In vivo, assembly of axonemal dyneins into cilia is a multi-step process. We show that Chlamydomonas ODA10 encodes an axonemal protein required for a late step in outer arm dynein assembly. Once dynein arms have assembled, they can be extracted and the ODA10p protein is no longer required for high-affinity binding onto axonemal binding sites.
Abstract
Assembly of outer dynein arms (ODAs) requires multiple steps and involves multiple proteins in addition to dynein subunits. The Chlamydomonas ODA10, ODA5, and ODA8 loci genetically interact and are hypothesized to function as an axonemal accessory complex, but only ODA5p was previously characterized. We positionally cloned ODA10 and identified the gene by rescuing an oda10 mutant with a hemagglutinin-tagged cDNA. ODA10 sequence predicts a conserved coiled-coil protein homologous to mouse ccdc151. ODA10p is present in cytoplasm and flagella, remains axonemal after detergent treatment, and is extracted with 0.6 M NaCl. Both outer arm dynein and ODA10p rebound to the axonemes when desalted extracts are mixed with oda10-mutant axonemes. Sucrose gradient separation of these extracts shows that ODA10p sediments near the top of the gradient, not with 23S outer dynein arm proteins. Unexpectedly, dynein and ODA10p fractions are able to bind individually to oda10 axonemes. ODA10p is present on oda8-mutant flagella at wild-type levels. However, ODA10p does not assemble into oda5 flagella and is absent from oda5 cytoplasm, suggesting a necessity of ODA5p for stability of ODA10p in vivo. The results suggest that ODA10p does not function as a part of a traditionally defined docking complex.
INTRODUCTION
The cilium is a highly conserved organelle in eukaryotic cells. Mammals have cilia in almost all cells for sensory and/or motility purposes, and disruptions of ciliary function are the foundation of a group of pleiotropic diseases collectively called ciliopathies that affect developmental, sensory, reproductive, and respiratory functions (Hildebrandt et al., 2011). To treat and accommodate patients who are affected by ciliopathies, it is crucial to understand the mechanism of ciliary assembly.
Dyneins are highly conserved minus end–directed, microtubule-associated motor complexes that have AAA ATPase domains. Axonemal dyneins are essential for proper functioning of motile cilia. Defects in dynein assembly disrupt motility and are one cause of human primary ciliary dyskinesia (PCD), which is characterized by infertility, respiratory tract infections, left–right asymmetry malformation, and hydrocephalus. Axonemal dyneins can be divided into subfamilies based on their structural and functional differences (Gokhale et al., 2012; Kamiya, 2012). Multiple unique inner dynein arms are distributed, each with 96-nm periodicity, and mutations in components of inner dynein arms typically affect flagellar waveform. In contrast, one (Chlamydomonas) or two (human) types of outer dynein arm are found along each doublet microtubule with 24-nm periodicity, and mutations in components of outer dynein arms reduce beat frequency. By far the most common genetic defects associated with PCD in humans are mutations that disrupt the assembly of outer dynein arms (Hildebrandt et al., 2011).
Assembly of the outer dynein arm complex has been extensively studied in Chlamydomonas reinhardtii. In Chlamydomonas, outer dynein arms consist of three heavy, two intermediate, and multiple light chains. In addition to their core components, outer dynein arms require a docking complex (DC) that facilitates their attachment onto microtubules (Takada and Kamiya, 1994; Takada et al., 2002; Koutoulis et al., 1997; Casey et al., 2003), cytoplasmic chaperones that assist preassembly of dynein complexes (Freshour et al., 2007; Omran et al., 2008; Duquesnoy et al., 2009; Mitchison et al., 2012), and an intraflagellar transport (IFT) adaptor protein specific for transportation of outer arm complexes (Ahmed and Mitchell, 2005; Ahmed et al., 2008).
Other gene products required for assembly of outer dynein arms (ODAs) in Chlamydomonas may form an accessory complex (AC) composed of three subunits: ODA5p, ODA8p, and ODA10p (Kamiya, 1988; Fowkes and Mitchell, 1998; Wirschell et al., 2004). Previous work on the oda5, oda8, and oda10 mutants and biochemical characterization of ODA5p supported the existence of a complex containing all three gene products that is localized on doublet microtubules. Genetic evidence for this complex comes from cytoplasmic complementation analysis in temporary dikaryons, quadraflagellate diploid cells that form when two haploid gametes fuse during mating. When two mutants that do not affect the same complex or pathway form a temporary dikaryon, a wild-type version of each protein is provided by its mating partner in the fused cytoplasm. Three groups of oda mutants were defined by the lack of cytoplasmic complementation seen when temporary dikaryons were formed between any two mutants in a group (Kamiya, 1988). One group was later discovered to include subunits of the ODA complex itself (Fowkes and Mitchell, 1998) and cytoplasmic preassembly chaperones required for complex formation (Omran et al., 2008; Mitchison et al., 2012). A second group included oda1 and oda3, which were later shown to encode subunits of the doublet-associated DC (Takada et al., 2002). A third group (oda5, oda8, and oda10) defines the proposed AC (Kamiya, 1988; Fowkes and Mitchell, 1998; Wirschell et al., 2004).
ODA5p is the only gene defining this AC that has been characterized. Consistent with its proposed function as the subunit of a doublet-associated AC, ODA5p is an axonemal coiled-coil protein, distantly homologous to two of the three DC subunits. Assembly of ODA5p in flagella is dependent on ODA10p but independent of ODA8p, the DC, or axonemal dynein complexes (Wirschell et al., 2004). In oda5, oda8, and oda10 mutant cells, dynein subunits are preassembled in the cytoplasm but do not assemble in flagella (Fowkes and Mitchell, 1998). However, outer dynein arms that are extracted from wild-type flagella not only can rebind to oda5, oda8, and oda10 axonemes, but they can also rescue beat frequency up to 52 Hz in vitro (Sakakibara and Kamiya, 1989). Further, purified 12S and 18S dynein fractions are also able to rebind to oda5 axonemes and rescue beat frequency (Takada and Kamiya, 1994). These rebinding studies suggest that dynein may rebind in the absence of the AC and appear to be in conflict with previous models that proposed a role for the AC as a second docking complex for binding of outer dynein arms in flagella (Fowkes and Mitchell, 1998; Wirschell et al., 2004). Furthermore, although outer dynein arm complexes form in the oda5 cytoplasm (Fowkes and Mitchell, 1998) and these complexes can rescue the motility of flagella in vivo in dikaryons with subunit-defective strains (Kamiya, 1988), their cytoplasmic abundance is reduced compared with wild-type or DC-mutant strains (Fowkes and Mitchell, 1998). This result suggests a unique role for ODA5p in the cytoplasm instead of, or in addition to, a role as part of an accessory docking complex in the flagellum.
Here we characterize the ODA10 protein and described its role in assembly of outer dynein arms. Our data support a new model in which ODA5, ODA8, and ODA10 proteins modify outer dynein arms into a form that binds with high affinity to axonemal binding sites.
RESULTS
Positional cloning of ODA10 locus
Oda10 was previously mapped near pf3 on chromosome 8 (parental ditype to nonparental ditype to tetratype = 132:0:5; Harris et al., 1987), and the pf3 locus was physically mapped to scaffold 29 of the Chlamydomonas genomic sequence version 3, but this genomic region remains incompletely assembled in current versions of the Chlamydomonas genome (Wirschell et al., 2013). We further mapped 67 progeny from a cross between oda10-1 and strain S1D2 (CC-2290). Linkage data were verified with previously characterized molecular markers: PSBQ, MCA1, and LI818 (Rymarquis et al., 2005). The results identified a general candidate region for ODA10 between markers LI818 and MCA1 on chromosome 8. The linkage between pf3 and ODA10 was genetically verified to be ∼1 cM, which corresponds to a physical distance of ∼100 kb (Rymarquis et al., 2005). Based on these linkage data, two candidate regions were identified within ∼100 kb of the predicted location of pf3 locus. Using an additional marker within one candidate region, V3S294, we mapped ODA10 between V3S294 and MCA1 (see Supplemental Table S1 for marker details). A candidate gene in this interval, C_290012 in JGI Chlamydomonas Genome Version 4, was selected for further analysis based on its homology to genes found only in organisms and tissues with motile cilia. A marker, ODA10-6, near the candidate gene was tested and showed no recombination with the ODA10 locus in all 67 test progeny. These results strongly support that C_290012 was the ODA10 locus.
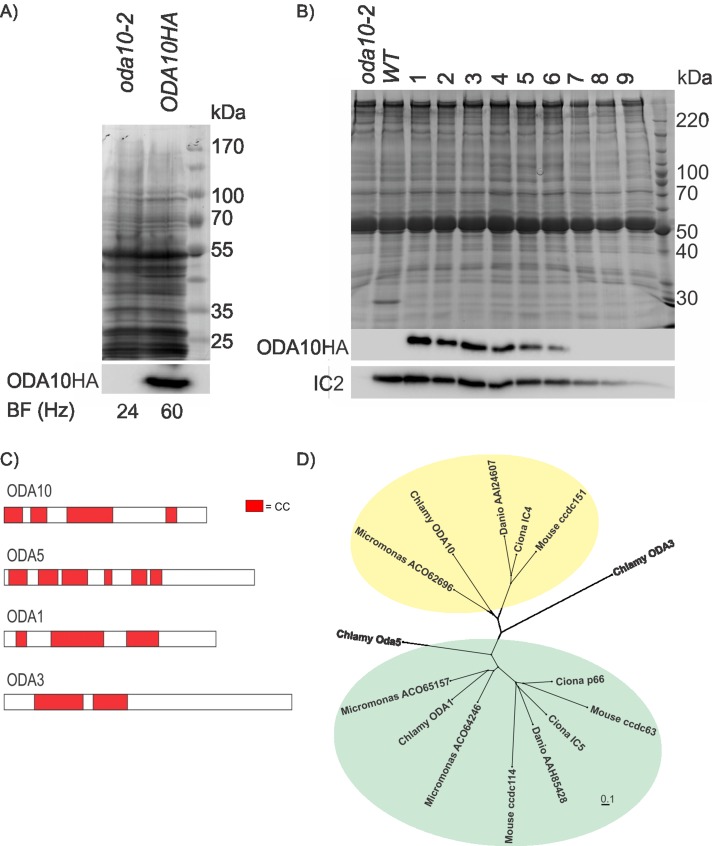
A BLAST search with C_290012 identified two cDNA clones in the Kazusa database (Asamizu et al., 1999). One of these clones (AV640669) was confirmed to be a full-length cDNA of candidate gene C_290012. To express this cDNA in C. reinhardtii, we cloned the predicted coding region into a C. reinhardtii expression vector that adds three hemagglutinin (HA) epitopes at the C terminus. When transformed into oda10-2 cells this construct rescued the beat frequency of oda10-2(V87.2) mutant (∼24 Hz) to that of wild type (∼60 Hz; Figure 1A) and restored assembly of outer dynein arms to wild-type levels as detected by Western blotting (Figure 1B). Comparison of multiple independent transformants showed a correlation between the amount of ODA10HAp and the amount of outer arm dynein in flagella. We conclude that gene C_290012 on chromosome 8 is the ODA10 locus, and the level of outer dynein arm assembly is dependent on the amount of ODA10p present in the flagella.
FIGURE 1:
ODA10p is a conserved coiled-coil protein. (A) A Coomassie-stained gel (top) and Western blot (bottom), probed with an antibody against HA epitope, of equally loaded (by cell number) whole-cell samples of oda10-2,ODA10HA and oda10-2 cells show expression of ODA10HAp in a strain with rescued beat frequency. (B) A Coomassie-stained gel (top) and western blot (bottom), probed with antibodies against HA epitope and IC2, of flagellar samples of mutant (oda10-2), wild type (137c), and rescued transformants (oda10-2,ODA10HA 1-9) show restored assembly of outer dynein arms to wild-type levels in transformant cells expressing the highest levels of ODA10HAp. (C) The distribution of predicted coiled-coil domains of ODA1p, ODA3p, ODA5p, and ODA10p. (D) All homologues of ODA10p and of three Chlamydomonas ODA10p paralogues (ODA5p, ODA1p [ = DC2], ODA3p [ = DC1]) in an alga (Micromonas), a basal chordate (Ciona), a fish (Danio), and a mammal (Mus) were aligned, and an unrooted tree was generated to display relative sequence similarities. Green background highlights homologues of ODA1p; yellow background highlights homologues of ODA10p. Protein sequences used in the alignments that are not given in the figure were Q8CDV6 (ccdc63), AAI60348 (ccdc114), AAH57069 (ccdc151), NP_001027646 (Ciona p66), NP_001072015 (Ciona IC4), NP_001071888 (Ciona IC5), AAC49732 (ODA3), AAK72125 (ODA1), AAS10183 (ODA5), and AGW18228 (ODA10).
ODA10p is a conserved coiled-coil protein
ODA10p is predicted to be a 56-kDa protein with pI of 7.93 and a high probability to form multiple α-helical coiled coils (Figure 1C). Direct BLASTp comparison of ODA10p to predicted Chlamydomonas proteins did not reveal strong similarity to any other proteins; however the overall predicted structural similarity suggested that ODA10p might share some evolutionary relationship to other predicted coiled-coil proteins important for dynein assembly. These include another protein genetically associated with the AC (ODA5p) and two proteins in the DC (ODA1p = DC2 and ODA3p = DC1). The distribution of coiled-coil domains in these proteins is shown in Figure 1C. All homologues of these four Chlamydomonas proteins (selected by BLASTp with a cutoff of 1E-10) in an alga (Micromonas), a basal chordate (Ciona), a fish (Danio), and a mammal (Mus) were aligned, and an unrooted tree was generated to display relative sequence similarities (Figure 1D). A single close homologue of ODA10p appeared in each genome. The other Chlamydomonas AC protein, ODA5p, appears to be distantly related to Chlamydomonas DC protein ODA1p (E value of 3E-22), and two ODA1p homologues were also apparent in the Micromonas, Ciona, and Mus genomes. Branch order suggests that the metazoan homologues arose before the divergence of chordates but after the divergence of metazoa from their common ancestor with algae and that one ODA1p homologue may have been lost in the Danio lineage. In contrast, no close homologues of ODA3p appear in any of these genomes, suggesting that it might be a recent invention in the Chlamydomonas lineage.
Biochemical fractionation of ODA10p
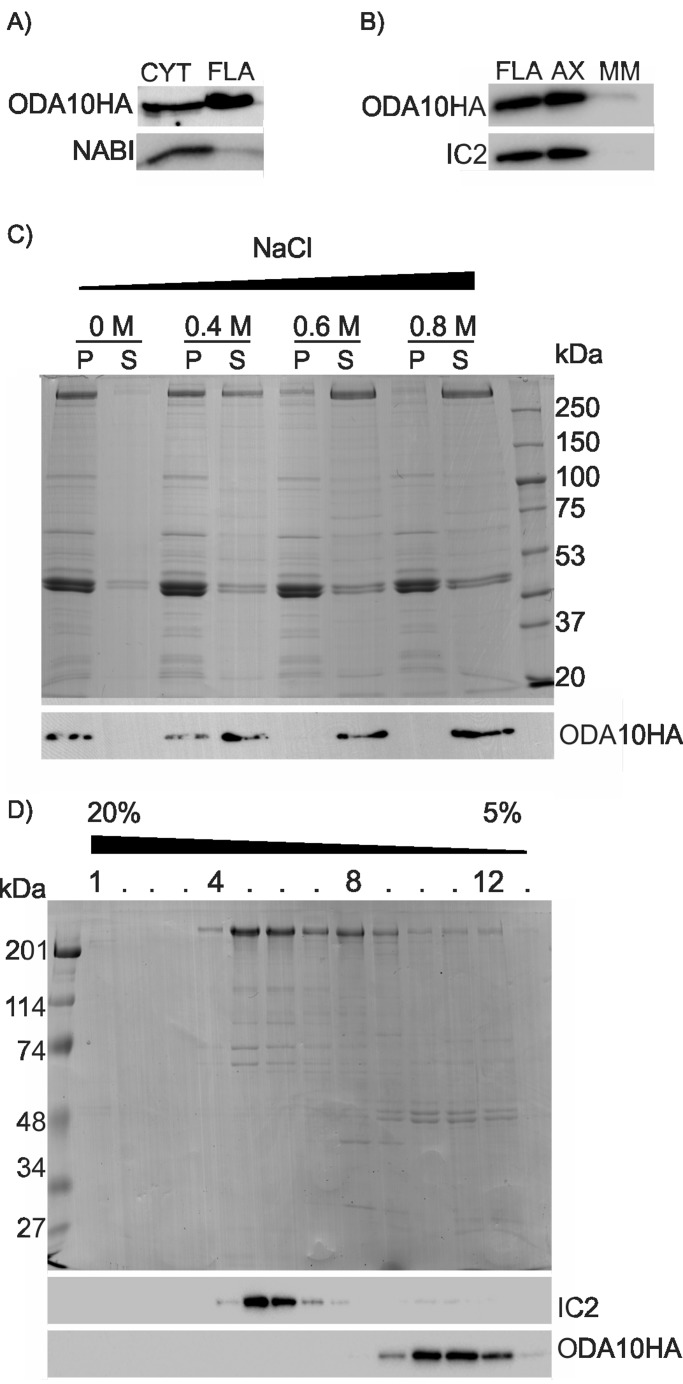
To determine the localization of ODA10p, cytoplasmic extract, flagella, axoneme, and membrane matrix fractions of oda10-2,ODA10HA cells were prepared at 1:1 stoichiometric ratios, and the abundance of ODA10p was detected with Western blots using an antibody against the HA epitope. ODA10p was present in cytoplasm and flagella at ∼1:1 ratio (Figure 2A) and stayed in the axonemal microtubule-associated fraction after treatment of flagella with detergent to remove membranes and soluble matrix (Figure 2B). When oda10-2,ODA10HA axonemes were extracted with NaCl, ODA10p was extracted quantitatively at concentrations >0.4 M NaCl (Figure 2C), similar to the concentration of NaCl required to extract outer dynein arms from axonemes.
FIGURE 2:
ODA10p is a salt-extractable axonemal protein that does not cofractionate with dynein. (A) Western blots of cytoplasmic extract and flagellar samples from equal numbers of cells, probed with antibodies against HA (ODA10p) and NAB1 (a cytoplasmic protein used as a control for cell body contamination in the flagellar fraction), show ODA10p distribution in cytoplasm and flagella. (B) Western blots of flagellar, detergent-resistant axonemal, and detergent-soluble membrane and matrix fractions, probed with antibodies against HA and IC2, show that ODA10p remains microtubule associated, similar to IC2. (C) A Coomassie-stained gel (top) and Western blot of soluble and insoluble fractions of wild-type axonemes after exposure to increasing salt concentrations, probed with an antibody against HA, show extraction of ODA10p >0.4 M NaCl. (D) A Coomassie-stained gel (top) and Western blot (bottom) of sucrose gradient fractions of a 0.6 M NaCl extract of ODA10HA axonemes. Outer row dynein (IC2) does not cosediment with ODA10p (ODA10HA).
Similar to Chlamydomonas docking complex, the Ciona ODA10p orthologue (IC4) copurifies with outer row dyneins on sucrose gradients (Hozumi et al., 2006). Because ODA10p not only showed some evolutionary relationship with docking complex proteins (Figure 1D), but was also hypothesized to have a similar function (Wirschell et al., 2004), we tested whether Chlamydomonas ODA10p cofractionates with the 23S dynein fraction, which has intact outer row dyneins together with docking complexes (Takada et al., 2002). Fractionation of dialyzed high-salt extract of ODA10HA axonemes by sucrose gradient centrifugation shows that ODA10HAp cosediments with tubulin at the top of the gradient (∼6 S), whereas outer row dynein sediments at ∼23 S, represented by its IC (IC2; Figure 2D).
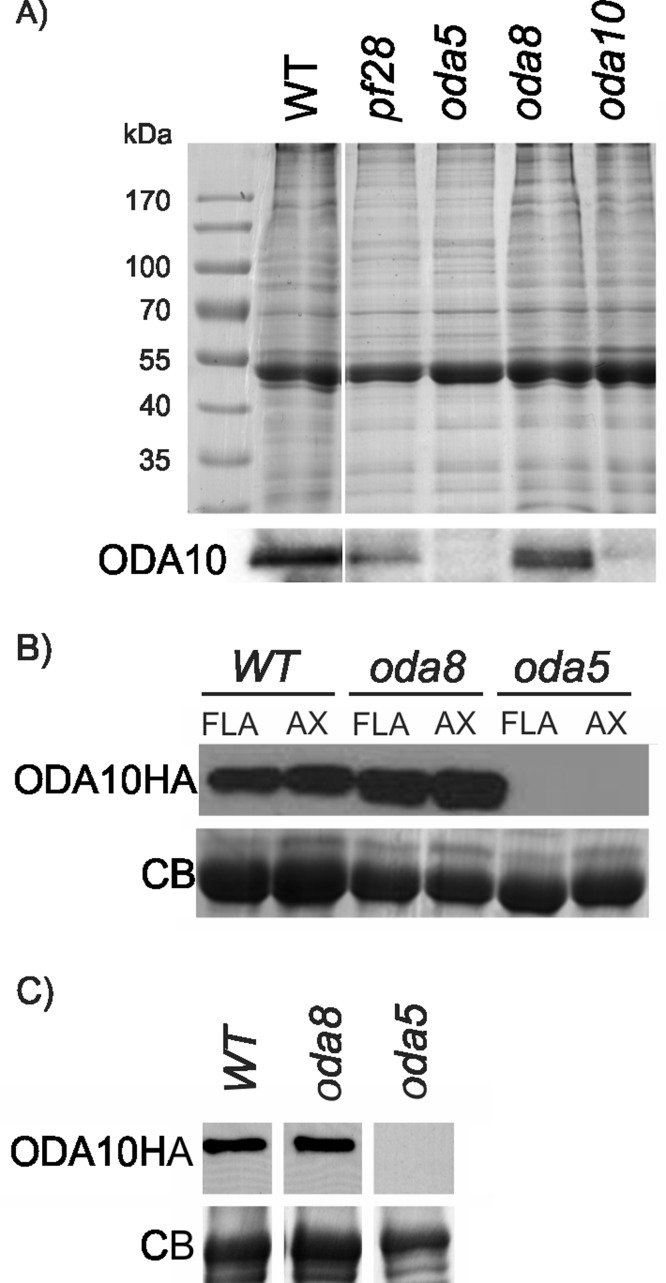
Effects of oda5 and oda8 mutations on ODA10p
To test the effect of other outer dynein arm mutants on assembly of ODA10p, we probed flagellar samples of pf28, oda5, oda8, and oda10-1 for the presence of ODA10p using a polyclonal antibody against endogenous ODA10p. The results show that flagellar assembly of ODA10p was significantly reduced in oda5 but not in oda8 or dynein heavy-chain mutant pf28 (Figure 3A). To confirm the effect of oda5 and oda8 on assembly of ODA10p, we probed samples of oda5,oda10 and oda8,oda10 double-mutant cells expressing an HA-tagged ODA10p (designated here as oda5,ODA10HA and oda8,ODA10HA) for the presence of ODA10p with the more sensitive antibody against the HA epitope. A Western blot with equally loaded flagellar samples of oda10,ODA10HA, oda5,ODA10HA, and oda8,ODA10HA cells showed that ODA10p did not assemble in oda5-mutant flagella, whereas ODA10p did assemble to wild-type levels in flagella of oda8-mutant cells (Figure 3B). To elucidate the reason for lack of assembly of ODA10p in oda5 mutants, we tested the level of ODA10p in the cytoplasm. The results showed an almost undetectable amount of ODA10p in the cytoplasm of oda5, whereas cytoplasmic abundance of ODA10p in oda8 was maintained at wild-type levels (Figure 3C). Thus ODA10p is unstable in the absence of ODA5p, whereas its stability in cytoplasm and assembly in flagella were not affected by lack of ODA8p. Because ODA complexes fail to assemble in either pf28 or oda8 flagella, the absence of outer dynein arms did not alter assembly of ODA10p.
FIGURE 3:
ODA10p is unstable in oda5 mutant. (A) A Coomassie-stained gel (top) and Western blot (bottom) of flagellar samples from the indicated strains, probed with a polyclonal antibody against ODA10p, show that ODA10p is missing from oda5 as well as oda10 flagella. (B) Flagellar and axonemal samples of oda8,ODA10HA oda5,ODA10HA and oda10,ODA10HA were probed with antibody against the HA epitope. The tubulin region of a Coomassie-stained gel (bottom) serves as a loading control. (C) A Western blot (top) of deflagellated cell body samples of oda10,ODA10HA oda8,ODA10HA and oda5,ODA10HA probed for ODA10HAp. The amount of ODA10p in cytoplasm of oda5 was greatly reduced but was near wild type in oda8. A portion of the Coomassie-stained gel serves as a loading control.
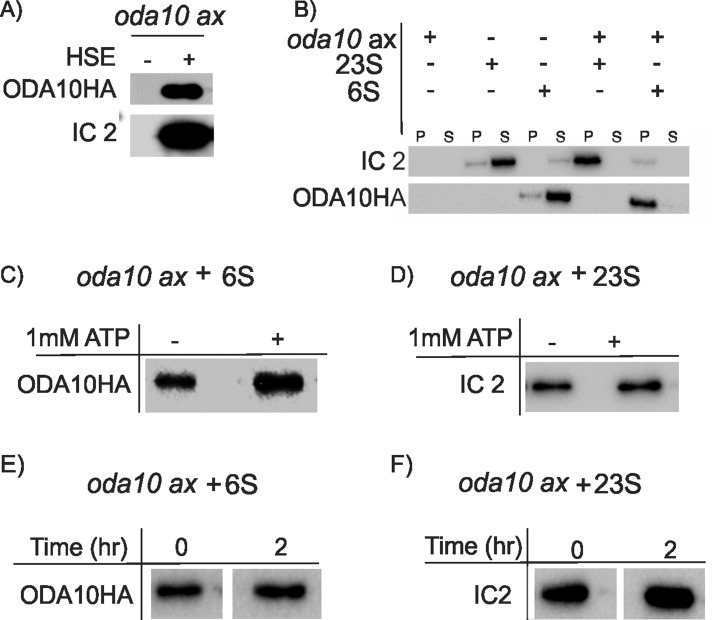
ODA10p and 23S dynein stably bind to oda10 axonemes independently in vitro
Sakakibara and Kamiya (1989) showed that a dialyzed high-salt extract of wild-type axonemes was sufficient to restore wild-type motility to axonemes of oda cell models, including oda10. We tested the ability of ODA10p to rebind to oda10 axonemes under these conditions. If ODA10p cannot rebind to the axonemes, then we could conclude that ODA10p is unnecessary for functional dynein binding in vitro. Mixing a dialyzed salt extract containing axonemal dynein with oda10 axonemes shows that dynein and ODA10HAp both rebound to oda10 axonemes in vitro (Figure 4A). To see whether ODA10p is necessary for dynein binding, we fractionated extracts by sucrose gradient centrifugation under conditions that preserve a 23S dynein fraction, which has been shown to functionally rescue oda1 axonemes in vitro (Takada et al., 1992). Binding experiments with independently pooled 23S and 6S fractions show that ODA10p and outer dynein arms each binds independently onto oda10 axonemes in vitro (Figure 4B). Therefore ODA10p is not required for the initial binding of outer dynein arms onto oda axonemes in vitro. Addition of 1 mM ATP did not affect binding of either ODA10p or outer dynein arms to oda10-mutant axonemes (Figure 4, C and D), ruling out dynein binding only by ATP-sensitive motor domain interaction with doublet microtubules (Haimo et al., 1979; Haimo and Fenton, 1988). To further test the stability of outer dynein arm and ODA10p axoneme association, we resuspended axonemes with bound proteins in buffer alone, incubated them on ice, and repelleted them. Neither ODA10p nor outer dynein arms dissociated from oda10 axonemes even after 2 h in buffer alone (Figure 4, E and F). Thus outer dynein arms extracted from wild-type axonemes can rebind to oda10 axonemes at high affinity in the absence of ODA10p.
FIGURE 4:
ODA10p and outer arm dynein stably assemble onto oda10 axonemes independently in vitro. Western blots probed with anti-IC2 and anti-HA detect proteins bound in vitro to oda10 axonemes. (A) oda10 axonemes were incubated with buffer alone (–) or a dialyzed HSE of wild-type axonemes from ODA10HA cells (+) for 1 h, and pellet fractions were examined. Both IC2 and ODA10HAp are present in the pellet. (B) oda10 axonemes were incubated on ice for 1 h with separate sucrose gradient fractions of outer row dyneins (∼23 S) or ODA10HAp (∼6 S) or buffer alone. Both pellet (P) and supernatant (S) were analyzed. (C, D) The binding experiment in B was repeated in presence of 1 mM ATP, and pellets were probed for ODA10HA or IC2. ODA10HA and outer dynein arms pellet with axonemes, regardless of presence of ATP. (E, F) oda10 axonemes previously incubated with either 6S (E) or 23S (F) fractions were pelleted, and half of each sample was resuspended in fresh buffer, incubated on ice for 2 h, and pelleted again. Equal amounts of ODA10HAp (E) and IC2 (F) are associated with oda10 axonemes before (0) and after (2) incubation.
DISCUSSION
Assembly of ODA motor complexes appears to be a complicated, multistep process that involves chaperoning steps in the cytoplasm (Mitchison et al., 2012), IFT-based transport (Ahmed et al., 2008), and specific docking sites on doublet microtubules (Takada and Kamiya, 1994; Takada et al., 2002; Wakabayashi et al., 2001). An additional step involving a hypothesized AC composed minimally of ODA5p, ODA8p, and ODA10p is required for assembly of outer dynein arms, but the specific role of the AC in assembly is yet to be defined.
ODA10p is a coiled-coil protein with weak similarity to ODA5p and DC proteins ODA1p and ODA3p. A single ODA10p orthologue occurs in the genomes of most organisms with motile cilia, including a diatom (Thalassiosira) that lacks inner dynein arms, whereas no orthologues are found in a moss (Physcomitrella) that lacks outer dynein arms, supporting a conserved role for ODA10p in outer dynein arm assembly or function. In contrast, clear orthologues of ODA5p were not detected in organisms more distantly related than Volvox, suggesting that the ODA5 gene may have been created relatively recently from a duplication of ODA1 within the Chlorophyte algae lineage. Independent duplications of ODA1 homologues have occurred in the Prasinophyte algae (Micromonas) and the Metazoa (Ciona, Mus), as seen in Figure 1. It will be interesting to determine whether any of these gene duplications also created a gene product functionally equivalent to ODA5p and thus interacting genetically with ODA10p. Alternatively, these gene duplication events might have created proteins that interact with each other to create a docking complex equivalent to the Chlamydomonas DC, which contains a heterodimer between coiled-coil proteins ODA1p and ODA3p (Wakabayashi et al., 2001), or proteins with tissue-specific roles, as suggested by the cilia-specific expression of ccdc114 and testis-specific expression of ccdc63 in humans (Onoufriadis et al., 2013). Evidence from purification of Ciona sperm outer dynein arms (Hozumi et al., 2006) shows that the Ciona ODA10p orthologue (IC4) copurifies with two Ciona ODA1p orthologues (IC5 and p66), but only one of these, IC5, is present at 1:1 stoichiometry with IC4. In Ciona IC4/IC5 may therefore be AC subunits equivalent to Chlamydomonas ODA10/ODA5.
Purification of outer dynein arm complexes from a wide range of organisms has not presented a consistent pattern of copurifying intermediate chains. Sea urchin ODA complexes typically have three ICs. Two of these are WD-repeat proteins homologous to Chlamydomonas IC1 and IC2, which are the only universal components of all outer dynein arms, and the third is a hybrid thioredoxin/nucleoside diphosphate kinase (TNDK) that copurifies with ODAs from the sea urchin Anthocidaris, the trout Oncorhynchus, the ascidian Halocynthia, and the mussel Mytilus (Ogawa et al., 1996), as well as the ascidian Ciona (Hozumi et al., 2006), but is not present in ODA fractions of Chlamydomonas (Pfister and Witman, 1984) or of ciliates such as Paramecium (Walczak et al., 1993). Database searches indicate that this TNDK-type IC is present in the genomes of a choanozoan (Monosiga) and a fungus (Batrachochytrium) in addition to the metazoa listed earlier but not in any other branches of eukaryotes and thus appears to be an Opisthokont invention. No other bands typically copurify with ODA complexes from ciliates, Chlamydomonas, sea urchin sperm, or mussel gill cilia. However, one or two additional unidentified bands are seen in fish sperm dynein fractions (Ogawa et al., 1996; Ogawa and Inaba, 2006), the three coiled-coil proteins already discussed cofractionate with Ciona sperm dynein (Hozumi et al., 2006), and under select buffer conditions and fractionation methods the coiled-coil DC1 and DC2 proteins copurify with Chlamydomonas outer arms (Takada et al., 2002). On the basis of the copurification of an ODA10p homologue with ODA complexes from Ciona sperm, we speculate that Chlamydomonas ODA10p also interacts directly with outer dynein arms but that this interaction is relatively weak and not maintained under typical extraction and purification conditions.
ODA10p has only some of the properties expected of a subunit of an accessory docking complex. In Chlamydomonas, ODA10p remains associated with flagella after detergent treatment and is extractable in half-molar salt solutions, but unlike the DC, ODA10p fails to copurify with outer dynein arms (Figure 2). In addition, the flagellar assembly of ODA10p is independent of the assembly of outer dynein arms, the docking complex, or the presence of the ODA8 gene product, consistent with the model of the AC as an independent axonemal complex (Wirschell et al., 2004). The instability of ODA10p in the oda5 mutant suggests the existence of a mutually dependent coiled-coil dimer between ODA10p and ODA5p, which may be similar to the dimer between mutually dependent coiled-coil proteins DC1 and DC2 in the docking complex (Wakabayashi et al., 2001).
Surprisingly, our functional tests do not support the model of ODA10p as the subunit of a complex involved in anchoring dynein onto axonemal microtubules. Our observation that dynein extracted from wild-type axonemes can rebind to oda10 axonemes at high affinity and in an ATP-insensitive manner, even though ODA10p is not present (Figure 4), combined with previous experiments showing that fractions containing only 12S and 18S dynein are sufficient to reconstitute functional outer dynein arms onto oda5 axonemes (Takada and Kamiya, 1994), shows that dynein that has already assembled once onto doublets in vivo retains its high-affinity binding state for doublet docking sites after salt extraction and no longer requires ODA5p and ODA10p to rebind. In contrast, experiments with DC mutants such as oda1 and oda3 show that the DC is required for outer dynein arm binding in vitro (Takada and Kamiya, 1994; Takada et al., 2002). We therefore hypothesize that the AC is an essential complex for the initial in vivo assembly of outer dynein arms but is not needed for dynein function postassembly. Because assembled outer dynein arm complexes can be immunoprecipitated from cytoplasmic extracts of oda5, oda8, and oda10 cells (Fowkes and Mitchell, 1998), the AC is also not essential for the initial cytoplasmic assembly steps, but further tests are needed to see whether the dynein that assembles in an oda10 cytoplasm is fully competent to bind to axonemes in vitro. The essential function of the AC thus could occur in the cytoplasm, during IFT transport, or at a late step of final association of dynein complexes with axonemal binding sites.
MATERIALS AND METHODS
Strains and culture conditions
The following strains of Chlamydomonas reinhardtii were obtained from the Chlamydomonas Genetics Center (University of Minnesota, Minneapolis, MN): wild-type 137c,mt- (CC-124), 137c,mt+ (CC-125), S1D2,mt- (CC-2290), mutant strains pf28,mt- (CC-1877) (an allele of oda2; Mitchell and Rosenbaum, 1985), oda1,mt+ (CC-2228), oda5,mt+(CC-2236), oda8,mt+(CC-2242), oda10-1,mt+ (CC-2246), and oda10-1,mt-(CC-2247) (Kamiya, 1988); strains oda5,ODA5HA (CC-4030) and insertional mutants oda10-2,mt- and oda10-2,mt-,arg7 (Wirschell et al., 2004) were obtained from George Witman (University of Massachusetts Medical School, Worcester, MA). oda5,oda10, oda5,oda8, oda8,oda10, oda5,oda10,ODA10HA, and oda8,oda10,ODA10HA were constructed in our laboratory. Strains with oda phenotype, oda5,oda10, oda8,oda10, and oda5,oda8, were selected from nonparental ditype tetrads of the crosses between oda5 and oda10, oda8 and oda10, and oda5 and oda8, respectively. The double mutant strains were crossed with oda10,ODA10HA to create oda5,oda10,ODA10HA and oda8,oda10,ODA10HA from nonparental ditype tetrads. oda mutant strains that expressed ODA10HA were selected using whole-cell Western blots probed with anti-HA antibody (see Antibodies and immunodetection for details). oda10-2,ODA10HA was created by cotransformation of HA-tagged ODA10 cDNA with ARG2 gene plasmid pJD67 into an arginine-dependent oda10-2,arg7 strain. Mating and tetrad analyses were performed according to standard protocols (Harris, 1989; Dutcher, 1995).
Antibodies and immunodetection
Antibodies used include rat monoclonal anti-HA epitope 3F10 (Roche, Indianapolis, IN), NAB1 (Agrisera AB, Vannas, Sweden), anti-ODA-IC2 (C11.4; Mitchell and Rosenbaum, 1986), and rabbit polyclonal anti-ODA10. Rabbit polyclonal anti-ODA10 was created by subcloning a cDNA region encoding an N-terminal fragment of ODA10p into the pMAL-p2 vector (New England BioLabs, Ipswich, MA) to make bacterially expressed SDS–PAGE-purified maltose-binding fusion proteins of ODA10p (amino acids 41–133), which was used to immunize rabbits for antibody production (Covance, Berkeley, CA). Antibodies were affinity purified against glutathione S-transferase fusion protein of the full-length ODA10 protein (Supplemental Figure S1). For Western blots, horseradish peroxidase–labeled secondary antibodies were detected using Super Signal West Dura Extend Duration (Pierce Chemical, Rockford, IL). The chemiluminescent signals were detected using either radiography or Chemidoc MP Imager (Bio-Rad, Hercules, CA). The blots were quantified using ImageJ (National Institutes of Health, Bethesda, MD) or Chemidoc software (Bio-Rad).
Positional cloning
ODA10 locus was positionally cloned using previously described amplification polymorphisms (Rymarquis et al., 2005). To do linkage analysis between markers and the oda10 locus, we isolated DNA from 67 fast-swimming recombinant progeny from a cross between oda10-1 and polymorphic strain S1D2. We initially used known markers on chromosome 8 and then designed markers to map additional loci on chromosome 8 contigs predicted by the Chlamydomonas genome assemblies 3.0 and 4.0. Markers PBSQ, LI818, and MCA1 from a molecular mapping kit (Rymarquis et al., 2005) were used to locate candidate regions for ODA10 between LI818 and MCA1. An additional marker, V3S294 on scaffold 29 of version 3 of the C. reinhardtii map (www.jgi.doe.gov/; Merchant et al., 2007), was designed to amplify between markers LI818 and MCA1 and used to map ODA10 between V3S294 and MCA1. Amplification probe ODA10-6 was selected to identify a locus near candidate gene C_290012. V3S294 and ODA10-6 were designed by the Primer-BLAST Primer designing tool, using standard procedures (Rymarquis et al., 2005). Information regarding the markers is shown in Supplemental Table S1. Amplification was carried at 94°C for 1 min, the chosen temperature range or annealing temperature for 1 min, 72°C for 1 min (40 cycles), and 72°C for 10 min. NEB Taq polymerase with either NEB buffer or Advantage buffer SA, or Advantage Buffer with Advantage Polymerase (Clontech, Mountain View, CA), was used as noted.
cDNA cloning
Candidate gene model C_290012 on scaffold 29 on version 3 of the C. reinhardtii map was used to identify two expressed sequence tag (EST) sequences (AV640669 and AV640714) deposited by the Kazusa DNA Research Institute (Asamizu et al., 1999). The full insert of EST AV640669 clone HCL020c08_r was sequenced, and sequences were deposited at the National Center for Biotechnology Information (Accession No. KF442967). When this sequence was compared with the predicted mRNA encoded by C_290012 (XM_00169594), it extended one exon and identified five more exons that were not predicted by Joint Genome Institute (JGI) version 3 or 4 of the Chlamydomonas genome. The coding region in this cDNA has two plausible start codons within the same reading frame. The second start codon was selected for generating expression constructs, based on alignment of predicted translation products from homologues in closely related algae (Volvox, Micromonas). Restriction sites NdeI and NruI were inserted, respectively, at the 5′ and 3′ ends of this predicted coding region, using a QuikChange II XL Site-directed Mutagenesis kit (Invitrogen, Carlsbad, CA). The coding region was cloned between NdeI and NruI sites in expression vector pGenDlinkerHA (Duquesnoy et al., 2009).
Transformations
DNA was introduced into Chlamydomonas using the glass bead method as described previously (Kindle et al., 1989). Tagged ODA10 expression plasmids were cotransformed with pJD67 plasmid into oda10-2,arg7,mt-(51b) mutant. Individual transformant colonies were screened for rescued phenotype in liquid under a light microscope. Cotransformation rates were 4.7–6.8%. Expression levels of tagged ODA10p in rescued strains were determined using anti-HA Western blots on whole-cell samples.
Protein extraction and fractionation
Flagella and flagellar fractions were prepared as described previously (Ahmed and Mitchell, 2005). High-salt extract of axonemes of oda10,ODA10HA prepared with 0.6 M NaCl were dialyzed against 500× volume HMDEK (30 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 5 M MgSO4, 1 mM dithiothreitol [DTT], 0.5 mM K acetate, pH 7.4) overnight at 4°C with one buffer change after the first hour. The high-salt extract was frozen in liquid nitrogen and kept at −75ºC until use. The dialyzed dynein extracts were separated in sucrose gradient as described before (Takada et al., 1994). Cytoplasmic extracts were made as described previously (Ahmed and Mitchell, 2005). Broken cells were pelleted in a microcentrifuge for 15 min, and supernatants were spun again for 30 min. The cytoplasmic extracts were either used immediately or frozen in liquid nitrogen and stored at −75°C until use.
Binding and dissociation assay
Axonemes were used within 24 h of preparation and were incubated with either crude dynein extract or pooled sucrose gradient fractions at ∼1:1 stoichiometric ratio (by cell count) on ice for 1 h, with or without 1 mM ATP. These mixtures were spun in a microcentrifuge at top speed for 5 min. The pellet was resuspended in fresh HMDEK. For dissociation assay, the resuspended pellets were incubated on the ice for the noted length of time and microcentrifuged at top speed for 5 min. The pellets were resuspended in HMDEK and mixed with equal amount of 2× SDS sample buffer. Supernatants were concentrated by acetone precipitation and resuspended in 1XSDS-SB, so that the final volumes of pellets and corresponding supernatants were equal.
Motility analysis
Swimming speed was measure using videomicroscopy (30 frames/s) of at least 50 cells under a Zeiss Axioskop light microscope (Carl Zeiss, Jena, Germany) with dark-field illumination through a 635-nm cutoff filter. Motility paths were tracked and average swimming speeds were measured using CellTrak 1.5 software (Motion Analysis, Santa Rosa, CA). Beat frequency was measured with stroboscopic illumination.
Computational analysis
The BLAST server at www.ncbi.nlm.nih.gov/BLAST (Altschul et al., 1997) and www.genome.jgi.doe.gov (Grigoriev et al., 2012) were used to search for homologous sequences. PairCoil (Berger et al., 1995) was used to determine whether a protein has a high probability of forming coiled coils, and COILS (Lupas et al., 1991) was used to determine regions of high coiled-coil probability. Antigenicity of the peptide was determined using web.expasy.org/protparam/(Gasteiger et al., 2005). Multiple sequence alignments of protein sequences selected with BLASTp (scores <1E-10) in the genomes of an alga (Micromonas), a basal chordate (Ciona), a fish (Danio), and a mammal (Mus) using Chlamydomonas ODA1, ODA3, ODA5, or ODA10 as bait were generated by MAFFT using MEGA5 (Tamura et al., 2011). An unrooted tree was generated from the alignment by the PHYLIP neighbor-joining method with a Jones–Taylor–Thornton distance matrix using SeaView (Gouy et al., 2010).
Supplementary Material
Acknowledgments
We thank the Kazusa DNA Research Institute for kindly providing cDNA plasmids, the G. Witman laboratory for a gift of the oda10-2 strain, and Mary Porter and Maureen Wirschell for communication regarding the project. We thank Judy Freshour, Brandon Smith, and Pauruv Desai for comments and technical support. This work was supported by National Institutes of Health Grant R01 044228 to D.R.M.
Abbreviations used:
- AC
accessory complex
- DC
docking complex
- ODA
outer dynein arm
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-06-0310) on October 2, 2013.
REFERENCES
- Ahmed NT, Gao C, Lucker BF, Cole DG, Mitchell DR. ODA16 aids axonemal outer row dynein assembly through an interaction with the intraflagellar transport machinery. J Cell Biol. 2008;183:313–322. doi: 10.1083/jcb.200802025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed NT, Mitchell DR. ODA16p, a Chlamydomonas flagellar protein needed for dynein assembly. Mol Biol Cell. 2005;16:5004–5012. doi: 10.1091/mbc.E05-07-0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asamizu E, Nakamura Y, Sato S, Fukuzawa H, Tabata S. A large scale structural analysis of cDNAs in a unicellular green alga, Chlamydomonas reinhardtii. I. Generation of 3433 non-redundant expressed sequence tags. DNA Res. 1999;6:369–373. doi: 10.1093/dnares/6.6.369. [DOI] [PubMed] [Google Scholar]
- Berger B, Wilson DB, Wolf E, Tonchev T, Milla M, Kim PS. Predicting coiled coils by use of pairwise residue correlations. Proc Natl Acad Sci USA. 1995;92:8259–8263. doi: 10.1073/pnas.92.18.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey DM, Inaba K, Pazour GJ, Takada S, Wakabayashi K, Wilkerson CG, Kamiya R, Witman GB. DC3, the 21-kDa subunit of the outer dynein arm-docking complex (ODA-DC), is a novel EF-hand protein important for assembly of both the outer arm and the ODA-DC. Mol Biol Cell. 2003;14:3650–3663. doi: 10.1091/mbc.E03-01-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duquesnoy P, et al. Loss-of-function mutations in the human ortholog of Chlamydomonas reinhardtii ODA7 disrupt dynein arm assembly and cause primary ciliary dyskinesia. Am J Hum Genet. 2009;85:890–896. doi: 10.1016/j.ajhg.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher SK. Mating and tetrad analysis in Chlamydomonas reinhardtii. Methods Cell Biol. 1995;47:531–540. doi: 10.1016/s0091-679x(08)60857-2. [DOI] [PubMed] [Google Scholar]
- Fowkes ME, Mitchell DR. The role of preassembled cytoplasmic complexes in assembly of flagellar dynein subunits. Mol Biol Cell. 1998;9:2337–2347. doi: 10.1091/mbc.9.9.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freshour J, Yokoyama R, Mitchell DR. Chlamydomonas flagellar outer row dynein assembly protein ODA7 interacts with both outer row and I1 inner row dyneins. J Biol Chem. 2007;282:5404–5412. doi: 10.1074/jbc.M607509200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the ExPASy server. In: JM Walker., editor. The Proteomics Protocols Handbook. Totowa, NJ: Humana Press; 2005. pp. 571–607. [Google Scholar]
- Gokhale A, Wirschell M, Sale W, Mitchell DR. Dynein motility in cilia and flagella. In: Hirose K, Amos LA, editors. Handbook of Dynein. Singapore: Pan Stanford; 2012. pp. 203–244. [Google Scholar]
- Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Grigoriev IV, et al. The genome portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res. 2012;40:D26–D32. doi: 10.1093/nar/gkr947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimo LT, Fenton RD. Interaction of Chlamydomonas dynein with tubulin. Cell Motil. Cytoskeleton. 1988;9:129–139. doi: 10.1002/cm.970090205. [DOI] [PubMed] [Google Scholar]
- Haimo LT, Telzer BR, Rosenbaum JL. Dynein binds to and crossbridges cytoplasmic microtubules. Proc Natl Acad Sci USA. 1979;76:5759–5763. doi: 10.1073/pnas.76.11.5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EH. The Chlamydomonas Source Book: A Comprehensive Guide to Biology and Laboratory Use. San Diego, CA: Academic Press; 1989. [DOI] [PubMed] [Google Scholar]
- Harris EH, Boynton JE, Gillham NW. Chlamydomonas nuclear gene loci. In: O'Brien SJ, editor. Genetic Maps. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1987. pp. 258–265. [Google Scholar]
- Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N Engl J Med. 2011;364:1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hozumi A, Satouh Y, Makino Y, Toda T, Ide H, Ogawa K, King SM, Inaba K. Molecular characterization of Ciona sperm outer arm dynein reveals multiple components related to outer arm docking complex protein 2. Cell Motil. Cytoskeleton. 2006;63:591–603. doi: 10.1002/cm.20146. [DOI] [PubMed] [Google Scholar]
- Kamiya R. Mutations at twelve independent loci result in absence of outer dynein arms in Chlamydomonas reinhardtii. J Cell Biol. 1988;107:2253–2258. doi: 10.1083/jcb.107.6.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya R. Functional diversity of axonemal dyneins. In: Hirose K, Amos LA, editors. Handbook of Dynein. Singapore: Pan Stanford; 2012. pp. 267–284. [Google Scholar]
- Kindle KL, Schnell RA, Fernandez E, Lefebvre PA. Stable nuclear transformation of Chlamydomonas using the Chlamydomonas gene for nitrate reductase. J Cell Biol. 1989;109:2589–2601. doi: 10.1083/jcb.109.6.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutoulis A, Pazour GJ, Wilkerson CG, Inaba K, Sheng H, Takada S, Witman GB. The Chlamydomonas reinhardtii ODA3 gene encodes a protein of the outer dynein arm docking complex. J Cell Biol. 1997;137:1069–1080. doi: 10.1083/jcb.137.5.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van DM, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Merchant SS, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DR, Rosenbaum JL. A motile Chlamydomonas flagellar mutant that lacks outer dynein arms. J Cell Biol. 1985;100:1228–1234. doi: 10.1083/jcb.100.4.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DR, Rosenbaum JL. Protein-protein interactions in the 18S ATPase of Chlamydomonas outer dynein arms. Cell Motil Cytoskeleton. 1986;6:510–520. doi: 10.1002/cm.970060510. [DOI] [PubMed] [Google Scholar]
- Mitchison HM, et al. Mutations in axonemal dynein assembly factor DNAAF3 cause primary ciliary dyskinesia. Nat Genet. 2012;44:381–382. doi: 10.1038/ng.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa K, Inaba K. Ap58: A novel in situ outer dynein arm-binding protein. Biochem Biophys Res Commun. 2006;343:385–390. doi: 10.1016/j.bbrc.2006.02.157. [DOI] [PubMed] [Google Scholar]
- Ogawa K, Takai H, Ogiwara A, Yokota E, Shimizu T, Inaba K, Mohri H. Is outer arm dynein intermediate chain 1 multifunctional. Mol Biol Cell. 1996;7:1895–1907. doi: 10.1091/mbc.7.12.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omran H, et al. Ktu/PF13 is required for cytoplasmic pre-assembly of axonemal dyneins. Nature. 2008;456:611–616. doi: 10.1038/nature07471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onoufriadis A, et al. Splice-site mutations in the axonemal outer dynein arm docking complex gene CCDC114 cause primary ciliary dyskinesia. Am J Hum Genet. 2013;92:88–98. doi: 10.1016/j.ajhg.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister KK, Witman GB. Subfractionation of Chlamydomonas 18 S dynein into two unique subunits containing ATPase activity. J Biol Chem. 1984;259:12072–12080. [PubMed] [Google Scholar]
- Rymarquis LA, Handley JM, Thomas M, Stern DB. Beyond complementation. Map-based cloning in Chlamydomonas reinhardtii. Plant Physiol. 2005;137:557–566. doi: 10.1104/pp.104.054221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara H, Kamiya R. Functional recombination of outer dynein arms with outer arm-missing flagellar axonemes of a Chlamydomonas mutant. J Cell Sci. 1989;92:77–83. [Google Scholar]
- Takada S, Kamiya R. Functional reconstitution of Chlamydomonas outer dynein arms from alpha-beta and gamma subunits: requirement of a third factor. J Cell Biol. 1994;126:737–745. doi: 10.1083/jcb.126.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S, Sakakibara H, Kamiya R. Three-headed outer arm dynein from Chlamydomonas that can functionally combine with outer-arm-missing axonemes. J Biochem. 1992;111:758–762. doi: 10.1093/oxfordjournals.jbchem.a123832. [DOI] [PubMed] [Google Scholar]
- Takada S, Wilkerson CG, Wakabayashi K, Kamiya R, Witman GB. The outer dynein arm-docking complex: composition and characterization of a subunit (oda1) necessary for outer arm assembly. Mol Biol Cell. 2002;13:1015–1029. doi: 10.1091/mbc.01-04-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi K, Takada S, Witman GB, Kamiya R. Transport and arrangement of the outer-dynein-arm docking complex in the flagella of Chlamydomonas mutants that lack outer dynein arms. Cell Motil Cytoskeleton. 2001;48:277–286. doi: 10.1002/cm.1015. [DOI] [PubMed] [Google Scholar]
- Walczak CE, Marchese-Ragona SP, Nelson DL. Immunological comparison of 22S, 19S, and 12S dyneins from Paramecium cilia. Cell Motil Cytoskeleton. 1993;24:17–28. doi: 10.1002/cm.970240103. [DOI] [PubMed] [Google Scholar]
- Wirschell M, et al. The nexin-dynein regulatory complex subunit DRC1 is essential for motile cilia function in algae and humans. Nat Genet. 2013;45:262–268. doi: 10.1038/ng.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirschell M, Pazour G, Yoda A, Hirono M, Kamiya R, Witman GB. Oda5p, a novel axonemal protein required for assembly of the outer dynein arm and an associated adenylate kinase. Mol Biol Cell. 2004;15:2729–2741. doi: 10.1091/mbc.E03-11-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.