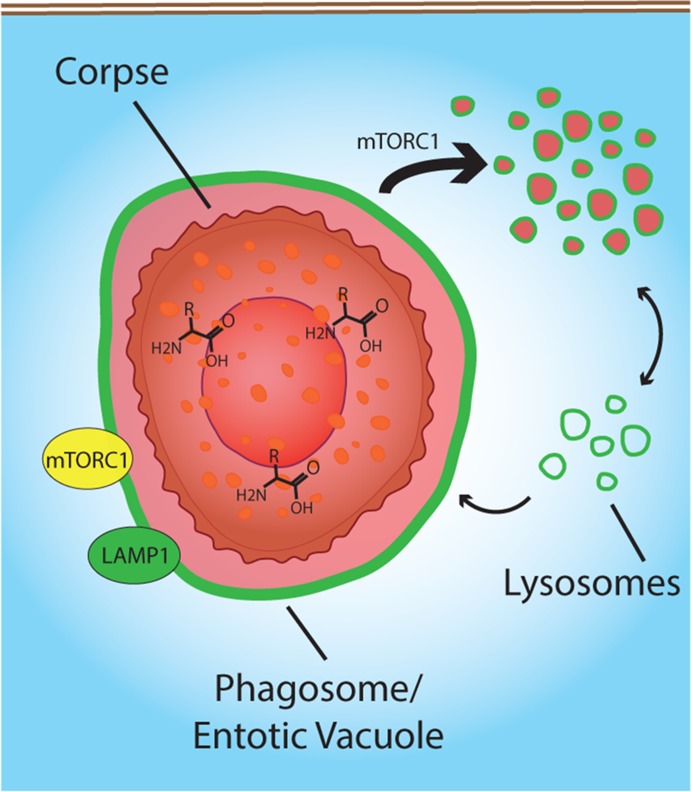
Phagosomes and entotic vacuoles harboring engulfed cells undergo an mTOR-regulated process of vacuolar fission that distributes engulfed contents throughout lysosome networks. Amino acid recovery from engulfed cells activates mTORC1 and rescues engulfing cells from the effects of amino acid starvation.
Abstract
Macroendocytic vacuoles formed by phagocytosis, or the live-cell engulfment program entosis, undergo sequential steps of maturation, leading to the fusion of lysosomes that digest internalized cargo. After cargo digestion, nutrients must be exported to the cytosol, and vacuole membranes must be processed by mechanisms that remain poorly defined. Here we find that phagosomes and entotic vacuoles undergo a late maturation step characterized by fission, which redistributes vacuolar contents into lysosomal networks. Vacuole fission is regulated by the serine/threonine protein kinase mammalian target of rapamycin complex 1 (mTORC1), which localizes to vacuole membranes surrounding engulfed cells. Degrading engulfed cells supply engulfing cells with amino acids that are used in translation, and rescue cell survival and mTORC1 activity in starved macrophages and tumor cells. These data identify a late stage of phagocytosis and entosis that involves processing of large vacuoles by mTOR-regulated membrane fission.
INTRODUCTION
The elimination of dying cells by phagocytosis is fundamental to the development and homeostasis of multicellular organisms (Elliott and Ravichandran, 2010). Failure to engulf or properly degrade apoptotic cells leads to tissue damage and inflammation and can cause developmental defects and autoimmune disease (Elliott and Ravichandran, 2010). Like phagocytosis, entosis is a form of cell engulfment, but entosis targets live cells rather than dead cells, and whereas phagocytosis occurs in normal development, the “cell-in-cell” structures that form by entosis are primarily found in human tumors (Overholtzer et al., 2007). Entosis is proposed to be a tumor-suppressive cell death mechanism (Florey et al., 2011) but may also contribute to genomic changes that promote tumor progression (Krajcovic et al., 2011; Krajcovic and Overholtzer, 2012).
The vacuoles formed by phagocytic or entotic cell engulfments undergo sequential steps of maturation leading to the fusion of lysosomes. Lysosomal hydrolytic enzymes degrade dead cells during the final stages of phagocytosis (Han and Ravichandran, 2011) or execute the death of live engulfed cells during entosis (Florey et al., 2011). The events that control phagosome maturation upstream of lysosome fusion are well described and include sequential lipid phosphorylations and protein recruitments, highlighted by the transition from Rab-5– to Rab-7–positive stages (Kinchen and Ravichandran, 2008; Fairn and Grinstein, 2012). The autophagy protein light chain 3 (LC3) is also targeted to apoptotic cell phagosomes and is believed to play a role in maturation, potentially by promoting membrane–membrane fusion (Florey et al., 2011; Florey and Overholtzer, 2012; Martinez et al., 2011). Like phagosomes, entotic vacuoles also recruit LC3, in a manner dependent on autophagy proteins such as Atg5 and Atg7 (Florey et al., 2011). For entosis, autophagy proteins regulate lysosome fusion and cell death (Florey et al., 2011), whereas during phagocytosis, autophagy proteins facilitate corpse degradation (Florey et al., 2011; Martinez et al., 2011).
Although the overall sequence of maturation events that controls lysosome fusion is well described (Kinchen and Ravichandran, 2008; Flannagan et al., 2012), by contrast the subsequent stages that follow the degradation of engulfed cargo after lysosome fusion are poorly understood. Here we investigate the maturation of phagosomes and entotic vacuoles post lysosome fusion and identify a late stage involving nutrient recovery from engulfed cells and mammalian target of rapamycin (mTOR)-regulated vacuolar fission.
RESULTS
Engulfing cells recover amino acids from internalized cells and reactivate mTOR complex 1
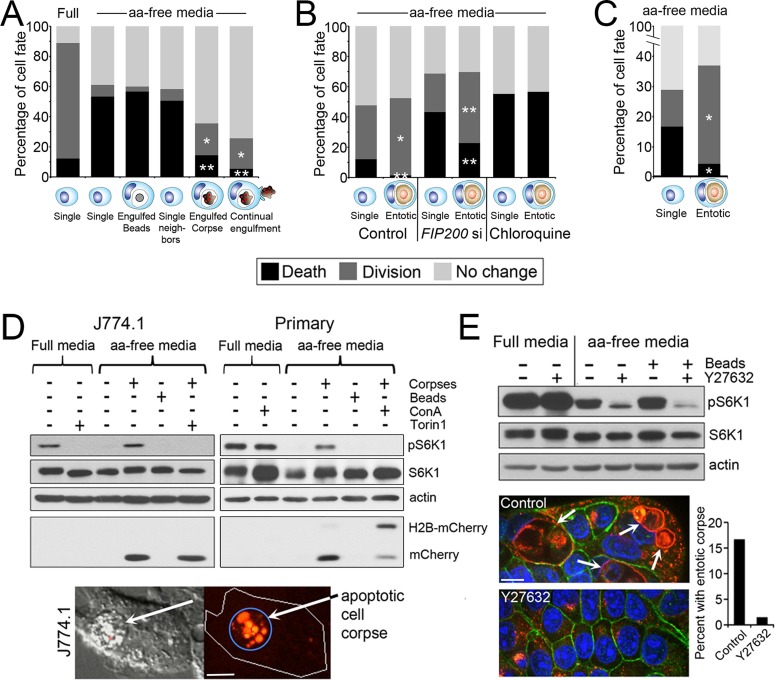
We hypothesized that entosis might provide engulfing cells with nutrients after the degradation of internalized cells that could, like nutrient recovery by autophagy, promote cell survival during starvation. To investigate whether cell engulfment mechanisms can generally promote the survival of starved cells, we first examined whether phagocytosis of apoptotic cells by macrophages could rescue the effects of amino acid starvation. Compared to full media conditions, J774.1 mouse macrophages cultured in the absence of amino acids ceased proliferating and readily underwent cell death (Figure 1A). These effects of amino acid withdrawal were rescued by the engulfment of apoptotic cell corpses but not uncoated latex beads or beads coated with the “eat-me” engulfment signal phosphatidylserine (PS), suggesting that phagocytes can recover amino acids from engulfed cells (Figure 1A and Supplemental Figure S1, A and B). Like macrophages with engulfed apoptotic corpses, MCF7 breast tumor cells and immortalized MCF10A mammary epithelial cells with engulfed entotic cell corpses also exhibited increased cell survival and proliferation in amino acid–free media compared with their neighbors (Figure 1, B and C). Even under conditions of autophagy inhibition, by the small interfering RNA (siRNA)–mediated knockdown of the Ulk kinase complex member FIP200, as described (Florey et al., 2011), starvation-induced apoptosis was significantly inhibited, and proliferation was enhanced by the digestion of a cell corpse (Figure 1B and Supplemental Figure S1C). Entotic cells had no advantage over their neighbors when treated with the lysosome inhibitor chloroquine, suggesting that lysosome function is required for entotic cells to gain a nutrient advantage, presumably by lysosomal digestion of corpses and export of amino acids (Figure 1B).
FIGURE 1:
Cell engulfment rescues the effects of amino acid deprivation. (A) Engulfment of apoptotic corpses rescues macrophages from amino acid deprivation–induced cell death. Fates of J774.1 macrophages cultured in full and amino acid (aa)–free media in the presence or absence of apoptotic corpses, determined by time-lapse microscopy for 48 h. Single, single cells, n = 90 for full and 90 for aa-free media; engulfed beads, cells that engulfed from one to five latex beads, n = 60; single neighbors, single cells within same microscopic fields as corpse-engulfing cells, n = 168; engulfed corpse, cells with one or two corpses engulfed before start of time lapse, n = 90; continual engulfment, cells supplied with corpses that were engulfed continuously throughout the time lapse, n = 74. *p < 0.02, **p < 0.001 (when compared with single cells in aa-free media; chi-squared). Data are from at least three independent experiments. (B) Entosis rescues MCF10A cells from the effects of amino acid deprivation. Fates of MCF10A cells (single) and MCF10A cells with an entotic cell corpse (entotic) in aa-free media time lapsed for 48 h (control and FIP200 siRNA [FIP200 si]–treated cells) or 18 h (chloroquine-treated cells). Control single cells, n = 360; control entotic, n = 137; FIP200si single, n = 558; FIP200si entotic, n = 179; chloroquine single, n = 92; chloroquine entotic, n = 37. *p < 0.002, **p < 0.001 (chi-squared). Data are from at least three independent experiments. (C) Entotic MCF-7 cells (n = 192) harboring an entotic corpse are rescued from cell death and proliferation arrest compared with single control cells (n = 567) in aa-free media. p < 0.001 (chi-squared). Cells were examined for 48 h by time-lapse microscopy. Data are from at least three independent experiments. (D) mTORC1 is reactivated in aa-free media by corpse digestion in J774.1 macrophages (left blots) and primary bone marrow–derived macrophages (right blots). Western blots show restoration of phosphorylated S6-kinase threonine 389 (pS6K) by apoptotic corpse engulfment but not latex bead engulfment in macrophages cultured in aa-free media. pS6K restoration is blocked by treatment with the mTOR inhibitor Torin1 and the lysosome inhibitor ConA. Torin1 was added to cultures 1 h before cell lysis; ConA was added for the duration of the experiment. Untreated macrophages digest the corpse-specific marker H2B-mCherry into free mCherry protein, which is inhibited by ConA treatment. Images show apoptotic corpse expressing H2B-mCherry (red fluorescence, arrow) engulfed by J774.1 macrophage. Bar, 10 μm. (E) mTORC1 is reactivated by entosis. Western blots show higher levels of pS6K in MCF-7 cells cultured in aa-free media under control conditions, with 15% of cells harboring entotic corpses (quantified in graph; representative of two independent experiments), compared with entosis-inhibited conditions with Y-27632 treatment. Images show entotic cell corpses (white arrows) in control cultures, which are absent from Y-27632–treated cultures. Immunofluorescence staining for Lamp1 (red) and β-catenin (green) and 4′,6-diamidino-2-phenylindole (DAPI)–stained nuclei (blue). Bar, 10 μm.
To examine whether the engulfment and degradation of cell corpses could restore amino acid signaling, we investigated whether mTOR complex 1 (mTORC1) activity, which requires amino acids (Hara et al., 1998; Kim et al., 2002), could be restored by cell engulfment in starved cells. Whereas J774.1 macrophages cultured in amino acid–free media displayed a loss of phosphorylation of threonine 389 of S6-ribosomal protein kinase (pS6K), a known substrate of mTORC1 (Pearson et al., 1995), the engulfment of apoptotic corpses by starved macrophages restored pS6K in an mTOR inhibitor (Torin1)–sensitive manner (Figure 1D). Unlike apoptotic corpses, the engulfment of uncoated or PS-coated latex beads did not restore pS6K, suggesting that the degradation of corpses, rather than simply their engulfment, could underlie reactivation of mTORC1 (Figure 1D and Supplemental Figure S1D). Phosphorylation of S6K was not increased by phagocytosis when macrophages were cultured in full media, demonstrating that the phagocytic uptake of apoptotic corpses does not generally increase mTORC1 signaling (Supplemental Figure S2). Similar to J774.1 mouse macrophages, primary mouse bone marrow–derived macrophages also reactivated mTORC1 in amino acid–free media when fed with apoptotic corpses but not with latex beads (Figure 1D). mTORC1 reactivation was blocked by treatment with the lysosome acidification inhibitor concanamycin A (ConA), which inhibited corpse degradation, as shown by the rescue of full-length, corpse-derived H2B-mCherry protein (Figure 1D), suggesting that the phagocytic engulfment and lysosomal degradation of apoptotic cell corpses, but not latex beads, reactivates mTORC1 signaling in macrophages, which is consistent with the recovery of amino acids from engulfed cells. Indeed we found that macrophages incorporate amino acids derived from engulfed corpses into protein synthesized by cells either in the absence or presence of exogenous amino acids supplied in media, demonstrating that engulfing cells recover amino acids from apoptotic corpses (Supplemental Figure S3).
To examine whether entosis, like phagocytosis, could reactivate mTORC1, we cultured MCF7 breast tumor cells under conditions permissive or restrictive for entosis in the absence of amino acids and examined levels of pS6K by Western blotting. Treatment of MCF7 cells with the Rho-kinase inhibitor Y-27632, a known blocker of entosis (Overholtzer et al., 2007), reduced pS6K in cells cultured in the absence of amino acids and prevented entosis while having no effect on pS6K in cells cultured in full media (Figure 1E). Unlike entotic cell corpses, the engulfment of latex beads had no effect on pS6K, suggesting that corpse degradation, rather than engulfment, may reactivate mTORC1. These data suggest that entosis, like phagocytosis, can supply cells with amino acids that signal to activate mTORC1.
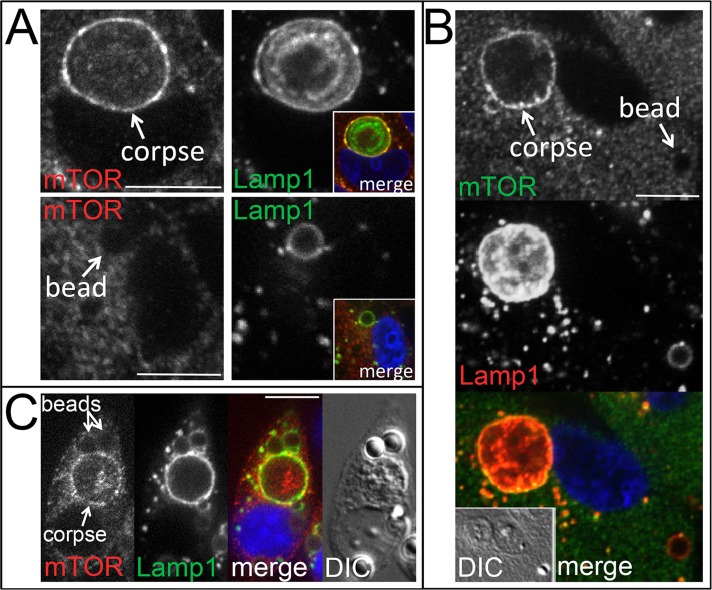
Amino acids are known to signal to mTORC1 through Rag GTPases that recruit mTORC1 to lysosomal membranes where the upstream activator Rheb resides (Sancak et al., 2008). Given that activated mTORC1 localizes to lysosomes, we examined whether mTOR would localize to phagosome and entotic vacuole membranes when reactivated by corpse degradation in amino acid–free media. Indeed, endogenous mTOR localized prominently to the lysosomal vacuoles harboring degrading entotic corpses but not to lysosomal vacuoles harboring latex beads, which did not reactivate mTORC1 (Figure 2A). Of interest, when cells were allowed to engulf both neighboring cells and latex beads, mTOR localized to the lysosomal vacuoles harboring degrading cells and not to those with engulfed beads (Figure 2B), even though RagC GTPase localized to both compartments (Supplemental Figure S4). Similarly, mTOR localized to phagosomes harboring degrading apoptotic cells in macrophages but not to those with uncoated or PS-coated latex beads, even within the same cell (Figure 2C and Supplemental Figure S1E), demonstrating compartment-specific reactivation of mTORC1, which is consistent with a recently proposed inside-out model of signaling to mTORC1 by amino acids from within lysosomes (Zoncu et al., 2011).
FIGURE 2:
mTOR is recruited to lysosomal vacuoles harboring engulfed cells but not latex beads. (A) Top, mTOR (left) is recruited to corpse-containing entotic vacuole (arrow) (MCF10A), where it colocalizes with Lamp1 (right); inset, colocalization of immunostained mTOR (red) and Lamp1 (green). DAPI-stained nucleus is shown in blue. Bottom, mTOR does not recruit to bead-containing vacuole (left, arrow), which is marked by Lamp1 (right) (MCF10A); inset, merged image with DAPI-stained nucleus in blue. All are confocal microscopic images. Bars, 10 μm. (B) mTOR localizes to corpse-containing entotic vacuole (labeled corpse with arrow; 70% were positive for mTOR, n = 37) but not to bead-containing lysosomal vacuole (bead, arrow; 2.3% positive for mTOR, n = 84) in the same MCF10A cell. Top and middle, confocal images of immunofluorescence for mTOR and Lamp1; bottom, merge with DAPI-stained nucleus (blue); inset, DIC. (C) mTOR localizes to apoptotic cell phagosomes (52% positive for mTOR, n = 54) but not latex bead phagosomes (1.9% positive for mTOR, n = 154) in J774.1 macrophages. Confocal microscopic images show macrophage with an engulfed apoptotic corpse and two beads, as indicated, stained for mTOR and Lamp1 by immunofluorescence. Right, merged image with DAPI-stained nucleus (blue) and DIC.
Phagosomes and entotic vacuoles undergo a late maturation phase involving fission
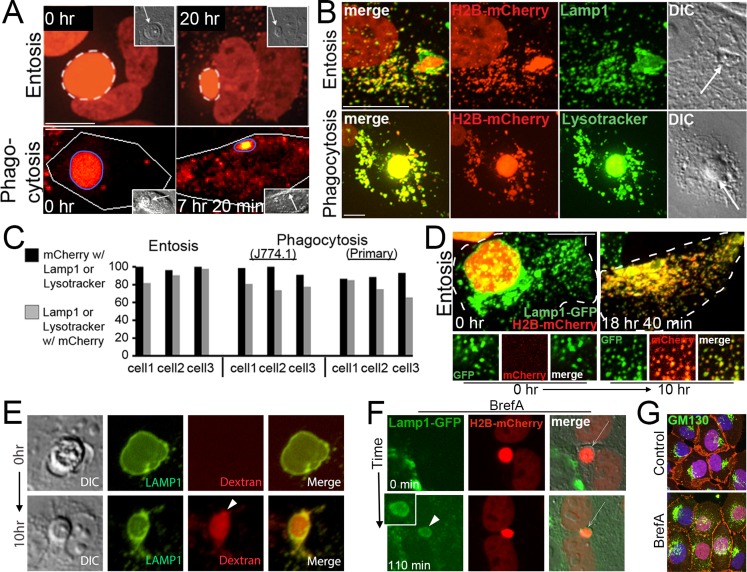
In our time-lapse analyses of the degradation of engulfed entotic or apoptotic cell corpses, we noted that mCherry fluorescence, derived from the corpse-specific H2B-mCherry marker, filled engulfing cells with puncta as corpses were degraded and vacuoles shrank in size (Figure 3A and Supplemental Videos S1 and S2). mCherry-labeled puncta colocalized with the lysosomal membrane protein Lamp1 in entotic cells and LysoTracker in macrophages, suggesting that entotic vacuole or phagosome contents were distributed throughout engulfing cells in lysosomes (Figure 3B). Indeed, the majority of Lamp1-positive vesicles in entotic cells colocalized with mCherry fluorescence, and conversely, nearly all mCherry-positive vesicles were Lamp1 positive (Figure 3C). Similarly, the majority of LysoTracker-positive vesicles in macrophages were labeled with mCherry, and nearly all mCherry puncta were labeled with LysoTracker (Figure 3C), suggesting that at late stages of corpse degradation, the luminal contents of phagosomes and entotic vacuoles are dispersed throughout the lysosome network of engulfing cells. Remarkably, time-lapse imaging of an entotic engulfing cell expressing Lamp1–green fluorescent protein (GFP) harboring an H2B-mCherry–expressing engulfed corpse demonstrated the redistribution of mCherry fluorescence from entotic vacuoles into the Lamp1-labeled lysosome network (Figure 3D and Supplemental Video S3). These data identify a late stage of entosis and phagocytosis that is associated with fission of vacuoles that redistributes a tracer of the lumen into the lysosome network of engulfing cells.
FIGURE 3:
Phagosomes and entotic vacuoles undergo fission. (A) mCherry fluorescence from an entotic corpse expressing H2B-mCherry (top) or an apoptotic corpse expressing H2B-mCherry (bottom) appears as puncta in the cytoplasm of engulfing cells (right) as entotic vacuoles (white dashed circle) and phagosomes (blue circle) shrink over time. Images from time-lapse analysis show mCherry fluorescence (red); insets, DIC; arrows indicate corpses. Note that entotic engulfing cell in top is binucleate and also expresses H2B-mCherry. Bars, 10 μm. See Supplemental Videos S1 and S2. (B) mCherry puncta colocalize with Lamp1-GFP in entotic cells (top) and LysoTracker in primary macrophages (bottom). Maximum projection images of mCherry fluorescence (red), Lamp1-GFP and LysoTracker green (green), and merged and DIC images as indicated; arrows indicate cell corpses. Bars, 10 μm. (C) Quantification of colocalization between mCherry and Lamp1 or LysoTracker in individual phagocytic or entotic engulfing cells. Black bars, percentage of mCherry puncta that colocalize with Lamp1 or LysoTracker; gray bars, percentage of Lamp1 or LysoTracker vesicles that colocalize with mCherry. Percentages for three individual engulfing cells for entosis (MCF10A) or phagocytosis (J774.1 and primary macrophages) are shown. For entosis, cell 1, n = 124 vesicles (black bar) and 137 vesicles (gray bar); cell 2, n = 132, 137; and cell 3, n = 131, 125. For J774.1 phagocytosis, cell 1, n = 66, 83; cell 2, n = 54, 76; and cell 3, n = 55, 67. For phagocytosis with primary macrophages, cell 1, n = 96, 100; cell 2, n = 174, 171; cell 3, n = 157, 143. (D) Corpse-derived mCherry fluorescence redistributes from the entotic vacuole to the lysosome network of the engulfing cell. Maximum projections from confocal time-lapse analysis of a Lamp1-GFP–expressing MCF10A cell with an engulfed entotic corpse expressing H2B-mCherry. Note that Lamp1-GFP–labeled lysosomes acquire red fluorescence over time as entotic vacuole undergoes fission. Top, merged green and red fluorescence of whole cell outlined with hatched white line. Bottom, individual and merged fluorescent channels at time 0 and 10 h for an area of cell cytoplasm. Bar, 10 μm. See Supplemental Video S3. (E) Entotic vacuole, labeled with Lamp1-GFP (green), accumulates 10-kDa red fluorescent dextran (arrowhead) from media over the course of 10 h as the vacuole shrinks in size. Also see Supplemental Figure S5a. (F) An entotic vacuole (arrow) in brefeldin A–treated cells fuses with Lamp1-GFP–labeled lysosomes (green) after polyethylene glycol (PEG)–initiated cell fusion. Arrowhead indicates Lamp1-GFP accumulation at entotic vacuole. Time indicates minutes after cell fusion. Also see Supplemental Figure S5, b and c. (G) Brefeldin A treatment as in F disrupts the Golgi, as shown by GM130 immunostaining (green) in mixed cultures used for cell fusion in F. β-Catenin immunostaining and H2B-mCherry are shown in red. Note that the H2B-mCherry–expressing cells used in this experiment do not express Lamp1-GFP (green), so cells with red nuclei show only GM130 immunofluorescence in the green channel, whereas cells without red nuclei show GM130 immunofluorescence and Lamp1-GFP fluorescence in the green channel.
Vacuoles undergoing fission also exhibit continual lysosome fusion
The redistribution of mCherry fluorescence from phagosomes and entotic vacuoles into lysosome networks prompted consideration of the mechanism of how preexisting lysosomes (see Supplemental Video S3) acquire vacuolar contents. We reasoned that lysosomes could undergo continual fusion/fission with corpse-containing vacuoles, which would rapidly disperse mCherry throughout lysosome networks as corpses are digested. To examine whether vacuoles undergo continual fusion with the endocytic pathway even as they shrink, we first added red fluorescent dextran to media as a tracer of the endocytic pathway and examined whether entotic corpse-containing vacuoles of different sizes, representing different stages of shrinkage, would accumulate red fluorescence. After adding dextran to media, we found that both large and small entotic vacuoles accumulated red fluorescence, consistent with continual fusion of endosomes to vacuoles during shrinkage (Figure 3E and Supplemental Figure S5A). To examine whether lysosomes undergo continual fusion to vacuoles, we devised a strategy in which cells with Lamp1-GFP–labeled lysosomes could be fused, by adding polyethylene glycol to cultures, to neighboring cells harboring corpse-containing vacuoles at different stages of shrinkage and the fusion of Lamp1-GFP–labeled vesicles to entotic vacuoles could be examined over time. Remarkably, after cell fusion, Lamp1-GFP fluorescence rapidly accumulated at corpse-containing vacuoles of different sizes (Supplemental Figure S5B and Supplemental Video S4). Lamp1-GFP accumulation at vacuoles was not inhibited by treatment of cells with brefeldin A, which disrupts the Golgi network, suggesting that Lamp1-GFP recruitment is not due to trafficking from the Golgi but instead occurs by lysosome fusion (Figure 3, F and G, Supplemental Figure S5C, and Supplemental Video S5). Together these data support a model in which continual fusion/fission of lysosomes with shrinking vacuoles underlies the rapid accumulation of a tracer of the vacuole lumen into lysosome networks.
mTORC1 regulates phagosome and entotic vacuole fission
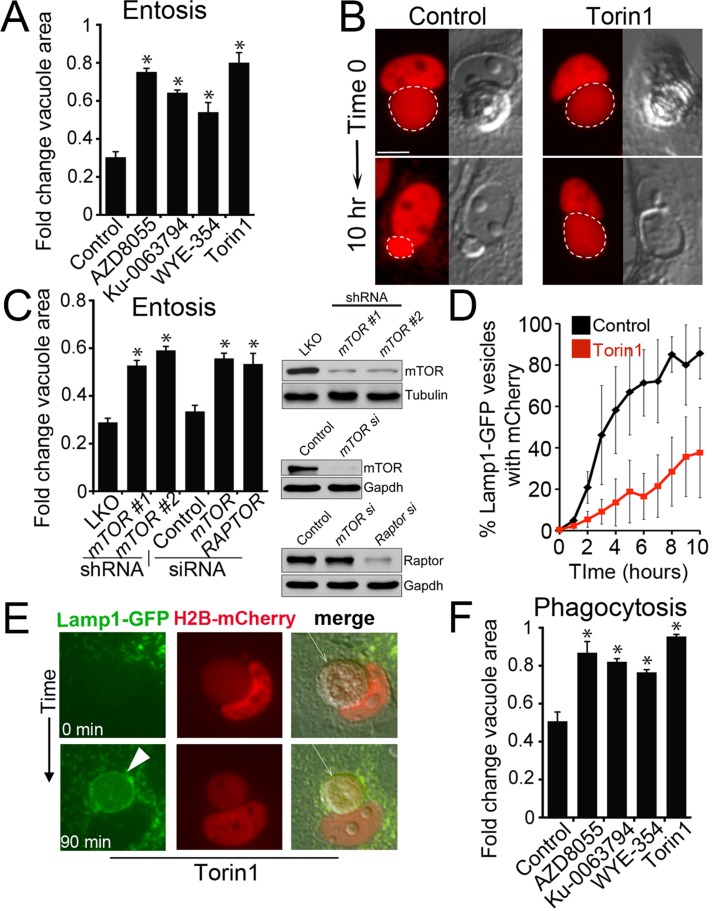
mTOR was recently shown to regulate the tubulation of lysosomes during autophagy that resets lysosome networks from autolysosomes (Yu et al., 2010), but how the much larger lysosomal vacuoles produced by the cell engulfment mechanisms phagocytosis and entosis are processed is unknown. Because entotic vacuoles also displayed tubulation during fission (Supplemental Video S6), we speculated that mTOR, which was recruited to phagosome and entotic vacuole membranes, might also play a role in fission of these vacuoles. To examine the requirement of mTOR activity for entotic vacuole fission, we treated cells with inhibitors of mTOR kinase activity (Thoreen et al., 2009; Liu et al., 2012) and monitored corpse-containing cells by time-lapse microscopy. Whereas control vacuoles underwent rapid shrinkage associated with corpse degradation, vacuoles in mTOR-inhibited cells shrank slowly, even as corpses degraded (Figure 4, A and B, and Supplemental Videos S7 and S8). mTOR inhibition slowed vacuole shrinkage when cells were cultured in full media or amino acid–free media (Supplemental Figure S6). Similarly, siRNA or short hairpin RNA (shRNA)–mediated knockdown of mTOR, as well as the mTORC1 component Raptor (Kim et al., 2002), slowed vacuole shrinkage (Figure 4C). The accumulation of mCherry fluorescence in lysosome networks was also delayed by inhibition of mTOR, consistent with a role of mTOR in vacuole fission (Figure 4D). Of interest the inhibition of mTOR had no effect on the recruitment of Lamp1-GFP to vacuoles (Figure 4E, Supplemental Figure S5D, and Supplemental Video S9), suggesting that mTOR regulates vacuole fission and does not play a role in lysosome fusion. Like entotic vacuoles, phagosomes in macrophages harboring apoptotic cells were slowed for shrinkage when cells were treated with inhibitors of mTOR, consistent with a requirement of mTOR activity for rapid phagosome fission (Figure 4F). Together these data demonstrate that mTOR regulates a program of vacuolar fission that is associated with the degradation of engulfed cells.
FIGURE 4:
Entotic vacuole and phagosome fission is regulated by mTOR. (A) Inhibitors of mTOR slow entotic vacuole shrinkage. Fold change of vacuole area after 10 h for entotic vacuoles (MCF10A cells) measured by time-lapse microscopy of control and mTOR inhibitor–treated cells cultured in full media. Error bars, SEM for n = 3 independent experiments. Total cell number analyzed for control, n = 57; AZD8055, n = 55; Ku-0063794, n = 55; WYE-354, n = 51; Torin1, n = 55. *p < 0.02 (Student's t test). (B) mTOR inhibition slows vacuole fission. Control entotic vacuole (left) undergoes rapid shrinkage, producing mCherry-labeled puncta as corpse degrades, whereas mTOR-inhibited cell (right) is delayed for fission, leaving a large vacuole as the corpse degrades. Images show mCherry fluorescence (red) and DIC. Dashed line, entotic vacuoles marked by mCherry. Bar, 10 μm. See Supplemental Videos S7 and S8. (C) shRNA- and siRNA-mediated knockdowns of mTOR and Raptor slow entotic vacuole shrinkage. Fold change of vacuole area after 10 h for entotic vacuoles (MCF10A cells) determined by time-lapse microscopy of control and mTOR or Raptor-knockdown cells. Western blots to the right of graph show protein knockdowns. Error bars, SEM for three independent experiments. Total cell number analyzed for pLKO, n = 121; mTOR #1 shRNA, n = 89; mTOR #2 shRNA, n = 131; control (nontargeting siRNA), n = 93; mTOR siRNA, n = 87; Raptor siRNA, n = 97. *p < 0.02 (Student's t test). (D) Torin1 treatment slows the redistribution of corpse-derived mCherry from the entotic vacuole to the lysosome network of engulfing cells. Quantification of percentage of Lamp1-GFP–labeled lysosomes with mCherry fluorescence in four individual control and Torin1-treated cells, examined by confocal microscopy and quantified in every z-plane at 0.5-μm intervals through the entire cell every hour for 10 h. Error bars, SD. After 10 h Torin1-treated cells have significantly less mCherry fluorescence in lysosome networks (p < 0.01; Student's t test). (E) An entotic vacuole (arrow) in Torin1-treated cells fuses with Lamp1-GFP–labeled lysosomes (green) after PEG-initiated cell fusion. Arrowhead, Lamp1-GFP accumulation at entotic vacuole. Time indicates minutes after cell fusion. Also see Supplemental Figure S5D and Supplemental Video S9. (F) mTOR inhibition slows phagosome shrinkage in full media. Fold change of phagosome area after 2 h. Error bars, SEM for three independent experiments. Total cell number analyzed for control, n = 50, AZD8055; n = 47, Ku-0063794, n = 30; WYE-354, n = 38; Torin1, n = 54. *p < 0.02 (Student's t test).
DISCUSSION
Although much is known regarding the complex mechanisms that control phagosome maturation up to the point of lysosome fusion (Kinchen and Ravichandran, 2008), by contrast surprisingly little is known about the final stages of degradation of engulfed cargo, including export of digested components and processing of vacuole membranes. Here we describe a late stage of phagosome and entotic vacuole maturation involving fission that both reduces the size of phagosomes and entotic vacuoles and redistributes degraded cargo into the lysosome network of phagocytes in an mTOR-regulated manner (Figure 5).
FIGURE 5:
Model for phagosome and entotic vacuole fission. The degradation of cell corpses after phagocytosis or entosis recruits mTORC1 (yellow) to Lamp1-positive (green) vacuole membranes. mTORC1 regulates vacuolar fission, which redistributes the luminal contents of phagosomes and entotic vacuoles into the lysosome network of engulfing cells. mTORC1-regulated fission (thick arrow) accelerates vacuole shrinkage by overcoming a constant rate of lysosome fusion (thin arrow). Amino acid export from phagosomes and entotic vacuoles likely provides the survival advantage of engulfing cells in amino acid–free media.
We found that Lamp1-GFP–labeled lysosomes continue to fuse with vacuoles even as they undergo shrinkage (Figure 3E and Supplemental Figure S5). Continual fusion/fission between lysosomes and the shrinking vacuole would be predicted to disperse vacuolar contents throughout lysosome networks, as we demonstrate occurs during vacuolar shrinkage. These cycles of fusion and fission are reminiscent of similar activities previously reported between late endosomes and lysosomes (Bright et al., 2005). Of interest, entotic vacuoles remain positive for Rab-7, a late endosome marker, at all sizes, representing different stages of shrinkage, suggesting that vacuoles are in fact hybrid late endosome/lysosome organelles (Supplemental Figure S7). Like late endosomes, large vacuoles such as phagosomes and entotic vacuoles may therefore continually fuse with lysosomes and also undergo fission in order to maintain lysosome network homeostasis.
In our working model for phagosome/entotic vacuole shrinkage, mTOR activity may facilitate an increased rate of vacuolar fission that overrides a constant rate of lysosome fusion in order to rapidly shrink vacuoles with degraded contents (Figure 5). Given that mTOR inhibition slows vacuole shrinkage but does not lead to vacuole growth despite continual lysosome fusion, which is mTOR independent (Figure 4E and Supplemental Figure S5D), we hypothesize that basal, mTOR-independent fission of vacuoles balances continual lysosome fusion and mTOR recruitment to vacuoles increases fission, by an as-yet-unidentified mechanism, to facilitate vacuolar shrinkage (Figure 5). In this model, the degradation of engulfed proteins into amino acids may serve as a convenient mechanism to initiate vacuolar shrinkage by recruiting mTORC1. Indeed, the specific localization of mTOR to phagosome and entotic vacuole membranes and not to latex-bead–containing lysosomal membranes, even within the same cells, demonstrates a potential role for the luminal contents of phagosomes or entotic vacuoles in specification of the sites of mTOR reactivation. Whereas amino acid signaling to mTORC1 was recently shown to involve leucine binding to leucyl-tRNA synthetase in the cytosol (Han et al., 2012) or glutaminolysis (Duran et al., 2012), our data appear to support another recently proposed model in which amino acids from within lysosomal compartments recruit mTORC1 by inducing a conformation change to the vacuolar H(+)-adenosine triphosphatase (v-ATPase), which binds to the mTORC1 recruiting complex Ragulator (Sancak et al., 2010; Zoncu et al., 2011).
The fission of vesicles from phagosomes was previously described to recycle transmembrane proteins, including phagocyte receptors, to the plasma membrane (Bajno et al., 2000; Botelho et al., 2000; Beron et al., 2001; Lee et al., 2005). Recycling fission regulated by Rab-11 maintains vacuolar size by balancing membrane addition due to vesicle fusion with membrane removal and is required to maintain sufficient levels of phagocyte receptors at the plasma membrane to support continued phagocytosis (Chen et al., 2010; Silver and Harrison, 2011). By contrast to recycling fission, the late stage of fission described here shrinks phagosomes and entotic vacuoles and is associated with the terminal stages of degradation of engulfed cargo. The regulation of phagosome fission by mTOR is reminiscent of the terminal stages of autophagy, in which mTOR-dependent membrane tubulation from large autolysosomes re-forms lysosome networks in a program termed autophagic lysosome reformation (ALR; Yu et al., 2010). TORC1 in yeast has also been shown to mediate vacuole fission, which may contribute to changes in vacuole size upon nutrient restriction and induction of autophagy (Michaillat et al., 2012). Like autolysosomes in mammalian cells, entotic vacuoles exhibit tubulation during fission, but unlike ALR, the complete blockage of tubulation with nocodazole has no effect on entotic vacuole shrinkage, suggesting that tubulation is not required overall to shrink large vacuoles (Supplemental Figure S8). Moreover, whereas mTOR activity is required for tubulation during ALR, treatment of cells with the mTOR inhibitor Torin1 has no effect on tubulation from entotic vacuoles, despite slowing vacuolar shrinkage (Supplemental Figure S8B and Figure 4A). In addition, whereas the lysosomes produced by ALR are initially devoid of luminal contents (Yu et al., 2010), the vesicles produced by phagosome and entotic vacuole fission harbor luminal contents, as evidenced by mCherry fluorescence (Supplemental Figure S8A). The mTOR-regulated substrates that control either ALR or vacuole shrinkage remain to be identified, and their discovery may shed light on whether mTOR facilitates lysosome network homeostasis in these different pathways by a common mechanism.
The data presented here demonstrate that phagocytosis and entosis can rescue cells from death induced by amino acid deprivation. Although we cannot rule out that signaling initiated by phagocytosis plays a role in the recruitment and reactivation of mTORC1 or the rescue of engulfing cells from starvation-induced cell death, the blockage of mTORC1 reactivation by lysosome inhibition suggests that corpse degradation is required for restored mTORC1 activity, potentially by supplying amino acids to engulfing cells. We also note that neighboring cells in the cell fate analyses that did not engulf apoptotic corpses did not exhibit a survival advantage, suggesting that secreted factors in media are unlikely to account for rescue from the effects of starvation. We found that macrophages incorporate amino acids derived from engulfed corpses into protein synthesized by cells either in the absence or presence of exogenous amino acids supplied in media (Supplemental Figure S3), which demonstrates that engulfing cells do recover amino acids from apoptotic corpses. For entosis, the rescue of engulfing cells from the effects of nutrient deprivation suggests that cannibalistic cell behavior, which is found in human cancers (Overholtzer and Brugge, 2008), could, like autophagy, provide cells with nutrients to sustain cell viability and support proliferation to contribute to tumor progression. For heterotypic cannibalism it was shown previously that the ingestion of live T-cells by metastatic melanoma cells could similarly rescue from cell death induced by starvation (Lugini et al., 2006). For homotypic cell cannibalism, in which neighboring tumor cells ingest each other, the death of some tumor cells, which is predicted to slow tumor growth, is balanced by the recovery of nutrients that could endow engulfing cells survival advantages that, like changes in cell ploidy (Krajcovic et al., 2011), may promote tumor progression.
MATERIALS AND METHODS
Cell culture and reagents
MCF10A cells were cultured as described (Florey et al., 2011). J774.1 mouse macrophages (American Type Culture Collection [ATCC], Manassas, VA) and MCF-7 cells (Lombardi Cancer Center at Georgetown University, Washington, DC) were cultured in DMEM plus 10% heat-inactivated fetal bovine serum (FBS) with penicillin/streptomycin. Primary mouse bone marrow–derived macrophages were prepared and cultured as described (Shree et al., 2011). U937 cells (ATCC) were cultured in RPMI-1640 plus 10% heat-inactivated FBS with penicillin/streptomycin. Amino acid–free medium was prepared by dialyzing heat-inactivated FBS (for J774.1 and primary macrophages) or horse serum (for MCF10A) for 4 h, followed by overnight at 4°C in phosphate-buffered saline (PBS) in MWCO 3500 dialysis tubing (21-152-9; Fisherbrand, Pittsburgh, PA) and addition to base media prepared without amino acids to 10% final. Cells expressing the H2B-mCherry nuclear marker or Lamp1-GFP were prepared by transducing cells with retroviruses made with the pBabe-H2B-mCherry and pRetro-Lamp1-GFP constructs, as described (Florey et al., 2011). Cell lines expressing fluorescent markers were selected by flow cytometry. Cells were treated with Y-27632 (Tocris Bioscience) at 10 μM, concanamycin A (Sigma, St. Louis, MO) at 100 nM, latrunculin B (L5288; Sigma) at 2.5 μM, Torin-1 (Tocris Bioscience, Bristol, UK) at 500 nM, AZD-8055 (Selleck Chemicals, Houston, TX) at 500 nM, WYE-354 (Selleck Chemicals) at 1 μM, Ku-0063794 (Selleck Chemicals) at 1 μM, Chloroquine (Sigma) at 12.5 μM, nocodazole (M1404; Sigma) at 1 μM, and brefeldin A (B7651; Sigma) at 10 μg/ml. Inhibitors were added to cultures ∼30 min before the start of biological assays unless indicated otherwise.
Time-lapse microscopy
Cells were cultured on glass-bottom dishes (MatTek, Ashland, MA), and time-lapse microscopy was performed in 37°C and 5% CO2 live-cell incubation chambers, as described (Florey et al., 2011). Fluorescence and differential interference contrast (DIC) images were acquired every 10–15 min for 48 h using a Nikon Ti-E inverted microscope attached to a CoolSNAP charge-coupled device camera (Photometrics, Tucson, AZ) and NIS Elements software (Nikon, Melville, NY). Cell fates, including cell death, division, and no change, were quantified manually using Elements software. Individual engulfing cells, or three randomly chosen neighboring cells per field, or three single cells per field, were analyzed and assigned a single fate. Phagosome and entotic vacuole areas were determined by mCherry fluorescence and quantified manually using Elements software. For entosis, time zero for vacuole shrinkage assays was determined as the time point when mCherry protein diffused out of the nucleus of a cell corpse and filled the vacuole. For phagocytosis, vacuole shrinkage assays were performed on phagocytic events that occurred during the time lapse, and time zero was determined as the time point when mCherry diffused from apoptotic corpses to fill the vacuole.
Phagocytosis assays
We plated 1 × 105 J774.1 or 4 × 105 primary bone marrow–derived mouse macrophages onto tissue culture plastic (for Western blotting) or glass-bottom dishes (for live-cell imaging) (MatTek) and cultured them in full medium for 24 h (for primary macrophages) or full medium with 200 U/ml interferon-γ for 48 h (for J774.1), as described (Florey et al., 2011). After 24–48 h, cells were washed three times in PBS and overlaid with 2 × 106 apoptotic cell corpses or latex beads in full or amino acid–free media. Apoptotic corpses were prepared by ultraviolet irradiation of H2B-mCherry–expressing U937 cells, as described (Florey et al., 2011), and were harvested by centrifugation after overnight incubation. Macrophages were lysed for Western blotting 48 h after addition of corpses, and unengulfed apoptotic corpses were removed before lysis by washing three times in PBS.
Immunofluorescence
The following antibodies were used for immunofluorescence (IF): anti-mTOR (2983; Cell Signaling), anti-Lamp1 (for MCF10A and MCF-7 cells; BD555798), anti-Lamp1 (for mouse macrophages; BD553792), and anti-RagC (9480; Cell Signaling, Danvers, MA). IF was performed on cells cultured on glass-bottom dishes (MatTek), essentially as described (Overholtzer et al., 2007). For staining of mTOR and Lamp1 in MCF10A cells, fixation was performed in 1:1 methanol:acetone at −20°C for 5 min. For IF of endogenous Lamp1- and mCherry-containing vesicles, MFC10A cells were fixed in 2% glutaraldehyde for 15 min at room temperature, followed by 1:1 methanol:acetone for 5 min at −20°C. For IF on J774 mouse macrophages, cells were fixed in 1:1 methanol:acetone at −20°C for 5 min. LysoTracker staining was performed by adding 50 nM LysoTracker Green (Invitrogen, Carlsbad, CA) to culture media for 30 min before imaging live cells. Colocalization of Lamp1 and mCherry vesicles, or LysoTracker Green and mCherry vesicles, was performed by manual counting of vesicles in z-stack confocal images using Volocity software (Perkin Elmer, Waltham, MA). Confocal microscopy was performed with the Ultraview Vox spinning-disk confocal system (Perkin Elmer) equipped with a Yokogawa CSU-X1 spinning-disk head and an electron-multiplying charge-coupled device camera (Hamamatsu C9100-13) coupled to a Nikon Ti-E microscope.
Western blotting
Cells were lysed in ice-cold RIPA buffer, and SDS–PAGE and Western blotting were performed as described (Florey et al., 2011), using Anti-mTOR (2983; Cell Signaling), anti-Raptor (2280; Cell Signaling), anti–S6-Kinase (9202; Cell Signaling), anti–phospho-S6-kinase threonine 389 (9234; Cell Signaling), anti-mCherry (ab125096; Abcam), anti-actin (A1978; Sigma), anti-Tubulin (3873; Cell Signaling), anti–glyceraldehyde-3-phosphate dehydrogenase (sc25778; Santa Cruz Biotechnology, Dallas, TX), and anti-LC3B (L7543; Sigma) antibodies.
siRNA and shRNA
siGenome SMART pool siRNAs against human FIP200, mTOR, and RAPTOR, as well as Control 2 nontargeting siRNAs, were obtained from Dharmacon (Pittsburgh, PA). MCF10A cells were seeded in a six-well plate at 6 × 104 per well and transfected with 100 nM siRNA using Oligofectamine (Invitrogen). Cells were assayed 48 h posttransfection. shRNA constructs targeting mTOR in the pLKO.1 vector were acquired from Addgene (plasmids 1855 and 1856; Sarbassov et al., 2005). Cell pools expressing the mTOR shRNAs were assayed 72 h after transduction. Control cells were transduced with the empty LKO.1 vector.
Entosis assays
MCF-7 cells were plated overnight onto glass-bottom dishes (Mattek) in the presence or absence of Y-27632 to block entosis. Cultures were switched to amino acid–free media the next day, in the presence or absence of Y-27632 and latex beads, and cultured for 24 h before lysis and analysis by Western blotting. Parallel plates were stained by immunofluorescence to quantify the percentage of cells with entotic corpses, identified by Lamp1 immunostaining and confocal microscopy.
PS-coated beads
Streptavidin-coated 6-μm microspheres (24158; Polysciences, Warrington, PA) were incubated with biotin-phosphatidylserine (L-31B16; Echelon) in PBS for 1 h under constant rolling at room temperature. Annexin–fluorescein isothiocyanate (Invitrogen) staining was performed according to the manufacturer's protocol.
Dextran labeling
To follow the fusion of endosomes with entotic vacuoles using fluorescent dextran as an endocytic tracer, we plated MCF10A-Lamp1-GFP cells onto glass coverslip dishes overnight and then added red fluorescent 10-kDa dextran (D1817; Invitrogen) to growth media at 100 μg/ml concentration, followed by time-lapse imaging of cells with entotic vacuoles of different sizes representing different stages of shrinkage. Ten of 10 entotic vacuoles imaged for 10 h acquired red dextran from the culture media.
Cell fusion assay
To examine the fusion of Lamp1-GFP–labeled lysosomes to entotic vacuoles, we plated MCF10A cells expressing Lamp1-GFP onto glass coverslips at a 1:1 ratio with MCF10A cells expressing H2B-mCherry. The next day, cells with an H2B-mCherry–labeled entotic corpse adjacent to Lamp1-GFP–expressing cells were identified and the stage positions marked, followed by the initiation of cell fusion by treatment of cells with a 1:1 polyethylene glycol (P3640; Sigma):serum-free growth medium mixture for 2.5 min in the tissue culture hood. After washing at least three times in PBS, cells were placed back onto the microscope, and cell fusions were imaged by time-lapse microscopy.
[35S]cysteine/methionine-labeled apoptotic cell engulfment
We labeled 2 × 106 U937 cells with 1.1 mCi of 35S labeling mix (NEG772007MC; Perkin Elmer) in 10 ml of labeling medium (81% RPMI-1640 without Cys/Met/l-Glut [7513; Sigma], 9% dialyzed FBS, 9% RPMI-1640, and 1% FBS) for 24 h. Radiolabeled U937 corpses were centrifuged and washed twice with PBS to remove 35S labeling medium. Filtered medium was prepared by collecting supernatant from apoptotic corpses after a 24-h incubation, followed by centrifugation and filtration through a 0.45-μm filter. GFP immunoprecipitation was performed using a GFP-Trap kit (ChromoTek) according to the manufacturer's protocol. Macrophages were lysed for Western blotting 24 h after addition of corpses, and unengulfed apoptotic corpses were removed before lysis by washing three times in PBS.
Statistics
The indicated p values were obtained using Student's t test or the chi-squared test, as indicated.
Supplementary Material
Acknowledgments
This work was supported by National Cancer Institute Grants CA177697 (M.O.) and CA148967 (J.A.J.) the Louis V. Gerstner, Jr. Young Investigators Fund (M.O.), and the Benjamin Friedman Research Fund (M.O). We thank members of the Overholtzer laboratory for critical reading of the manuscript.
Abbreviations used:
- aa
amino acid
- ALR
autophagic lysosome reformation
- ConA
concanamycin A
- DIC
differential interference contrast
- FIP200
focal adhesion kinase family interacting protein of 200 kDa
- Gapdh
glyceraldehyde-3-phosphate dehydrogenase
- GFP
green fluorescent protein
- GM130
Golgi matrix protein of 130 kDa
- IF
immunofluorescence
- Lamp1
lysosomal-associated membrane protein 1
- LC3
microtubule-associated protein 1 light chain 3
- mTOR
mammalian target of rapamycin
- mTORC1
mammalian target of rapamycin complex 1
- PS
phosphatidylserine
- pS6K1
phosphorylated threonine 389 of ribosomal protein S6 kinase 1
- Raptor
regulatory-associated protein of mTOR
- Rheb
ras homologue enriched in brain
- shRNA
short hairpin RNA
- siRNA
small interfering RNA
- S6K1
ribosomal protein S6 kinase 1
- Ulk
Unc-51-like kinase
- v-ATPase
vacuolar-type H(+)-adenosine triphosphatase
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-07-0408) on October 2, 2013.
REFERENCES
- Bajno L, Peng XR, Schreiber AD, Moore HP, Trimble WS, Grinstein S. Focal exocytosis of VAMP3-containing vesicles at sites of phagosome formation. J Cell Biol. 2000;149:697–706. doi: 10.1083/jcb.149.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beron W, Mayorga LS, Colombo MI, Stahl PD. Recruitment of coat-protein-complex proteins on to phagosomal membranes is regulated by a brefeldin A-sensitive ADP-ribosylation factor. Biochem J. 2001;355:409–415. doi: 10.1042/0264-6021:3550409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botelho RJ, Hackam DJ, Schreiber AD, Grinstein S. Role of COPI in phagosome maturation. J Biol Chem. 2000;275:15717–15727. doi: 10.1074/jbc.M910068199. [DOI] [PubMed] [Google Scholar]
- Bright NA, Gratian MJ, Luzio JP. Endocytic delivery to lysosomes mediated by concurrent fusion and kissing events in living cells. Curr Biol. 2005;15:360–365. doi: 10.1016/j.cub.2005.01.049. [DOI] [PubMed] [Google Scholar]
- Chen D, Xiao H, Zhang K, Wang B, Gao Z, Jian Y, Qi X, Sun J, Miao L, Yang C. Retromer is required for apoptotic cell clearance by phagocytic receptor recycling. Science. 2010;327:1261–1264. doi: 10.1126/science.1184840. [DOI] [PubMed] [Google Scholar]
- Duran RV, Oppliger W, Robitaille AM, Heiserich L, Skendaj R, Gottlieb E, Hall MN. Glutaminolysis activates Rag-mTORC1 signaling. Mol Cell. 2012;47:349–358. doi: 10.1016/j.molcel.2012.05.043. [DOI] [PubMed] [Google Scholar]
- Elliott MR, Ravichandran KS. Clearance of apoptotic cells: implications in health and disease. J Cell Biol. 2010;189:1059–1070. doi: 10.1083/jcb.201004096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairn GD, Grinstein S. How nascent phagosomes mature to become phagolysosomes. Trends Immunol. 2012;33:397–405. doi: 10.1016/j.it.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Flannagan RS, Jaumouille V, Grinstein S. The cell biology of phagocytosis. Annu Rev Pathol. 2012;7:61–98. doi: 10.1146/annurev-pathol-011811-132445. [DOI] [PubMed] [Google Scholar]
- Florey O, Kim SE, Sandoval CP, Haynes CM, Overholtzer M. Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat Cell Biol. 2011;13:1335–1343. doi: 10.1038/ncb2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florey O, Overholtzer M. Autophagy proteins in macroendocytic engulfment. Trends Cell Biol. 2012;22:374–380. doi: 10.1016/j.tcb.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han CZ, Ravichandran KS. Metabolic connections during apoptotic cell engulfment. Cell. 2011;147:1442–1445. doi: 10.1016/j.cell.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JM, Jeong SJ, Park MC, Kim G, Kwon NH, Kim HK, Ha SH, Ryu SH, Kim S. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell. 2012;149:410–424. doi: 10.1016/j.cell.2012.02.044. [DOI] [PubMed] [Google Scholar]
- Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- Kinchen JM, Ravichandran KS. Phagosome maturation: going through the acid test. Nat Rev Mol Cell Biol. 2008;9:781–795. doi: 10.1038/nrm2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajcovic M, et al. A non-genetic route to aneuploidy in human cancers. Nat Cell Biol. 2011;13:324–330. doi: 10.1038/ncb2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajcovic M, Overholtzer M. Mechanisms of ploidy increase in human cancers: a new role for cell cannibalism. Cancer Res. 2012;72:1596–1601. doi: 10.1158/0008-5472.CAN-11-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WL, Kim MK, Schreiber AD, Grinstein S. Role of ubiquitin and proteasomes in phagosome maturation. Mol Biol Cell. 2005;16:2077–2090. doi: 10.1091/mbc.E04-06-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, et al. Kinome-wide selectivity profiling of ATP-competitive mammalian target of rapamycin (mTOR) inhibitors and characterization of their binding kinetics. J Biol Chem. 2012;287:9742–9752. doi: 10.1074/jbc.M111.304485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugini L, et al. Cannibalism of live lymphocytes by human metastatic but not primary melanoma cells. Cancer Res. 2006;66:3629–3638. doi: 10.1158/0008-5472.CAN-05-3204. [DOI] [PubMed] [Google Scholar]
- Martinez J, Almendinger J, Oberst A, Ness R, Dillon CP, Fitzgerald P, Hengartner MO, Green DR. Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc Natl Acad Sci USA. 2011;108:17396–17401. doi: 10.1073/pnas.1113421108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaillat L, Baars TL, Mayer A. Cell-free reconstitution of vacuole membrane fragmentation reveals regulation of vacuole size and number by TORC1. Mol Biol Cell. 2012;23:881–895. doi: 10.1091/mbc.E11-08-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overholtzer M, Brugge JS. The cell biology of cell-in-cell structures. Nat Rev Mol Cell Biol. 2008;9:796–809. doi: 10.1038/nrm2504. [DOI] [PubMed] [Google Scholar]
- Overholtzer M, Mailleux AA, Mouneimne G, Normand G, Schnitt SJ, King RW, Cibas ES, Brugge JS. A nonapoptotic cell death process, entosis, that occurs by cell-in-cell invasion. Cell. 2007;131:966–979. doi: 10.1016/j.cell.2007.10.040. [DOI] [PubMed] [Google Scholar]
- Pearson RB, Dennis PB, Han JW, Williamson NA, Kozma SC, Wettenhall RE, Thomas G. The principal target of rapamycin-induced p70s6k inactivation is a novel phosphorylation site within a conserved hydrophobic domain. EMBO J. 1995;14:5279–5287. doi: 10.1002/j.1460-2075.1995.tb00212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Shree T, Olson OC, Elie BT, Kester JC, Garfall AL, Simpson K, Bell-McGuinn KM, Zabor EC, Brogi E, Joyce JA. Macrophages and cathepsin proteases blunt chemotherapeutic response in breast cancer. Genes Dev. 2011;25:2465–2479. doi: 10.1101/gad.180331.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver KE, Harrison RE. Kinesin 5B is necessary for delivery of membrane and receptors during FcgammaR-mediated phagocytosis. J Immunol. 2011;186:816–825. doi: 10.4049/jimmunol.1002161. [DOI] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465:942–946. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.