Abstract
Objective
Cytochrome P450 (CYP) enzymatic activity, which is influenced by CYP genetic polymorphism, is known to affect the inter-individual variation in the efficacy and tolerability of antidepressants in major depressive disorder (MDD). Escitalopram is metabolized by CYP2D6, and recent studies have reported a correlation between clinical outcomes and CYP2D6 genetic polymorphism. The purpose of this study was to determine the relationship between the CYP2D6 P34S polymorphism (C188T, rs1065852) and the efficacy of escitalopram treatment in Korean patients with MDD.
Methods
A total of 94 patients diagnosed with MDD were recruited for the study and their symptoms were evaluated using the 21-item Hamilton Depression Rating scale (HAMD-21). The association between the CYP2D6 P34S polymorphism and the clinical outcomes (remission and response) was investigated after 1, 2, 4, 8, and 12 weeks of escitalopram treatment using multiple logistic regression analysis and χ2 test.
Results
The proportion of P allele carriers (PP, PS) in remission status was greater than that of S allele homozygotes (SS) after 8 and 12 weeks of escitalopram treatment. Similarly, P allele carriers exhibited a greater treatment response after 8 and 12 weeks of escitalopram treatment than S allele homozygotes.
Conclusion
Our results suggest that the P allele of the CYP2D6 P34S polymorphism is a favorable factor in escitalopram treatment for MDD, and that the CYP2D6 P34S polymorphism may be a good genetic marker for predicting escitalopram treatment outcomes.
Keywords: CYP2D6 polymorphism, Major depressive disorder, Escitalopram, Treatment response
INTRODUCTION
Major depressive disorder (MDD) is characterized by heterogeneity of etiology and involves both genetic and environmental factors.1 Information on the effect of neurobiological factors on the treatment of MDD is limited. MDD patients are treated with a series of antidepressants depending on the severity and recurrence of the disease. Although numerous antidepressants are available for the treatment of MDD, a significant proportion of patients do not exhibit a response in enough dose and duration of antidepressant treatment, and experience tolerability or adverse effects of antidepressants that can result in discontinuation of treatment.2,3 The expectation that accurate individualization of antidepressant selection may optimize clinical outcomes has led psychopharmacogenetics to target personalized medicine, which can maximize treatment response and minimize the possibility of nonresponse, side effects, or dangerous adverse events in antidepressant treatment, based on patients' genetic variabilities.4
The cytochrome P450 (CYP) system in the liver is responsible for the first phase of the metabolism of numerous ingested chemicals, and CYP enzymes convert these substances into electrophilic intermediates, which are then conjugated by phase II enzymes to yield hydrophilic derivatives that can be excreted.5 Escitalopram is a selective serotonin reuptake inhibitor which is widely used in the treatment of MDD and anxiety disorders.6 Escitalopram is metabolized by CYP2D6, 2C19, and 3A4.7 Escitalopram is first metabolized via N-demethylation to S-desmethylcitalopram by CYP3A4 and 2C19.8 Then, this metabolite is further demethylated, mainly by CYP2D6, to form S-didemethylcitalopram.9 Escitalopram and S-desmethylcitalopram are the active forms of the metabolites with antidepressant effects, whereas S-didemethylcitalopram is an inactive form.8 Based on CYP enzymatic activity, patients taking escitalopram exhibit various plasma concentrations of escitalopram and its metabolites S-desmethylcitalopram and Sdidemethylcitalopram.10
CYP2D6 polymorphism is one of the most important genetic polymorphisms because of its influence on the enzymatic activity of CYP2D6, which is responsible for the metabolism of numerous antidepressants.11 Based on the metabolic activity of the CYP2D6 polymorphism, 4 phenotypes have been identified: extensive metabolizers (EMs), intermediate metabolizers (IMs), poor metabolizers (PMs), and ultrarapid metabolizers (UMs).12,13 PMs often exhibit significantly higher plasma drug concentrations. Therefore, PMs may have greater susceptibility to adverse effects, whereas UMs usually show subtherapeutic plasma concentrations resulting in decreased efficacy of antidepressants.14 In addition to the higher susceptibility of PMs to adverse effects of antidepressants, recent studies reported that PMs exhibit significantly lower treatment responses than EMs.15-18
The genetic polymorphism of CYP enzymes is not altered by medication. therefore, these can be bio-markers for predicting enzymatic activities related to the metabolism of escitalopram. Furthermore, a recent study suggested an association between genetic polymorphism of CYP enzymes and treatment response to escitalopram.19
There are 80 identified variations of the CYP2D6 gene among the CYP genetic superfamily.20 Ethnicity is a factor in the occurrence of CYP2D6 variability. For example, the allele frequency of CYP2D6*10 in Asians is about 50%,21 whereas it is only 1-2% in Caucasians.12 Differences in the allele frequency of CYP2D6*10 are also observed among Asian populations: 38.1% in Japanese 22 and 51.3% in Chinese.23 It has been reported that in Koreans the CYP2D6 alleles *1, *2, *5, *10, and *41 occur with a frequency exceeding 1%.24 Especially, a CYP2D6 genotyping study in the Korean population indicated that CYP2D6*10 was the most frequent allele in Koreans, with a frequency of 45%.24 The CYP2D6*10 allele includes the P34S (rs1065852) and S486T (rs1135840) mutations. The P34S mutation is a single nucleotide polymorphism (SNP) in *4, *10, *14A, *36, and many other alleles of CYP2D6,25 and is known to eliminate the important proline-rich ("PPGP") sequence near the amino terminus essential for the folding of the P450 enzyme.26,27 The P34S mutation of CYP2D6*10 makes the enzymatic activity very unstable, and it usually leads to reduced affinity for substrates.28 The relationship between the P34S mutation and drug metabolism by the CYP2D6 enzyme has urged recent pharmacogenetic studies to focus on the influence of this SNP on the clinical outcomes of several psychotropic treatments.19,29
We postulated that CYP2D6 P34S polymorphism is a most powerful marker for detecting CYP2D6*10 allele and other CYP2D6 alleles with P34S polymorphism are negligible in the study, because other alleles with P34S polymorphism have low frequency in Korean population, not exceeding 1%.24 Therefore, we chose the CYP2D6* 10 P34S polymorphism as a genetic marker for predicting escitalopram treatment outcomes in this study.
Although numerous studies have suggested that inter-individual variations in clinical outcomes and tolerability in the treatment of MDD are influenced by CYP2D6 polymorphism, few studies have focused on the CYP2D6*10 polymorphism and treatment response to escitalopram. The purpose of this study was to investigate the relationship between the CY P2D6*10 P34S polymorphism (C188T, rs1065852) and the efficacy of escitalopram treatment in Korean patients with MDD.
METHODS
Subjects and clinical assessments
A total of 94 patients diagnosed with MDD were recruited from Korea University Anam Hospital and enrolled from March 2009 through June 2011. Using the Structured Clinical Interview for DSM-IV Axis I disorders (SCID-I) and the Korean version of the Diagnostic Interview for Genetic Studies (KDIGS), trained psychiatrists examined all the subjects. The severity of depression was evaluated using the 21-item Hamilton Depression Rating (HAMD-21) scale. All subjects enrolled had a minimum score of 18 on the HAMD-21 scale. Exclusion criteria for enrollment were: 1) primary or co-morbid diagnoses of schizophrenia, schizoaffective disorder, bipolar disorder, dementia or MDD with psychotic features, based on DSM-IV criteria within the previous 6 months, 2) serious or unstable medical illness, 3) personal or family history of substance abuse/dependence. All subjects were at least 18 years of age. Patients already on other psychotropic medication were permitted a 2-week washout period. Demographic data, medical history, and laboratory data of all the subjects were documented. During the study treatment period, all subjects received escitalopram (Lexapro®; Lundbeck, Copenhagen, Denmark) at a daily dose of 5-40 mg, and psychotropic drugs, such as antipsychotics and mood stabilizers, were not permitted. In accordance with the Declaration of Helsinki, all subjects gave informed consent to participate in the study, and the study protocol was approved by ethics review board of from Korea University Anam Hospital. We evaluated clinical symptoms using the HAMD-21 scale and the Clinical global impression scale-Severity (CGI-S) at baseline and after 1, 2, 4, 8, and 12 weeks of treatment. Response was defined as a reduction of 50% or more in the HAMD-21 score compared to baseline, and remission status was defined as a HAMD-21 total score of 7 points or less.30 The side-effects profile during treatment was assessed using the Udvalg for Kliniske Undersogelser (UKU) Side Effect Rating Scale (UKU-SERS).31 To evaluate specific clusters of adverse effects, UKU-SERS items were grouped as psychic side effects (items 1.1, 1.2, 1.3, 1.4, 1.5, 1.6, and 1.10), sleep symptoms (items 1.7, 1.8, and 1.9), extrapyramidal symptoms (items 2.1, 2.2, 2.3, 2.4, 2.5, and 2.6), gastro-intestinal symptoms (items 3.4, 3.5, and 3.6), autonomic side effects (items 3.1, 3.2, 3.3, 3.7, 3.8, 3.9, 3.10, and 3.11), skin reactions (items 4.1a, 4.1b, 4.1c, 4.1d, 4.1e, 4.2, 4.3, and 4.4), weight changes (items 4.5 and 4.6), hormonal side effects (items 4.7, 4.8, 4.9, and 4.10), sexual side effects (4.11, 4.12, 4.13, 4.14, 4.15, and 4.16), and headache (items 4.17a, 4.17b, and 4.17c).
DNA analysis
We extracted DNA from peripheral blood of the subjects and polymerase chain reaction (PCR) was performed using the sense primer 5'-CCA TTT GGT AGT GAG GCAGGT AT-3' and antisense primer 5'-CAC CAT CCATGT TTG CTT CTG GT-3'. Each amplification mixture contained 50 ng of DNA, 3 µL of 10× PCR buffer, 2.5 µL of 2.5 mM dNTP, 10 pmol of each primer, and 0.5 µL of Taq polymerase (5 U/µL; Takara, Kyoto, Japan). We amplified the samples using a thermocycler (Perkin-Elmer, Boston, MA, USA) using the following conditions: an initial 7 minutes at 94℃, followed by 35 cycles of 45 seconds at 94℃, 45 seconds at 56℃, and 45 seconds at 72℃. The amplification reaction was terminated at 4℃ after a final 5 minutes extension at 72℃. The amplified DNA was digested with the restriction endonuclease HphI (New England BioLabs, Ipswich, MA, USA), which cleaves at site 188C. The digested DNA was electrophoresed on 3% agarose gel and stained with ethidium bromide to detect the 213- and 58-bp fragments corresponding to the 188C (34Pro) allele and the 122-, 101-, and 58-bp fragments corresponding to the 188T (34Ser) allele.
Statistical analysis
An efficacy analysis was conducted on the "intention-to-treat" group. The intention-to-treat group included all subjects who received escitalopram at least once and who had at least one efficacy measurement. Hardy-Weinberg equilibrium was examined using the χ2 test. The genetic association of the CYP2D6 P34S polymorphism with treatment efficacy was analyzed using multiple logistic regression controlling for sex and age as covariates. In allele frequency analysis, χ2 test was used. All statistical analyses were performed using SPSS version 18.0 (SPSS Inc., Chicago, IL, USA).
RESULTS
Subject clinical characteristics and Hardy-Weinberg equilibrium for the CYP2D6 P34S polymorphism
The intention-to-treat group consisted of 94 subjects. χ2 test analysis of genotype frequencies confirmed that the subjects were in Hardy-Weinberg equilibrium (χ2=3.45, p=0.063). Of the 94 patients initially recruited, 56 patients completed the 12-week study and 38 patients withdrew because of a failure to draw blood, lack of efficacy, personal conflict or other personal decision, loss to treatment or adverse events during the 12 weeks of treatment.
The mean age, onset age, family history of MDD and baseline HAMD-21 and CGI-S scores are presented in Table 1. There were no significant differences in these demographic data among the genotypes. The gender distribution during the study period did not differ between the 3 genotypes. The baseline HAMD-21 and CGI-S scores also did not differ among the genotypes.
Table 1.
Demographic characteristics of the major depressive disorder intention-to-treat group
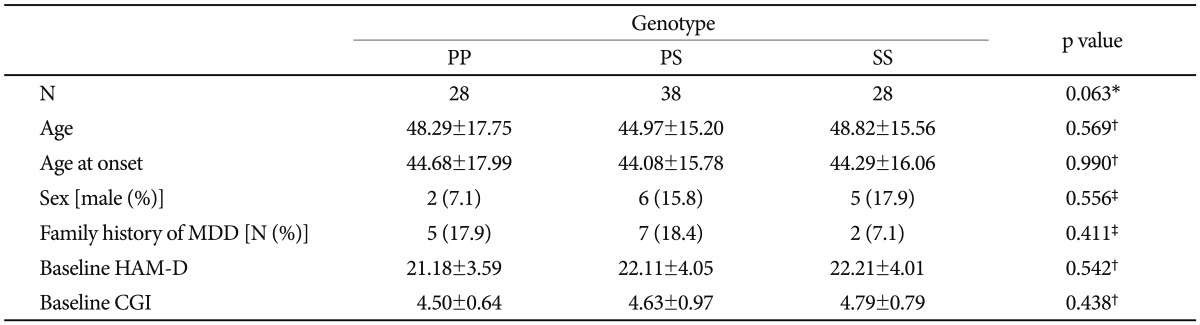
*p value for Hardy-Weinberg equilibrium, †genotype comparisons were performed using ANOVA, ‡genotype comparisons were performed using Fisher's exact test. MDD: major depressive disorder, HAM-D: Hamilton Depression Rating Scale, CGI: Clinical Global Impression, ANOVA: analysis of variance
Association between remission status and the CYP2D6 P34S polymorphism in escitalopram treatment
We investigated the association between the CYP2D6 P34S polymorphism and remission status after 1, 2, 4, 8, and 12 weeks of escitalopram treatment using multiple logistic regression. The correlations between the CYP2D6 P34S polymorphism and remission status in escitalopram treatment are presented in Table 2. A significant association was detected after 8 and 12 weeks of treatment (p=0.049 and 0.011, respectively) in a recessive model, which compared P allele carriers (PP homozygotes and PS heterozygotes) with SS homozygotes. The proportion of SS homozygotes in remission was lower than that of P allele carriers after 8 and 12 weeks of treatment. There were also significant differences in the remission status among the genotypes in the co-dominant (PP homozygotes vs. PS heterozygotes vs. SS homozygotes) after 8 and 12 weeks of treatment (p=0.044 and 0.013, respectively). However, in dominant models which compared S allele carrier with PP homozygotes, there were no significant relationship between the polymorphism and remission status. In the allele frequency analysis using χ2 test, there were significant differences among the genotypes after 4, 8, and 12 weeks of treatment (p=0.029, 0.032, and 0.007, respectively). Our results support the postulation that P allele carriers tend to enter remission in escitalopram treatment.
Table 2.
Association analysis of the CYP2D6 P34S polymorphism with remission status in escitalopram treatment of MDD patients
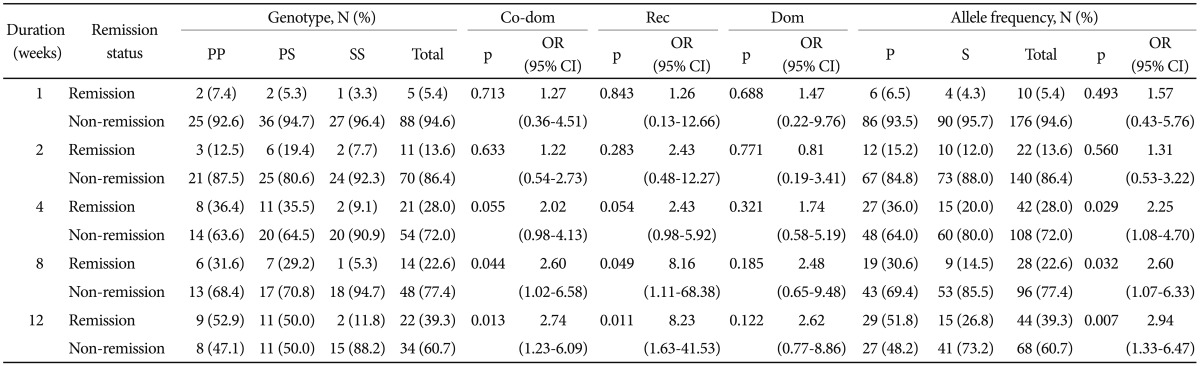
Data were obtained by logistic regression after controlling for age and sex as covariates. In allele frequency analysis, χ2 test was used. MDD: major depressive disorder, Co-dom: co-dominant model (SS vs. PS vs. PP), Rec: recessive model (SS vs. PP+PS), Dom: dominant model (SS+PS vs. PP), OR: odds ratio, CI: confidence interval
Association between treatment response and the CYP2D6 P34S polymorphism in escitalopram treatment
As shown in Table 3, a significant association was observed between the genotype and treatment response after 8 and 12 weeks of treatment (p=0.030 and p<0.001, respectively) by logistic regression in the recessive model. The proportions of S allele homozygotes in responders were lower than that of P allele carriers after 8 weeks and 12 weeks of treatment. A significant correlation was also observed after 8 and 12 weeks of treatment (p=0.031 and 0.001, respectively) in the co-dominant model. In allele frequency analysis, the proportions of P allele in responders were significantly larger than those of S alleles after 8 weeks and 12 weeks of treatment (p=0.011 and p<0.001, respectively).
Table 3.
Association analysis of the CYP2D6 P34S polymorphism with treatment response to escitalopram in MDD patients
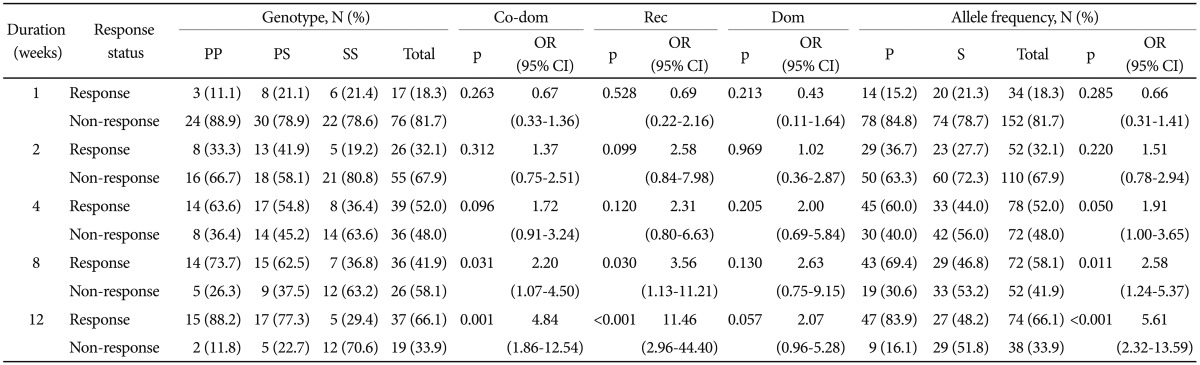
Data were obtained by logistic regression after controlling for age and sex as covariates. In allele frequency analysis, χ2 test was used. MDD: major depressive disorder, Co-dom: co-dominant model (SS vs. PS vs. PP), Rec: recessive model (SS vs. PP+PS), Dom: dominant model (SS+PS vs. PP), OR: odds ratio, CI: confidence interval
Association of the CYP2D6 P34S polymorphism with escitalopram-induced side effects in escitalopram treatment
To evaluate the correlation between the CYP2D6 P34S polymorphism and escitalopram-induced adverse effects, UKU-SERS was used. The adverse effects on psychic symptoms, sleep symptoms, extrapyramidal symptoms, gastro-intestinal symptoms, autonomic symptoms, skin reactions, hormonal problems, sexual dysfunction, and headache were investigated by multiple logistic regression analysis. An impact of the CYP2D6 P34S polymorphism on medication-induced adverse effects was not observed for all the symptom categories of UKU-SERS (data not shown).
DISCUSSION
The involvement of CYP2D6 in the metabolism of several antidepressants, including escitalopram, is well recognized.32,33 The metabolic activity of CYP2D6 exhibited inter-individual variability, ranging from complete absence (PMs) to overexpression of activity (UMs) in population-based CYP2D6 polymorphism studies.12,24 With respect to pharmacokinetics, the influence of the CYP2D6 genotype on several antidepressants has been well documented,34-37 but reports on the impact of the CYP2D6 genotype on clinical outcome are limited.38
In this study, we found a significant association between remission status and the CYP2D6 P34S polymorphism in P allele carriers after 8 and 12 weeks of escitalopram treatment in a recessive and co-dominant model. There was also a significant correlation between treatment response and P allele carriers after 8 and 12 weeks of treatment. Additionally, a significant relationship was observed between allele frequencies of the CYP2D6 P34S polymorphism and remission status and treatment response. These results suggest that the P allele of the CYP2D6 P34S polymorphism is a favorable factor in escitalopram treatment for MDD. Demographic data analysis revealed no differences in the baseline HAMD-21 and CGI-S scores between genotypes, suggesting that CYP2D6 P34S is not a risk factor for the development of MDD. This finding is consistent with the results of a previous study, which showed that the occurrence of MDD was not influenced by CYP2D6 polymorphism.39
Although an association between the CYP2D6 P34S polymorphism and the clinical response to antidepressants was reported by previous pharmacogenetic studies,40,41 there has been no consistent explanation for the effects of the CYP2D6*10 polymorphism on antidepressant action. Recently, Tsai et al.19 reported that CYP2D6 polymorphism, including the *10 allele, influenced escitalopram treatment outcomes in MDD patients, based on gene dosage analyses. This study indicated that IMs with CYP2D6 polymorphism exhibited a significantly higher frequency of remission from MDD during an 8-week treatment with escitalopram. However, Serretti et al.42 reported that the CYP2D6 genes are not associated with response and remission in a sample of depressive patients treated with various antidepressants, including escitalopram. These contradictory results may be explained by the differences in the study designs. In the study of Serretti et al.,22 antidepressants were divided into SSRI, TCA, NRI, SNRI, and NaSSA, and the efficacy of treatment was evaluated based on the genetic polymorphisms of CYP1A2, CYP2C9, CYP2C19, and CYP2D6. Problems of stratification in the study was pointed out by a recent report,29 because only 1 of 247 subjects received escitalopram in the study. Consistent with the results of our present study, our previous study on CYP2D6 P34S polymorphism and treatment response to mirtazapine, which showed that CYP2D6 P34S polymorphism was significantly associated with remission status, and that the P allele of the CYP2D6 P34S polymorphism is a favorable factor for a response to mirtazapine treatment in Koreans with major depression.29
There has been a discussion on not only the influence of CYP2D6 polymorphism on treatment outcomes, but also on the correlation between CYP2D6 polymorphism and druginduced adverse effects. Several recent studies on pharmacogenetic evaluation of antidepressant-induced adverse effects reported that CYP2D6 PMs tend to exhibit an increase in concentration-dependent adverse events due to slow elimination of antidepressants from the body.17,18 Furthermore, Chou et al.43 suggested that the total duration of hospital stay was longer for patients in the CYP2D6 PM group due to a higher number of adverse drug effects. However, our analysis of the CYP2D6 P34S polymorphism and the medication-related adverse effects could not reveal the correlation between them in any categories of adverse effect on UKU-SERS.
Based on the results of previous studies, altered CYP2D6 metabolic activity modulated by CYP2D6 polymorphism is associated with a generally less favorable treatment outcome in patients treated with antidepressants that are CYP2D6 substrates.18 It has been reported that CYP2D6 PMs have greater susceptibility to adverse effects44,45 and lower treatment response15,16 to antidepressants than EMs due to higher plasma drug concentrations. On the other hand, reduced antidepressant efficacy in CYP2D6 UMs was attributed to insufficient plasma drug concentrations to produce the desired therapeutic effects with the usual doses.46,47 The frequency of CYP2 D6*10 is relatively high in Asian IMs, and lower catalytic activities toward various psychotropic drugs, including antidepressants, were observed in patients with CYP2D6*10/*10 or CYP2D6*10/*null genotypes.25 Based on the aforementioned reports and the results of our study, we postulate that SS allele homozygotes of the CYP2D6 P34S polymorphism show low efficacy of treatment for MDD due to their escitalopram metabolizer phenotype. In this study, it remains unclear whether patients with the homozygous SS allele are PMs, UMs, or IMs due to lack of plasma drug concentration measurements. However, studies on the association between drug concentration levels and treatment outcomes in MDD have yielded inconsistent results. In one study with a naturalistic clinical study design, treatment response was not associated with plasma concentrations or influenced by CYP2D6 polymorphism, above or below the lower limit of a presumed therapeutic range of antidepressants.48 In another study,19 an influence of CYP2D6 polymorphism on escitalopram efficacy was observed, but there were no significant differences in the serum concentrations of escitalopram and its metabolites S-desmethylcitalopram and S-didemethylcitalopram among the allelic combinations of CYP2D6 polymorphism, including the CYP2 D6*10 allele. Therefore, further studies that take into consideration both the genotype of the polymorphism and plasma drug concentrations as a phenotype are required to demonstrate a direct correlation between plasma drug concentration and the polymorphism and to elucidate the influence of the CYP2D6 P34S polymorphism on the efficacy and tolerability of escitalopram.
To our knowledge, this is first study to evaluate the association between CYP2D6 polymorphism and efficacy of escitalopram, focusing on a single locus of the CYP2D6 P34S polymorphism. However, this study has several limitations; First, we investigated only 1 SNP of the CYP2D6 polymorphism, not allelic combinations of all CYP2D6 polymorphisms detected in the Korean population. For a thorough evaluation, gene screening and association studies of whole CYP2D6 genetic polymorphisms are required to identify the genetic markers associated with treatment response to escitalopram. However, the CYP2D6 P34S polymorphism is an important genetic marker, indicating CYP2D6*10 allele which is not only the most frequent allele in the Korean population,24 but also in the Asian population.21 Therefore, an investigation of the causal relationship between single SNP and efficacy of escitalopram, focusing on the CYP2D6 P34S polymorphism, will also be meaningful.
Second, relatively small sample sizes at baseline for each genotype groups and small number of completers for 12-week escitalopram treatment schedule make nothing to be concluded strongly in this study. In addition, there were performance of multiple comparisons in the analyses, therefore, our suggestions in this study can not be free from the risk of type I error.
Third, we could not determine the patients' CYP2D6 metabolizer phenotype, which was considered an important factor in previous pharmacogenetic studies on CYP2D6 polymorphism and antidepressant efficacy, because of a lack of plasma escitalopram concentration measurements in our study. Therefore, the correlation between treatment response to escitalopram and enzymatic activity of CYP2D6 remains unclear. With respect to population stratification, bias cannot be excluded from our results. However, given that the Korean population has a relatively high degree of genetic homogeneity, such stratification bias is unlikely in our subjects. In this study, individual subjects received escitalopram at a dose range of 5-40 mg, not the same dose, during the 12-week trial. However, we did not control the dose of escitalopram as a covariate in the statistical analysis of the association between polymorphism genotypes and treatment outcomes. Thus, there is a possibility that the variation in the escitalopram dose influenced the treatment response. Finally, we could not detect an influence of the CYP2D6 P34S polymorphism on adverse effects of escitalopram in our study. Few studies have examined the association between CYP2D6 polymorphism and tolerability of antidepressants. We expect that future studies will pay more attention to this topic.
In summary, we found that CYP2D6 P34S was associated with remission status and treatment response in escitalopram treatment. We postulate that CYP2D6 P34S is a genetic marker for predicting the clinical outcome of escitalopram treatment, and the P allele is a favorable factor in escitalopram treatment in Korean patients with major depression.
Acknowledgments
This study was supported by a grant of the Korean Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (A120004) and by a grant of the Korean Health 21 R & D Project, Ministry of Health and Welfare, Republic of Korea (HI10C2020).
References
- 1.Duffy A, Grof P, Robertson C, Alda M. The implications of genetics studies of major mood disorders for clinical practice. J Clin Psychiatry. 2000;61:630–637. doi: 10.4088/jcp.v61n0906. [DOI] [PubMed] [Google Scholar]
- 2.Kelly K, Posternak M, Alpert JE. Toward achieving optimal response: understanding and managing antidepressant side effects. Dialogues Clin Neurosci. 2008;10:409–418. doi: 10.31887/DCNS.2008.10.4/kkelly. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khan A, Khan SR, Walens G, Kolts R, Giller EL. Frequency of positive studies among fixed and flexible dose antidepressant clinical trials: an analysis of the food and drug administration summary basis of approval reports. Neuropsychopharmacology. 2003;28:552–557. doi: 10.1038/sj.npp.1300059. [DOI] [PubMed] [Google Scholar]
- 4.Hamdani N, Gorwood P. Role of genetics in psychiatric treatment. Presse Med. 2008;37:902–911. doi: 10.1016/j.lpm.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 5.Nebert DW. Polymorphisms in drug-metabolizing enzymes: what is their clinical relevance and why do they exist? Am J Hum Genet. 1997;60:265–271. [PMC free article] [PubMed] [Google Scholar]
- 6.Baldwin DS, Reines EH, Guiton C, Weiller E. Escitalopram therapy for major depression and anxiety disorders. Ann Pharmacother. 2007;41:1583–1592. doi: 10.1345/aph.1K089. [DOI] [PubMed] [Google Scholar]
- 7.Rao N. The clinical pharmacokinetics of escitalopram. Clin Pharmacokinet. 2007;46:281–290. doi: 10.2165/00003088-200746040-00002. [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi K, Chiba K, Yagi T, Shimada N, Taniguchi T, Horie T, et al. Identification of cytochrome P450 isoforms involved in citalopram N-demethylation by human liver microsomes. J Pharmacol Exp Ther. 1997;280:927–933. [PubMed] [Google Scholar]
- 9.von Moltke LL, Greenblatt DJ, Giancarlo GM, Granda BW, Harmatz JS, Shader RI. Escitalopram (S-citalopram) and its metabolites in vitro: cytochromes mediating biotransformation, inhibitory effects, and comparison to R-citalopram. Drug Metab Dispos. 2001;29:1102–1109. [PubMed] [Google Scholar]
- 10.Reis M, Lundmark J, Bengtsson F. Therapeutic drug monitoring of racemic citalopram: a 5-year experience in Sweden, 1992-1997. Ther Drug Monit. 2003;25:183–191. doi: 10.1097/00007691-200304000-00007. [DOI] [PubMed] [Google Scholar]
- 11.Bertilsson L. Metabolism of antidepressant and neuroleptic drugs by cytochrome p450s: clinical and interethnic aspects. Clin Pharmacol Ther. 2007;82:606–609. doi: 10.1038/sj.clpt.6100358. [DOI] [PubMed] [Google Scholar]
- 12.Sachse C, Brockmoller J, Bauer S, Roots I. Cytochrome P450 2D6 variants in a Caucasian population: allele frequencies and phenotypic consequences. Am J Hum Genet. 1997;60:284–295. [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Barcelo M, Chow LY, Chiu HF, Wing YK, Lee DT, Lam KL, et al. Genetic analysis of the CYP2D6 locus in a Hong Kong Chinese population. Clin Chem. 2000;46:18–23. [PubMed] [Google Scholar]
- 14.Lee MS. Role of genetic polymorphisms related to neurotransmitters and cytochrome P-450 enzymes in response to antidepressants. Drugs Today (Barc) 2007;43:569–581. doi: 10.1358/dot.2007.43.8.1130447. [DOI] [PubMed] [Google Scholar]
- 15.Lobello KW, Preskorn SH, Guico-Pabia CJ, Jiang Q, Paul J, Nichols AI, et al. Cytochrome P450 2D6 phenotype predicts antidepressant efficacy of venlafaxine: a secondary analysis of 4 studies in major depressive disorder. J Clin Psychiatry. 2010;71:1482–1487. doi: 10.4088/JCP.08m04773blu. [DOI] [PubMed] [Google Scholar]
- 16.Mulder H, Wilmink FW, Beumer TL, Tamminga WJ, Jedema JN, Egberts AC. The association between cytochrome P450 2D6 genotype and prescription patterns of antipsychotic and antidepressant drugs in hospitalized psychiatric patients: a retrospective follow-up study. J Clin Psychopharmacol. 2005;25:188–191. doi: 10.1097/01.jcp.0000155832.79777.b5. [DOI] [PubMed] [Google Scholar]
- 17.Rau T, Wohlleben G, Wuttke H, Thuerauf N, Lunkenheimer J, Lanczik M, et al. CYP2D6 genotype: impact on adverse effects and nonresponse during treatment with antidepressants-a pilot study. Clin Pharmacol Ther. 2004;75:386–393. doi: 10.1016/j.clpt.2003.12.015. [DOI] [PubMed] [Google Scholar]
- 18.D'Empaire I, Guico-Pabia CJ, Preskorn SH. Antidepressant treatment and altered CYP2D6 activity: are pharmacokinetic variations clinically relevant? J Psychiatr Pract. 2011;17:330–339. doi: 10.1097/01.pra.0000405363.95881.01. [DOI] [PubMed] [Google Scholar]
- 19.Tsai MH, Lin KM, Hsiao MC, Shen WW, Lu ML, Tang HS, et al. Genetic polymorphisms of cytochrome P450 enzymes influence metabolism of the antidepressant escitalopram and treatment response. Pharmacogenomics. 2010;11:537–546. doi: 10.2217/pgs.09.168. [DOI] [PubMed] [Google Scholar]
- 20.Commitee N. The Human Cytochrome P450 (CYP) Allele. [Accessed March 14, 2013]. Available at: http://www.cypalleles.ki.se/cyp2d6.htm.
- 21.Ingelman-Sundberg M. Pharmacogenetics: an opportunity for a safer and more efficient pharmacotherapy. J Intern Med. 2001;250:186–200. doi: 10.1046/j.1365-2796.2001.00879.x. [DOI] [PubMed] [Google Scholar]
- 22.Nishida Y, Fukuda T, Yamamoto I, Azuma J. CYP2D6 genotypes in a Japanese population: low frequencies of CYP2D6 gene duplication but high frequency of CYP2D6*10. Pharmacogenetics. 2000;10:567–570. doi: 10.1097/00008571-200008000-00010. [DOI] [PubMed] [Google Scholar]
- 23.Ji L, Pan S, Marti-Jaun J, Hanseler E, Rentsch K, Hersberger M. Single-step assays to analyze CYP2D6 gene polymorphisms in Asians: allele frequencies and a novel*14B allele in mainland Chinese. Clin Chem. 2002;48:983–988. [PubMed] [Google Scholar]
- 24.Lee SY, Sohn KM, Ryu JY, Yoon YR, Shin JG, Kim JW. Sequence-based CYP2D6 genotyping in the Korean population. Ther Drug Monit. 2006;28:382–387. doi: 10.1097/01.ftd.0000211823.80854.db. [DOI] [PubMed] [Google Scholar]
- 25.Sakuyama K, Sasaki T, Ujiie S, Obata K, Mizugaki M, Ishikawa M, et al. Functional characterization of 17 CYP2D6 allelic variants (CYP2D6.2, 10, 14A-B, 18, 27, 36, 39, 47-51, 53-55, and 57) Drug Metab Dispos. 2008;36:2460–2467. doi: 10.1124/dmd.108.023242. [DOI] [PubMed] [Google Scholar]
- 26.Johansson I, Oscarson M, Yue QY, Bertilsson L, Sjoqvist F, Ingelman-Sundberg M. Genetic analysis of the Chinese cytochrome P4502D locus: characterization of variant CYP2D6 genes present in subjects with diminished capacity for debrisoquine hydroxylation. Mol Pharmacol. 1994;46:452–459. [PubMed] [Google Scholar]
- 27.Zhou SF. Polymorphism of human cytochrome P450 2D6 and its clinical significance: Part I. Clin Pharmacokinet. 2009;48:689–723. doi: 10.2165/11318030-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 28.Heim MH, Meyer UA. Evolution of a highly polymorphic human cytochrome P450 gene cluster: CYP2D6. Genomics. 1992;14:49–58. doi: 10.1016/s0888-7543(05)80282-4. [DOI] [PubMed] [Google Scholar]
- 29.Jeon SO, Lee HY, Ham BJ, Kang RH, Jeong YJ, Lee MS. CYP2D6 P34S polymorphism and mirtazapine responses in Koreans with major depression. Mol Cell Toxicol. 2009;5:346–353. [Google Scholar]
- 30.Frank E, Prien RF, Jarrett RB, Keller MB, Kupfer DJ, Lavori PW, et al. Conceptualization and rationale for consensus definitions of terms in major depressive disorder. Remission, recovery, relapse, and recurrence. Arch Gen Psychiatry. 1991;48:851–855. doi: 10.1001/archpsyc.1991.01810330075011. [DOI] [PubMed] [Google Scholar]
- 31.Kim JH, Choi SW, Joe SH, Ha TH, Yoo HJ, Choi JE, et al. Reliability and validity of the Korean version of UKU-SERS-Pat in patients with bipolar disorder. Nord J Psychiatry. 2008;62:496–502. doi: 10.1080/08039480801985229. [DOI] [PubMed] [Google Scholar]
- 32.Bertilsson L, Dahl ML, Dalen P, Al-Shurbaji A. Molecular genetics of CYP2D6: clinical relevance with focus on psychotropic drugs. Br J Clin Pharmacol. 2002;53:111–122. doi: 10.1046/j.0306-5251.2001.01548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirchheiner J, Brosen K, Dahl ML, Gram LF, Kasper S, Roots I, et al. CYP2D6 and CYP2C19 genotype-based dose recommendations for antidepressants: a first step towards subpopulation-specific dosages. Acta Psychiatr Scand. 2001;104:173–192. doi: 10.1034/j.1600-0447.2001.00299.x. [DOI] [PubMed] [Google Scholar]
- 34.Preskorn S, Patroneva A, Silman H, Jiang Q, Isler JA, Burczynski ME, et al. Comparison of the pharmacokinetics of venlafaxine extended release and desvenlafaxine in extensive and poor cytochrome P450 2D6 metabolizers. J Clin Psychopharmacol. 2009;29:39–43. doi: 10.1097/JCP.0b013e318192e4c1. [DOI] [PubMed] [Google Scholar]
- 35.Ueda M, Hirokane G, Morita S, Okawa M, Watanabe T, Akiyama K, et al. The impact of CYP2D6 genotypes on the plasma concentration of paroxetine in Japanese psychiatric patients. Prog Neuropsychopharmacol Biol Psychiatry. 2006;30:486–491. doi: 10.1016/j.pnpbp.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 36.LLerena A, Dorado P, Berecz R, Gonzalez AP, Peñas-LLedó EM. Effect of CYP2D6 and CYP2C9 genotypes on fluoxetine and norfluoxetine plasma concentrations during steady-state conditions. Eur J Clin Pharmacol. 2004;59:869–873. doi: 10.1007/s00228-003-0707-y. [DOI] [PubMed] [Google Scholar]
- 37.Ohara K, Tanabu S, Ishibashi K, Ikemoto K, Yoshida K, Shibuya H. CYP2D6*10 alleles do not determine plasma fluvoxamine concentration/dose ratio in Japanese subjects. Eur J Clin Pharmacol. 2003;58:659–661. doi: 10.1007/s00228-002-0529-3. [DOI] [PubMed] [Google Scholar]
- 38.Zhou SF. Polymorphism of human cytochrome P450 2D6 and its clinical significance: part II. Clin Pharmacokinet. 2009;48:761–804. doi: 10.2165/11318070-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 39.Bijl MJ, Luijendijk HJ, van den Berg JF, Visser LE, van Schaik RH, Hofman A, et al. Association between the CYP2D6*4 polymorphism and depression or anxiety in the elderly. Pharmacogenomics. 2009;10:541–547. doi: 10.2217/pgs.09.9. [DOI] [PubMed] [Google Scholar]
- 40.Gerstenberg G, Aoshima T, Fukasawa T, Yoshida K, Takahashi H, Higuchi H, et al. Relationship between clinical effects of fluvoxamine and the steady-state plasma concentrations of fluvoxamine and its major metabolite fluvoxamino acid in Japanese depressed patients. Psychopharmacology (Berl) 2003;167:443–448. doi: 10.1007/s00213-003-1430-1. [DOI] [PubMed] [Google Scholar]
- 41.Murphy GM, Jr, Kremer C, Rodrigues HE, Schatzberg AF. Pharmacogenetics of antidepressant medication intolerance. Am J Psychiatry. 2003;160:1830–1835. doi: 10.1176/appi.ajp.160.10.1830. [DOI] [PubMed] [Google Scholar]
- 42.Serretti A, Calati R, Massat I, Linotte S, Kasper S, Lecrubier Y, et al. Cytochrome P450 CYP1A2, CYP2C9, CYP2C19 and CYP2D6 genes are not associated with response and remission in a sample of depressive patients. Int Clin Psychopharmacol. 2009;24:250–256. doi: 10.1097/YIC.0b013e32832e5b0d. [DOI] [PubMed] [Google Scholar]
- 43.Chou WH, Yan FX, de Leon J, Barnhill J, Rogers T, Cronin M, et al. Extension of a pilot study: impact from the cytochrome P450 2D6 polymorphism on outcome and costs associated with severe mental illness. J Clin Psychopharmacol. 2000;20:246–251. doi: 10.1097/00004714-200004000-00019. [DOI] [PubMed] [Google Scholar]
- 44.Shams ME, Arneth B, Hiemke C, Dragicevic A, Muller MJ, Kaiser R, et al. CYP2D6 polymorphism and clinical effect of the antidepressant venlafaxine. J Clin Pharm Ther. 2006;31:493–502. doi: 10.1111/j.1365-2710.2006.00763.x. [DOI] [PubMed] [Google Scholar]
- 45.Zourkova A, Ceskova E, Hadasova E, Ravcukova B. Links among paroxetine-induced sexual dysfunctions, gender, and CYP2D6 activity. J Sex Marital Ther. 2007;33:343–355. doi: 10.1080/00926230701385589. [DOI] [PubMed] [Google Scholar]
- 46.Kawanishi C, Lundgren S, Agren H, Bertilsson L. Increased incidence of CYP2D6 gene duplication in patients with persistent mood disorders: ultrarapid metabolism of antidepressants as a cause of nonresponse. A pilot study. Eur J Clin Pharmacol. 2004;59:803–807. doi: 10.1007/s00228-003-0701-4. [DOI] [PubMed] [Google Scholar]
- 47.Breil F, Verstuyft C, Orostegui L, Buhl C, Alvarez JC, Chouinard G, et al. Non-response to consecutive antidepressant therapy caused by CYP2D6 ultrarapid metabolizer phenotype. Int J Neuropsychopharmacol. 2008;11:727–728. doi: 10.1017/S1461145708008742. [DOI] [PubMed] [Google Scholar]
- 48.Grasmader K, Verwohlt PL, Rietschel M, Dragicevic A, Muller M, Hiemke C, et al. Impact of polymorphisms of cytochrome-P450 isoenzymes 2C9, 2C19 and 2D6 on plasma concentrations and clinical effects of antidepressants in a naturalistic clinical setting. Eur J Clin Pharmacol. 2004;60:329–336. doi: 10.1007/s00228-004-0766-8. [DOI] [PubMed] [Google Scholar]


