Cortical NuMA is essential for regulating spindle orientation and recruiting dynein/dynactin to the cell cortex. NuMA stability and spindle orientation activity require interactions with 4.1 family proteins in metaphase. In anaphase, loss of NuMA phosphorylation results in its cortical association in an LGN- and 4.1-independent manner.
Abstract
The epidermis is a multilayered epithelium that requires asymmetric divisions for stratification. A conserved cortical protein complex, including LGN, nuclear mitotic apparatus (NuMA), and dynein/dynactin, plays a key role in establishing proper spindle orientation during asymmetric divisions. The requirements for the cortical recruitment of these proteins, however, remain unclear. In this work, we show that NuMA is required to recruit dynactin to the cell cortex of keratinocytes. NuMA's cortical recruitment requires LGN; however, LGN interactions are not sufficient for this localization. Using fluorescence recovery after photobleaching, we find that the 4.1-binding domain of NuMA is important for stabilizing its interaction with the cell cortex. This is functionally important, as loss of 4.1/NuMA interaction results in spindle orientation defects, using two distinct assays. Furthermore, we observe an increase in cortical NuMA localization as cells enter anaphase. Inhibition of Cdk1 or mutation of a single residue in NuMA mimics this effect. NuMA's anaphase localization is independent of LGN and 4.1 interactions, revealing two distinct mechanisms responsible for NuMA cortical recruitment at different stages of mitosis. This work highlights the complexity of NuMA localization and reveals the importance of NuMA cortical stability for productive force generation during spindle orientation.
INTRODUCTION
Robust regulation of spindle orientation is essential for driving asymmetric cell divisions and plays a critical role during many morphogenetic processes throughout tissue development and homeostasis (Poulson and Lechler, 2012). During the development of the mammalian epidermis, mitotic spindle orientation in the proliferative basal cells is crucial not only for dictating daughter cell fate, but also for initiating stratification of the entire tissue (Smart, 1970; Lechler and Fuchs, 2005). During symmetric divisions that serve to increase the surface area of the epidermis, spindles align parallel to the underlying basement membrane and generate two identical daughter cells, which both inherit progenitor fates. In asymmetric divisions, however, the mitotic spindle orients perpendicular to the basement membrane, so that one daughter is displaced into a new cell layer, where it will ultimately undergo terminal differentiation.
Progenitor cells in the mammalian epidermis use evolutionarily conserved cortical machinery to orient their mitotic spindles during asymmetric cell divisions (Lechler and Fuchs, 2005; Poulson and Lechler, 2010; Williams et al., 2011). This cortical machinery includes the polarity protein Par3, as well as mInscuteable, LGN, nuclear mitotic apparatus (NuMA), and dynein/dynactin. Loss of any one of these proteins, both in invertebrate models and in the mammalian epidermis, leads to defects in spindle orientation (Kraut et al., 1996; Schober et al., 1999; Bowman et al., 2006; Siller et al., 2006; Williams et al., 2011). Nevertheless, it remains unclear how these proteins are recruited to the cell cortex and whether their interactions with additional cortical proteins are required for spindle orientation activity. Previous work in Caenorhabditis elegans highlighted the importance of dynein/dynactin recruitment to the cell cortex, where this complex is believed to generate directional forces on astral microtubules to facilitate spindle rotation or displacement (Lu and Johnston, 2013; McNally, 2013). The asymmetry in forces has been postulated, in different cell types, to be due to either asymmetric localization of dynein/dynactin or asymmetric activation. In this study, we investigate the mechanism underlying spindle orientation establishment in keratinocytes isolated from mouse epidermis, which serve as a powerful culture model for studying this process in mammalian cells (Lechler and Fuchs, 2005).
Although keratinocytes show a clear polarization of dynein and dynactin to the cell cortex (Lechler and Fuchs, 2005), the precise mechanism underlying their cortical recruitment is a matter of debate. A previous study performed using Madin–Darby canine kidney cells proposed that LGN can directly recruit dynein/dynactin through interactions with the dynein heavy chain (Zheng et al., 2013). In other cases, however, it appears that NuMA is required for this localization through interactions between dynein/dynactin and the amino terminus of NuMA (Kotak et al., 2012).
In addition, the requirements for cortical localization of NuMA remain unclear. NuMA is a 230-kDa coiled-coil protein that harbors binding domains for several proteins within its carboxy terminus, including LGN. Although many studies using a variety of cell types have demonstrated a genetic requirement for LGN in cortical NuMA localization, there are certain cell types that require additional proteins for cortical NuMA recruitment (Bowman et al., 2006; Izumi et al., 2006; Siller et al., 2006; Williams et al., 2011). For example, localization of the NuMA homologue Mud in Drosophila neuroblasts requires not only LGN, but also Ran1 and Canoe (Speicher et al., 2008; Wee et al., 2011). The requirements for NuMA cortical localization in both the epidermis and cultured keratinocytes have not been addressed.
Although efficient cortical recruitment of NuMA, dynein, and dynactin is essential for spindle orientation, another critical aspect of this process is the successful tethering of these components to the cortex to facilitate their productive force generation on astral microtubules. There is evidence in C. elegans that without tethering to the F-actin–rich cortex, force generators are pulled into the cell on membrane invaginations rather than directing force on the mitotic spindle (Redemann et al., 2010). Nonetheless, no direct interactions or adaptors have been identified in the epidermis that link the spindle orientation machinery to the cell cortex.
In this work, we demonstrate that NuMA is required for dynein/dynactin localization to the keratinocyte cell cortex. NuMA's localization in turn depends on LGN, yet NuMA also requires members of the F-actin–binding 4.1 family of proteins for stabilization at the membrane and robust execution of spindle orientation. In addition, we observe an increase and expansion of cortical NuMA association in anaphase. This localization can be mimicked by pharmacological Cdk1 inhibition or mutation of a Cdk1 phosphorylation site in the carboxy terminus of NuMA. Unlike the cortical localization of NuMA in metaphase, this anaphase localization is independent of both LGN and 4.1. This study thus reveals that a collaboration between multiple distinct pathways is important for regulating NuMA's cortical targeting and stability throughout mitosis.
RESULTS
NuMA is required for cortical dynein/dynactin recruitment
Studies in different cell types suggest disparate mechanisms for recruitment of dynein/dynactin to the cell cortex during spindle orientation. Although NuMA is required for dynein/dynactin localization in symmetrically dividing HeLa cells (Kotak et al., 2012), recent reports suggest that LGN may also recruit dynein through interactions with dynein heavy chain (Kotak et al., 2012; Zheng et al., 2013). Complicating this is the fact that LGN is required for NuMA localization (Bowman et al., 2006; Izumi et al., 2006; Siller et al., 2006; Williams et al., 2011). Therefore, before we pursued the mechanism of NuMA localization in keratinocytes, we first sought to determine whether NuMA was, in fact, required for dynein/dynactin localization in these cells.
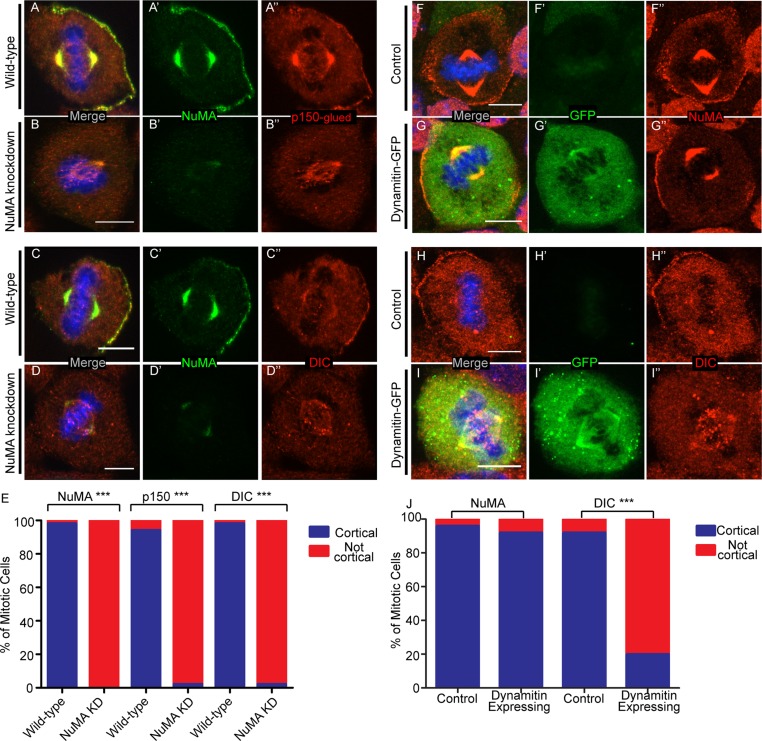
We began by using previously characterized NuMA shRNA constructs to knock down NuMA expression in cultured keratinocytes (Williams et al., 2011). We found significant loss of NuMA protein levels in NuMA shRNA knockdown samples when compared with control samples (Supplemental Figure S1A and Figure 1, A′–D′). We then examined the localization of p150glued (a dynactin component) and dynein intermediate chain (DIC), the dynein subunit that directly interacts with p150glued (Vaughan and Vallee, 1995). To quantitate the effect of NuMA loss on cortical localization of these components, we generated intensity line scans beginning at the cortex and leading into the cytoplasm to reveal variations in cortical and cytoplasmic fluorescence levels. Supplemental Figure S2 illustrates examples of these data plotted for individual cells and their corresponding average values. These data clearly demonstrate both the alterations in cortical protein recruitment and the extent of cell-to-cell variability. We found that the presence or absence of cortical localization was easily distinguishable, with “cortical” representing at least a 1.5-fold difference between cortical and cytoplasmic signal. Considering the ease of distinguishing “cortical” versus “not cortical” localization, we report our data throughout the remainder of this article by categorizing cells into one of these two groups. In addition, for a number of experiments we have included the intensity scans. Although both p150glued and DIC colocalized with NuMA at the cell cortex in wild-type cells, we did not detect cortical accumulation in NuMA-knockdown cells (Figure 1, A–E, and Supplemental Figure S2B). Although this does not rule out a complementary direct role for LGN in dynein/dynactin recruitment, these data indicate that NuMA is necessary for their localization in keratinocytes.
FIGURE 1:
NuMA recruits dynein/dynactin to the cell cortex of keratinocytes. (A–D) Immunofluorescence analysis of endogenous NuMA, p150glued, and DIC localization in wild-type and NuMA-knockdown mouse keratinocytes, as indicated. (E) Quantitation of cells with cortical NuMA, p150-glued, and DIC localization. n = 50 cells for each, p < 0.0001 for each. (F–I) Immunofluorescence analysis of endogenous NuMA and DIC localization in untransfected and dynamitin-GFP–transfected cells. (J) Quantitation of cells with cortical NuMA and DIC localization. n = 25 cells for each, p = 1 for NuMA, p < 0.0001 for DIC. Scale bars, 10 μm.
To determine whether NuMA specifically recruited dynactin and/or dynein, we disrupted the dynactin complex by overexpressing one subunit, p50 dynamitin. We found that most p50 dynamitin–transfected keratinocytes showed a loss of cortical p150glued localization when compared with control cells, thus confirming this protein's effect on dynactin localization (Burkhardt et al., 1997; Supplemental Figure S1, C–E). Furthermore, p50-dynamitin–transfected cells showed a significant defect in the recruitment of cortical DIC when compared with controls (Figure 1, H–J, and Supplemental Figure S2C), despite the fact that cortical NuMA was readily observed (Figure 1, F, G, and J). Together, these data demonstrate that NuMA cortical localization is independent of dynactin localization and that dynein cortical localization is dependent upon dynactin. Having established that NuMA is required for recruiting dynein/dynactin to the cell cortex, we proceeded to investigate how NuMA localization and stability are controlled.
NuMA cortical and spindle pole localizations are regulated by distinct cytoskeletal components
To begin to address how NuMA is targeted to the cell cortex in keratinocytes, we examined the cytoskeletal requirements for proper cortical targeting. In control cells (dimethyl sulfoxide treated), NuMA localized to spindle poles and to the cell cortex, as previously reported (Lechler and Fuchs, 2005; Supplemental Figure S3, A and E). Whereas NuMA localization to spindle poles was lost after treatment with the microtubule-depolymerizing drug nocodazole, NuMA cortical localization remained unperturbed (Supplemental Figure S3, B, D, and F). Consistent with a microtubule-independent recruitment, both full-length NuMA tagged with green fluorescent protein (GFP) and a NuMA deletion construct lacking the microtubule-binding domain localized to the cell cortex when transfected into keratinocytes (Supplemental Figure S3, I–K).
We next examined whether the F-actin cytoskeleton is required for NuMA localization by treating keratinocytes with the actin-depolymerizing drug latrunculin A. Although this treatment did not affect NuMA spindle pole localization, it resulted in a clear loss of its cortical localization (Supplemental Figure S3, C, G, and H). Taken together, these data suggest that the two prominent sites of NuMA localization are regulated by distinct cytoskeletal elements: spindle pole localization requires microtubules, whereas cortical localization requires F-actin.
To further understand the role that F-actin plays in cortical NuMA recruitment, we examined NuMA localization in cells treated with the myosin II inhibitor blebbistatin (Straight et al., 2003). There was no defect in either spindle pole or cortical localization of NuMA after this treatment (Supplemental Figure S3, L–N), indicating that NuMA localization is independent of actomyosin contractility and likely requires F-actin exclusively for structural stability. Similar to NuMA localization, cortical p150glued and DIC localizations were maintained upon treatment with nocodazole (Supplemental Figure S3, B, D, and F) or blebbistatin (Supplemental Figure S3, M–N) but were lost upon treatment with latrunculin A (Supplemental Figure S3, C and G).
LGN is necessary but not sufficient for cortical NuMA localization
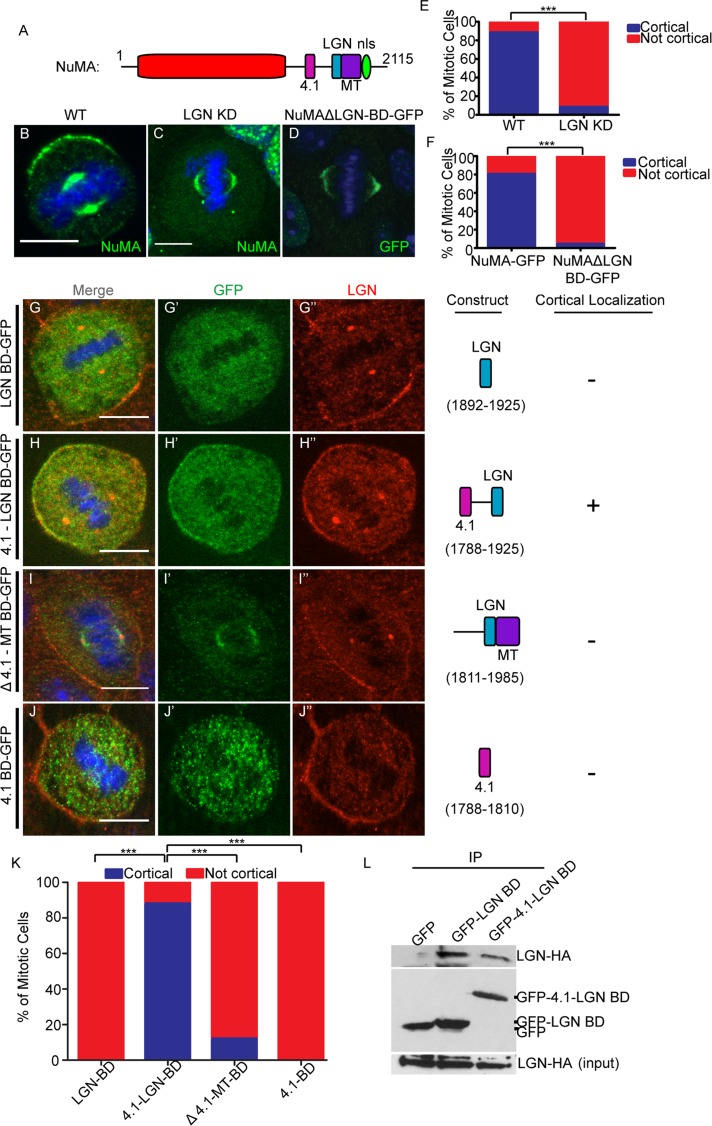
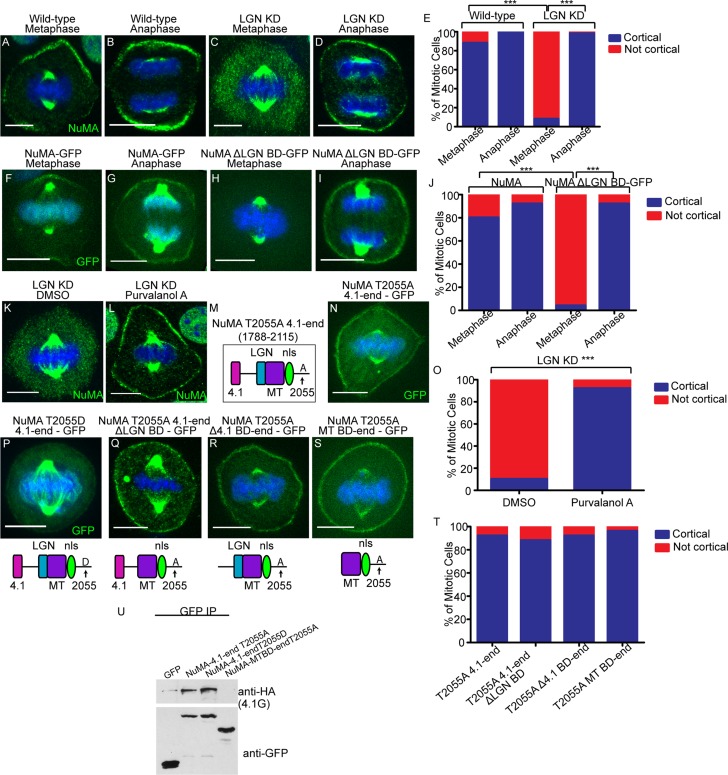
In addition to the cytoskeleton, other protein–protein interactions are required for cortical NuMA localization. NuMA has a number of motifs that mediate direct interactions with other proteins (Figure 2A). The best-characterized of these is LGN, which is essential for NuMA localization in a wide array of cell types, including keratinocytes (Bowman et al., 2006; Izumi et al., 2006; Siller et al., 2006; Williams et al., 2011). We first verified these findings using a previously published LGN short hairpin RNA (shRNA)–knockdown approach (Williams et al., 2011). LGN levels were greatly reduced in knockdown cells as compared with controls (Supplemental Figure S1B). In keratinocytes, NuMA was clearly and specifically lost at the cell cortex upon LGN knockdown; however, its spindle pole localization was not affected (Figure 2, C and E). Consistent with this result, a NuMA mutant lacking the LGN-binding domain (LGN BD) was also unable to localize to the cell cortex, despite its successful recruitment to spindle poles (Figure 2, D and F).
FIGURE 2:
LGN binding is necessary but not sufficient for cortical NuMA recruitment, which may require association with 4.1. (A) The characterized binding regions within the NuMA protein. (B, C) Immunofluorescence analysis of endogenous NuMA localization in wild-type (WT) and LGN-knockdown keratinocytes. (D) Localization of GFP-tagged NuMA lacking the LGN-binding domain (ΔLGN BD-GFP) in wild-type cells. (E) Quantitation of NuMA cortical localization in WT and LGN-knockdown cells. n = 50 cells, p < 0.001. (F) Quantitation of cortical NuMA-GFP and NuMAΔLGN-BD-GFP localization. n = 25 cells, p < 0.0001. (G–J) Various truncation constructs of NuMA (see Construct column) tagged to GFP were transfected into wild-type cells. The amino acids spanned in each construct are specified in the Construct column. Cells were stained for endogenous LGN, and subsequent immunofluorescence analysis was performed to compare localization of these constructs with respect to cortical LGN. The Cortical column indicates whether cortical localization was detected for each construct (+, presence in; –, absence from cortex). (K) Quantitation of cortical localization of NuMA deletion constructs, as indicated. n = 25 cells for each, p < 0.0001 when comparing the 4.1-LGN BD to either the LGN BD or Δ4.1-MT BD. Scale bars, 10 μm. (L) Immunoprecipitation of GFP-tagged LGN-BD– and 4.1-LGN BD–transfected keratinocytes. Lysates were probed with anti-HA antibodies to detect associated LGN-HA. Middle blot, amounts of GFP fusion proteins in the immunoprecipitates; bottom, levels of LGN-HA in the lysates.
To determine whether the LGN BD of NuMA was sufficient for cortical localization, we expressed a GFP-tagged fragment of NuMA containing the entire LGN BD, as previously determined biochemically (Du et al., 2001, 2002). We first confirmed that this fragment was able to interact with LGN when transfected into keratinocytes (Figure 2L). This fragment, however, was unable to localize to the cell cortex (Figure 2, G and K, and Supplemental Figure S2D). Owing to this unexpected result, we generated a larger fragment of NuMA that contained the 4.1-binding domain (4.1 BD), which is amino terminal to the LGN BD (Du et al., 2002; Krauss et al., 2004). Whereas the addition of the 4.1 BD did not significantly increase the amount of LGN binding (Figure 2L), it was sufficient to rescue cortical NuMA localization (Figure 2, H and K, and Supplemental Figure S2D). In addition, we generated a construct that contained all sequences carboxy terminal to the 4.1 BD, and this fragment did not localize to the cell cortex (Figure 2, I and K). Similarly, the 4.1 BD alone was unable to localize to the cell cortex (Figure 2, J and K). Therefore, in the context of these short carboxy-terminal–domain fragments of NuMA (which do not oligomerize as the full-length protein does), LGN binding was necessary but not sufficient for NuMA cortical localization, which required the addition of the 4.1-binding domain.
NuMA/4.1 interactions are required for NuMA cortical stability and spindle orientation integrity
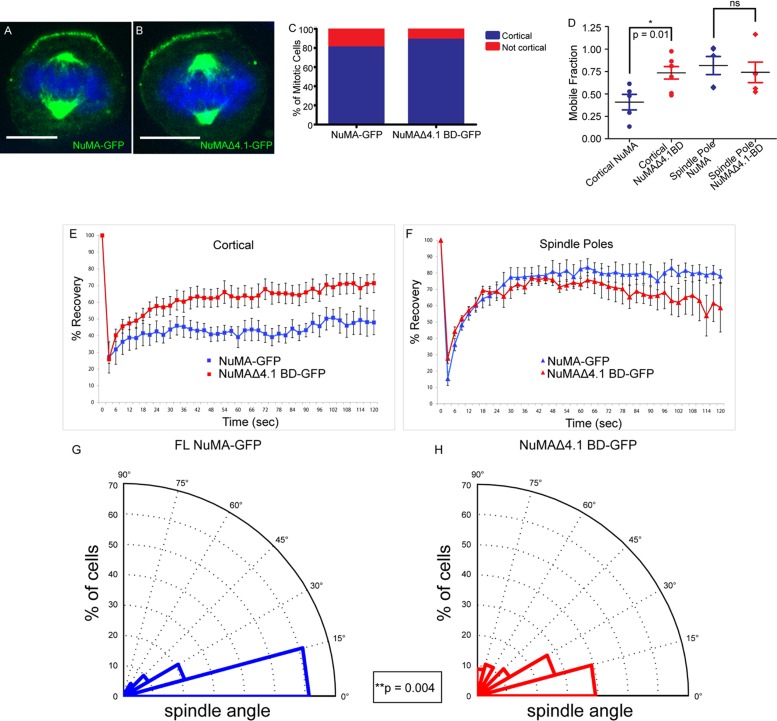
The 4.1 family of proteins contains both a FERM domain and a spectrin-actin–binding domain that allows them to link the actin cytoskeleton to the cell cortex (Hoover and Bryant, 2000). Our results suggested that the 4.1 family was an attractive candidate for recruiting cortical NuMA. However, directly testing its role in NuMA localization proved difficult because multiple 4.1 family members (and multiple isoforms of each) are expressed in skin (unpublished data). In addition, loss of 4.1 proteins renders membranes fragile (Baines, 2010), suggesting that any observed effects on protein localization and spindle orientation could be secondary and due to effects beyond direct NuMA recruitment. To test whether 4.1 binding was necessary for cortical NuMA recruitment, we monitored the ability of a full-length NuMA mutant that lacks the 4.1 BD to target to the cell cortex. To our surprise, we found that this mutant localized normally to both the cell cortex and the spindle poles (Figure 3, A–C). Therefore, in the context of the full-length protein, the 4.1 BD is not essential for localization (despite its requirement in smaller fragments).
FIGURE 3:
4.1/NuMA interactions are required for cortical NuMA stability and mitotic spindle orientation. (A, B) Full-length NuMA-GFP and NuMAΔ4.1 BD-GFP constructs were transfected into wild-type cells, and immunofluorescence analysis was performed to determine localization. (C) Quantitation of cortical accumulation of GFP-tagged NuMA and NuMAΔ4.1 BD. n = 25 cells for each, p = 0.7. (D) Dot plot showing the distribution of mobile fractions of cortical and spindle pole NuMA-GFP for both WT and NuMAΔ4.1 BD. p (cortical) = 0.01; p (spindle poles) = 0.63. (E, F) FRAP analysis of full-length NuMA-GFP (FL) vs. a GFP-tagged NuMA construct lacking the 4.1-binding domain (Δ4.1 BD) transfected into wild-type cells. Recovery profiles of bleached GFP cortical signal (E) and spindle pole signal (F) from both constructs over 2 min. (G, H) Analysis of mitotic spindle alignment with the center of the cortical NuMA-GFP crescent in FL and NuMAΔ4.1 BD-GFP–transfected wild-type cells plated on 100 μM laminin. Radial histograms illustrate the distribution of spindle angles from both constructs. Histograms are grouped into 15° bins, with 0° representing a spindle aligned with the center of the NuMA crescent. n = 62 for NuMA-GFP, n = 56 for NuMAΔ4.1 BD-GFP; p = 0.004 using a Kolmogorov–Smirnov test. Scale bars, 10 μm.
Although 4.1 interactions are not required for full-length NuMA localization, there may be a functional significance for the NuMA-4.1 interaction in spindle orientation. One possibility is that oligomeric NuMA is able to target to the cell cortex via LGN (and/or additional interactions) and that 4.1 stabilizes NuMA at the cortex. Because stability is likely to be important for NuMA's ability to generate productive forces at the cell cortex, we performed fluorescence recovery after photobleaching (FRAP) analysis to directly measure NuMA turnover. NuMAΔ4.1 BD–GFP demonstrated a marked increase in mobility at the cortex (80% mobile fraction) compared with full-length NuMA (50% mobile fraction; Figure 3, D and E). This change in mobility was specific to the cell cortex, as the turnover at the spindle poles was not significantly different in the mutant compared with the full-length construct (Figure 3, D and F). These results demonstrate that whereas the 4.1-binding domain may not be required for NuMA localization, this interaction clearly plays a significant role in enhancing NuMA stability at the cortex. These experiments may underestimate the role of 4.1 interactions in stabilizing NuMA at the cortex, as they were performed in the background of endogenous wild-type NuMA.
To determine whether the differences in cortical stability are important for robust spindle orientation, we sought to quantify the ability of GFP-tagged full-length NuMA and NuMAΔ4.1 BD to align the mitotic spindle. To this end, we measured the spindle angle relative to the center of the cortical NuMA crescent in metaphase keratinocytes. For this analysis, we discovered that wild-type keratinocytes were unable to properly align their spindles unless plated on coverslips precoated with laminin or fibronectin (Supplemental Figure S4). These proteins are critical basement membrane constituents in the skin that interact with αβ1 integrins. β1 Integrin is required for proper spindle orientation in the epidermis, and here we show that a related requirement exists in vitro (Lecher and Fuchs, 2007).
Wild-type keratinocytes that were plated on laminin and transfected with full-length NuMA displayed a strong alignment between the spindle and the cortical NuMA crescent, thus demonstrating a robust ability to orient the mitotic spindle (Figure 3G). NuMAΔ4.1 BD-GFP–transfected cells, on the other hand, showed a significant impairment in spindle orientation (Figure 3H). We noted both a substantial decrease in the number of spindles highly aligned (within 15°) with the center of the NuMA crescent and an increase in misaligned spindles. Therefore, the 4.1-binding domain of NuMA is critical for maintaining NuMA's robust spindle-orienting abilities. These data suggest that stability of cortical NuMA is required for productive force generation in order to successfully drive robust spindle orientation.
The 4.1-binding domain of NuMA is essential for stretch-induced spindle reorientation
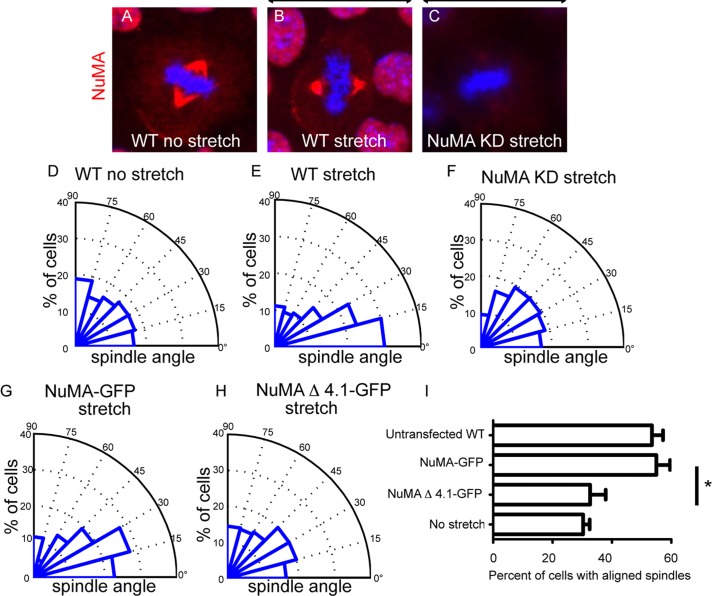
In certain cell types, mitotic spindle alignment is dictated by cell geometry and/or external forces applied to the cell (Fink et al., 2011). To determine whether keratinocytes realigned their spindles in response to external forces, we grew these cells on flexible substrates that we then subjected to a sustained 25% uniaxial stretch for 90 min. We found that cells under these conditions oriented their mitotic spindles along the stretch axis (Figure 4, A, B, D, and E). Mitotic spindles aligned with cortical NuMA crescents upon stretch, suggesting that NuMA might play a role in stretch-induced spindle reorientation (Figure 4B). To determine whether this was the case, we examined the ability of NuMA-knockdown cells to reorient their spindles in response to stretch. These cells demonstrated a significant deficiency in this activity (Figure 4, C and F). We thus conclude that keratinocytes reorient their mitotic spindles in response to stretch in an NuMA-dependent manner. We then asked whether this activity was also impaired in cells expressing a NuMA construct containing the 4.1 BD deletion. Similar to our observations in unstretched cells, we found a significant effect of NuMAΔ4.1 BD–GFP expression on spindle reorientation. Whereas cells expressing the full-length NuMA-GFP construct oriented well upon stretch, those expressing the 4.1 BD deletion were impaired in this activity (Figure 4, G–I). Therefore, using two distinct assays, we demonstrated a role for the 4.1 BD in potentiating NuMA's function in spindle orientation. Of note, all quantitations for both of these assays were performed on metaphase cells.
FIGURE 4:
Mitotic spindle alignment with stretch requires NuMA's 4.1-binding domain. (A–C) Representative images of endogenous NuMA (red) in unstretched cells (A), stretched WT cells (B), and stretched NuMA-knockdown cells (C). Arrows illustrate the direction of stretch applied along the horizontal axis. (D–F) Radial histograms quantitating the spindle orientation relative to the stretch axis, as indicated. Histograms are grouped into 15° bins, with 0° representing a spindle aligned parallel to the stretch axis. Cells were stretched 25% for 90 min. (G, H) Radial histogram for WT cells transfected with (G) NuMA-GFP with applied stretch and (H) NuMAΔ4.1-GFP with applied stretch. All radial histograms display cumulative data from >60 cells from three different experiments for each condition. (I) Quantification of the percentage of mitotic cells that align with the stretch axis (mitotic angle <30°). All conditions represent stretched cells except where “no stretch” is specified. Mean + SEM for three trials for each condition. p < 0.03. Scale bar, 10 μm.
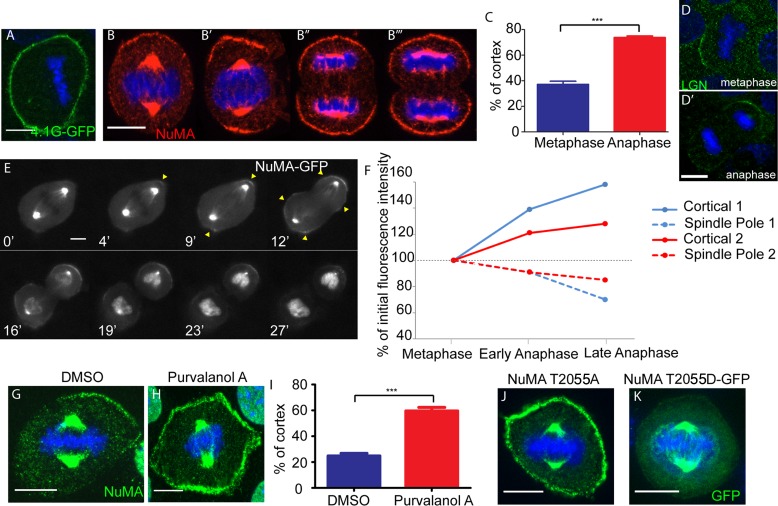
Cortical NuMA is enriched upon anaphase onset
Unlike LGN and NuMA, 4.1G was not highly polarized in cultured keratinocytes, instead showing uniform localization around the entire cell cortex (Figure 5A). It is important to note, however, that we have not tested all isoforms of 4.1 family members and therefore cannot rule out that some of these may be polarized in a manner similar to NuMA. Although NuMA shows a polarized localization in many cultured keratinocytes, we also noted bipolar and nonpolarized localizations in some cells. In fixed cells, we consistently noted stronger cortical signals (and less polarized signals) in anaphase cells (Figure 5, B and C). This cortical enrichment was specific to NuMA, as other components of the machinery, such as LGN, did not increase and expand as dramatically across the cortex in anaphase (Figure 5D), similar to reports in other cell types (Kiyomitsu and Cheeseman, 2013). We examined this more closely by taking time-lapse movies of NuMA-GFP dynamics in mitotic primary keratinocytes isolated from transgenic mice that express NuMA-GFP in the epidermis (Poulson and Lechler, 2010). Once again, we observed a notable increase in cortical NuMA as mitotic cells entered anaphase (Figure 5, E and F, and Supplemental Movie S1). In addition, we consistently noted that the spindle pole NuMA signal diminished as cortical signal increased (Figure 5, E and F). Therefore NuMA's localization at the cell cortex is enhanced substantially in anaphase, which may play an important role in stabilizing the spindle during cell division, as a recent report suggested (Kiyomitsu and Cheeseman, 2013).
FIGURE 5:
NuMA cortical expansion occurs during anaphase onset and Cdk1 inactivation. (A) 4.1G tagged to GFP was transfected into wild-type cells, and immunofluorescence analysis was performed to determine localization. (B) Immunofluorescence analysis of endogenous NuMA localization in wild-type cells during different stages of mitosis. The same exposure time was used for image acquisition of B–B′′′. (C) Quantitation of percentage of the cortex that NuMA covers in metaphase and late anaphase. Mean + SEM. n = 50 cells, p < 0.0001. (D, D′) Localization of endogenous LGN in metaphase- and anaphase-stage keratinocytes. (E) Frames taken from a time-lapse movie imaged in wide field of a dividing NuMA-GFP primary cell isolated from a K14: NuMA-GFP transgenic mouse (see Supplemental Movie S1). Time is displayed in minutes (′) at the bottom left of each frame. (F) Graph of cortical NuMA-GFP and spindle pole NuMA-GFP intensities quantified from time-lapse movies of two dividing NuMA-GFP primary cells. Cortical 1 and Spindle Pole 1 are from the same cell (shown in E), and Cortical 2 and Spindle Pole 2 are from a second cell. (G, H) Immunofluorescence analysis of endogenous NuMA in wild-type cells treated with DMSO or 100 μM purvalanol A to inhibit cyclin-dependent kinase activity. (I) Quantitation of percentage of the cortex that NuMA spans in DMSO- or purvalanol A-treated cell. Mean + SEM. n = 50, p < 0.0001. (J) A full-length NuMA construct with a T2055A mutation (to prevent phosphorylation at that residue) was transfected into wild-type cells, and immunofluorescence analysis of NuMA was performed to determine localization. (K) A full-length NuMA construct with a GFP-tagged phosphomimetic T2055D mutation was transfected into wild-type cells, and immunofluorescence analysis was performed to determine localization. Scale bars, 10 μm.
Cdk1 phosphorylation regulates NuMA localization
To investigate how NuMA might be regulated at the metaphase-to-anaphase transition, we began by examining the role of Cdk1. Considering the known role for cyclin B degradation and Cdk1 inactivation in driving anaphase onset (Murray et al., 1989), we first tested the effect of Cdk1 inhibition on NuMA localization in metaphase cells. On treating cells with the Cdk1 inhibitor purvalanol A, we saw a significant increase and expansion in the cortical localization of endogenous NuMA as compared with vehicle-treated controls (Figure 5, G–I). These data are consistent with Cdk1 inactivation in anaphase promoting the cortical localization of NuMA. Further investigation, however, is required to determine whether local control of phosphorylation during metaphase occurs to maintain a cortical pool of NuMA.
Consistent with our Cdk1 findings, previous data demonstrated that phosphorylation of NuMA can control its localization (Compton and Luo, 1995). Although these studies were performed before a cortical role for NuMA during asymmetric cell divisions had been established, they convincingly showed that loss of a Cdk1 phosphorylation site at NuMA threonine residue 2055 (initially reported as residue 2040, which is equivalent to residue 2055 in the 2115–amino acid–long isoform of NuMA used in this study) resulted in increased cortical NuMA association. We thus evaluated the ability of a T2055A NuMA mutant to localize to the cell cortex in keratinocytes and were able to reproduce the earlier observations that this mutant localized robustly and uniformly to the cell cortex (Figure 5J and Supplemental Figure S2E). To further assess how phosphorylation affects NuMA cortical localization, we generated a NuMA T2055D mutant that mimics constitutive phosphorylation. When transfected into keratinocytes, this construct localized strongly to the spindle poles with minimal detectable cortical localization (Figure 5K and Supplemental Figure S2E). Taken together, this evidence demonstrates that the phosphorylation of threonine residue T2055 is important for regulating NuMA's subcellular localization in keratinocytes.
The increase and expansion of cortical NuMA in anaphase could rely on the same binding partners as earlier in mitosis or could be mediated by distinct mechanisms. The fact that we did not see increased LGN localization at the cortex at anaphase onset suggested that distinct mechanisms might mediate metaphase and anaphase NuMA cortical recruitment. To directly test whether the same machinery was responsible for recruiting cortical NuMA in metaphase and anaphase, we reexamined NuMA localization in wild-type and LGN-knockdown keratinocytes. Although NuMA localization was lost from the cortex in metaphase LGN KD cells as previously discussed, cortical NuMA returned to wild-type levels in anaphase (Figure 6, A–E, and Supplemental Figure S2F). Similarly, whereas the NuMAΔLGN-GFP protein was not recruited to the cortex in metaphase, its cortical localization was rescued to wild-type levels in anaphase (Figure 6, F–J). This suggested that in anaphase, NuMA associates with the cortex in an LGN-independent manner. To determine whether Cdk1 inhibition could rescue the metaphase localization of NuMA in LGN-knockdown keratinocytes, we treated these cells with purvalanol A or DMSO (control). Cdk1 inhibition was sufficient to restore the metaphase localization of NuMA in LGN-depleted cells (Figure 6, K, L, and O).
FIGURE 6:
Anaphase localization of NuMA is independent of LGN and 4.1 binding. (A–D) Immunofluorescence analysis of endogenous NuMA localization in wild-type and LGN-knockdown keratinocytes in metaphase and anaphase, as indicated. (E) Quantitation of cells with cortical NuMA accumulation, as indicated. n = 50 cells, p < 0.0001 for NuMA localization in WT vs. LGN KD cells in metaphase. (F–I) Epifluorescence of NuMA-GFP and NuMAΔLGN–BD-GFP transfected keratinocytes in metaphase and anaphase, as indicated. (J) Quantitation of cells with cortical NuMA accumulation, as indicated. n = 25 cells, p < 0.0001 for NuMA-GFP vs. NuMAΔLGN BD-GFP cortical localization in metaphase. (K, L) Immunofluorescence analysis of endogenous NuMA localization in LGN-knockdown keratinocytes treated with DMSO or 100 μM purvalanol A for 3 min. (M) NuMA construct that harbors the T2055A mutation and spans from the carboxy-terminal 4.1-binding domain to the end of the protein. (N) A GFP-tagged version of the NuMA fragment diagrammed in M was expressed in wild-type cells, and immunofluorescence analysis was performed to determine localization. (O) Quantitation of cells from K and L with cortical NuMA accumulation, as indicated. n = 50 cells, p < 0.0001. (P–S) GFP epifluorescence of NuMA constructs as indicated. The domain structure of each NuMA fragment is diagrammed beneath its corresponding image. (T) Quantitation of the ability of NuMA deletion/mutant constructs from N and Q–S to target to the cell cortex in metaphase. n = 25 cells, all p > 0.05. (U) Immunoprecipitation of GFP-tagged NuMA constructs/mutants with 4.1G-HA. Top and bottom, amount of 4.1G-HA and GFP fusion proteins in the immunoprecipitates, respectively.
To further isolate the region(s) that were required for NuMA's anaphase cortical targeting, we first generated a carboxy-terminal construct of NuMA harboring the T2055A mutation and found that it localized well to the cell cortex (Figure 6, M and N). As with the full-length protein, mutating T2055 to an aspartate residue dramatically decreased cortical accumulation of the protein (Figure 6P). We then deleted the LGN-binding domain, the 4.1-binding domain, or both in tandem to determine whether either of these (or both redundantly) played a role in anaphase localization of NuMA. To our surprise, all of these constructs were able to localize to the cell cortex as robustly as the control (Figure 6, Q–T, and Supplemental Figure S2G). This demonstrates that in keratinocytes, the anaphase localization of NuMA is independent of both its LGN- and 4.1-binding domains.
To ensure that phosphorylation was not controlling 4.1 interactions, we performed biochemical experiments to test the ability of the T2055A and T2055D mutants to bind to 4.1G-hemagglutinin (HA). We did not detect a significant difference in their binding (Figure 6U). However, we found that the smallest construct (MTBD-end-T2055A) that localizes well to the cell cortex (and lacks both the LGN and 4.1 BDs) does not interact with 4.1G, ruling out an additional binding site in the extreme carboxy terminus (Figure 6U). These data suggest that in anaphase keratinocytes, NuMA cortical localization is independent of both LGN and 4.1 family members and is, instead, dependent on Cdk1 inactivation. Taken together, these findings reveal a profound distinction between the mechanisms that underlie cortical NuMA recruitment during metaphase and anaphase.
DISCUSSION
Regulation of spindle orientation is required for both asymmetric cell divisions and many aspects of morphogenesis. Although a core set of proteins, including LGN, NuMA, and dynein/dynactin, is important for spindle orientation in many cell types studied, several aspects of their localization and function remain poorly understood. In this work, we identified the 4.1-interacting domain of NuMA as an important mediator of spindle orientation. Although not required for cortical NuMA recruitment, NuMA/4.1 interactions play an essential role in stabilizing NuMA at the cortex to ensure robust spindle positioning. In addition, we showed how posttranslational modifications of NuMA differentially affect its localization throughout mitosis, a mechanism that functions independently of LGN and 4.1 binding.
Recent reports proposed distinct ways for recruiting dynein/dynactin to the cell cortex: a NuMA-dependent mechanism and an LGN-dependent mechanism (Kotak et al., 2012; Zheng et al., 2013). Because LGN is required for NuMA localization, it is somewhat difficult to exclude a direct role for LGN in dynein/dynactin recruitment. However, we demonstrated that under conditions in which LGN is at the cell cortex but NuMA is absent, there is no targeting of dynein/dynactin. Whereas NuMA is essential for dynein/dynactin recruitment, we cannot rule out the possibility that LGN plays an important role in regulating stability and/or activity of the dynein/dynactin complex at the membrane. Furthermore, we showed that dynactin is the likely proximal complex recruited by NuMA, as disruption of dynactin prevents dynein association with the cortex.
In mitotic keratinocytes, NuMA localizes to two distinct subcellular regions—the spindle poles and the cell cortex (Lechler and Fuchs, 2005). The mechanisms of localization for these two NuMA pools are distinct, as disruption of microtubules causes loss of spindle pole NuMA, whereas F-actin disruption selectively affects cortical localization. However, whereas microtubules and the microtubule-binding domain of NuMA are not required for localization, they may play a regulatory role in controlling the levels of NuMA at the cortex. Indeed, at least two studies found that microtubules are important for disassembly of asymmetric cell division machinery at the cell cortex (Werts et al., 2011; Zheng et al., 2013). This is consistent with our observations in time-lapse movies, in which puncta of NuMA can be seen streaming from the cortex into the spindle pole/reforming nucleus at the end of anaphase (Supplemental Movie S1).
Of importance, we found a novel role for the 4.1-binding domain of NuMA in regulating NuMA's cortical behavior and spindle orientation. Despite the finding that 4.1 interactions are not essential for localization of the full-length NuMA construct harboring a 4.1 BD deletion, we showed that 4.1-binding interactions play a critical role in maintenance of NuMA cortical stability and spindle orientation. Although LGN interactions are required for NuMA localization, they are not sufficient for robust spindle orientation in the absence of 4.1 interactions. The effect of 4.1 on cortical NuMA stability is likely due to the presence of increased binding sites for cortical association and the ability to link NuMA to the F-actin cytoskeleton. We suggest that the stabilizing role of 4.1 is critical for proper spindle orientation because of its ability to facilitate robust cortical NuMA tethering, which is required for productive force generation by dynein/dynactin complexes on astral microtubules. Future experiments to address this, as well as the consequences of loss of NuMA/4.1 interactions in the tissue context, are necessary. Owing to the presence of multiple 4.1 family members in the epidermis, we have not been able to directly address the role of this protein family. However, loss of these proteins may have pleiotropic effects on the cell cortex that could cause spindle orientation defects in multiple ways. By deleting the 4.1 BD of NuMA, we directly address the role of 4.1/NuMA interactions rather than 4.1’s role in establishing membrane domains. While this study was in revision, Kiyomitsu and Cheeseman (2013) published data showing that 4.1/NuMA interactions are important for anaphase-specific localization of NuMA in symmetrically dividing HeLa cells. Our work shows that in asymmetrically dividing keratinocytes, this interaction also stabilizes NuMA in metaphase. It is possible that this also occurs in HeLa and other cell types; however, this will prove to be more difficult to assess because these cells do not orient to a polarized landmark.
During the course of this work we also identified an increase in cortical association of NuMA during anaphase. This is likely due to loss of Cdk1 phosphorylation because 1) pharmacological inhibition of Cdk1 in metaphase cells resulted in robust recruitment of NuMA to the cell cortex, and 2) mutation of a Cdk1 phosphorylation site in the carboxy terminus of NuMA resulted in uniform cortical localization. Strikingly, this localization is independent of LGN, which is essential for the metaphase cortical localization of NuMA. It is also independent of 4.1 interactions. Although a recent article reported similar findings on the role of Cdk1 phosphorylation in NuMA localization, this was believed to occur through 4.1 interactions in HeLa cells (Kiyomitsu and Cheeseman, 2013). Our data clearly demonstrate that 4.1 is not required for the anaphase interactions in keratinocytes because 1) the phosphorylation status of NuMA did not affect its ability to interact with 4.1, and 2) loss of the 4.1-binding domain, both alone and in combination with the LGN-binding domain, did not disrupt metaphase/anaphase localization of a nonphosphorylatable form of NuMA. This suggests that phosphorylation regulates an additional molecular interaction that alters the relative stability of NuMA at the spindle poles and cell cortex. In conclusion, this study augments our understanding of the regulatory mechanisms throughout mitosis that are required for efficient recruitment, stabilization, and function of cortical NuMA during spindle positioning, which are all essential for driving successful asymmetric divisions and downstream developmental programs.
MATERIALS AND METHODS
Lentiviral production and gene knockdown
shRNA constructs for mouse NuMA (NM_133947.2-1676s1c1) and LGN (NM_029522.1-1617s1c1) with a pLKO.1 lentiviral backbone were ordered from the Broad Institute's Mission TRC-1 mouse library, as previously described (Williams et al., 2011). For production of lentivirus, 293FT cells were grown at 37°C with 7.5% CO2 on 10-cm plates in DMEM and 10% fetal bovine serum (without antibiotics) until they reached 50–70% confluency. Cells were then transfected using calcium phosphate with the following vectors: pLKO.1 NuMA siRNA (Sigma), pMDL g/pRRE, pRSV-Rev, and pMD2.G (Addgene). Five hours after transfection, new growth medium was added and cells were grown for 48 h. The 293FT medium containing the viral particles was put through a 0.45-μm filter (VWR) and centrifuged through an Amicon Ultra Centrifuge filter at 4000 rpm for 15 min.
For subsequent retroviral transduction, wild-type mouse keratinocytes were infected with the concentrated lentiviral-packaged shRNA particles. Virus was diluted in 0.05 mM Ca2+-containing medium with the addition of 6 μg/ml Polybrene and added to cells at 30–50% confluency. After 24 h, the medium was replaced, and the cells were given an additional 24 h to recover before addition of 2 μg/ml puromycin (Amresco) to select for cells expressing the shRNA construct. Medium was changed every 3 d over a 5- to 14-d period before cells were passaged. Gene knockdown was confirmed by immunofluorescence and Western blot analysis.
DNA constructs
All constructs used in this study were cloned into the pEGFP-C1 vector (Clontech), with the exception of NuMAΔMT BD–GFP, NuMAΔLGN BD–GFP, and NuMAΔ4.1 BD–GFP, which were cloned into the pEGFP-N1 vector (Clontech).
Cell culture
All keratinocytes were grown at 37°C with 7.5% CO2 in media containing 0.05 mM Ca2+. NuMA- and LGN-knockdown cell lines were maintained in media supplemented with 2 μg/ml puromycin (Amresco). Before imaging, cells to be fixed were plated on glass coverslips, and cells for live imaging were plated on 35-mm MatTek glass-bottom dishes (no. 1.5). On reaching ∼80% confluency, all cells were treated for 16 h in 2 mM thymidine (Sigma-Aldrich) and released into fresh media containing 0.4 mM Ca2+ for 1.5 h before fixation or live imaging. For drug treatments, 10 μM nocodazole (Sigma-Aldrich), 40 nM latrunculin A (Enzo Life Sciences), 100 μM blebbistatin (Sigma-Aldrich) and the corresponding concentration of DMSO for each were incubated with cells for 10 min before fixation. Purvalanol A (Tocris Bioscience) at 100 μM concentration and DMSO were incubated with cells for 3 min before fixation. For spindle orientation experiments, cells were plated on glass coverslips coated with 100 μM laminin (Invitrogen), 100 μM fibronectin (Life Technologies), or 100 μM collagen (Sigma-Aldrich).
Immunofluorescence staining and analysis
Cells were fixed for 3 min in −20°C methanol. Primary antibodies used in this study include chicken α-GFP (Abcam), rabbit α-NuMA (Abcam), mouse α-p150glued (BD Biosciences), mouse α-DIC (Millipore), and guinea pig α-LGN. Secondary antibodies include Alexa Fluor 488 (Invitrogen) and Rhodamine Red-X (Jackson ImmunoResearch Laboratories). A phosphate-buffered saline solution containing 90% glycerol and 2.5 mg/ml p-phenylenediamine (Sigma-Aldrich) was used to mount samples. Images were collected using a Zeiss motorized Axio Imager Z1 fluorescence microscope with Apotome attachment, an AxioCam MRm camera, a 63×/1.4 numerical aperture (NA) Plan Apochromat objective, Zeiss Immersol 518F oil, and AxioVision Digital Image Processing Software. Photoshop (Adobe) and ImageJ software were used for postacquisition processing.
FRAP analysis
Mouse keratinocytes were plated on 35-mm MatTek glass-bottom dishes (no. 1.5). On reaching ∼60% confluency, cells were transfected with either full-length NuMA-GFP or NuMAΔ4.1 BD–GFP using TransIT-LT1 transfection reagent (Mirus). Keratinocytes were washed and supplemented with fresh media 8 h after transfection. Cells were treated with 2 mM thymidine for 16 h and released from thymidine for 1.5 h. Cells were mounted on a 37°C stage within a 5% CO2 chamber and left to equilibrate for 10 min. Imaging was performed using a Zeiss LSM 710 confocal scanning light microscope with a 63×/1.4 NA oil immersion objective and a pinhole size of 1 airy unit. Enhanced GFP was excited using an argon 488-nm laser line and emission gated between 493 and 598 nm. FRAP experiments were performed using the regions, bleaching, and time series modules from the Zeiss ZEN software. After a region of interest was defined, 75% laser power at the appropriate wavelength was used for three iterations to bleach the GFP signal. After bleaching, images within the same focal plane were acquired every 3 s to monitor fluorescence recovery. The Zen software recorded the mean fluorescence intensity of each region of interest for every time point. A separate region of interest was drawn outside of the bleached cell to be used as a background control. The percentage of recovery was determined by first normalizing fluorescence intensities to background intensities and then normalizing each time point to the initial intensity reading. The mobile fraction was determined as mf = (Imax − I0)/(I − I0) as previously described (Shen et al. 2008), where Imax is defined as the maximum fluorescence intensity recorded postbleaching, and I0 is defined as the initial fluorescence intensity immediately after bleaching. Statistical analysis was performed using a Student's t test.
Time-lapse imaging
Primary NuMA-GFP cells isolated from K14: NuMA-GFP mouse backskin were plated on MatTek glass-bottom dishes (no. 1.5) coated with 100 μM laminin (Invitrogen). Cells were treated with 2 mM thymidine for 16 h, washed, and then reintroduced into fresh media for 1.5 h before imaging. Movies were acquired using a Leica DMI6000 inverted microscope, a 63×/1.4 NA Plan Apochromat objective, Leica immersion oil, and an OrcaER camera (Hamamatsu Photonics). The imaging chamber was maintained at 37°C with 5% CO2. Simple PCI software was used for image acquisition (eCommerce Solutions), and Photoshop and ImageJ software were used for postacquisition processing.
Stretch experiments
Stretch experiments were performed similar to those previously described (Ray et al., 2013). Briefly, cells were plated onto laminin-coated polydimethylsiloxane substrates. Cells were released from thymidine block into 0.5 mM Ca2+-containing media 1 h before stretching. Cells were then stretched for 90 min with a 25% static stretch. After stretch, cells were fixed in methanol and imaged as described.
Protein interactions
Keratinocyte extracts were prepared by scraping transfected cells into buffer (50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 100 mM KCl, 1 mM MgCl2, 1 mM ethylene glycol tetraacetic acid, 1 mM dithiothreitol, 1% Triton X-100, pH 7.4, with protease inhibitors). After a 5-min centrifuge at 4°C, soluble extracts were added to GFP-Trap beads (Chromogen) and incubated on a rotator for 30–60 min. After extensive washing with lysis buffer, proteins were eluted from the beads in sample buffer and subjected to Western blotting. Rabbit anti-LGN (ProteinTech Group) was used for Western blotting.
Statistical analysis
Fisher's exact test was used for all statistical analyses, with the exception of FRAP data, which were analyzed with a Student's t test, and the polar histograms of spindle orientation, which were analyzed with the Kolmogorov–Smirnov test.
Supplementary Material
Acknowledgments
We thank Duane Compton and Sharon Krauss for reagents and members of the Lechler lab for comments on the manuscript. This work was supported by grants from the National Institutes of Health (R01AR055926, R03AR056045), the Sidney Kimmel Foundation, and the March of Dimes to T.L.
Abbreviations used:
- CDK1
cyclin-dependent kinase 1
- DIC
dynein intermediate chain
- FRAP
fluorescence recovery after photobleaching
- NuMA
nuclear mitotic apparatus
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-05-0277) on October 9, 2013.
*These authors contributed equally to the manuscript.
†Present address: University of North Carolina School of Medicine, Chapel Hill, NC 27514.
REFERENCES
- Baines AJ. The spectrin-ankyrin-4.1-adducin membrane skeleton: adapting eukaryotic cells to the demands of animal life. Protoplasma. 2010;244:99–131. doi: 10.1007/s00709-010-0181-1. [DOI] [PubMed] [Google Scholar]
- Bowman SK, Neumuller RA, Novatchkova M, Du Q, Knoblich JA. The Drosophila NuMA homolog Mud regulates spindle orientation in asymmetric cell division. Dev Cell. 2006;10:731–742. doi: 10.1016/j.devcel.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Burkhardt JK, Echeverri CJ, Nilsson T, Vallee RB. Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J Cell Biol. 1997;139:469–484. doi: 10.1083/jcb.139.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton DA, Luo C. Mutation of the predicted p34cdc2 phosphorylation sites in NuMA impair the assembly of the mitotic spindle and block mitosis. J Cell Sci. 1995;108:621–633. doi: 10.1242/jcs.108.2.621. [DOI] [PubMed] [Google Scholar]
- Du Q, Stukenberg PT, Macara IG. A mammalian partner of Inscuteable binds NuMA and regulates mitotic spindle organization. Nat Cell Biol. 2001;3:1069–1075. doi: 10.1038/ncb1201-1069. [DOI] [PubMed] [Google Scholar]
- Du Q, Taylor L, Compton DA, Macara IG. LGN blocks the ability of NuMA to bind and stabilize microtubules. A mechanism for mitotic spindle assembly regulation. Curr Biol. 2002;12:1928–1933. doi: 10.1016/s0960-9822(02)01298-8. [DOI] [PubMed] [Google Scholar]
- Fink J, et al. External forces control mitotic spindle positioning. Nat Cell Biol. 2011;13:771–778. doi: 10.1038/ncb2269. [DOI] [PubMed] [Google Scholar]
- Hoover KB, Bryant PJ. The genetics of the protein 4.1 family: organizers of the membrane and cytoskeleton. Curr Opin Cell Biol. 2000;12:229–234. doi: 10.1016/s0955-0674(99)00080-0. [DOI] [PubMed] [Google Scholar]
- Izumi Y, Ohta N, Hisata K, Raabe T, Matsuzaki F. Drosophila Pins-binding protein Mud regulates spindle-polarity coupling and centrosome organization. Nat Cell Biol. 2006;8:586–593. doi: 10.1038/ncb1409. [DOI] [PubMed] [Google Scholar]
- Kiyomitsu T, Cheeseman IM. Cortical dynein and asymmetric membrane elongation coordinately position the spindle in anaphase. Cell. 2013;154:391–402. doi: 10.1016/j.cell.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S, Busso C, Gonczy P. Cortical dynein is critical for proper spindle positioning in human cells. J Cell Biol. 2012;199:97–110. doi: 10.1083/jcb.201203166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss SW, Lee G, Chasis JA, Mohandas N, Heald R. Two protein 4.1 domains essential for mitotic spindle and aster microtubule dynamics and organization in vitro. J Biol Chem. 2004;279:27591–27598. doi: 10.1074/jbc.M402813200. [DOI] [PubMed] [Google Scholar]
- Kraut R, Chia W, Jan LY, Jan YN, Knoblich JA. Role of inscuteable in orienting asymmetric cell divisions in Drosophila. Nature. 1996;383:50–55. doi: 10.1038/383050a0. [DOI] [PubMed] [Google Scholar]
- Lechler T, Fuchs E. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature. 2005;437:275–280. doi: 10.1038/nature03922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechler T, Fuchs E. Desmoplakin: an unexpected regulator of microtubule organization in the epidermis. J Cell Biol. 2007;176:147–154. doi: 10.1083/jcb.200609109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu MS, Johnston CA. Molecular pathways regulating mitotic spindle orientation in animal cells. Development. 2013;140:1843–1856. doi: 10.1242/dev.087627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally FJ. Mechanisms of spindle positioning. J Cell Biol. 2013;200:131–140. doi: 10.1083/jcb.201210007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AW, Solomon MJ, Kirschner MW. The role of cyclin synthesis and degradation in the control of maturation promoting factor activity. Nature. 1989;339:280–286. doi: 10.1038/339280a0. [DOI] [PubMed] [Google Scholar]
- Poulson ND, Lechler T. Robust control of mitotic spindle orientation in the developing epidermis. J Cell Biol. 2010;191:915–922. doi: 10.1083/jcb.201008001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulson ND, Lechler T. Asymmetric cell divisions in the epidermis. Int Rev Cell Mol Biol. 2012;295:199–232. doi: 10.1016/B978-0-12-394306-4.00012-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Foote HP, Lechler T. beta-Catenin protects the epidermis from mechanical stresses. J Cell Biol. 2013;202:45–52. doi: 10.1083/jcb.201212140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redemann S, Pecreaux J, Goehring NW, Khairy K, Stelzer EH, Hyman AA, Howard J. Membrane invaginations reveal cortical sites that pull on mitotic spindles in one-cell C. elegans embryos. PLoS One. 2010;5:e12301. doi: 10.1371/journal.pone.0012301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober M, Schaefer M, Knoblich JA. Bazooka recruits Inscuteable to orient asymmetric cell divisions in Drosophila neuroblasts. Nature. 1999;402:548–551. doi: 10.1038/990135. [DOI] [PubMed] [Google Scholar]
- Siller KH, Cabernard C, Doe CQ. The NuMA-related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nat Cell Biol. 2006;8:594–600. doi: 10.1038/ncb1412. [DOI] [PubMed] [Google Scholar]
- Smart IH. Variation in the plane of cell cleavage during the process of stratification in the mouse epidermis. British J Dermatol. 1970;82:276–282. doi: 10.1111/j.1365-2133.1970.tb12437.x. [DOI] [PubMed] [Google Scholar]
- Speicher S, Fischer A, Knoblich J, Carmena A. The PDZ protein Canoe regulates the asymmetric division of Drosophila neuroblasts and muscle progenitors. Curr Biol. 2008;18:831–837. doi: 10.1016/j.cub.2008.04.072. [DOI] [PubMed] [Google Scholar]
- Straight AF, Cheung A, Limouze J, Chen I, Westwood NJ, Sellers JR, Mitchison TJ. Dissecting temporal and spatial control of cytokinesis with a myosin II inhibitor. Science. 2003;299:1743–1747. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- Vaughan KT, Vallee RB. Cytoplasmic dynein binds dynactin through a direct interaction between the intermediate chains and p150Glued. J Cell Biol. 1995;131:1507–1516. doi: 10.1083/jcb.131.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee B, Johnston CA, Prehoda KE, Doe CQ. Canoe binds RanGTP to promote Pins(TPR)/Mud-mediated spindle orientation. J Cell Biol. 2011;195:369–376. doi: 10.1083/jcb.201102130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werts AD, Roh-Johnson M, Goldstein B. Dynamic localization of C. elegans TPR-GoLoco proteins mediates mitotic spindle orientation by extrinsic signaling. Development. 2011;138:4411–4422. doi: 10.1242/dev.070979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SE, Beronja S, Pasolli HA, Fuchs E. Asymmetric cell divisions promote Notch-dependent epidermal differentiation. Nature. 2011;470:353–358. doi: 10.1038/nature09793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Wan Q, Liu J, Zhu H, Chu X, Du Q. Evidence for dynein and astral microtubule-mediated cortical release and transport of Galphai/LGN/NuMA complex in mitotic cells. Mol Biol Cell. 2013;24:901–913. doi: 10.1091/mbc.E12-06-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.