17, and L35 synthesis, suggesting that cytokinesis was inhibited. However, single elongated cells were also observed after repression of L2 (Figure 3), presumably formed from the mother cells with elongated buds and indicating that cytokinesis may not be completely arrested but instead slowed significantly. Chitin was present at the “bulge” of the cell in the L35 strain, perhaps signifying that the bulge formed as a result of separating the previous bud from the mother.
The repression of r-protein synthesis also affected DNA organization and distribution in a variety of ways. Many small, scattered DAPI-stained bodies were observed in several strains (S1, S5, S19, S20, L5, L17, and L18; Figure 3), perhaps due to continued replication of mitochondrial DNA. Smaller nuclei were seen in the S20 strain, indicating that the organization of the chromosomes may be altered in these cells. The absence of visible nuclei in some cells in the chains after repression of L17 synthesis (Figure 3) suggests that nuclear segregation could be distorted. Finally, DNA is detectable in some bud necks after repression of S5, S20, and L5 synthesis, implying that the transport of DNA into the bud may also take longer than normal after depletion of these proteins.
Formation of chains in Pgal-RPL17
We were interested in watching the development of abnormal cell morphology and chose to follow the formation of cell chains in the Pgal-RPL17 strain. A culture was grown in galactose medium and shifted to glucose medium. After 2 h, an aliquot was placed on a microscope slide, and two different pairs of cells were chosen for continuous observation (Figure 4). As a control, a cell from Pgal-RPL17 growing in galactose was also observed. Photographs were taken at the indicated times after the beginning of microscopy. Both cells in each of the pairs from the glucose culture formed new buds at opposite ends relative to points of attachment from the previous division. After ∼4 h (6 h after the shift to glucose medium), the buds had grown to a size that approximated “normal daughter size,” but neither the new daughters nor the original mother cells separated from each other. Note that DAPI staining of L17 cells chains clearly showed a nucleus in most but not all cells, and the chitin staining showed bud scars forming at all cell–cell interfaces (Figure 3). Thus it appears that mitosis between most mother–daughter pairs in the chains may have been completed (although nondisjunction cannot be excluded), but cytokinesis did not follow. In contrast, the galactose-grown control cells went through budding and created a small pile of normal cells (Figure 4).
FIGURE 4:
Rgal-RPL17 was grown in galactose medium until an OD600 of ∼0.6 (∼107 cells/ml) and shifted to glucose medium for 2 h. Samples of the culture were prepared for microscopy of live cells, and two pairs of connected cells of approximately equal size were chosen for continuous observation for ∼4 h. Pictures were taken at the indicated times (hours) after the beginning of microscopy.
DISCUSSION
The signaling that connects ribosome biogenesis with the cell cycle and other cellular processes is poorly understood. To establish the basic parameters for this network, we inhibited the synthesis of 54 of the 80 individual r-proteins in haploid S. cerevisiae and followed the effects using flow cytometry and light microscopy analyses. Because inhibition of the synthesis of each of the 54 r-proteins prevented colony formation and disturbed the formation of functional ribosomes (Ferreira-Cerca et al., 2005; Pöll et al., 2009), with ensuing reduction of protein synthesis capacity, we anticipated a uniform response independent of which protein gene was repressed. To our surprise, abolishing the supply of individual r-proteins generates protein-specific responses.
We found three classes of changes in cell cycle progression, depending on the protein whose synthesis is repressed (Figure 1, B–D and F). First, for 31 proteins (22 from the 40S and nine from the 60S subunit), the cell cycle distribution shifted from 2N toward 1N, indicating that progression through G1 phase is prolonged or arrested. A similar phenotype was observed in strains deleted for genes for nonessential ribosomal proteins (Hoose et al., 2012). Second, for nine 60S subunit proteins, we observed accumulation of mother cells with two or three buds or chains of cells that collectively contain >2N equivalents of DNA (3N or trail), suggesting a delay or arrest at some point between late G2 phase and cytokinesis (referred to as G2/M). A similar “checkpoint” was recently reported in response to a combination of inhibition of 40S and 60S r-protein synthesis in human cells (Fumagalli et al., 2012). Third, for 15 proteins (four from the 40S and 11 from the 60S subunit) no significant cell cycle response was observed.
We also observed three types of morphological change. First, for many proteins we observed an increase in cell volume (Figure 1, B and C, and Supplemental Figure S2). Preliminary observations suggest that this may be related to the accumulation of large vacuoles (Md.S. and L.L., unpublished data), similar to vacuoles described by Bernstein et al. (2007) upon repression of the synthesis of proteins from the small subunit processome. Second, in some strains, bud morphology was changed to extremely long, almost cylindrical, buds, indicating that the normal switch from polar to isotropic cell wall growth (Lew and Reed, 1993) fails to occur. Third, distorted bud site selection was observed in most of the strains analyzed for this feature. Screens for protein haploinsufficiency in diploid yeast previously indicated links between ribosome protein synthesis and bud site selection and cell morphology (Ni and Snyder, 2001), but this is the first report of a systematic analysis of the effect of repressed r-protein synthesis.
The protein specificity of the effect on cell cycle and bud morphology demonstrates that the effect on cell cycle does not stem simply from lack of ribosomes but instead from an insufficient supply of specific proteins. Furthermore, there is little correlation between the strains in the cell cycle and bud morphology classifications. Specifically, the L4, L7, and L18 strains exhibit both the G2/M and the long-bud phenotype, whereas the L3, L17, L28, L35, L37, and L40 strains also generate the G2/M phenotype but do not exhibit formation of elongated buds. Furthermore, the L2, L8, L9, and L19 strains also show long buds but are either N or G1 with respect to flow cytometry. We also note that distorted bud site selection appears to be a general phenotype, not associated with a particular cell cycle phenotype. We conclude that even though each of the phenotypes we observed was specific to the protein whose synthesis was repressed, there does not seem to be a “master” signal that coordinately generates all the phenotypes we described. This argument is strengthened by the fact that the phenotypes described here are not correlated with the previously reported disturbances of rRNA processing using the same set of strains we used (Ferreira-Cerca et al., 2005; Pöll et al., 2009). In the latter case, four different phenotypes with respect to the processing of the 5′ end of 5.8S rRNA and cleavage in the Internal Transcribed Spacer 2 (ITS2) were noted after repression of 40S r-protein genes (Ferreira-Cerca et al., 2005). In contrast, repression of most 40S r-protein genes resulted in a single phenotype (G1 arrest). Regarding the 60S r-protein genes, we observed the G2/M phenotype after repressing expression of the L3, L4, L7, L17, L18 L28, L35, L37, and L40 genes, but repression of these genes does not generate the same rRNA processing phenotype. Depletion of L3, L4, L7, or L18 causes a relative increase of immature compared with mature 5′ ends of 5.8S rRNA, whereas reduced synthesis of L17 and L28 results in a decrease in the same ratio (Pöll et al., 2009). Furthermore, the phenotype of an increased ratio between immature and mature ends was also seen for L8, L16, L32, and L33, whereas these strains belong to the G1 or no-change groups in our study.
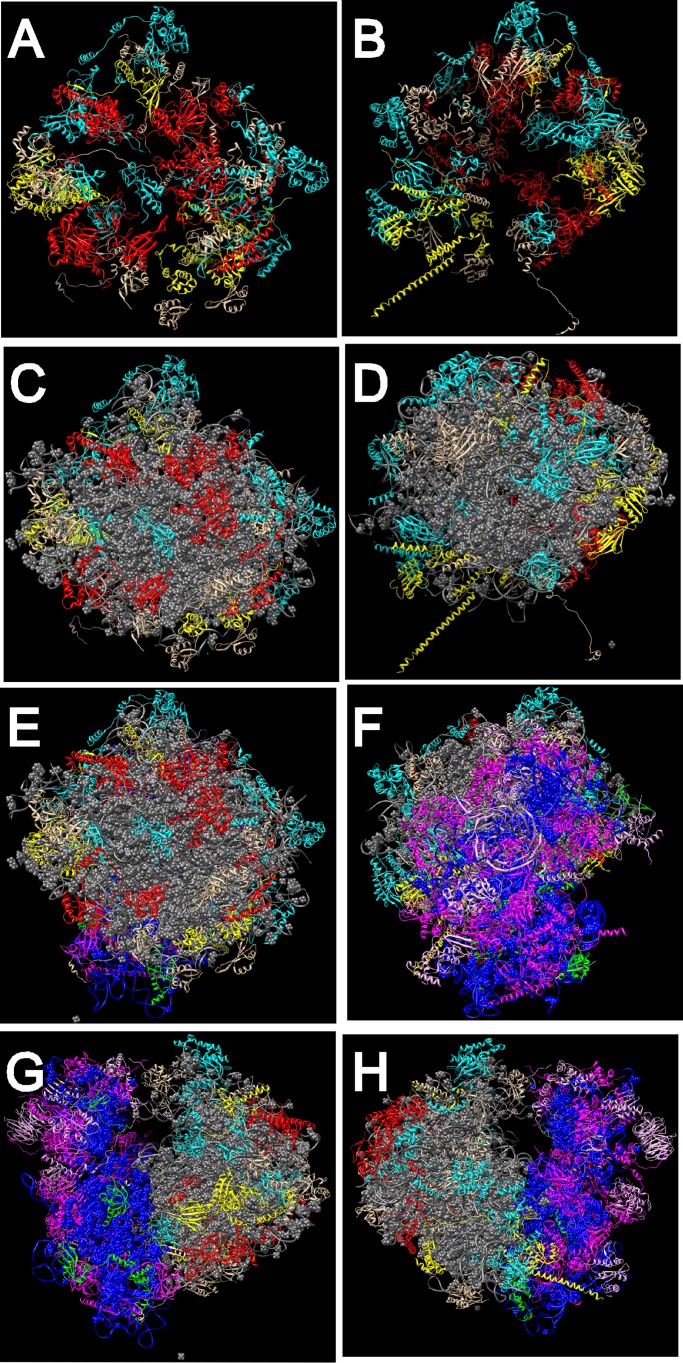
In contrast to the incomplete overlap in the classifications just discussed, there is an intriguing correlation between the cell cycle phenotypes and the locations in the ribosome of the proteins whose reduced synthesis induces these phenotypes (Figure 5). Proteins linked to the G2/M response all map to the solvent side of the 60S subunit. On the other hand, proteins whose cessation of synthesis generates the G1 response are either 40S proteins or 60S proteins on the interface side, forming a ring around the 25S rRNA domain that connects to the 40S subunit. Of interest, the same subunit specificity may also apply to the few ribosomal assembly factors that have been tested for effects on the cell cycle. Cessation of the synthesis of a number of proteins from the small subunit processome results in G1 arrest (Bernstein and Baserga, 2004; Bernstein et al., 2007), whereas repression of the synthesis of the 60S-specific factor Nop15 or Rrp14 generates accumulation of cells during the late part of the cell cycle (Oeffinger and Tollervey, 2003; Oeffinger et al., 2007; Yamada et al., 2007). The differential effects of disrupting the 40S and 60S pathways may also apply to mammalian cells. Conditional deletion of the RPS6 gene in liver cells prevents the transition from G1 to S phase (Thomas et al., 2000; Fumagalli et al., 2009), but combined disruption of the synthesis of one protein from each subunit activates a G2/M checkpoint (Fumagalli et al., 2012) that is akin to our observations in yeast.
FIGURE 5:
Grouping of proteins on the model for the S. cerevisiae ribosome. (A, B) The 60S subunit proteins (rRNA omitted) viewed from the solvent side (A) or the interface side (B). (C, D) The 60S proteins (rRNA included) viewed from the solvent side (C) or interface side (D). (E, F) The 80S ribosome viewed from the solvent side of the 60S subunit (E) or the solvent side of the 40S subunit (F). (G, H) The 80S ribosome viewed from the P-stalk side (G) or the L1 stalk side (H). Proteins are color coded according to the phenotype generated in response to repression of their synthesis. Red, 60S proteins with G2/M phenotype; yellow, 60S proteins with G1 phenotype; magenta, 40S proteins with G1 phenotype; turquoise, 60S proteins with no phenotype; green, 40S proteins with no phenotype; gray, 25S, 5.8S, and 5S rRNA; blue, 18S rRNA. Models were rendered with Chimera using Protein Data Base files 3U5B, 3U5C, 3U5D, and 3U5E. Phenotypes generated upon repression of the synthesis of specific proteins are listed in Figure 1, D and F; see also Figure 1, B and C.
The separation into topological domains required for advancement through G1 and G2/M suggests that progression through different parts of the cell cycle depends on the integrity of distinct regions of the ribosome. There are several, potentially overlapping models that could account for this. First, the mature ribosome, a precursor of the ribosome, or a preassembly neighborhood of ribosomal components (Zhang et al., 2007) could serve as a reservoir for proteins involved in control of the cell cycle. The solvent side of the 60S subunit or pre-60S might interact with nonribosomal proteins required for progression through late stages of the cell cycle, whereas the interface side of the 60S and the 40S subunit or precursor particles might interact with nonribosomal proteins required for completion of the G1 phase. Such interactions might involve steps in both the nucleus and the cytoplasm because the G2/M group includes r-proteins that join the preribosome early as well as late in the assembly pathway. Thus L4 and L17 (equivalent to the L4 and L22 bacterial r-proteins, respectively) are expected to bind to the nascent ribosome in the nucleus because they are known as “early binding proteins” in bacteria (Röhl and Nierhaus, 1982), whereas L40 is added to the preribosome after its export to the cytoplasm (Fernandez-Pevida et al., 2012). We speculate that cell cycle proteins important for late step(s) of the cell cycle could attach to the 60S solvent side in the nucleus and be released in the cytoplasm upon maturation of the ribosome.
A second possibility is that repressed synthesis of individual r-proteins results in the generation of “specialized” ribosomes with different affinities for translation-regulatory factors and individual messengers. This model would predict that factors stimulating the synthesis of proteins promoting passage through G1 or G2/M bind to separate domains of the ribosome. Alternatively, specialized ribosomes could simply be error-prone. In this regard, we note that all proteins associated with the G1 phenotype are on the 40S subunit (containing the mRNA-decoding center) and on the 60S interface side. Although none of these 60S proteins has been implicated in translation accuracy, it is probable that the loss of a protein on the 60S interface side would affect the rRNA intersubunit bridges, which are known to be important for translation accuracy (Liang et al., 2007).
It is well established that surveillance mechanisms in both the nucleus and the cytoplasm normally degrade incorrectly assembled ribosomal particles (e.g., Dez et al., 2006; McIntosh et al., 2011; Karbstein, 2013) and prevent precursor ribosomes from commencing mRNA translation (Strunk et al., 2012). In fact, most rRNA synthesized during the repression of the r-protein genes in the strains used here is degraded (Ferreira-Cerca et al., 2005; Pöll et al., 2009; (A.B., J.M.Z., and L.L., unpublished data). However, a few nascent precursor ribosomes might escape the surveillance mechanisms, resulting in the formation of small numbers of ribosomes with an incomplete set of r-proteins, which may change their propensity for translating different groups of cell cycle proteins (Xue and Barna, 2012). This model is compatible with the growth of an extra set of ribs in mice lacking the gene for r-protein L38 (Kondrashov et al., 2011). The finding of 60S ribosomal subunits lacking r-protein L1 (McIntosh et al., 2011) is also compatible with this model.
Third, several reports suggest that cell cycle proteins could directly affect ribosome metabolism in other ways. The Cdc14 phosphatase, which is part of the mitosis exit network, is located in the nucleolus throughout the cell cycle, except during anaphase, when it is released into the nucleoplasm and subsequently into the cytoplasm (Bloom et al., 2011). The proximity between Cdc14 and rRNA genes might contribute to a link between ribosome biogenesis and cell cycle progression. It has also been reported that the cell cycle Cdc48 ATPase is part of a complex required for the degradation of dysfunctional mutant large ribosomal subunits (Fujii et al., 2012).
A fourth possibility is that some r-proteins and ribosome assembly factors are bifunctional (Warner and McIntosh, 2009). This appears to be the case for Nop7, also known as Yph1, a protein that has been identified as a component of both the 60S precursor ribosomal particle and the origin of DNA replication particle (Du and Stillman, 2002). Observations made with higher eukaryotes further support the concept of a connection between ribosome biogenesis and cell division. Certain r-proteins in mammalian cells inhibit the Mdm2 ubiquitin ligase and thereby affect the stability of p53 (Bursac et al., 2012; Morgado-Palacin et al., 2012; Suzuki et al., 2012). In addition, Alliegro et al. (2012) described a structure called the nucleolinus, which is associated with the nucleolus and may be necessary for formation of the cell division apparatus.
A final point is that our results may be relevant to understanding cell fate and development in higher eukaryotes. Many mutations that disturb normal ribosome biogenesis in metazoans are known to result in abnormal development and a variety of congenital diseases called ribosomopathies. These include Diamond–Blackfan anemia, X-linked dyskeratosis congenita, Shwachman–Diamond syndrome, and other cancers (Lipton and Ellis, 2010; Farrar et al., 2011; Wang et al., 2012; Ruggero, 2013). The molecular mechanisms responsible for these connections remain enigmatic but appear to involve changes in the nucleolus resulting from a distortion of normal ribosome biogenesis (Narla and Ebert, 2010; Narla et al., 2011). Recent experiments show that a significant fraction of human ribosome assembly factors are homologous to those identified in yeast, but human cells also have a significant number of assembly factors with no yeast counterparts (Tafforeau et al., 2013). Yeast may thus prove to be a good model for some ribosomopathies but not others (reviewed in Woolford and Baserga, 2013). We expect that our observation of signaling between ribosome biogenesis and cell cycle control and cell morphology will provide useful models for understanding some, but certainly not all, human ribosomopathies.
MATERIALS AND METHODS
Strains and growth conditions
The strains used are listed in Supplemental Table S1. In all strains except the BY4741 control strain, the expression of a single r-protein is under control of the galactose-inducible promoter. The Pgal-RPL4B strain (see the Supplemental Material) was constructed in our lab by standard protocols. The Pgal-RPL4B plasmid was constructed from pGILDA (Clonetech) by recombineering (Lee et al., 2001). All other strains were generous gifts from Philipp Milkereit (University of Regensburg, Regensburg, Germany) and John Woolford (Carnegie Mellon University, Pittsburgh, PA; Ferreira-Cerca et al., 2005; Pöll et al., 2009; J. Woolford, personal communication). In brief, all galactose-controlled genes except for RPL7 and RPL18 are on centromer plasmids (low copy). The RPL7 and RPL18 galactose-controlled genes are in the chromosome at their cognate positions. RPL4 repression was tested in two strains carrying the Pgal-RPL4A gene in the chromosome or the Pgal-RPL4B gene on a plasmid (see the Supplemental Material). In all other cases only one paralogue (where two exist) was used. The identity of the r-protein under Pgal control in all strains used was verified in our lab by PCR amplification of DNA between a primer in the galactose promoter and another in the structural gene for the r-protein under galactose control (Md.S. and L.L., unpublished results). Supplemental Table S1 provides complete genotypes of all strains used.
Cultures were grown asynchronously in 1% yeast extract, 2% peptone, 2% galactose (YPGal) at 30˚C until mid–log phase (OD600 ∼ 0.8–1.0, corresponding to ∼[1.5–2] × 107 cells/ml) and then shifted (1:10 dilution) into 1% yeast extract, 2% peptone, 2% glucose (YPD). Cells were harvested 16 h after the shift of media. Cultures were diluted as necessary with prewarmed media to keep the OD600 <1.0 using a Hitachi U1100 spectrophotometer (Hitachi High-Technologies Corporation, Japan).
Bright-field microscopy
Cells (1 OD600 unit corresponding to ∼2 × 107 cells) were collected by centrifugation in a Biofuge (Sorvall Heraeus) at 13,000 rpm for 1 min, resuspended in 100 μl of YPGal or YPD medium, and sonicated using the Branson Sonifier 450 at power setting 3 for 5 s to break up clumps of cells. Cells were viewed under a bright-field microscope (Nikon Microphot-SA) at 40× magnification, and pictures were taken using the SPOT RT camera. The fraction of cells with one, two, or thee buds was counted in a hemocytometer. At least 100 cells were counted for each strain. To image the sorted cells collected from the FACS, approximately 5000 cells were prepared for microscopy as described, except that they were reconstituted with 100 μl of phosphate-buffered saline (PBS) solution. Cell suspensions were viewed through an Imagining Retiga 4000R fluorescence microscope and imaged by an Olympus BX41 camera.
Confocal microscopy
Cells (1 OD600 unit) were collected and washed with PBS. The cell pellet was resuspended in 1 ml of 70% cold ethanol on a rocker for 2 h at 4°C. A final concentration of 100 ng/ml DAPI (Sigma-Aldrich, St. Louis, MO) or 100 ng/ml calcofluor (Sigma-Aldrich) was added to the cells, which were then incubated for 30 min in the dark at room temperature (Lieberman, 2004). Cells were washed, reconstituted with 100 μl of PBS, and sonicated before being viewed under a Leica TCS SP5 confocal microscope equipped with LAS AF Lite Software using the 63× oil immersion lens. The filter was set at 380 nm for the excitation and 425 nm for the emission.
Flow cytometry
Cells (1 OD600 unit) were collected and fixed with 1 ml of cold 70% EtOH and placed on a rocker at 4°C overnight. The cells were washed with 1 ml of 50 mM Tris-HCl (pH 7.5) and resuspended in 450 μl of the same buffer. The cells were then supplemented with 50 μl of 10 mg/ml RNaseA in water (Sigma-Aldrich; stock solution was heated at 100°C for 15 min to inactivate any DNases), incubated at 37°C for 2–4 h, washed with 1 ml PBS twice, and resuspended in 500 μl of PBS. Propidium iodide (PI; 15 μg/ml; Sigma-Aldrich) was added, and samples were subsequently kept in the dark. Cells were stored for ∼24 h at 4°C (the maximal storage time for the stained cells was 1 wk), sonicated, and vortexed. Fifty microliters of the PI-stained cells was added to 450 μl of PBS and passed through an Epics XL (Beckman Coulter, Ft. Collins, CO) or a Cyan ADP flow cytometer (Beckman Coulter) and interrogated by a 488-nm excitation wavelength; emitted light was collected using a 630-nm band pass filter. The flow rate was set at 200 cells/s, with a total number of ∼20,000 cells collected. Data for distribution of DNA content per cell and forward light scattering were analyzed using Summit Software (Beckman Coulter).
Cell sorting was performed on a MoFlo Flow Cytometer (Beckman Coulter). Cells were fixed, labeled with propidium iodide, and analyzed as described. Aggregates were eliminated using a Pulse Width gate. Gates were set around the N, 2N, and 3N peaks in the PI histogram, and cells within those gates were collected and inspected by microscopy. Imaging flow cytometry was performed on an Amnis ImageStreamX imaging flow cytometer (Amnis Corporation, Seattle, WA) at 60× magnification. Focused cells were identified and gated using the Gradient RMS Brightfield feature. The 1N, 2N, and 3N gates were set on the corresponding peaks using an intensity histogram of the propidium iodide, and cells within each gate were visualized.
Supplementary Material
Acknowledgments
We thank Susan Fretz for help with strain construction and Philipp Milkereit and John Woolford for strains. We also thank John Woolford for helpful discussions. We gratefully acknowledge our departmental colleagues (Suzanne Rosenberg, Stephen Miller, Michelle Starz-Gaiano, Rachel Brewster, and Chere Petty) for use of and help with flow cytometer and microscopes. We thank the National Heart, Lung and Blood Institute at the National Institutes of Health for use of the Flow Cytometry Core Facility. This work was supported by National Science Foundation Grants MCB0349443 and 0920578 to J.M.Z. and L.L. and National Science Foundation Major Research Instrumentation Grant DBI-0722569 to D.B. and T.G. Finally, we acknowledge the use of the UCSF Chimera Package supported by National Institute of General Medical Sciences Grant P41-GM103311.
Abbreviation used:
- r-protein
ribosomal protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-02-0097) on October 9, 2013.
Present addresses: *EBSCO, Ipswich, MA 01938.
†Department of Therapeutic Radiology, Yale University, New Haven, CT 06520.
REFERENCES
- Alliegro MC, Hartson S, Alliegro MA. Composition and dynamics of the nucleolinus, a link between the nucleolus and cell division apparatus in surf clam (Spisula) oocytes. J Biol Chem. 2012;287:6702–6713. doi: 10.1074/jbc.M111.288506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein KA, Baserga SJ. The small subunit processome is required for cell cycle progression at G1. Mol Biol Cell. 2004;15:5038–5046. doi: 10.1091/mbc.E04-06-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein KA, Bleichert F, Bean JM, Cross FR, Baserga SJ. Ribosome biogenesis is sensed at the Start cell cycle checkpoint. Mol Biol Cell. 2007;18:953–964. doi: 10.1091/mbc.E06-06-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom J, Cristea IM, Procko AL, Lubkov V, Chait BT, Snyder M, Cross FR. Global analysis of Cdc14 phosphatase reveals diverse roles in mitotic processes. J Biol Chem. 2011;286:5434–5445. doi: 10.1074/jbc.M110.205054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursac S, et al. Mutual protection of ribosomal proteins L5 and L11 from degradation is essential for p53 activation upon ribosomal biogenesis stress. Proc Natl Acad Sci USA. 2012;109:20467–20472. doi: 10.1073/pnas.1218535109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derenzini M. The AgNORs. Micron. 2000;31:117–120. doi: 10.1016/s0968-4328(99)00067-0. [DOI] [PubMed] [Google Scholar]
- Derenzini M, Trere D, Pession A, Montanaro L, Sirri V, Ochs RL. Nucleolar function and size in cancer cells. Am J Pathol. 1998;152:1291–1297. [PMC free article] [PubMed] [Google Scholar]
- Dez C, Houseley J, Tollervey D. Surveillance of nuclear-restricted pre-ribosomes within a subnucleolar region of Saccharomyces cerevisiae. EMBO J. 2006;25:1534–1546. doi: 10.1038/sj.emboj.7601035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du YC, Stillman B. Yph1p, an ORC-interacting protein: potential links between cell proliferation control, DNA replication, and ribosome biogenesis. Cell. 2002;109:835–848. doi: 10.1016/s0092-8674(02)00773-0. [DOI] [PubMed] [Google Scholar]
- Farrar JE, et al. Abnormalities of the large ribosomal subunit protein, Rpl35a, in Diamond-Blackfan anemia. Blood. 2008;112:1582–1592. doi: 10.1182/blood-2008-02-140012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrar JE, Vlachos A, Atsidaftos E, Carlson-Donohoe H, Markello TC, Arceci RJ, Ellis SR, Lipton JM, Bodine DM. Ribosomal protein gene deletions in Diamond-Blackfan anemia. Blood. 2011;118:6943–6951. doi: 10.1182/blood-2011-08-375170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Pevida A, Rodriguez-Galan O, Diaz-Quintana A, Kressler D, de la Cruz J. Yeast ribosomal protein l40 assembles late into precursor 60 s ribosomes and is required for their cytoplasmic maturation. J Biol Chem. 2012;287:38390–38407. doi: 10.1074/jbc.M112.400564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira-Cerca S, Poll G, Gleizes PE, Tschochner H, Milkereit P. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol Cell. 2005;20:263–275. doi: 10.1016/j.molcel.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Ferreira-Cerca S, Poll G, Kuhn H, Neueder A, Jakob S, Tschochner H, Milkereit P. Analysis of the in vivo assembly pathway of eukaryotic 40S ribosomal proteins. Mol Cell. 2007;28:446–457. doi: 10.1016/j.molcel.2007.09.029. [DOI] [PubMed] [Google Scholar]
- Fujii K, Kitabatake M, Sakata T, Ohno M. 40S subunit dissociation and proteasome-dependent RNA degradation in nonfunctional 25S rRNA decay. EMBO J. 2012;31:2579–2589. doi: 10.1038/emboj.2012.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita J. Cold shock response in mammalian cells. J Mol Microbiol Biotechnol. 1999;1:243–255. [PubMed] [Google Scholar]
- Fumagalli S, Di Cara A, Neb-Gulati A, Natt F, Schwemberger S, Hall J, Babcock GF, Bernardi R, Pandolfi PP, Thomas G. Absence of nucleolar disruption after impairment of 40S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat Cell Biol. 2009;11:501–508. doi: 10.1038/ncb1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli S, Ivanenkov VV, Teng T, Thomas G. Suprainduction of p53 by disruption of 40S and 60S ribosome biogenesis leads to the activation of a novel G2/M checkpoint. Genes Dev. 2012;26:1028–1040. doi: 10.1101/gad.189951.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamalinda M, Jakovljevic J, Babiano R, Talkish J, de la Cruz J, Woolford JL., Jr Yeast polypeptide exit tunnel ribosomal proteins L17, L35 and L37 are necessary to recruit late-assembling factors required for 27SB pre-rRNA processing. Nucleic Acids Res. 2013;41:1965–1983. doi: 10.1093/nar/gks1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoose SA, Duran C, Malik I, Eslamfam S, Shasserre SC, Downing SS, Hoover EM, Dowd KE, Smith R, 3rd, Polymenis M. Systematic analysis of cell cycle effects of common drugs leads to the discovery of a suppressive interaction between gemfibrozil and fluoxetine. PLoS One. 2012;7:e36503. doi: 10.1371/journal.pone.0036503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakovljevic J, Ohmayer U, Gamalinda M, Talkish J, Alexander L, Linnemann J, Milkereit P, Woolford JL., Jr Ribosomal proteins L7 and L8 function in concert with six A(3) assembly factors to propagate assembly of domains I and II of 25S rRNA in yeast 60S ribosomal subunits. RNA. 2012;18:1805–1822. doi: 10.1261/rna.032540.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner L, Melnikov S, de Loubresse NG, Ben-Shem A, Iskakova M, Urzhumtsev A, Meskauskas A, Dinman J, Yusupova G, Yusupov M. Crystal structure of the 80S yeast ribosome. Curr Opin Struct Biol. 2012;22:759–767. doi: 10.1016/j.sbi.2012.07.013. [DOI] [PubMed] [Google Scholar]
- Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, Tyers M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbstein K. Quality control mechanisms during ribosome maturation. Trends Cell Biol. 2013;23:242–250. doi: 10.1016/j.tcb.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kief DR, Warner JR. Coordinate control of synthesis of ribosomal ribonucleic acid and ribosomal proteins during nutritional shift-up in Saccharomyces cerevisiae. Mol Cell Biol. 1981;1:1007–1015. doi: 10.1128/mcb.1.11.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch AL, Ehrenfeld E. The size and shape of bacteria by light scattering measurements. Biochim Biophys Acta. 1968;165:262–273. doi: 10.1016/0304-4165(68)90054-8. [DOI] [PubMed] [Google Scholar]
- Kondrashov N, Pusic A, Stumpf CR, Shimizu K, Hsieh AC, Xue S, Ishijima J, Shiroishi T, Barna M. Ribosome-mediated specificity in hox mRNA translation and vertebrate tissue patterning. Cell. 2011;145:383–397. doi: 10.1016/j.cell.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamouille S, Derynck R. Cell size and invasion in TGF-beta-induced epithelial to mesenchymal transition is regulated by activation of the mTOR pathway. J Cell Biol. 2007;178:437–451. doi: 10.1083/jcb.200611146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- Lempiainen H, Uotila A, Urban J, Dohnal I, Ammerer G, Loewith R, Shore D. Sfp1 interaction with TORC1 and Mrs6 reveals feedback regulation on TOR signaling. Mol Cell. 2009;33:704–716. doi: 10.1016/j.molcel.2009.01.034. [DOI] [PubMed] [Google Scholar]
- Lew DJ, Reed SI. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Liu Q, Fournier MJ. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol Cell. 2007;28:965–977. doi: 10.1016/j.molcel.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Lieberman H. Cell Cycle Checkpoint Control Protocols. Totowa, NJ: Humana Press; 2004. [Google Scholar]
- Lipton JM, Ellis SR. Diamond Blackfan anemia 2008–2009: broadening the scope of ribosome biogenesis disorders. Curr Opin Pediatr. 2010;22:12–19. doi: 10.1097/MOP.0b013e328334573b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maaløe O, Kjeldgaard NO. Control of Macromolecular Synthesis. New York: Benjamin; 1966. [Google Scholar]
- Mayer C, Grummt I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene. 2006;25:6384–6391. doi: 10.1038/sj.onc.1209883. [DOI] [PubMed] [Google Scholar]
- McIntosh KB, Bhattacharya A, Willis IM, Warner JR. Eukaryotic cells producing ribosomes deficient in Rpl1 are hypersensitive to defects in the ubiquitin-proteasome system. PLoS One. 2011;6:e23579. doi: 10.1371/journal.pone.0023579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montanaro L, Trere D, Derenzini M. Changes in ribosome biogenesis may induce cancer by down-regulating the cell tumor suppressor potential. Biochim Biophys Acta. 2012;1825:101–110. doi: 10.1016/j.bbcan.2011.10.006. [DOI] [PubMed] [Google Scholar]
- Morgado-Palacin L, Llanos S, Serrano M. Ribosomal stress induces L11- and p53-dependent apoptosis in mouse pluripotent stem cells. Cell Cycle. 2012;11:503–510. doi: 10.4161/cc.11.3.19002. [DOI] [PubMed] [Google Scholar]
- Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115:3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narla A, Hurst SN, Ebert BL. Ribosome defects in disorders of erythropoiesis. Int J Hematol. 2011;93:144–149. doi: 10.1007/s12185-011-0776-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L, Snyder M. A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol Biol Cell. 2001;12:2147–2170. doi: 10.1091/mbc.12.7.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M, Fatica A, Rout MP, Tollervey D. Yeast Rrp14p is required for ribosomal subunit synthesis and for correct positioning of the mitotic spindle during mitosis. Nucleic Acids Res. 2007;35:1354–1366. doi: 10.1093/nar/gkl824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M, Tollervey D. Yeast Nop15p is an RNA-binding protein required for pre-rRNA processing and cytokinesis. EMBO J. 2003;22:6573–6583. doi: 10.1093/emboj/cdg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöll G, Braun T, Jakovljevic J, Neueder A, Jakob S, Woolford JL, Jr, Tschochner H, Milkereit P. rRNA maturation in yeast cells depleted of large ribosomal subunit proteins. PLoS One. 2009;4:e8249. doi: 10.1371/journal.pone.0008249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röhl R, Nierhaus KH. Assembly map of the large subunit (50S) of Escherichia coli ribosomes. Proc Natl Acad Sci USA. 1982;79:729–733. doi: 10.1073/pnas.79.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado IV, Kressler D, de la Cruz J. Functional analysis of Saccharomyces cerevisiae ribosomal protein Rpl3p in ribosome synthesis. Nucleic Acids Res. 2007;35:4203–4213. doi: 10.1093/nar/gkm388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D. Translational control in cancer etiology. Cold Spring Harb Perspect Biol 5. 2013:a012336. doi: 10.1101/cshperspect.a012336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaechter M, Maaloe O, Kjeldgaard NO. Dependency on medium and temperature of cell size and chemical composition during balanced grown of Salmonella typhimurium. J Gen Microbiol. 1958;19:592–606. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- Slaughter BD, Smith SE, Li R. Symmetry breaking in the life cycle of the budding yeast. Cold Spring Harb Perspect Biol. 2009;1:a003384. doi: 10.1101/cshperspect.a003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunk BS, Novak MN, Young CL, Karbstein K. A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell. 2012;150:111–121. doi: 10.1016/j.cell.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Kogo R, Kawahara K, Sasaki M, Nishio M, Maehama T, Sasaki T, Mimori K, Mori M. A new PICTure of nucleolar stress. Cancer Sci. 2012;103:632–637. doi: 10.1111/j.1349-7006.2012.02219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafforeau L, Zorbas C, Langhendries JL, Mullineux ST, Stamatopoulou V, Mullier R, Wacheul L, Lafontaine DL. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of pre-rRNA processing factors. Mol Cell. 2013;51:539–551. doi: 10.1016/j.molcel.2013.08.011. [DOI] [PubMed] [Google Scholar]
- Thomas BC, Chamberlain J, Engelke DR, Gegenheimer P. Evidence for an RNA-based catalytic mechanism in eukaryotic nuclear ribonuclease P. RNA. 2000;6:554–562. doi: 10.1017/s1355838200991477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G, Siegmann M, Gordon J. Multiple phosphorylation of ribosomal protein S6 during transition of quiescent 3T3 cells into early G1, and cellular compartmentalization of the phosphate donor. Proc Natl Acad Sci USA. 1979;76:3952–3956. doi: 10.1073/pnas.76.8.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Luo Y, Hong Y, Peng J, Lo L. Ribosome biogenesis factor Bms1-like is essential for liver development in zebrafish. J Genet Genomics. 2012;39:451–462. doi: 10.1016/j.jgg.2012.07.007. [DOI] [PubMed] [Google Scholar]
- Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins. Mol Cell. 2009;34:3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford JL Jr, Baserga SJ. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013;195:643–681. doi: 10.1534/genetics.113.153197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S, Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Biol. 2012;13:355–369. doi: 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H, Horigome C, Okada T, Shirai C, Mizuta K. Yeast Rrp14p is a nucleolar protein involved in both ribosome biogenesis and cell polarity. RNA. 2007;13:1977–1987. doi: 10.1261/rna.553807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza D, Ghavidel A, Heitman J, Schultz MC. Rapamycin induces the G0 program of transcriptional repression in yeast by interfering with the TOR signaling pathway. Mol Cell Biol. 1998;18:4463–4470. doi: 10.1128/mcb.18.8.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Harnpicharnchai P, Jakovljevic J, Tang L, Guo Y, Oeffinger M, Rout MP, Hiley SL, Hughes T, Woolford JL., Jr Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes Dev. 2007;21:2580–2592. doi: 10.1101/gad.1569307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.