Characterization of the cell cycle–regulated transcripts in U2OS cells yielded 1871 unique genes. FOXM1 targets were identified via ChIP-seq, and novel targets in G2/M and S phases were verified using a real-time luciferase assay. ChIP-seq data were used to map cell cycle transcriptional regulators of cell cycle–regulated gene expression in U2OS cells.
Abstract
We identify the cell cycle–regulated mRNA transcripts genome-wide in the osteosarcoma-derived U2OS cell line. This results in 2140 transcripts mapping to 1871 unique cell cycle–regulated genes that show periodic oscillations across multiple synchronous cell cycles. We identify genomic loci bound by the G2/M transcription factor FOXM1 by chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) and associate these with cell cycle–regulated genes. FOXM1 is bound to cell cycle–regulated genes with peak expression in both S phase and G2/M phases. We show that ChIP-seq genomic loci are responsive to FOXM1 using a real-time luciferase assay in live cells, showing that FOXM1 strongly activates promoters of G2/M phase genes and weakly activates those induced in S phase. Analysis of ChIP-seq data from a panel of cell cycle transcription factors (E2F1, E2F4, E2F6, and GABPA) from the Encyclopedia of DNA Elements and ChIP-seq data for the DREAM complex finds that a set of core cell cycle genes regulated in both U2OS and HeLa cells are bound by multiple cell cycle transcription factors. These data identify the cell cycle–regulated genes in a second cancer-derived cell line and provide a comprehensive picture of the transcriptional regulatory systems controlling periodic gene expression in the human cell division cycle.
INTRODUCTION
Examining the periodic expression patterns of the human cell cycle using genomic approaches can provide a complete picture of one of the most tightly regulated processes in the life of a cell. This knowledge allows, in turn, the examination of how different regulators of the cell cycle machinery interact and affect the timing of cell cycle progression. This is especially important, as perturbations in cell cycle progression can lead to apoptosis or cancer.
The cell cycle has been studied extensively at the molecular level, and transcriptional programs have been measured and analyzed using microarray technology in budding yeast (Cho et al., 1998; Spellman et al., 1998), fission yeast (Rustici et al., 2004; Oliva et al., 2005; Peng et al., 2005), bacteria (Laub et al., 2000), primary human fibroblasts (Cho et al., 1998; Iyer et al., 1999; Bar-Joseph et al., 2008), mouse fibroblasts (Ishida et al., 2001), Arabidopsis (Menges et al., 2002, 2005), the human keratinocyte cell line HaCaT (Pena-Diaz et al., 2013), and HeLa cells (Crawford and Piwnica-Worms, 2001; Whitfield et al., 2002; Sadasivam et al., 2012). The analysis of cell cycle–regulated gene expression in fibroblasts and HeLa cells suggests both commonality and cell-type specificity in the cell cycle–regulated gene expression programs of human cells. Here we provide the full complement of cell cycle–regulated genes in a fourth human cell line, the commonly used osteosarcoma cell line U2OS. In addition, to shed light on cell cycle regulation, we determine the genome-wide binding patterns of FOXM1 and analyze publicly available data for additional cell cycle transcriptional regulators, thus providing an interpretive framework for the periodic gene expression program in U2OS cells.
DNA replication and mitosis are two major cellular events whose transcriptional programs must be precisely coordinated and executed in order for a cell to successfully divide. To ensure that these two events are successfully completed in an orderly manner, there are regulatory mechanisms, ranging from transcriptional control (e.g., RB and pocket protein sequestration of E2Fs) to protein degradation (e.g., anaphase-promoting complex), that assure that a cell progresses forward in the process and reversal does not occur. These processes, in turn, have their own systems of control and regulation. Transcription factors provide one aspect of this control, regulating the timing and levels of gene expression. One of these transcription factors, the G1/S-phase regulator E2F1, has been well studied, whereas the corresponding transcription factor for mitotic gene expression, FOXM1, has been less well studied (reviewed in Wierstra and Alves, 2007; Alvarez-Fernandez and Medema, 2013).
At the beginning of and throughout S phase, the E2F family of transcription factors regulates the expression of genes involved in initiation and continued replication of DNA (Johnson et al., 1994; Slansky and Farnham, 1996; Helin, 1998). As cells progress through S phase, the MuvB core complex (LIN9, LIN37, LIN52, LIN54, and RBBP4) dissociates from the DREAM complex (TFDP1, p130 [RBL2], E2F4, and the MuvB core complex) and binds to B-Myb (the B-Myb-MuvB complex). This complex then binds to the promoters of PLK1 and CCNB1, two genes expressed in G2 and mitosis, as well as a number of other mitotic cell cycle genes (Sadasivam et al., 2012). In late S phase, the transcription factor FOXM1 is recruited to these promoters by the B-Myb-MuvB complex before B-Myb dissociates from the complex and is degraded by the proteasome (Litovchick et al., 2007).
FOXM1 activates expression of genes critical for the proper progression of mitosis, including CCNB1, PLK1, CDC25B, CDC25C, AURKB, and BIRC5 (Kalinichenko et al., 2003; Kim et al., 2005; Laoukili et al., 2005; Wonsey and Follettie, 2005; Alvarez-Fernandez et al., 2010; Nakamura et al., 2010; Bonet et al., 2012; Down et al., 2012; Sadasivam et al., 2012). In HeLa cells, FOXM1 mRNA is cell cycle regulated, with peak expression in G2 (Whitfield et al., 2002). The FOXM1 protein is also phosphorylated in M phase. These include multiple phosphorylation events involving multiple cyclin-CDK complexes, including cyclin D1/Cdk4 (Wierstra and Alves, 2006b, 2008), cyclin E/Cdk2 (Wierstra and Alves, 2006a, 2008), cyclin A/Cdk2 (Wierstra and Alves, 2006a, 2008; Laoukili et al., 2008), cyclin A/Cdk1 (Wierstra and Alves, 2006a), and cyclin B1/Cdk1 (Major et al., 2004; Chen et al., 2009), as well as phosphorylation by PLK1 (Chen et al., 2009). These modifications culminate in the destruction of FOXM1 protein during mitosis by the APC/C (Laoukili et al., 2008; Park et al., 2008). FOXM1 expression is often increased in different types of cancer, such as squamous cell carcinoma (Calvisi et al., 2009; Chen et al., 2009; Gemenetzidis et al., 2009; Waseem et al., 2010; Hui et al., 2012; Teh et al., 2013), rhabdomyosarcoma (Wan et al., 2012), hepatocellular carcinoma (Liu et al., 2012; Xia et al., 2012), pancreatic cancer (Huang et al., 2012), glioblastoma (Liu et al., 2006), breast cancer (Madureira et al., 2006; Ahmad et al., 2010, 2011; Kwok et al., 2010; Bergamaschi et al., 2011; Wang and Gartel, 2011; Park et al., 2012), and leukemia (Nakamura et al., 2010). FOXM1 has also been shown to drive expression of MMP2 and MMP9, which are implicated in metastases and tumor cell invasion (Wang et al., 2008; Ahmad et al., 2010; Chen et al., 2011; Lok et al., 2011; Ahmed et al., 2012; Lynch et al., 2012; Xue et al., 2012).
Although FOXM1 is perhaps the best-studied forkhead box protein involved in cell cycle control, there are a number of others that have roles in cell cycle progression. FOXK1 and FOXJ3 are required for cell cycle progression in U2OS cells, as determined in both live-cell luminescence assays (Grant et al., 2012) and genome-wide small interfering RNA (siRNA) screens (Mukherji et al., 2006; Kittler et al., 2007). In addition, there are a number of other FOX genes that result in cell cycle phenotypes when knocked down in siRNA screens, including FOXA2, FOXA1, and FOXL2 (Mukherji et al., 2006; Kittler et al., 2007).
Here we identify the cell cycle–regulated genes in U2OS cells after synchronization using either a double-thymidine block or a thymidine–nocodazole block. We identify 2140 transcripts mapping to 1871 unique cell cycle–regulated genes and analyze FOXM1 genomic loci by genome-wide chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq). We show that FOXM1 is bound to many genes involved in the G2- to M-phase transition, as well as a number of genes involved in DNA replication. Inclusion of ChIP-seq data for E2F1, 4, and 6 from the Encyclopedia of DNA Elements (ENCODE) project, FOXK1 (Grant et al., 2012), and the DREAM complex (Sadasivam et al., 2012) provides a comprehensive picture of the transcriptional regulatory system controlling periodic gene expression in the human cell division cycle.
RESULTS
We identified the full complement of genes periodically expressed across multiple synchronous cell division cycles in the commonly studied human U2OS osteosarcoma-derived cell line (Ponten and Saksela, 1967; Laoukili et al., 2005). This extends and contrasts with periodic, cell cycle–regulated gene expression data from three additional human cell lines: HeLa cells (Whitfield et al., 2002; Sadasivam et al., 2012), primary foreskin fibroblasts (Cho et al., 2001; Bar-Joseph et al., 2008), and HaCaT cells (Pena-Diaz et al., 2013).
Identification of periodically expressed transcripts
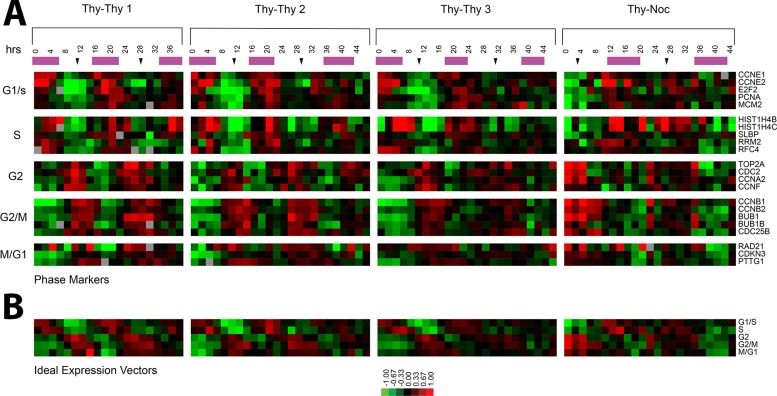
Two complementary methods were used to obtain synchronous populations of U2OS cells. The first is a thymidine–thymidine (Thy-Thy) synchronization that arrests cells at the G1/S boundary, and the second is a thymidine–nocodazole (Thy-Noc) synchronization that arrests cells in mitosis. Each of these methods provides the best synchrony in the cell cycle phases that immediately follow the arrest. Thy-Thy provides the best synchrony for the G2 and M phases, and Thy-Noc provides the best synchrony in G1/S and S phases. Three Thy-Thy synchronization time courses were performed, which sampled the population at 2-h intervals for 38 h (Thy-Thy 1) or 46 h (Thy-Thy 2 and Thy-Thy 3; Figure 1; all figures are available in blue/yellow color format in Supplemental Figure S10). The Thy-Thy time courses measured gene expression across 2.5 synchronous cell cycles, traversing two complete cycles of S phase and mitosis, and into a third round of DNA replication. A fourth time course was performed using the Thy-Noc synchronization protocol, and time points were collected at 2-h intervals for 44 h, which resulted in two full cycles of mitosis and S phase (Figure 1). Gene expression was measured genome-wide using these two complementary methods across four independent time courses, resulting in the analysis of multiple synchronous cell cycles.
FIGURE 1:
Periodic expression of well-characterized cell cycle genes. (A) Expression profiles of known periodically expressed genes for each of the four time courses. The purple bars indicate S phase, and black arrows indicate mitosis as estimated by flow cytometry or Western blots. (B) The idealized vector for each phase is the average expression of the known genes shown in A.
The degree of synchrony in each cell cycle time course was monitored by flow cytometry analysis of DNA content by propidium iodide staining (Supplemental Figure S1). In addition, cell lysates were prepared and Western blots performed for well-characterized cell cycle–regulated proteins (CCNB1 and FOXM1; unpublished data). All data used in this study were from experiments that showed clear cell cycle synchrony using these two measures.
Periodically expressed genes were identified as previously described (Whitfield et al., 2002). We collected and isolated total RNA from synchronous U2OS cells every 2 h for a minimum of 38 h and determined the expression of all genes in the genome using Agilent 4 × 44,000–element DNA microarrays (41,000 probes representing 19,637 unique Entrez GeneIDs). Each time course was analyzed independently.
To verify our ability to detect periodic cell cycle–regulated genes, we selected a set of known cell cycle–regulated genes from the literature previously shown to have peak expression in each of the five major phases of the cell cycle (G1/S, S, G2, G2/M, and M/G1; Figure 1). Expression of these genes in each time course is shown with DNA replication and mitosis indicated (Figure 1). For G1/S, we examined CCNE1, CCNE2, E2F2, PCNA, and MCM2. For S phase, we selected genes involved in DNA replication or the packaging of newly replicated DNA, including HIST1H4B, HIST1H4C, SLBP, RRM2, and RFC4. G2 was represented by TOP2A, CDC2, CCNA2, and CCNF. G2/M included five genes required for mitosis: CCNB1, CCNB2, BUB1, BUB1B, and CDC25B. Following on the observation from Whitfield et al. (2002) that a set of genes showed peak expression during mitosis into G1, we selected three genes for the M/G1 transition: RAD21, CDKN3, and PTTG1. These genes were averaged to generate an idealized expression vector for each cell cycle phase (Figure 1B).
A Fourier transform (Whitfield et al., 2002) was used to generate a periodicity score for each gene. The periodicity score quantifies how periodic a gene is across each of the four time courses and is the magnitude of the sine and cosine components of the Fourier analysis (see Materials and Methods). The periodicity score for each gene was summed across the four time courses and then scaled by the highest correlation to one of five idealized vectors of the known cell cycle genes for each cell cycle phase (G1/S, S, G2, G2/M, M/G1; Figure 1; Spellman et al., 1998; Whitfield et al., 2002). Table 1 gives examples of genes and their periodicity score (Supplemental Table S1).
TABLE 1:
Sample periodicity scores from selected cell cycle–regulated genes.
| Rank | Gene | Periodicity score | Maximum fold change |
|---|---|---|---|
| 1 | SHISA3 | 27.0 | 7 |
| 2 | PIF1 | 26.5 | 4.5 |
| 4 | KIF20A | 23.5 | 6 |
| 7 | PLK1 | 22.5 | 3 |
| 11 | CCNB1 | 19.5 | 2.5 |
| 14 | CCNE1 | 19.0 | 5.5 |
| 23 | CENPE1 | 16.8 | 5 |
| 161 | PCNA | 9.15 | 3.5 |
| 964 | CIT | 4.90 | 1.5 |
| 1162 | BRCA1 | 4.50 | 2 |
| 1288 | PRIM1 | 4.28 | 2.5 |
| 2557 | E2F1 | 3.12 | 2 |
The maximum fold change is the largest difference in peak-to-trough ratio for each gene rounded down to the nearest 0.5 fold change.
To establish a cutoff for a cell cycle–regulated gene, we randomized the data either by rows or both rows and columns and repeated the analysis on the randomized data to estimate a false discovery rate (FDR). We selected a periodicity score of 2.65, which resulted in 3568 periodically expressed probes with estimated FDR of 1.29% when randomizing by rows and columns and 3.67% when randomizing by rows only. To estimate the false-negative rate (FNR), we used a list of known cell cycle–regulated genes (Whitfield et al., 2002, Table 2). We determined the FNR at this cutoff to be 11%.
TABLE 2:
Enrichment of transcription factor–binding loci by cell cycle phase.
| Transcription factor | G1/S | S | G2 | G2/M | M/G1 | Minimum p | Minimum p phase |
|---|---|---|---|---|---|---|---|
| E2F1 (HeLa) | 1.20E-06 | 0.16 | 0.70 | 0.41 | 1.00 | 1.20E-06 | G1/S |
| E2F6 (K562) | 0.0010 | 0.20 | 0.59 | 1.00 | 0.36 | 0.0010 | G1/S |
| E2F6 (K562b) | 0.0018 | 0.18 | 0.91 | 1.00 | 0.12 | 0.0018 | G1/S |
| E2F6 (HeLa) | 0.29 | 0.42 | 1.00 | 0.69 | 0.46 | 0.29 | G1/S |
| FOXM1 | 0.79 | 0.53 | 1.09E-40 | 8.79E-33 | 0.00054 | 1.09E-40 | G2 |
| E2F4 (HeLa) | 1.43E-05 | 0.05 | 9.71E-16 | 5.51E-09 | 0.66 | 9.71E-16 | G2 |
| E2F4 (K562b) | 9.73E-07 | 0.00076 | 2.78E-10 | 1.56E-07 | 0.66 | 2.78E-10 | G2 |
As in the prior analysis of HeLa cells, we found that a portion of the probes display a sinusoidal pattern in the first cell cycle but do not repeat in the second cell cycle (Whitfield et al., 2002). These genes are likely induced by the synchronization procedure. To filter out these probes, we calculated an autocorrelation score for each gene in each time course. The autocorrelation determines whether the gene expression ratio at a given time is a good predictor of the ratio one cell cycle later. Therefore genes that oscillate and thus repeat their expression values over multiple cell cycles receive high autocorrelations, whereas genes that do not oscillate receive low or negative autocorrelations. Altogether, 690 probes with negative autocorrelations were removed, leaving 2878 probes.
As a final filtering step, we removed 48 probes that showed systematic bias associated with date of hybridization. These probes had no clear periodic expression and are almost certainly systematic artifacts from microarray hybridization. To objectively remove genes with “date-biased” expression patterns, we projected the spectrum for each of the 2878 probes onto the first two principal components and organized by k-means clustering. Forty-eight probes that displayed expression patterns that correlated with date of hybridization clustered together (Supplemental Figure S2A). Those 48 probes that grouped together with alternating high and low expression levels, correlated with the hybridization batch (Supplemental Figure S2, A and B), were removed, leaving 2830 cell cycle–regulated probes corresponding to 2140 Entrez GeneIDs, with 1871 of these being unique (Supplemental Table S2).
Overview of periodic gene expression in U2OS cells
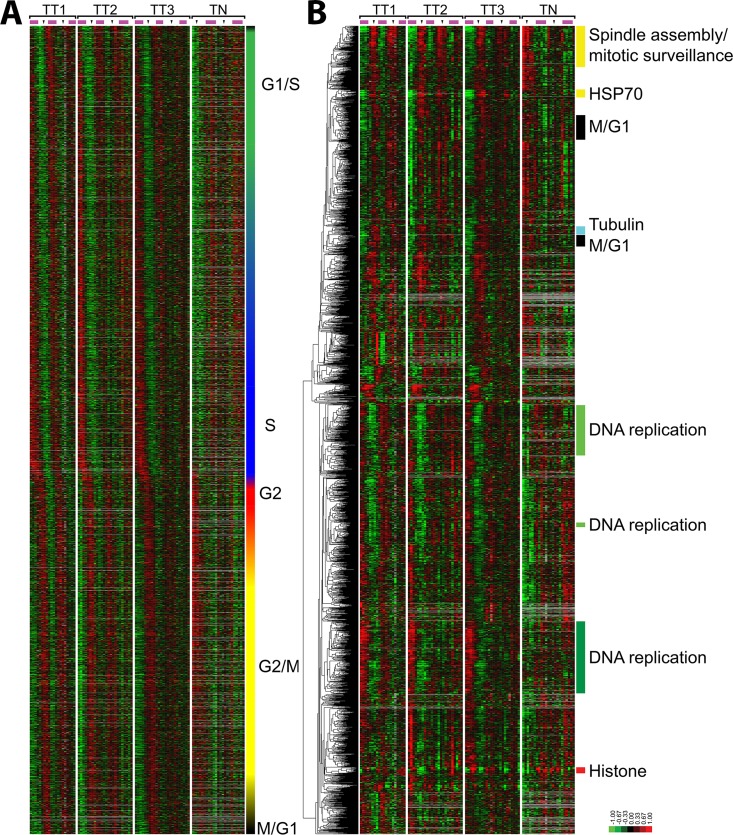
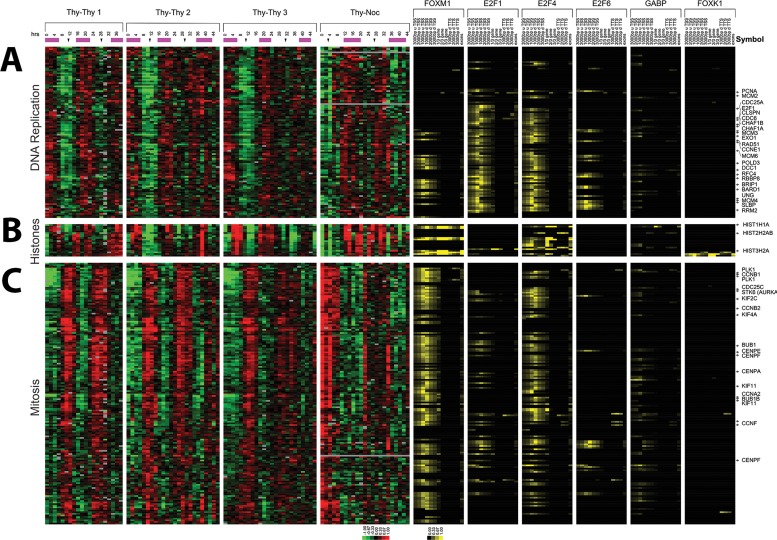
To display the selected genes in the order of their timing of peak expression within the cell division cycle, we organized genes by their phase of peak expression determined by Fourier analysis (Figure 2A). The alternating pattern of high and low gene expression levels shows persistent waves of gene expression across multiple synchronous cell cycles. The phase of peak expression of each gene was assigned based on peak correlation to the idealized phase expression profiles shown in Figure 1. This method assigns the probes to G1/S (1018 probes; 702 unique genes), S phase (546 probes; 355 unique genes), G2 phase (390 probes; 278 unique genes), G2/M (598 probes; 392 unique genes), and M/G1 (278 probes; 144 unique genes; Figure 2A).
FIGURE 2:
Periodically expressed genes in the U2OS cell cycle. (A) The 2830 probes that show periodic expression in U2OS cells, ordered by the point of their peak expression as calculated from their sine and cosine values in the Fourier transform. Phase assignments were performed using the correlation coefficients with the ideal vectors defined in Figure 1. The color bar on the right indicates the phase of peak expression (G1/S, green; S, blue; G2, red; G2/M, yellow; and M/G1, black); blended colors indicate interspersed phase assignments. (B) The 2830 periodically expressed probes from U2OS cells were ordered by average linkage hierarchical clustering. The enriched biological processes of select clusters is indicated.
The expression program in U2OS cells has two distinct waves of expression—one that occurs in G1 and S phases, and a second that occurs in G2 and M phases. We displayed the distribution of genes in the two phases by clustering the correlation values for each gene to the five idealized expression vectors (Supplemental Figure S3). Genes assigned to G1/S or S phase had similar correlation scores to the idealized G1/S and S-phase vectors, whereas genes that were classified as G2, G2/M, or M/G1 had high correlations to the G2 and G2/M vectors. Considerable overlap is observed among genes of the G1/S and S-phase transition and between the genes of the G2 and G2/M transition. Thus the boundaries between G1/S and S phase and between G2 and M phase are necessarily somewhat arbitrary, as genes assigned to G1/S can be immediately adjacent to genes assigned to S phase. In contrast, there is a distinct break between the G1- and S-phase genes and the G2- and M-phase genes. We analyzed the same distributions in HeLa cells, where we did not observe a distinct break between the two waves of expression (Supplemental Figure S3). This suggests that the distinct break observed here may be specific to U2OS cells and reflect specific mutations or biological changes in this cell line.
To organize the genes in an unbiased manner, we hierarchically clustered all cell cycle–regulated probes (Eisen et al., 1998). The Database for Annotation Visualization and Integrated Discovery (DAVID; Dennis et al., 2003) was used to identify enriched cellular processes in each cluster. Genes involved in mitosis (M phase, p = 6.6 × 10−42), including cyclins A2, B1, B2, and F, primarily fell into one large cluster, whereas genes involved in DNA replication separated into three large clusters, each with weaker but still significant levels of enrichment for S-phase processes (DNA replication, p = 1.4 × 10−10; DNA metabolic process, 1.4 × 10−6). The first cluster of S-phase genes includes four minichromosome maintenance proteins (MCM 2, 3, 4, and 10), PCNA, CDT1, CHAF1A, CHAF1B, E2F2, and E2F8. The second cluster of DNA replication genes includes RMI1, DSCC1, and MCM6. The third and final DNA replication cluster includes two more E2F genes, E2F1 and E2F7, PLK3, RMI2, CDC45L, RBBP8, DHFR, BRIP1, PRIM1, RRM2, and RFC4. A small but distinct cluster was found completely comprising histone genes (nucleosome assembly, p = 4.9 × 10−23). There was also a small cluster of genes containing primarily heat shock proteins (labeled the HSP70 cluster), as well as the HSP70-binding protein BAG3, which has antiapoptotic properties (Takayama et al., 1999; Doong et al., 2000; Romano et al., 2003a, b).
The overall patterns of gene expression reflect the fundamental biological processes necessary to duplicate a cell, such as DNA replication, DNA packaging, formation of the mitotic spindle, and mitosis. It includes not only the proteins required to perform these processes (e.g., MCM proteins, the histones, proteins at the DNA replication fork, nucleotide biosynthesis, nucleosome assembly and mitotic genes), but also key regulators of those genes (e.g., E2F transcription factors, cyclins, and key kinases such as PLK). Each of these is coordinately regulated in a tightly controlled manner, with peak expression at their required points in the cell cycle and then reduced expression until they are needed the following cell cycle.
Cell cycle regulation by FOXM1
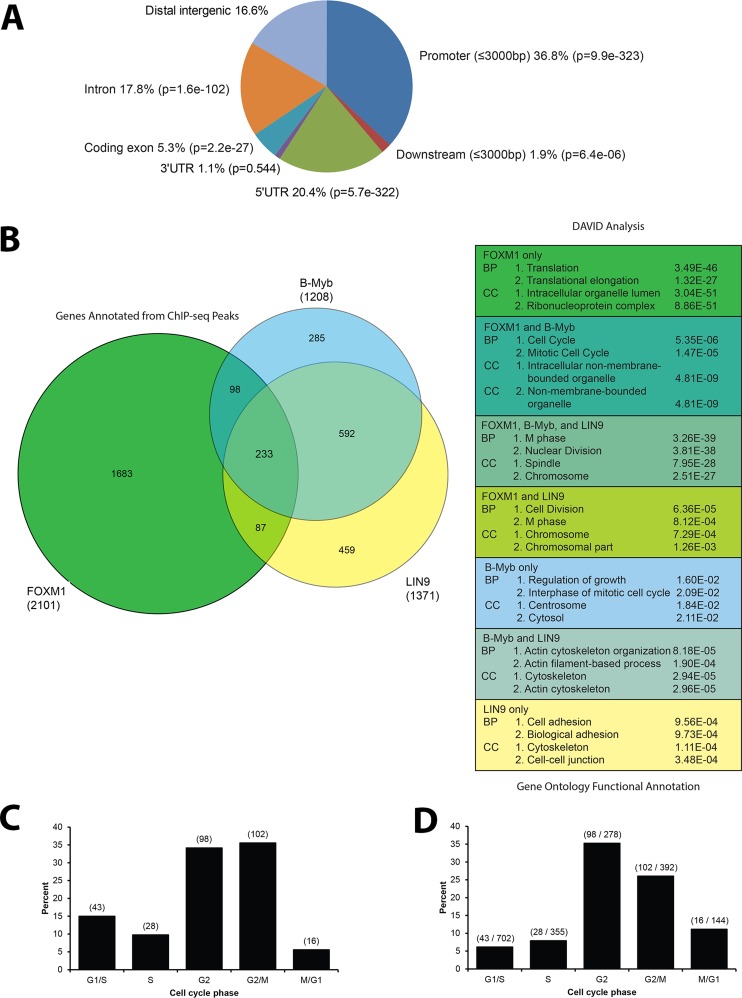
To understand the network of transcription factors controlling cell cycle–regulated gene expression and better understand the role of FOXM1 in the transcriptional program that regulates mitosis, we performed ChIP-seq for endogenous FOXM1 in asynchronous HeLa cells. IPs were performed in duplicate using an antibody against the endogenous FOXM1 (C20; Santa Cruz Biotechnology) and sequenced independently. As a conservative estimate of FOXM1 binding, we focused the analysis on those ChIP-seq regions found in both ChIP-seq assays. The first sequencing analysis (run A) gave 17.1 million sequence reads, with 8.4 million reads mapped (Hg 19), and the second sequencing analysis (run B) gave 17.0 million sequence reads, with 8.4 million mapped. Using the MACS module in Cistrome (Zhang et al., 2008; Liu et al., 2011; www.cistrome.com), we found 5727 peaks in A and 2849 peaks in B, with 2215 regions identified in both; these regions could be associated with 2367 unique genes by the Gene Centered Annotation (GCA) module in Cistrome (Shin et al., 2009; Figure 4 and Supplemental Figure S4). We refer to these intersecting regions as FOXM1 genomic loci. Of the genomic loci, 36.8% were in promoter regions within 3000 base pairs upstream from the transcription start site, and 1.9% of the genomic loci were within 3000 base pairs downstream of the transcription start site. Of the genomic loci, 20.4% were in the 5′-untranslated region (5′UTR) of target genes and 1.1% were in the 3′UTR. Genomic loci in exonic regions accounted for 5.3%, and 17.8% of the loci were in intronic regions. Distal intergenic regions accounted for 16.6% of the ChIP-seq genomic loci (Figure 3A).
FIGURE 4:
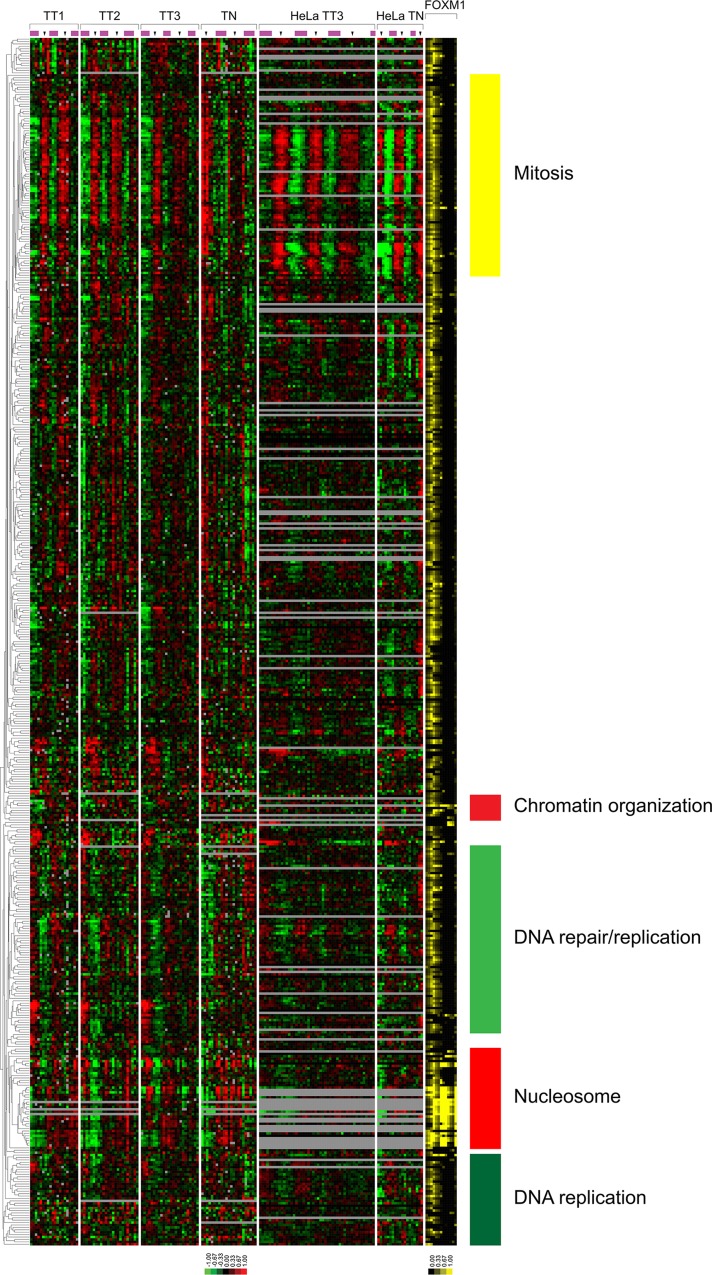
The expression profiles of genes bound by FOXM1. The clustered U2OS and HeLa cell expression profiles of the 501 genes bound by FOXM1 that are cell cycle regulated in U2OS cells. FOXM1 transcription factor binding is shown as percentage coverage of the UCSC genome browser gene model as defined by GCA for each gene (see Supplemental Figure S6 for more detail).
FIGURE 3:
FOXM1 ChIP-seq indicates a role for FOXM1 in the cell cycle and translation. (A) CEAS analysis of the overlapping FOXM1 ChIP-seq analysis shows enrichment in promoters, downstream regions, 5′UTRs, and coding exons relative to background. (B) Venn diagram showing the overlapping gene targets of FOXM1, B-Myb, and LIN9 (b-Myb and LIN9 results from Sadasivam et al., 2012). There is enrichment in cell cycle Gene Ontology (GO) terms for biological process (BP) and cellular component (CC) in the overlapping gene lists for FOXM1/B-Myb, FOXM1/B-Myb/LIN9, and FOXM1/LIN9. FOXM1 genes that do not overlap with B-Myb or LIN9 show an enrichment in the GO term translation. (C) Bar graph showing the percentage of FOXM1 targets per cell cycle phase. (D) Bar graph showing the percentage of genes in each phase that are bound by FOXM1.
Recently it was shown using ChIP-reChIP that B-Myb and FOXM1 cooccupy the promoters of genes involved in mitosis (PLK1, CCNB1, and AURKA; Sadasivam et al., 2012). Similar results were found using ChIP-reChIP of LIN9 followed by FOXM1. We compared our FOXM1 ChIP-seq results with the ChIP-seq results of Sadasivam et al. (2012; Figure 3B). The gene targets found in all three ChIP-seq experiments were enriched for genes involved in mitosis (DAVID, M phase, p = 3.26 × 10−39). There was also enrichment for cell cycle–related processes for the FOXM1/B-Myb overlap (Figure 3B; cell cycle, p = 5.35 × 10−06) as well as for the FOXM1/LIN9 overlap (cell division, p = 6.36 × 10−05). Of interest, after removal of the FOXM1 target genes from the B-Myb/LIN9 target list (i.e., target genes of all three transcription factors vs. targets of B-Myb and LIN9 but not FOXM1), the most enriched biological process was actin cytoskeleton organization (p = 8.18 × 10−05). The list of FOXM1 target genes only was enriched for the biological process of translation (p = 3.49 × 10−46) and translation elongation (p = 1.32 × 10−27; Figure 3B).
We display the expression of genes bound by FOXM1 in our ChIP-seq that were also cell cycle regulated (Figure 4), as well as those that were not cell cycle regulated (Supplemental Figure S4). To represent FOXM1 binding relative to gene models, we show the percentage coverage of different regions of each gene model as defined by GCA (Supplemental Figure S6). We then linked the genes for each FOXM1 ChIP-seq loci via Entrez GeneIDs to genes that are cell cycle regulated in U2OS cells. Of the 1871 unique cell cycle–regulated genes in U2OS cells, 287 showed evidence of FOXM1 occupancy at their promoters. Because FOXM1 is known to drive the expression of G2/M phase genes, we first examined the expression of known G2/M FOXM1 targets, AURKB, CCNB1, CCNB2, PLK1, and TOP2A, which all had FOXM1 bound in their promoters (Supplemental Figure S5). Of the 278 genes expressed in G2, 98 (35.2%, p < 0.001) were bound by FOXM1 in our ChIP-seq data. Of the 392 genes expressed in G2/M, 102 (26%, p < 0.001) were bound by FOXM1. Progressing through the cell cycle, there were 16 M/G1 genes bound by FOXM1 out of 144 (11.1%; not significantly enriched [NS]). Surprisingly, we found a number of G1- and S-phase genes that were bound by FOXM1, including TYMS, RMI1, and replication-dependent histone genes. FOXM1 binds to 6.1% of the genes expressed in G1/S (43 of 702; NS) and 7.8% of S-phase genes (28 of 355; NS).
Many FOXM1 target genes in different cell types have been reported (reviewed in Wierstra and Alves, 2007). Given that FOXM1 has roles in cell cycle progression, metastasis (through MMP2 and MMP9; Wang et al., 2008), and development in many cell types, we focused on FOXM1 targets in cell types that display epithelial morphology. In our ChIP-seq analysis we identified the promoters of all four previously published FOXM1 targets in HeLa cells (AURKA, PLK1, CCNB1, and RACGAP1; Sadasivam et al., 2012). Looking at genes previously shown to be bound by FOXM1 by ChIP in U2OS cells, we found five of nine published FOXM1 targets (Dai et al., 2007; Wang et al., 2007; Chen et al., 2009; Chetty et al., 2009; Ahmed et al., 2012).
Of the previously reported G2- or M-phase FOXM1 target genes that were also cell cycle regulated in HeLa cells, 32 of 39 (∼82%) were found in our analysis of U2OS cells, whereas only five of 16 (∼31%) of the published FOXM1 targets expressed in G1 or S phases were identified in our analysis in U20S cells (Supplemental Figure S6 and Supplemental Table S4). Most FOXM1 ChIP-seq target genes were cell cycle regulated in U2OS cells (27 of the top 30) and HeLa cells (28 of the top 30; Whitfield et al., 2002). Of the 12 published FOXM1 target genes not found in our ChIP-seq data, only three were cell cycle regulated: CDC25A in U2OS and HeLa, BRCA2 in U2OS only, and SKP2 in HeLa only (Supplemental Figure S7).
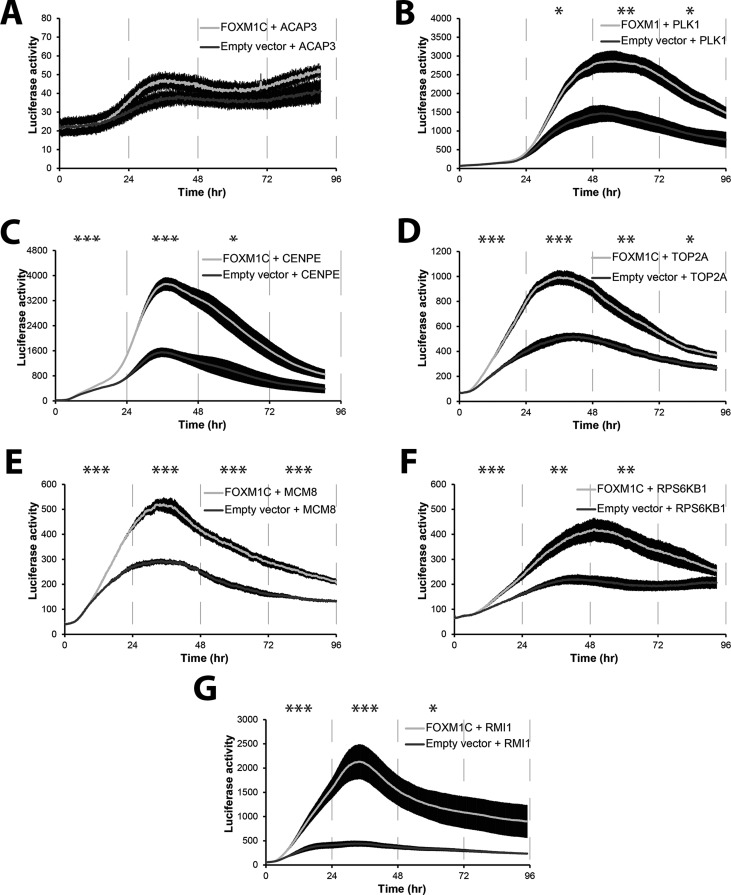
To determine whether FOXM1 can induce luciferase expression from promoters found in our ChIP-seq data, we used the LumiCycle, a luminometer that measures luciferase expression from live cells in real time (Grant et al., 2012). We cotransfected U2OS cells with either a Flag-tagged FOXM1C construct or the empty vector (pBABE-puro) and a promoter luciferase target. We tested the ability of FOXM1 to activate seven genes found among our ChIP-seq data. These included the well-characterized FOXM1 targets CENPE, PLK1, and TOP2A genes, with peak expression in G2 or G2/M phase. We also tested a subset of the novel FOXM1 targets identified in our screen: MCM8, RPS6KB1, and RMI1. As a negative control, we used the FOXK1 target ACAP3 (Grant et al., 2012), which contains a forkhead box cis-element but is not bound by FOXM1, as determined by our ChIP-seq analysis. As expected, the ACAP3 promoter showed no induction after transfection with FOXM1, whereas all other promoters were activated by exogenous FOXM1C expression (Figure 5). Given that both MCM8 and RMI1 are expressed in S phase, this demonstrates that FOXM1C can activate expression of S-phase genes, albeit at a lower level of induction. RPS6KB1 is not cell cycle regulated in HeLa cells but is in U2OS cells and can be overexpressed in cancers (Sinclair et al., 2003; Yamnik et al., 2009). RPS6KB1, a downstream target of mammalian target of rapamycin (mTOR), plays multiple roles in translation, including phosphorylation of eIF-4B, as well as phosphorylation of the negative regulator of eIF4A, PDCD4, targeting PDCD4 for destruction (reviewed in Fenton and Gout, 2011).
FIGURE 5:
FOXM1 overexpression activates the expression of a number of target genes found via ChIP-seq. (A) Neither FOXM1 (light line) nor the empty vector control (dark line) activates the ACAP3 promoter. ACAP3 is a known FOXK1 target (Grant et al., 2012) and is used as a negative control. (B–D) FOXM1 activates the promoters of PLK1, CENPE, and TOP2A, known FOXM1 targets (Figure 4 and Supplemental Figure S6). (E) FOXM1 activates the MCM8 promoter. (F) FOXM1 activates the promoter of RPS6KB1, which phosphorylates the 6S ribosomal protein. (G) The previously unknown FOXM1 target RMI1 promoter is activated by FOXM1 overexpression. Error bars, SEM. Time points were binned and averaged for every 24 h for a one-way analysis of variance. *p ≤ 0.05; **p ≤ 0.01, ***p ≤ 0.005.
Transcriptional regulators of the cell cycle gene expression program
To identify the transcriptional regulators of the genome-wide cell cycle gene expression program, we obtained ChIP-seq data from the ENCODE project for transcription factors E2F1, E2F4, E2F6, and GA-binding protein transcription factor, α subunit 60 kDa (GABPA [GABP]), each implicated in cell cycle control. We reanalyzed the data to identify ChIP-seq loci using MACS and associated loci with gene regions. We also included ChIP-seq data for FOXM1 and FOXK1 (Grant et al., 2012). Cell cycle gene expression data were collapsed to Entrez GeneIDs and merged with the Entrez GeneID–associated ChIP-seq loci. Gene expression and ChIP-seq data were coclustered and in the following are discussed in detail by biological process.
The DNA replication cluster includes genes involved in the process of DNA synthesis, including components of the prereplication complex (MCM2, 3, 4, and 6), nucleotide biosynthesis (RRM2), DNA replication (PCNA, CLSPN, EXO1, POLD3, RFC4, and DCC1), and DNA packaging (CHAF1A, CHAF1B, and SLBP). We also find genes involved in S-phase regulation (E2F1, CDC6, CDC25A, CCNE1, and RBBP8) and DNA repair (UNG, BARD1, BRIP1, and RAD51). The G1/S-phase transcriptional regulator E2F1 is bound to 67% (68 of 101) of the unique genes in this DNA replication cluster. E2F4, which is a member of the DREAM complex that represses the expression of S-phase genes during G0 (Litovchick et al., 2007), is bound to 60% of the genes in this cluster (61 of 101). Greater than half (56) of the genes in this cluster have both E2F1 and E2F4 bound (Figure 6A). Comparison with cell cycle–regulated genes in HeLa cells (Whitfield et al., 2002) shows that 42% (39 of 93) of those bound by E2F1 or E2F4 were regulated there.
FIGURE 6:
Biological process–specific gene clusters have coordinated transcription factor binding. (A) After the inclusion of FOXM1 ChIP-seq data as well as ENCODE data, hierarchical clustering shows that the genes in the U2OS S phase cluster are predominantly bound by E2F1 and E2F4. (B) The histone cluster shows E2F4 bound to more gene models than E2F1 or FOXM1. (C) Genes in the mitosis gene cluster are primarily bound by FOXM1 or E2F4. Transcription factor binding is shown as percentage of coverage of each portion of the gene model as defined by GCA (see Supplemental Figure S6 for more details).
Histone genes clustered together in a tight group likely due to their unique mechanism of regulation, which is primarily by posttranscriptional mechanisms controlled by histone SLBP (Wang et al., 1996; Whitfield et al., 2000). Consistent with this, only two of the genes in this cluster are bound by E2F1, but, surprisingly, 53% of the histone genes are bound by E2F4 (9 of 17 genes in the cluster) and 47% by FOXM1 (8 of 17). Given that E2F4 is known to repress S-phase genes in G0, it may play a similar role for the histone genes, whose expression is tightly restricted to S phase. Of these genes, 18% (3 of 17) were bound by FOXK1, recently shown to be involved in G1/S-phase gene activation (Figure 6B).
The mitosis cluster contains genes involved in the processes of DNA segregation and chromosome organization (KIF11, KIF2C, and KIF4A) and regulation of mitosis (PLK1, CCNB1, CCNB2, BUB1, STK6 [AURKA], CCNA2, CCNF, and CDC25C). Of the mitotic genes, 58% (68 of 118, p < 0.001) are bound by E2F4, likely as a component of the repressive DREAM complex; 76% (90 of 118, p < 0.001) are bound by FOXM1, and only 24% (28 of 118, NS) are bound by E2F1. Eleven percent (13 of 118, NS) are bound by E2F6 and 3% (4 of 118, NS) by FOXK1. GABPA binds to 35% (41 of 118, p < 0.001), albeit at a lower percentage of promoter coverage than FOXM1 or E2F4 (Figure 6C). Of the genes found in this cluster, 37% (44 of 118) are also cell cycle regulated in HeLa cells, corresponding to the “spindle assembly” and “mitotic surveillance” clusters (Whitfield et al., 2002).
Transcription factor binding by cell cycle phase
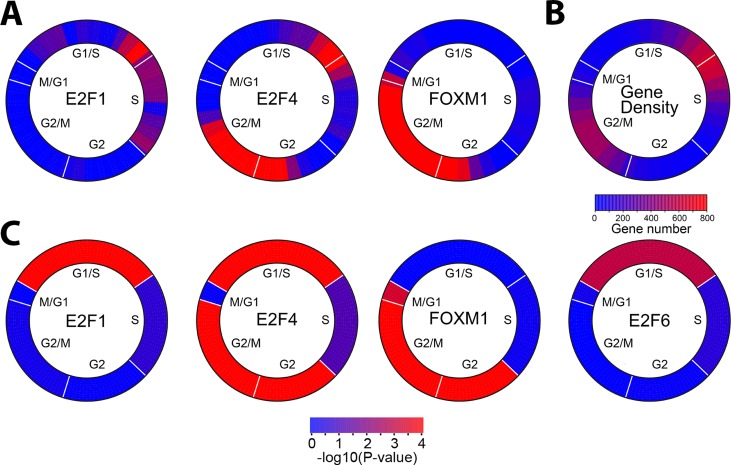
Transcription factor binding across the cell cycle was displayed as a function of the significance of enrichment (Fisher's exact test) of a transcription factor–binding site over a sliding window of phase-ordered genes (Figure 7A). This demonstrates that E2F1 binding shows significant enrichment in late G1/S phase of the cell cycle, whereas FOXM1 binding is significantly enriched at G2/M phase of the cell cycle. As expected, both show low enrichment of binding in the opposite phase. E2F4 shows biphasic binding, with enrichment in G1/S and G2/M phases. We also examined gene density by cell cycle phase and found that the regions with peak binding of these three transcription factors correspond to the highest gene density (Figure 7B).
FIGURE 7:
Transcription factor binding as a function of cell cycle phase. (A) Enrichment of transcription factor targets using a sliding window across the cell cycle indicates enrichment of E2F1, E2F4, and FOXM1 binding to genes that show peak expression at specific times during the cell cycle. Phase angles were calculated from the arc tangent of the Fourier analysis. Transcription factor binding enrichment was calculated using Fischer's exact test over a sliding window of 30° with a 10° overlap between neighboring windows. Cell cycle phase is indicated (B) Gene density in each sliding window was calculated for the expression of all the cell cycle–regulated genes in U2OS cells. (C) Enrichment of transcription factor targets in each cell cycle phase was calculated using estimated phase boundaries rather than a sliding window. The significance of transcription factor binding enrichment was calculated using Fischer's exact test. Transcription factor binding data were from HeLa cells, except for E2F6, which was from K562 cells. The p values for each transcription factor at each phase are given in Table 2.
To determine the specific cell cycle phase in which each transcription factor is enriched, we calculated enrichment using the phase assignments for each gene that we assigned using our idealized vectors (Figure 7C and Table 2). We find that E2F1 binding is significantly enriched in G1/S (p = 1.20 × 10−6). E2F enrichment is not significant in other cell cycle phases but is bound to 47% (16 of 34) of S-phase genes, 26% (16 of 60) of G2 genes, 27% (20 of 74) of G2/M genes, and 10% (2 of 20) M/G1 genes. E2F4 shows biphasic enrichment, with significant interactions with genes that peak at G1/S (p = 1.43 × 10−5), as well as with those that show peak expression at G2/M phase (p = 8.79 × 10−33; Table 2 and Figure 7C). E2F4 is bound to ∼53% (35 of 65) of G1/S, ∼44% (15 of 34) of S phase, and ∼28 (17 of 60) of G2 genes and ∼48% (36 of 74) G2/M phase targets. FOXM1-binding sites show significant enrichment for genes that peak in G2 (p = 1.09 × 10−40), G2/M (p = 8.79 × 10−33), and M/G1 (p = 5.4 × 10−4). FOXM1 is bound to a 73% (44 of 60) of G2 genes and 70% (52 of 74) of G2/M-phase genes but only ∼15% (10 of 65) of G1/S and ∼20% (7 of 34) of S-phase genes. E2F6 showed only weak enrichment for G1/S (p = 0.0010 and 0.0018) and only when using binding sites defined in K562 or K562b cells (Table 2 and Figure 7C). These data demonstrate a complex interplay of regulation between G1/S- and G2/M-phase transcription factors, which includes a biphasic binding by E2F4.
Identification of a common set of cell cycle–regulated genes
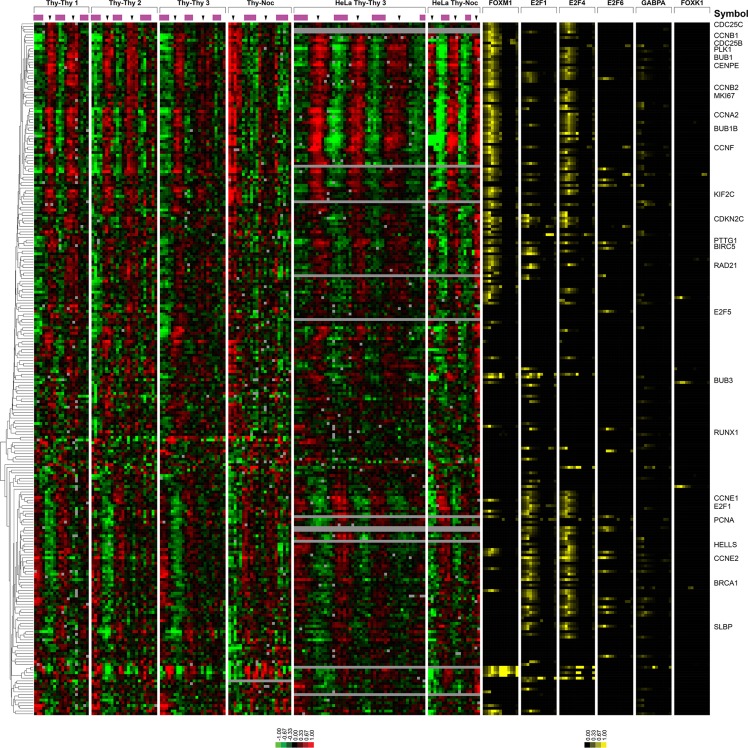
To identify the common cell cycle genes regulated across multiple cell types, we merged and compared the U2OS, HeLa, and ENCODE data (Figure 8). We compared the 1871 unique genes cell cycle regulated in U2OS cells to the 651 unique cell cycle-regulated genes in HeLa cells. We found that 253 of the 651 (38.8%) HeLa cell cycle–regulated genes were also cell cycle regulated in U2OS cells. These overlapping genes predominantly consisted of those involved in core processes of mitosis and S phase. Many of these genes are well known cell cycle genes, such as PLK1, CCNB1, E2F1, and CCNE2; however, there were a number of less well-characterized genes that are cell cycle regulated in all four cell types, including FLJ10156 (FAM64A), ARL6IP, DLG7 (DLGAP5), HMGB3, TROAP, SHCBP1, ANP32E, GPR126, HJURP, SLC38A2, and C14orf130 (UBR7), which may represent less well-studied proteins involved in essential biological processes required to duplicate a cell.
FIGURE 8:
Core cell cycle–regulated genes between HeLa cells and U2OS cells are bound by FOXM1 or E2F1. The 253 genes that show periodic regulation in both HeLa cells and U2OS cells have been clustered after the inclusion of the FOXM1 and FOXK1 ChIP-seq data and the ENCODE data for selected transcription factors. Transcription factor binding is shown as percentage of coverage of each portion of the gene model as defined by GCA (see Supplemental Figure S6 for more details).
DISCUSSION
We identified 1871 unique genes expressed in a cell cycle–dependent manner in the well-studied U2OS cancer cell line. This cell line was chosen because it is amenable to multiple synchronization methods, with a low percentage of noncycling cells. Each gene was assigned to a cell cycle phase: 56% had peak expression in G1 or S phase, whereas 35.8% had peak expression in G2 or M phase.
Comparison to other cell cycle expression data sets in mammalian cells
There have been a number of reports on the periodic expression of cell cycle genes in mammalian cells. These reports often used different cell lines/types, as well as different analysis methods. We focus on three studies that use human cells and either a double-thymidine or thymidine–nocodazole block.
Whitfield et al. (2002) published the cell cycle–regulated genes in HeLa cells. They reported 651 unique genes that were cell cycle regulated. Here we report 1871 unique genes that are cell cycle regulated in U2OS cells. Of these genes, 253 (38.8%) are cell cycle regulated in both cell lines when analyzed using the same analysis pipeline.
Genes cell cycle regulated in both studies include well-known genes that have peak expression in G1/S phase, such as E2F1, BARD1, CHAF1A, CHAF1B, as well as CCNE1 and CCNE2. This also holds true for genes involved in DNA replication with peak expression in S phase, including RRM2, PRIM1, RBBP8, and RFC2. U2OS genes peaking in mitosis have good overlap with mitosis genes in HeLa and foreskin fibroblasts, including PLK1, CCNB1, CDC25B, CDC25C, CENPA, CENPE, CENPF, and TOP2A. There are also overlapping genes that have peak expression during mitosis into G1, including CEP70, UBE2D3, BAIAP2, and PDGFA. Thus we identified a large number of previously known cell cycle–regulated genes, as well as a number of novel cell cycle–regulated genes in U2OS cells.
Although the methods for determining which genes are cell cycle regulated are very similar for both HeLa and U2OS, there are experimental differences that may affect the results. These include different array platforms and differences in sample preparation, sample collection methodologies, and time-point collection. There are also differences in the lengths of each time course and how many cell cycles were contained in each. HeLa cells have a cell cycle length of ∼14–15 h, so a 48-h time course can contain three complete cell cycles. The cell cycle length of U2OS cells is ∼18–20 h, and so a 48-h time course would be expected to have only ∼2.5 cell cycles.
Bar-Joseph et al. (2008) published the cell cycle–regulated genes in normal foreskin fibroblasts. We compared the U2OS data set to the cell cycle–regulated genes in their experiments (Bar-Joseph et al., 2008). Of the 480 genes identified as cell cycle regulated in foreskin fibroblasts, 228 (47.5%) were also regulated in U2OS cells. Like the U2OS data set, the samples in the foreskin fibroblast data set were collected every 2 h and arrayed on commercially available arrays (Affymetrix U133A 2.0 arrays; Bar-Joseph et al., 2008). Thus there are a number of potential reasons for the amount of overlap between the U2OS and the foreskin fibroblast data sets: cell type, transformed versus normal cells, arrays, and analysis methods.
This compares similarly to the analysis in Bar-Joseph et al. (2008), in which there was ∼40% overlap between the genes identified as regulated in HeLa cells and those cell cycle regulated in foreskin fibroblasts. Reanalysis of the HeLa data set with their analysis method shows that 362 of 481 genes (75.3%) overlapped (Bar-Joseph et al., 2008). Overlap of the genes identified as cell cycle regulated in U2OS, HeLa (Whitfield et al., 2002), and foreskin fibroblasts (Bar-Joseph et al., 2008) shows 142 cell cycle–regulated genes in all three cell types (Supplemental Table S3).
Despite the pairwise overlaps being in the 40% range, there are 142 genes that are cell cycle regulated in all three cell types. Many of these are involved in the core processes of either DNA replication or mitosis, which are tightly regulated and involve a discrete set of genes. These overlapping genes may be “core cell cycle” regulators that are critical for all cell types. Recently Pena-Diaz et al. (2013) reported the cell cycle–regulated genes in HaCaT human keratinocytes. They found that of 1249 Entrez genes cell cycle regulated in this cell line, 125 genes also were cell cycle regulated in HeLa and foreskin fibroblasts. This is similar to the number we report (142 genes) as cell cycle regulated in three different cell types. The inclusion of HaCat cells in our comparison of all cell types reduces the number of common periodic genes to 96 (Supplemental Figure S8 and Supplemental Table S3). These 96 genes represent genes involved in core cell cycle processes.
Many of the cell cycle–regulated genes in all three cell types have either E2F1 or FOXM1 bound at their promoters. This implies that these two transcription factors are responsible for the highly periodic expression patterns seen in either S phase (E2F1) or G2/M (FOXM1). Unfortunately, E2F1 does not bind the promoter of FOXM1, nor does FOXM1 bind the promoter of E2F1, so it appears that E2F1 does not directly induce FOXM1. This would imply that there is not a continuous transcription factor–based circuit that regulates the cell cycle; instead, there is an interplay of transcriptional activation, phosphorylation, degradation, and sequestration (among other methods) that controls cell cycle regulation.
It is becoming apparent that there may be cell type–dependent, cell cycle–regulated genes. As cell cycle–regulated genes are catalogued for different cell lines (or types), it will be interesting to determine which genes are cell line dependent and which genes are cell type dependent, and to determine with more precision which genes are invariant across all cycling cells. These results suggest that it may be informative to determine the differences in cell cycle gene expression between different cell types/lines of the same lineage, embryonic stem cells, or induced pluripotent stem cells to begin to determine the changes through developmental lineages.
Regulation of the cell cycle gene expression program
Previous reports showed that FOXM1 is required for proper progression through mitosis due to a requirement for the FOXM1 activation of critical mitosis genes (e.g., PLK1, CDC25B, and CCNB1; Laoukili et al., 2005; Wonsey and Follettie, 2005). Here, via FOXM1 ChIP-seq, we showed that not only are these genes activated by FOXM1, but in addition a number of other genes that are involved in proper mitotic progression are also activated by FOXM1. The number of genes that are direct FOXM1 targets implies that FOXM1 is a regulator of G2/M-phase transcription. In addition to binding a large number of G2/M-phase genes, FOXM1 binds a large number of targets that are involved with translation, a process that is ongoing throughout the cell cycle. Of interest, FOXM1 is not cell cycle regulated at the RNA level in U2OS cells, whereas it is regulated in HeLa cells. This is possibly due to cell type–specific methods of regulation or perhaps to U2OS cells having a relatively intact p53 pathway whereas HeLa cells do not, due to the presence of HPV E6 and E7 proteins (Gao et al., 2009). There is 43% overlap between the three S-phase clusters in HeLa cells and the S-phase cluster in U2OS cells.
Recently it was shown that FOXM1 binds to G2/M-phase gene promoters via CHR elements (Chen et al., 2013), which are also important to both DREAM and B-Myb-MuvB complex binding (Sadasivam et al., 2012). Similar to the results published here (Figure 3B), Chen et al. (2013) show that FOXM1 binds to many of the same promoters as LIN9 and B-MYB and that both FOXM1 ChIP-seq data sets are enriched for genes involved in mitosis. However, due to experimental differences, the data set presented here is also enriched for genes involved in translation that are not cell cycle regulated. Our data support the possibility that FOXM1 is weakly bound to a subset of genes expressed during S phase (Figures 6, A and B, and 7).
The genes cell cycle regulated in both HeLa and U2OS cells are generally bound by at least one of the cell cycle transcription factors. Of the 253 genes that are cell cycle regulated in U2OS and HeLa cells, 183 (∼72%) of the promoters are bound by at least one transcription factor, 70 (∼28%) of the promoters are bound by two transcription factors, and 63 (∼25%) of the promoters are bound by three or more. Thus each cell cycle gene may be regulated by the combinatorial effects of multiple cell cycle transcriptional factors. This would allow for very precise temporal regulation of cell cycle genes, as well as provide a high degree of redundancy in the system.
These data provide a catalogue of the cell cycle–regulated genes in U2OS cells and, along with associated transcription factor binding data, are available to anyone for any purpose. This will provide a resource for the scientific community, and the full data set is available from Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/query/acc.cgiat) at accession number GSE52100.
MATERIALS AND METHODS
Cell culture, synchronization, and RNA preparation
HeLa and U2OS cells were passaged in a 37°C humidified incubator in DMEM with 10% fetal bovine serum and 100 U of penicillin–streptomycin following standard protocols.
U2OS cells were synchronized using a double-thymidine protocol or a thymidine–nocodazole protocol. Briefly, 3.0 × 105 cells were plated in 16 ml of DMEM. After 24 h of growth, thymidine was added to a final concentration of 2.5 mM. After 18 h in thymidine media cells were washed twice with prewarmed CO2-equilibrated phosphate-buffered saline (PBS) and allowed to grow for 8 h in prewarmed CO2 equilibrated DMEM. Again thymidine was added to a final concentration of 2.5 mM for another 18 h. Cells were washed twice with PBS and released into DMEM. For the thymidine–nocodazole synchronization, U2OS cells were plated (5.0 × 105 cells) and allowed to grow for 24 h. Thymidine (2.5 mM) was added for 18 h before cells were washed twice with prewarmed CO2-equilibrated PBS before treatment with DMEM supplemented with 100 ng/ml nocodazole for 12 h. Floating cells were collected and spun down, washed twice with prewarmed CO2-equilibrated PBS, and resuspended in prewarmed CO2-equilibrated DMEM. Nonfloating cells were washed twice with prewarmed CO2-equilibrated PBS and released into prewarmed CO2-equilibrated DMEM, and the resuspended floating cells were added back to each plate. Cells were collected every 2 h for a minimum of 36 h using RNeasy Plus Mini Kit (Qiagen, Valencia, CA). Zero-hour samples were collected while cells were still in arrest conditions.
Synchrony was monitored via fluorescence-activated cell sorting (FACS) analysis of propidium iodide–labeled cells (DartLab, Geisel School of Medicine at Dartmouth College) and FOXM1 phosphorylation state or cyclin B1 expression via Western blots (see later description). Samples were collected for Western blot analysis using SDS–PAGE sample buffer.
Reference RNA was isolated from asynchronously growing U2OS cells using an RNeasy Plus Mini Kit. The same reference was used for all hybridization experiments.
Microarray hybridization and analysis
Microarrays were run as described previously (Grant et al., 2012). Briefly, cellular RNA was amplified and Cy3 (asynchronous U2OS RNA) or Cy5 (sample) labeled using the Quick-Amp Labeling kit (Agilent Technologies, Santa Clara, CA) following the manufacturer's protocol, except that the reaction volumes were reduced by one-half. Labeled cRNA was hybridized to Agilent Whole Human Genome Oligonucleotide arrays (4 × 44k) following the manufacturer's protocol. Microarrays were scanned using a GenePix 4000B scanner (Molecular Devices, Sunnyvale, CA). Spot pixel intensities were determined using GenePix Pro 5.1 software. Poor-quality spots were identified and flagged by hand and excluded from subsequent analysis. Arrays were stored in the University of North Carolina Microarray Database (Chapel Hill, NC; UMD). The full raw microarray data are available from the GEO at accession number GSE50988 (part of SuperSeries GSE52100).
Each time course was retrieved from the UMD independently from each other time course with the following conditions. Only spots with ratio of intensity over background of >1.5 were used. Genes missing >30% of their data were excluded from further analysis. Genes were normalized using Lowess normalization.
Identification of periodically expressed transcripts
Periodically expressed transcripts were identified using the same method as in Whitfield et al. (2002). Briefly, missing data were imputed using a k-nearest neighbors algorithm (Troyanskaya et al., 2001) using k = 12. Then each time course was centered by removing the first eigengene (Alter et al., 2000). Imputed data were removed from the data set as the last step of the analysis.
Rough estimates of the U2OS cell cycle were initially obtained from Western blot analysis of cell cycle–regulated phosphorylation of FOXM1 and FACS analysis for each time course. This estimate was then refined by performing a Fourier transform on each gene in each time course (Whitfield et al., 2002, Eqs. 1–3) with equally spaced values of time (every 15 min) for the estimated cell cycle length ±two hours.
An offset (φ; Whitfield et al., 2002, Eqs. 1 and 2) was determined for each time course relative to the first time course. The Fourier transform was then repeated for each time course using the following values of T and φ: Thy-Thy 1 (T = 17.65, φ = 0.0), Thy-Thy 2 (T = 18.6, φ = 0.0), Thy-Thy 3 (T = 18, φ = 0.0), and Thy-Noc (T = 23.95, φ = 2.3). The vectors for each data set were then summed and genes ranked by the magnitude of their combined vectors (C). To compensate for the imperfect match to sine or cosine curves, each gene was scaled by its correlation to an idealized vector. The ideal vector for each cell cycle phase (G1/S, S, G2, G2/M, and M/G1) was defined by the average expression profiles of the indicated genes in Figure 1. Using a standard Pearson correlation, each gene received a peak correlation score, which was its largest absolute value correlation with each of the ideal vectors. This peak correlation score was then used to scale each gene's C, generating a periodicity score for each gene.
Randomized data were then used to set a cutoff value for the minimum periodicity score to be considered cell cycle regulated. The data were randomized 10 times either within rows only or in rows and columns. The full analysis pipeline was performed for each of these randomizations using the same parameters as for the unrandomized data. We chose a minimum periodicity score of 2.65, which gave us 3568 genes with an initial false-positive rate of 3.67% when randomizing by rows only. Inclusion of the Thy-Noc time course resulted in improved false-positive and false-negative rates, despite having a lower degree of synchrony than the Thy-Thy time courses (Supplemental Figure S9)
To account for genes that received a high Fourier score but did not have a sinusoidal expression pattern throughout each time course, we calculated autocorrelation scores (Whitfield et al., 2002, Eq. 5). We calculated and summed the autocorrelation scores for each time course, leaving 2878 genes. To remove any genes that had an obvious date bias from technical issues during array hybridization, we found their power spectra using the Fourier transform of each time course. The date-biased genes were then removed by projecting the power spectra onto their first two principal components and clustered by k-means (k = 2; Supplemental Figure S2). Removing these genes gave us a final data set of 2830 probes. The 2830 probes correspond to 2140 Entrez GeneIDs with 1871 unique gene identifiers.
ChIP-seq and analysis
FOXM1 ChIP-seq was carried out as previously described (Lupien et al., 2008; Grant et al., 2012) using the FOXM1 antibody sc-502 (C20; Santa Cruz Biotechnology, Santa Cruz, CA). Briefly, asynchronous HeLa cells were fixed using 1% formaldehyde before sonication to produce DNA fragment lengths of 200–600 base pairs with a Bioruptor (Diagenode, Sparta, NJ). Anti-FOXM1 was bound to a mix of Protein A and Protein G Dynabeads (Life Technologies, Grand Island, NY) before an 18-h incubation at 4°C with the fragmented DNA. Bound DNA was washed and the cross-links reversed before DNA purification with a QIAquick PCR purification kit (Qiagen). DNA concentrations were measured using Quant-iT PicoGreen (Life Technologies). Library construction and sequencing for each ChIP-seq run were carried out independently at the High Throughput Sequencing Facility at the University of North Carolina (Chapel Hill, NC) using an Illumina Genome Analyzer II. Fastq files were mapped to the human genome using Bowtie (version 0.12) using the “best” flag to constrain alignments to those with the best read quality and fewest mismatches. The first ChIP-seq run resulted in 17.1 million sequence reads (8.4 mapped sequence reads), and the second ChIP-seq run resulted in 17.0 million reads (8.4 million mapped reads; human genome build Hg18). Enriched peaks were determined independently for each run using MACS, version 1.3 (run 1, mfold 32, p < 1.0 × 10−5; run 2, mfold 25, p < 1.0 × 10−5; Zhang et al., 2008). This resulted in 5727 peaks for the first run and 2849 peaks for the second. As a conservative estimate of FOXM1 binding, we analyzed the intersection of the sequences under the peaks that were found in both ChIP-seq runs, resulting in 2215 shared FOXM1 genomic loci. We then determined the distribution of the shared FOXM1 genomic loci using the cis-Regulatory Element Annotation System (CEAS; http://liulab.dfci.harvard.edu/CEAS/; Zhang et al., 2008; Shin et al., 2009) implemented in Cistrome (www.cistrome.com/). Raw ChIP-seq data and BED files are available from GEO at accession number GSE52098 (part of SuperSeries GSE52100).
Real-time luciferase assays
U2OS cells were plated at ∼20–25% density in 30-mm dishes and allowed to grow for 24 h. After 24 h, the growth medium was replaced with assay medium (Phenol red–free L15 [Life Technologies], 10% fetal bovine serum, 1% penicillin/streptomycin, 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid buffer, and 0.1 mM luciferin). Cells were then transfected with equal amounts of plasmid (typically 250 ng of each plasmid) using FuGENE 6 (Life Technologies) following the manufacturer's protocol. Tissue culture dishes were sealed with glass coverslips and silicone grease and transferred to the LumiCycle (Actimetrics, Wilmette, IL) at 36°C. Data analysis was performed with LumiCycle Analysis software (Actimetrics).
Western blots
Antibodies to FoxM1 C-20 (1:500) and cyclinB1 H-433 (1:2000) were purchased from Santa Cruz Biotechnology. Anti–glyceraldehyde-3-phosphate dehydrogenase was purchased from American Research Products (Belmont, MA). Western blots were run following standard protocols.
Plasmid construction
FOXM1 expression vectors, the ACAP3/CENTB5, and the RPS6KB1 promoter constructs have been described previously (Grant et al., 2012). We obtained commercially available promoter constructs for PLK1 (S119035), CENPE (S118567), TOP2A (S118760), and RMI1 (S113323) from Switchgear Genomics (Menlo Park, CA).
The FOXM1 target promoter construct, pGL3-MCM8, was cloned based on ChIP-seq loci as determined by MACS. Primers were designed using Primer 3 (Rozen and Skaletsky, 2000) to provide an amplicon length between 800 and 1000 base pairs. DNA fragments were amplified via PCR and cloned into Zero Blunt TOPO (Life Technologies) before being subcloned into pGL3-basic (Promega, Madison, WI) using standard methods. All plasmids were verified by sequencing (Molecular Biology and Proteomics Core Facility, Dartmouth College).
Functional annotation
Functional annotation of genes was performed using DAVID (Dennis et al., 2003; Huang da et al., 2009).
Cell cycle–wide binding profiles
We investigated the distribution of transcription factor target genes in the cell cycle. First, we identified a list of 2830 cell cycle probes in U2OS cells and sorted them according to their peak expression time in the cell cycle. Then we examined the enrichment of the target genes of a given transcription factor in each sliding window of the cell cycle. We used a window size of 30° with 10° overlap between neighboring windows. We used Fisher's exact test to determine the significance of enrichment of target genes for a transcription factor in each cell cycle window.
The target genes for E2F1, E2F4, and E2F6 in HeLa cells were determined from ChIP-seq data generated by the ENCODE project (Gerstein et al., 2012). The FOXM1 target genes were determined from the ChIP-seq presented here.
Supplementary Material
Acknowledgments
This work was supported by the V Foundation for Cancer Research, ACS-IRG 82-003-17, and National Institutes of Health Grants R01 CA130795, R01 HG004499, and R25 CA134286. C.C. is supported by the start-up funding package provided by the Geisel School of Medicine at Dartmouth College.
Abbreviations used:
- ChIP-seq
chromatin immunoprecipitation followed by high-throughput sequencing
- ENCODE
Encyclopedia of DNA Elements
- Thy-noc
thymidine-nocodazole
- Thy-thy
thymidine-thymidine
- SEM
standard error of the mean
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-05-0264) on October 9, 2013.
REFERENCES
- Ahmad A, Ali S, Wang Z, Ali AS, Sethi S, Sakr WA, Raz A, Rahman KM. 3,3’-Diindolylmethane enhances taxotere-induced growth inhibition of breast cancer cells through downregulation of FoxM1. Int J Cancer. 2011;129:1781–1791. doi: 10.1002/ijc.25839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad A, Wang Z, Kong D, Ali S, Li Y, Banerjee S, Ali R, Sarkar FH. FoxM1 down-regulation leads to inhibition of proliferation, migration and invasion of breast cancer cells through the modulation of extra-cellular matrix degrading factors. Breast Cancer Res Treat. 2010;122:337–346. doi: 10.1007/s10549-009-0572-1. [DOI] [PubMed] [Google Scholar]
- Ahmed M, et al. FoxM1 and its association with matrix metalloproteinases (MMP) signaling pathway in papillary thyroid carcinoma. J Clin Endocrinol Metab. 2012;97:E1–E13. doi: 10.1210/jc.2011-1506. [DOI] [PubMed] [Google Scholar]
- Alter O, Brown PO, Botstein D. Singular value decomposition for genome-wide expression data processing and modeling. Proc Natl Acad Sci USA. 2000;97:10101–10106. doi: 10.1073/pnas.97.18.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Fernandez M, Halim VA, Krenning L, Aprelia M, Mohammed S, Heck AJ, Medema RH. Recovery from a DNA-damage-induced G2 arrest requires Cdk-dependent activation of FoxM1. EMBO Rep. 2010;11:452–458. doi: 10.1038/embor.2010.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Fernandez M, Medema RH. Novel functions of FoxM1: from molecular mechanisms to cancer therapy. Front Oncol. 2013;3:30. doi: 10.3389/fonc.2013.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Joseph Z, Siegfried Z, Brandeis M, Brors B, Lu Y, Eils R, Dynlacht BD, Simon I. Genome-wide transcriptional analysis of the human cell cycle identifies genes differentially regulated in normal and cancer cells. Proc Natl Acad Sci USA. 2008;105:955–960. doi: 10.1073/pnas.0704723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergamaschi A, Christensen BL, Katzenellenbogen BS. Reversal of endocrine resistance in breast cancer: interrelationships among 14-3-3zeta, FOXM1, and a gene signature associated with mitosis. Breast Cancer Res. 2011;13:R70. doi: 10.1186/bcr2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonet C, Giuliano S, Ohanna M, Bille K, Allegra M, Lacour JP, Bahadoran P, Rocchi S, Ballotti R, Bertolotto C. Aurora B Is regulated by the mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK) signaling pathway and is a valuable potential target in melanoma cells. J Biol Chem. 2012;287:29887–29898. doi: 10.1074/jbc.M112.371682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvisi DF, et al. Forkhead box M1B is a determinant of rat susceptibility to hepatocarcinogenesis and sustains ERK activity in human HCC. Gut. 2009;58:679–687. doi: 10.1136/gut.2008.152652. [DOI] [PubMed] [Google Scholar]
- Chen CH, Chien CY, Huang CC, Hwang CF, Chuang HC, Fang FM, Huang HY, Chen CM, Liu HL, Huang CY. Expression of FLJ10540 is correlated with aggressiveness of oral cavity squamous cell carcinoma by stimulating cell migration and invasion through increased FOXM1 and MMP-2 activity. Oncogene. 2009;28:2723–2737. doi: 10.1038/onc.2009.128. [DOI] [PubMed] [Google Scholar]
- Chen YJ, Dominguez-Brauer C, Wang Z, Asara JM, Costa RH, Tyner AL, Lau LF, Raychaudhuri P. A conserved phosphorylation site within the forkhead domain of FoxM1B is required for its activation by cyclin-CDK1. J Biol Chem. 2009;284:30695–30707. doi: 10.1074/jbc.M109.007997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Muller GA, Quaas M, Fischer M, Han N, Stutchbury B, Sharrocks AD, Engeland K. The forkhead transcription factor FOXM1 controls cell cycle-dependent gene expression through an atypical chromatin binding mechanism. Mol Cell Biol. 2013;33:227–236. doi: 10.1128/MCB.00881-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Yuan K, Tao ZZ, Xiao BK. Deletion of forkhead Box M1 transcription factor reduces malignancy in laryngeal squamous carcinoma cells. Asian Pac J Cancer Prev. 2011;12:1785–1788. [PubMed] [Google Scholar]
- Chetty C, Bhoopathi P, Rao JS, Lakka SS. Inhibition of matrix metalloproteinase-2 enhances radiosensitivity by abrogating radiation-induced FoxM1-mediated G2/M arrest in A549 lung cancer cells. Int J Cancer. 2009;124:2468–2477. doi: 10.1002/ijc.24209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho RJ, et al. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol Cell. 1998;2:65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- Cho RJ, Huang M, Campbell MJ, Dong H, Steinmetz L, Sapinoso L, Hampton G, Elledge SJ, Davis RW, Lockhart DJ. Transcriptional regulation and function during the human cell cycle. Nat Genet. 2001;27:48–54. doi: 10.1038/83751. [DOI] [PubMed] [Google Scholar]
- Crawford DF, Piwnica-Worms H. The G(2) DNA damage checkpoint delays expression of genes encoding mitotic regulators. J Biol Chem. 2001;276:37166–37177. doi: 10.1074/jbc.M103414200. [DOI] [PubMed] [Google Scholar]
- Dai B, Kang SH, Gong W, Liu M, Aldape KD, Sawaya R, Huang S. Aberrant FoxM1B expression increases matrix metalloproteinase-2 transcription and enhances the invasion of glioma cells. Oncogene. 2007;26:6212–6219. doi: 10.1038/sj.onc.1210443. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Doong H, Price J, Kim YS, Gasbarre C, Probst J, Liotta LA, Blanchette J, Rizzo K, Kohn E. CAIR-1/BAG-3 forms an EGF-regulated ternary complex with phospholipase C-gamma and Hsp70/Hsc70. Oncogene. 2000;19:4385–4395. doi: 10.1038/sj.onc.1203797. [DOI] [PubMed] [Google Scholar]
- Down CF, Millour J, Lam EW, Watson RJ. Binding of FoxM1 to G2/M gene promoters is dependent upon B-Myb. Biochim Biophys Acta. 2012;1819:855–862. doi: 10.1016/j.bbagrm.2012.03.008. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton TR, Gout IT. Functions and regulation of the 70 kDa ribosomal S6 kinases. Int J Biochem Cell Biol. 2011;43:47–59. doi: 10.1016/j.biocel.2010.09.018. [DOI] [PubMed] [Google Scholar]
- Gao D, Inuzuka H, Korenjak M, Tseng A, Wu T, Wan L, Kirschner M, Dyson N, Wei W. Cdh1 regulates cell cycle through modulating the claspin/Chk1 and the Rb/E2F1 pathways. Mol Biol Cell. 2009;20:3305–3316. doi: 10.1091/mbc.E09-01-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemenetzidis E, et al. FOXM1 upregulation is an early event in human squamous cell carcinoma and it is enhanced by nicotine during malignant transformation. PLoS One. 2009;4:e4849. doi: 10.1371/journal.pone.0004849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant GD, Gamsby J, Martyanov V, Brooks L, 3rd, George LK, Mahoney JM, Loros JJ, Dunlap JC, Whitfield ML. Live-cell monitoring of periodic gene expression in synchronous human cells identifies forkhead genes involved in cell cycle control. Mol Biol Cell. 2012;23:3079–3093. doi: 10.1091/mbc.E11-02-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helin K. Regulation of cell proliferation by the E2F transcription factors. Curr Opin Genet Dev. 1998;8:28–35. doi: 10.1016/s0959-437x(98)80058-0. [DOI] [PubMed] [Google Scholar]
- Huang C, Qiu Z, Wang L, Peng Z, Jia Z, Logsdon CD, Le X, Wei D, Huang S, Xie K. A novel FoxM1-caveolin signaling pathway promotes pancreatic cancer invasion and metastasis. Cancer Res. 2012;72:655–665. doi: 10.1158/0008-5472.CAN-11-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hui MK, Chan KW, Luk JM, Lee NP, Chung Y, Cheung LC, Srivastava G, Tsao SW, Tang JC, Law S. Cytoplasmic forkhead box M1 (FoxM1) in esophageal squamous cell carcinoma significantly correlates with pathological disease stage. World J Surg. 2012;36:90–97. doi: 10.1007/s00268-011-1302-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida S, Huang E, Zuzan H, Spang R, Leone G, West M, Nevins JR. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol Cell Biol. 2001;21:4684–4699. doi: 10.1128/MCB.21.14.4684-4699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer VR, et al. The transcriptional program in the response of human fibroblasts to serum. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- Johnson DG, Ohtani K, Nevins JR. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 1994;8:1514–1525. doi: 10.1101/gad.8.13.1514. [DOI] [PubMed] [Google Scholar]
- Kalinichenko VV, Gusarova GA, Tan Y, Wang IC, Major ML, Wang X, Yoder HM, Costa RH. Ubiquitous expression of the forkhead box M1B transgene accelerates proliferation of distinct pulmonary cell types following lung injury. J Biol Chem. 2003;278:37888–37894. doi: 10.1074/jbc.M305555200. [DOI] [PubMed] [Google Scholar]
- Kim IM, Ramakrishna S, Gusarova GA, Yoder HM, Costa RH, Kalinichenko VV. The forkhead box m1 transcription factor is essential for embryonic development of pulmonary vasculature. J Biol Chem. 2005;280:22278–22286. doi: 10.1074/jbc.M500936200. [DOI] [PubMed] [Google Scholar]
- Kittler R, et al. Genome-scale RNAi profiling of cell division in human tissue culture cells. Nat Cell Biol. 2007;9:1401–1412. doi: 10.1038/ncb1659. [DOI] [PubMed] [Google Scholar]
- Kwok JM, Peck B, Monteiro LJ, Schwenen HD, Millour J, Coombes RC, Myatt SS, Lam EW. FOXM1 confers acquired cisplatin resistance in breast cancer cells. Mol Cancer Res. 2010;8:24–34. doi: 10.1158/1541-7786.MCR-09-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laoukili J, Alvarez M, Meijer LA, Stahl M, Mohammed S, Kleij L, Heck AJ, Medema RH. Activation of FoxM1 during G2 requires cyclin A/Cdk-dependent relief of autorepression by the FoxM1 N-terminal domain. Mol Cell Biol. 2008a;28:3076–3087. doi: 10.1128/MCB.01710-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laoukili J, Alvarez-Fernandez M, Stahl M, Medema RH. FoxM1 is degraded at mitotic exit in a Cdh1-dependent manner. Cell Cycle. 2008b;7:2720–2726. doi: 10.4161/cc.7.17.6580. [DOI] [PubMed] [Google Scholar]
- Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A, Clevers H, Medema RH. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7:126–136. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- Laub MT, McAdams HH, Feldblyum T, Fraser CM, Shapiro L. Global analysis of the genetic network controlling a bacterial cell cycle. Science. 2000;290:2144–2148. doi: 10.1126/science.290.5499.2144. [DOI] [PubMed] [Google Scholar]
- Litovchick L, Sadasivam S, Florens L, Zhu X, Swanson SK, Velmurugan S, Chen R, Washburn MP, Liu XS, DeCaprio JA. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol Cell. 2007;26:539–551. doi: 10.1016/j.molcel.2007.04.015. [DOI] [PubMed] [Google Scholar]
- Liu M, et al. FoxM1B is overexpressed in human glioblastomas and critically regulates the tumorigenicity of glioma cells. Cancer Res. 2006;66:3593–3602. doi: 10.1158/0008-5472.CAN-05-2912. [DOI] [PubMed] [Google Scholar]
- Liu T, et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 2011;12:R83. doi: 10.1186/gb-2011-12-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, et al. MicroRNA-135a contributes to the development of portal vein tumor thrombus by promoting metastasis in hepatocellular carcinoma. J Hepatol. 2012;56:389–396. doi: 10.1016/j.jhep.2011.08.008. [DOI] [PubMed] [Google Scholar]
- Lok GT, Chan DW, Liu VW, Hui WW, Leung TH, Yao KM, Ngan HY. Aberrant activation of ERK/FOXM1 signaling cascade triggers the cell migration/invasion in ovarian cancer cells. PLoS One. 2011;6:e23790. doi: 10.1371/journal.pone.0023790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch TP, Ferrer CM, Jackson SR, Shahriari KS, Vosseller K, Reginato MJ. Critical role of O-Linked beta-N-acetylglucosamine transferase in prostate cancer invasion, angiogenesis, and metastasis. J Biol Chem. 2012;287:11070–11081. doi: 10.1074/jbc.M111.302547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madureira PA, Varshochi R, Constantinidou D, Francis RE, Coombes RC, Yao KM, Lam EW. The forkhead box M1 protein regulates the transcription of the estrogen receptor alpha in breast cancer cells. J Biol Chem. 2006;281:25167–25176. doi: 10.1074/jbc.M603906200. [DOI] [PubMed] [Google Scholar]
- Major ML, Lepe R, Costa RH. Forkhead box M1B transcriptional activity requires binding of Cdk-cyclin complexes for phosphorylation-dependent recruitment of p300/CBP coactivators. Mol Cell Biol. 2004;24:2649–2661. doi: 10.1128/MCB.24.7.2649-2661.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M, de Jager SM, Gruissem W, Murray JA. Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J. 2005;41:546–566. doi: 10.1111/j.1365-313X.2004.02319.x. [DOI] [PubMed] [Google Scholar]
- Menges M, Hennig L, Gruissem W, Murray JA. Cell cycle-regulated gene expression in Arabidopsis. J Biol Chem. 2002;277:41987–42002. doi: 10.1074/jbc.M207570200. [DOI] [PubMed] [Google Scholar]
- Mukherji M, et al. Genome-wide functional analysis of human cell-cycle regulators. Proc Natl Acad Sci USA. 2006;103:14819–14824. doi: 10.1073/pnas.0604320103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Hirano I, Okinaka K, Takemura T, Yokota D, Ono T, Shigeno K, Shibata K, Fujisawa S, Ohnishi K. The FOXM1 transcriptional factor promotes the proliferation of leukemia cells through modulation of cell cycle progression in acute myeloid leukemia. Carcinogenesis. 2010;31:2012–2021. doi: 10.1093/carcin/bgq185. [DOI] [PubMed] [Google Scholar]
- Oliva A, Rosebrock A, Ferrezuelo F, Pyne S, Chen H, Skiena S, Futcher B, Leatherwood J. The cell cycle-regulated genes of Schizosaccharomyces pombe. PLoS Biol. 2005;3:e225. doi: 10.1371/journal.pbio.0030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YY, et al. FOXM1 mediates Dox resistance in breast cancer by enhancing DNA repair. Carcinogenesis. 2012;33:1843–1853. doi: 10.1093/carcin/bgs167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HJ, Costa RH, Lau LF, Tyner AL, Raychaudhuri P. Anaphase-promoting complex/cyclosome-CDH1-mediated proteolysis of the forkhead box M1 transcription factor is critical for regulated entry into S phase. Mol Cell Biol. 2008;28:5162–5171. doi: 10.1128/MCB.00387-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena-Diaz J, Hegre SA, Anderssen E, Aas PA, Mjelle R, Gilfillan GD, Lyle R, Drablos F, Krokan HE, Saetrom P. Transcription profiling during the cell cycle shows that a subset of Polycomb-targeted genes is upregulated during DNA replication. Nucleic Acids Res. 2013;41:2846–2856. doi: 10.1093/nar/gks1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, et al. Identification of cell cycle-regulated genes in fission yeast. Mol Biol Cell. 2005;16:1026–1042. doi: 10.1091/mbc.E04-04-0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponten J, Saksela E. Two established in vitro cell lines from human mesenchymal tumours. Int J Cancer. 1967;2:434–447. doi: 10.1002/ijc.2910020505. [DOI] [PubMed] [Google Scholar]
- Romano MF, et al. BAG3 protein controls B-chronic lymphocytic leukaemia cell apoptosis. Cell Death Differ. 2003a;10:383–385. doi: 10.1038/sj.cdd.4401167. [DOI] [PubMed] [Google Scholar]
- Romano MF, et al. BAG3 protein regulates cell survival in childhood acute lymphoblastic leukemia cells. Cancer Biol Ther. 2003b;2:508–510. doi: 10.4161/cbt.2.5.524. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Rustici G, Mata J, Kivinen K, Lio P, Penkett CJ, Burns G, Hayles J, Brazma A, Nurse P, Bahler J. Periodic gene expression program of the fission yeast cell cycle. Nat Genet. 2004;36:809–817. doi: 10.1038/ng1377. [DOI] [PubMed] [Google Scholar]
- Sadasivam S, Duan S, DeCaprio JA. The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes Dev. 2012;26:474–489. doi: 10.1101/gad.181933.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Liu T, Manrai AK, Liu XS. CEAS: cis-Regulatory Element Annotation System. Bioinformatics. 2009;25:2605–2606. doi: 10.1093/bioinformatics/btp479. [DOI] [PubMed] [Google Scholar]
- Sinclair CS, Rowley M, Naderi A, Couch FJ. The 17q23 amplicon and breast cancer. Breast Cancer Res Treat. 2003;78:313–322. doi: 10.1023/a:1023081624133. [DOI] [PubMed] [Google Scholar]
- Slansky JE, Farnham PJ. Introduction to the E2F family: protein structure and gene regulation. Curr Top Microbiol Immunol. 1996;208:1–30. doi: 10.1007/978-3-642-79910-5_1. [DOI] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S, Xie Z, Reed JC. An evolutionarily conserved family of Hsp70/Hsc70 molecular chaperone regulators. J Biol Chem. 1999;274:781–786. doi: 10.1074/jbc.274.2.781. [DOI] [PubMed] [Google Scholar]
- Teh MT, et al. Exploiting FOXM1-orchestrated molecular network for early squamous cell carcinoma diagnosis and prognosis. Int J Cancer 132, 2095–210. 2013 doi: 10.1002/ijc.27886. [DOI] [PubMed] [Google Scholar]
- Troyanskaya O, Cantor M, Sherlock G, Brown P, Hastie T, Tibshirani R, Botstein D, Altman RB. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17:520–525. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- Wan X, Yeung C, Kim SY, Dolan JG, Ngo VN, Burkett S, Khan J, Staudt LM, Helman LJ. Identification of FoxM1/Bub1b signaling pathway as a required component for growth and survival of rhabdomyosarcoma. Cancer Res. 2012;72:5889–5899. doi: 10.1158/0008-5472.CAN-12-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Banerjee S, Kong D, Li Y, Sarkar FH. Down-regulation of forkhead box M1 transcription factor leads to the inhibition of invasion and angiogenesis of pancreatic cancer cells. Cancer Res. 2007;67:8293–8300. doi: 10.1158/0008-5472.CAN-07-1265. [DOI] [PubMed] [Google Scholar]
- Wang IC, Chen YJ, Hughes DE, Ackerson T, Major ML, Kalinichenko VV, Costa RH, Raychaudhuri P, Tyner AL, Lau LF. FoxM1 regulates transcription of JNK1 to promote the G1/S transition and tumor cell invasiveness. J Biol Chem. 2008;283:20770–20778. doi: 10.1074/jbc.M709892200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Gartel AL. The suppression of FOXM1 and its targets in breast cancer xenograft tumors by siRNA. Oncotarget. 2011;2:1218–1226. doi: 10.18632/oncotarget.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZF, Whitfield ML, Ingledue TC, 3rd, Dominski Z, Marzluff WF. The protein that binds the 3’ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 1996;10:3028–3040. doi: 10.1101/gad.10.23.3028. [DOI] [PubMed] [Google Scholar]
- Waseem A, Ali M, Odell EW, Fortune F, Teh MT. Downstream targets of FOXM1: CEP55 and HELLS are cancer progression markers of head and neck squamous cell carcinoma. Oral Oncol. 2010;46:536–542. doi: 10.1016/j.oraloncology.2010.03.022. [DOI] [PubMed] [Google Scholar]
- Whitfield ML, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield ML, Zheng LX, Baldwin A, Ohta T, Hurt MM, Marzluff WF. Stem-loop binding protein, the protein that binds the 3’ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol Cell Biol. 2000;20:4188–4198. doi: 10.1128/mcb.20.12.4188-4198.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierstra I, Alves J. FOXM1c is activated by cyclin E/Cdk2, cyclin A/Cdk2, and cyclin A/Cdk1, but repressed by GSK-3alpha. Biochem Biophys Res Commun. 2006a;348:99–108. doi: 10.1016/j.bbrc.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Wierstra I, Alves J. Transcription factor FOXM1c is repressed by RB and activated by cyclin D1/Cdk4. Biol Chem. 2006b;387:949–962. doi: 10.1515/BC.2006.119. [DOI] [PubMed] [Google Scholar]
- Wierstra I, Alves J. FOXM1, a typical proliferation-associated transcription factor. Biol Chem. 2007;388:1257–1274. doi: 10.1515/BC.2007.159. [DOI] [PubMed] [Google Scholar]
- Wierstra I, Alves J. Cyclin E/Cdk2, P/CAF, and E1A regulate the transactivation of the c-myc promoter by FOXM1. Biochem Biophys Res Commun. 2008;368:107–115. doi: 10.1016/j.bbrc.2008.01.039. [DOI] [PubMed] [Google Scholar]
- Wonsey DR, Follettie MT. Loss of the forkhead transcription factor FoxM1 causes centrosome amplification and mitotic catastrophe. Cancer Res. 2005;65:5181–5189. doi: 10.1158/0008-5472.CAN-04-4059. [DOI] [PubMed] [Google Scholar]
- Xia L, Huang W, Tian D, Zhu H, Zhang Y, Hu H, Fan D, Nie Y, Wu K. Upregulated FoxM1 expression induced by hepatitis B virus X protein promotes tumor metastasis and indicates poor prognosis in hepatitis B virus-related hepatocellular carcinoma. J Hepatol. 2012;57:600–612. doi: 10.1016/j.jhep.2012.04.020. [DOI] [PubMed] [Google Scholar]
- Xue YJ, et al. Overexpression of FoxM1 is associated with tumor progression in patients with clear cell renal cell carcinoma. J Transl Med. 2012;10:200. doi: 10.1186/1479-5876-10-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamnik RL, Digilova A, Davis DC, Brodt ZN, Murphy CJ, Holz MK. S6 kinase 1 regulates estrogen receptor alpha in control of breast cancer cell proliferation. J Biol Chem. 2009;284:6361–6369. doi: 10.1074/jbc.M807532200. [DOI] [PubMed] [Google Scholar]
- Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.