Abstract
Proliferation and differentiation are tightly controlled during neural development. In the embryonic neural plate, primary neurogenesis is driven by the proneural pathway. Here we report the characterization of Maturin, a novel, evolutionarily conserved protein that is required for normal primary neurogenesis. Maturin is detected throughout the early nervous system, yet it is most strongly expressed in differentiating neurons of the embryonic fish, frog and mouse nervous systems. Maturin expression can be induced by the proneural transcription factors Neurog2, Neurod1, and Ebf3. Maturin overexpression promotes neurogenesis, while loss-of-function inhibits the differentiation of neuronal progenitors, resulting in neural plate expansion. Maturin knockdown blocks the ability of Neurog2, Neurod1, and Ebf3 to drive ectopic neurogenesis. Maturin and Pak3, are both required for, and can synergize to promote differentiation of the primary neurons in vivo. Together, our results suggest that Maturin functions during primary neurogenesis and is required for the proneural pathway to regulate neural differentiation.
Keywords: Pak3, primary neurogenesis, neuronal differentiation, neural plate
Introduction
Normal neural development requires a precise balance of proliferation and differentiation (Dehay and Kennedy, 2007; Donovan and Dyer, 2005; Salomoni and Calegari, 2010). Identifying the genes and signaling systems that regulate the differentiation of neuronal progenitors is essential, as premature or postponed neurogenesis can alter the number and types of neurons generated during development (Agathocleous and Harris, 2009; Donovan and Dyer, 2005; Dyer and Cepko, 2001; Guillemot, 2007; Hindley and Philpott, 2012). In Xenopus laevis the first progenitors to differentiate generate the primary neurons, which have served as a valuable model for identifying the genes and signaling systems driving vertebrate neurogenesis (Henningfeld et al., 2007). Shortly after gastrulation, the primary neurons differentiate along three, longitudinal, bilaterally symmetric stripes on either side of the embryonic midline and are detected by their expression of neural-specific, class II beta-tubulin (tubb2b) (Chitnis et al., 1995; Hartenstein, 1989; Oschwald et al., 1991).
Primary neurogenesis is driven by a cascade of proneural basic helix-loop-helix (bHLH) transcription factors that regulate the activity of downstream targets required for cell fate determination, cell cycle exit and the eventual differentiation of neuronal progenitors. Neurogenin 2 (neurog2; also known as X-ngnr-1), is the first proneural gene of the cascade and induces the expression of the bHLH transcription factors neuronal differentiation 1 (neurod1, also known as XNeuroD) and early B-cell factor 3 (ebf3, also known as Xcoe3) (Lee et al., 1995; Ma et al., 1996; Pozzoli et al., 2001). Neurog2, neurod1 and ebf3 are all expressed in the primary neurons, and when misexpressed, can induce ectopic neurons and drive their differentiation in vivo and in vitro (Lee et al., 1995; Ma et al., 1996; Pozzoli et al., 2001).
The p21-activated kinase 3 (Pak3) functions downstream of the proneural transcription factors and is required for cell cycle exit and differentiation of the primary neurons (Souopgui et al., 2002). Neurog2, neurod1 and ebf3 can all induce ectopic expression of pak3. Pak3 expression is detected in the primary neurons as they differentiate in the neural plate. Similarly, the cells of other neural tissues also express pak3 during differentiation. Pak3 function is required for primary neurons to exit the cell cycle and differentiate, since morpholino oligonucleotides that block pak3 translation inhibit neural differentiation and increase cell proliferation, resulting in neural plate expansion. Pak3 can be made constitutively active by artificial myristoylation, which targets the protein to cell membranes. Misexpression of myrPak3 results in cell cycle arrest and premature neuronal differentiation. Interestingly, non-myristoylated Pak3 was found to be functionally inactive, as it neither altered primary neurogenesis (tubb2b expression), nor embryonic development when misexpressed (Souopgui et al., 2002).
Here we report the identification of Maturin, an acidic, evolutionarily conserved protein that is required for normal primary neurogenesis. Maturin has no identifiable functional or structural domains, yet its primary amino acid sequence has been highly conserved in vertebrates. Xenopus laevis maturin is detected throughout the early nervous system, but is most highly expressed in differentiating neurons. A similar expression pattern is observed in both zebrafish and mouse embryos. Blocking Maturin function inhibits differentiation of the primary neurons, increases the number of proliferating neural progenitors, and results in neural plate enlargement. Conversely, Maturin overexpression promotes neural differentiation within the neural plate. The proneural pathway transcription factor(s) Neurog2, Neurod1 and Ebf3 can all induce maturin transcription and Maturin knockdown blocks neural differentiation initiated by Neurog2, Neurod1 and Ebf3. Maturin gain- and loss-of-function phenotypes mimic those of Pak3, and Maturin and Pak3 functions are both required for differentiation of the primary neurons. Maturin and the constitutively active myrPak3 can synergistically drive primary neurogenesis. Surprisingly, Maturin can also synergize with non-myristoylated Pak3. Our results suggest that Maturin and Pak3 are both required, and function synergistically in the neural plate to regulate normal primary neurogenesis.
Materials and methods
Animals
Xenopus embryos were obtained by in vitro fertilization following standard protocols and developmental stages were determined according to Nieuwkoop and Faber (Nieuwkoop and Faber, 1994). Wild-type AB and Tg(elav3l:GFP) zebrafish (Danio rerio) were obtained from the Zebrafish International Resource Center (ZIRC). Zebrafish embryos were staged as described (Kimmel et al., 1995). Swiss Webster E14 timed-pregnant females were purchased from Charles River Laboratories International (Wilmington, MA). The Committee for the Humane Use of Animals at SUNY Upstate Medical University approved all procedures.
Xenopus Maturin and ortholog analysis
The X. laevis (IMAGE: 5570100; pCMVSport6.Maturin) and mouse (IMAGE: 4535651; pCMVSport6.mouseMaturin) full-length cDNA clones was purchased from Open Biosystems (ThermoScientific, Huntsville, AL). Zebrafish Maturin (Accession: NM_001144806) was PCR amplified from 48hpf whole larvae cDNA using primers DrMaturinfor and DrMaturinrev (Supp. Table 1), TA cloned, and sequence verified. Putative Maturin orthologs were identified using BLASTP and/or BLASTN searches of GenBank and ENSEMBL databases. Protein sequences were aligned using the ClustalW algorithm and MegAlign ver. 10.1.2 (DNASTAR Inc., Madison, WI). Maturin is required for the differentiation (maturation) of neural progenitors, hence the name (first suggested by Andrea Viczian). The Human Gene Nomenclature Committee has approved the official gene name as: “maturin, neural progenitor differentiation regulator homolog (Xenopus)”, and the gene symbol as MTURN.
Plasmid construction
pCS2R.MattargetYFP was generated by amplification of YFP (primers YFPfor and YFPrev) and Mattarget (primers Mattargetfor and Mattargetrev)(Supp. Table 1). PCR products were TA cloned, then excised and ligated in frame into pCS2R. pCS2R.Pak3 was obtained by excising Pak3 from pBK-CMV.Pak3 and ligating into pCS2R. Construction of pCS2.Ngnr1, pCS2+MT.XNeuroD, pCS2.Xebf3, and pCS2.myrPak3 were previously described (Lee et al., 1995; Ma et al., 1996; Pozzoli et al., 2001; Souopgui et al., 2002). All plasmids and sequences are available upon request.
Reverse transcription PCR analysis
For analysis of maturin expression at different stages five eggs or whole embryos at the indicated stages were homogenized using Qiashredder (Qiagen, Valencia, CA), total RNA was extracted (RNeasy Mini Kit; Qiagen), and cDNA was synthesized using 500 ng of total RNA (Quantitect Reverse Transcription Kit; Qiagen) per manufacturers instructions. RT-PCR was performed using equal amounts of template cDNA. The primer pairs used were: Maturinfor and Maturinrev, Pax6for and Pax6rev, and H4for with H4rev (Supp. Table 1) (Zuber et al., 2003). For zebrafish maturin cloning, 30 embryos at 48hpf were homogenized in TRIzol reagent (Life technologies, Invitrogen, Grand Island, NY) to extract total RNA. Zebrafish cDNA was synthesized using 500 ng total RNA (M-MLV Reverse Transcriptase; Promega, Madison, WI) per manufacturer’s instructions.
Animal cap assay
Total RNA was isolated from animal caps (10 per condition) or two embryos per stage, reverse transcribed and RTPCR performed as previously described (Viczian et al., 2009; Zuber et al., 2003). The primers used were tubb2for, tubb2brev, ncam1for, ncam1rev, actc1for, actc1rev (Supp. Table 1).
Antisense morpholino oligonucleotides, RNA synthesis and microinjection
Embryos were injected into one ventral blastomere at the 8-cell stage with 30 ng of CoMO or MatMO2 with 200 pg nlsβ-Gal cRNA. Noggin (20 pg) was used as a positive control. Sox2 expression was detected by in situ hybridization as described below. Capped RNAs were synthesized in vitro from NotI linearized plasmids using the SP6 mMessage Machine kit (Ambion, Austin, TX). CoMO, Pak3MO and MatMO1/2 morpholino oligonucleotides were obtained from GeneTools (Philomath, OR; Supplementary Table 1). cRNAs and morpholinos were injected into one blastomere at the 2-, 4- or 8-cell stage. Embryos were devitellinized at stage 10, cultured to stage 15, and the relative size of the neural plate was determined using an optical micrometer.
Probes and whole mount in situ hybridization
Digoxigenin (DIG) or fluorescein (FL)-labelled antisense riboprobes were transcribed in vitro from pCMVSport6.Maturin (EcoRI, T7), pCMVSport6.mouseMaturin (BamHI, T7), pGEMTEZ.zebrafishMaturin (NdeI, T7), pBSKS+.tubb2b (BamHI, T3), pBSSK+.Sox2 (XbaI, T7), and pGEM-T Easy.Rax (NcoI, SP6) using RNA Polymerase Plus (Ambion) as previously described (Good et al., 1989; Mizuseki et al., 1998; Zuber et al., 2003). Fast Red (Roche) was used to visualize probes when section in situs were co-labeled with DAPI.
β-Gal staining and wholemount in situ hybridization was performed as described previously (Viczian et al., 2006; Zuber et al., 2003). In situ hybridization of tissue sections was performed as previously described (Chalmers et al., 2003; Regad et al., 2007; Viczian et al., 2006), except that stage 15 and 32 embryos were fixed in Dent’s fixative and embedded in 15% fish gelatin/15% for sectioning.
Immunohistochemistry and BrdU labeling
For immunohistochemistry sections were stained as previously described (Viczian et al., 2003), with the exception that blocking was done for 1hr. Phosphorylated Histone H3 wholemount immunostaining was carried out as previously described (Saka and Smith, 2001; Sive et al., 2000). For bromodeoxyuridine (BrdU) labeling, stage 32 embryos were incubated in 1 mM BrdU (Sigma, St. Louis, MO) in 0.1X Modified Marc’s Ringer (MMR) solution for 3 hours at 18°C. Stage 42 embryos were incubated in BrdU for 3 hr at RT. Primary antibodies were: anti-GFP polyclonal (1:500; Life Technologies, Madison, WI), anti-BrdU monoclonal (1:20, clone G3G4; DSHB, Iowa City, IA), anti-PCNA (1:250, Sigma, St. Louis, MO), anti-phophorylated Histone H3 (1:1,000, Millipore, Billerica, MA) and anti-acTUBB3B (TuJ1) (1:1,000, Covance, Princeton, NJ). Secondary antibodies were: goat anti-rabbit Alexa Fluor 488, goat anti-mouse Alexa Fluor 555, goat anti-mouse IgG2a Alexa fluor 555, goat anti-mouse IgG2a Alexa Fluor 488 (all at 1:500; Molecular Probes, Invitrogen, Eugene, OR), and goat anti-rabbit IgG-HRP (1:1,000, Millipore, Billerica, MA).
TUNEL staining
Wholemount TUNEL staining was performed as described (Hensey and Gautier, 1998). For imaging, embryos were dehydrated in 100% methanol and cleared in BBBA (2 parts Benzyl Benzoate: 1 part Benzyl alcohol). TUNEL counts were performed within an area of 0.138 mm2 on both sides of the midline of each embryo using Image J (NIH) plugins for cell counting and ROI managing.
Western blotting
Using standard protocols, 30 μg of total protein was loaded per lane, separated by SDS-PAGE and blotted onto a PVDF membrane (BioRad). Antibodies used were: polyclonal anti-GFP (1:1000; Molecular Probes), polyclonal anti-β-actin (1:1,000; Cell Signaling, Danvers, MA) and goat anti-rabbit HRP conjugate (1:1,000; Millipore, Billerica, MA).
Neural plate/eye field expansion, cell density and neural plate cell number
Sox2 and rax expression domains were determined by measuring the distance from the midline to lateral edge of expression and the average per cent change relative to the uninjected side ± s.e.m determined. Cell density was determined by counting the number of DAPI positive nuclei within a 100 μm × 200 μm area in five consecutive sections per embryo and averaging. Total neural plate cell number was calculated by counting all nuclei within the sox2 expression domain on 3 consecutive sections per embryo and averaging. All graphs and statistical analysis was performed using Graph Pad Prism version 5.0c (Graph Pad Software, La Jolla, CA). Two tailed Student’s t-tests were used and P ≤ 0.05 was considered significant.
Image acquisition and processing
Whole embryo images were taken using a Leica MZ16A fluorescence stereomicroscope with a MicroPublisher 3.3 RTV digital camera (Q-Imaging, Surrey, BC, Canada) and Q-Capture software version 3.1.2 (Q-Imaging, Surrey, BC, Canada). Section images were obtained with a DM6000 B upright microscope with motorized Z-focusing (Leica Microsystems, Bannockburn, IL) fitted with a Retiga-SRV camera (Q-Imaging) and Volocity Software version 6.2.1 (PerkinElmer). All images were prepared for publication using Photoshop and Illustrator CS6 version 13.0.4 (Adobe Systems Inc., San Jose, CA).
Results
Maturin is a novel, evolutionarily conserved vertebrate protein
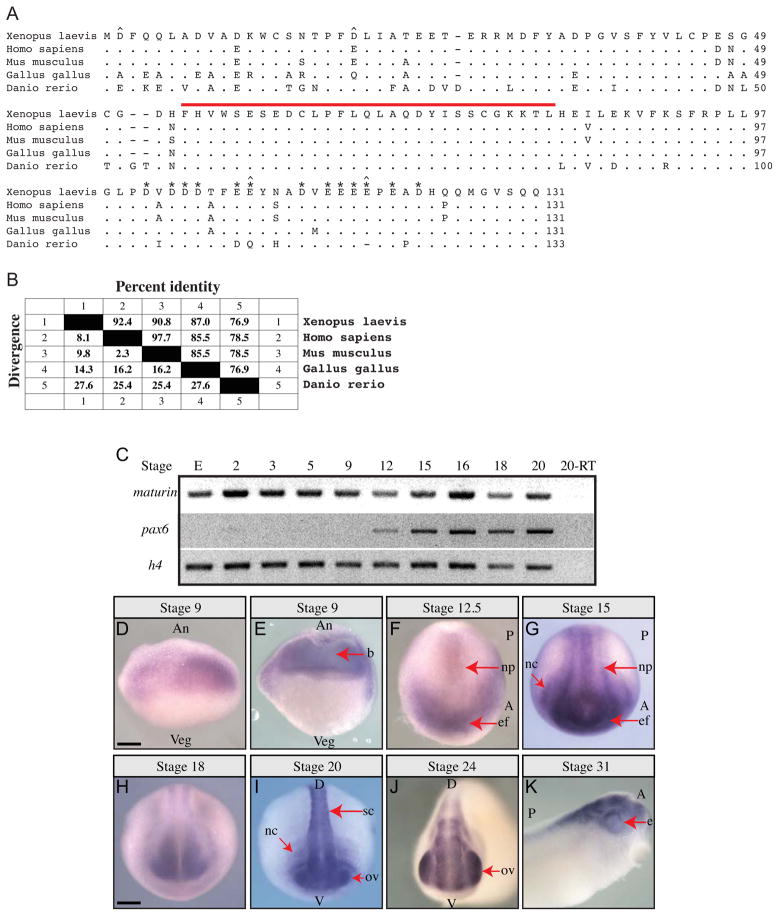
Maturin was identified in a screen designed to detect genes required for eye field specification ((Viczian et al., 2009) and M. E. Zuber, unpublished). The Xenopus laevis maturin cDNA codes for a predicted protein of 131 amino acids (a.a.) with a molecular weight of 15 kDa (Fig. 1A). A search of protein domain databases (including Prosite, InterProScan, NCBI Conserved Domain Database and Motif-Scan) failed to identify any known functional or structural motifs (Marchler-Bauer et al., 2011; Pagni et al., 2007; Quevillon et al., 2005; Sigrist et al., 2010). Putative signal peptide cleavage sites were not detected suggesting Maturin is not a secreted protein (Petersen et al., 2011). Maturin is an acidic protein with a predicted net charge of −21.5 at pH 7 (pI=3.9). Thirty of the 131 Maturin residues are either aspartic (D) or glutamic (E) acid, nearly half of which are located in a 22 residue acidic domain near the carboxy-terminus (Fig. 1A; asterisks).
Fig. 1.
Maturin predicted protein sequence, conservation and expression during Xenopus laevis embryonic development. (A) Maturin protein sequence alignment. Predicted amino acid sequence of X. laevis Maturin aligned with putative orthologs from other vertebrates. Periods indicate amino acid identity, while gaps (dashes) were inserted to optimize the alignment. The 29-residue Maturin Motif is indicated by a red over-line. Aspartic (D) and glutamic (E) amino acids in the acidic domain are labeled with asterisks. The four positions at which nonconservative changes of highly acidic amino acids have taken place are indicated with a caret (^). (B) Amino acid identity among vertebrate Maturins. Percent identity shared with X. laevis Maturin is shown. Genbank accession numbers are: X. laevis BC045253, H. sapiens NM_152793 (C7orf41), M. musculus BC042507 (2410066E13Rik), G. gallus XM_003640720.1, D. rerio NM_001144806. (C) Detection of maturin and pax6 transcripts using RT-PCR. Gene specific primers were used to detect maturin and pax6 transcripts in total RNA isolated from eggs (E) and whole embryos of the indicated developmental stage. Histone h4 was used as a loading control. (D–K) Maturin expression in developing embryos. Whole mount in situ hybridization was used to detect Xenopus laevis maturin expression. Developmental stage is indicated above each panel. (E) Stage 9 embryo cut open with a razor following whole mount in situ hybridization. An, animal; Veg, vegetal; b, blastocoel; A, anterior; P, posterior; np, neural plate; ef, eye field; nc, neural crest; D, dorsal; V, ventral; sc, spinal cord; ov, optic vesicle; e, eye. Scale bars = 400μm.
Whole genome scans indicated the presence of a single maturin gene in diploid vertebrate organisms. No significant homology with any other gene was detected (not shown). Despite the lack of any identifiable functional domain, Maturin has been highly conserved among vertebrate species (Fig. 1A). The X. laevis Maturin protein is 92.4% and 90.8% identical to the human (synonym C7ORF41) and mouse (synonym 2410066E13RIK) Maturin proteins, respectively (Fig. 1B). Less conservation is observed with the predicted chicken and zebrafish orthologs (Fig. 1B). Of the 30 highly acidic amino acids (aspartic and glutamic acid) in X. laevis Maturin, 19 are invariant (Fig. 1A). Of the remaining 11 variant positions, all but four (two in chicken, two in zebrafish) are conservative (D → E or E → D) substitutions (Fig. 1A, caret). We also identified a novel 29-residue region (a.a. 54–82 of X. laevis) with perfect conservation (Fig. 1A). Due to its high level of conservation this region was named the Maturin Motif (Fig. 1A; red over-line).
Maturin is expressed maternally and is enriched in neural tissues
By RT-PCR maturin was detected in the egg, indicating it is expressed maternally (Fig. 1C). In contrast to pax6 transcripts, which were only detected after the late blastula stage (stage 9), maturin was detected through the eye vesicle stage (stage 20).
Maturin transcript was first detected by whole mount in situ hybridization in blastula, with the most intense staining located in the animal half of the embryo (Fig. 1D and E). Expression extends into the marginal zone, but was not observed in other areas of the vegetal half of the embryo, nor in the outer epithelial cell layer of the blastocoel roof (Fig. 1E). At the small yolk plug stage (stage 12.5), expression was faint in central and posterior regions of the neural plate, yet robust in the anterior neural plate, including the eye field (Fig. 1F; ef). By the neurula stage (stage 15), expression became more intense in the posterior neural plate (Fig. 1G). Expression is also first detected in presumptive neural crest at these stages. At later developmental stages, transcripts appeared restricted to the developing nervous system (Fig. 1H–K). Expression was detected in the developing brain, spinal cord and optic vesicles at early and late tailbud stages (stages 24 to 31; Fig. 1J and K). Thus, maturin codes for a novel evolutionarily conserved vertebrate gene, which is expressed maternally, but is enriched in neural tissues.
Maturin is expressed in differentiating neurons of the vertebrate nervous system
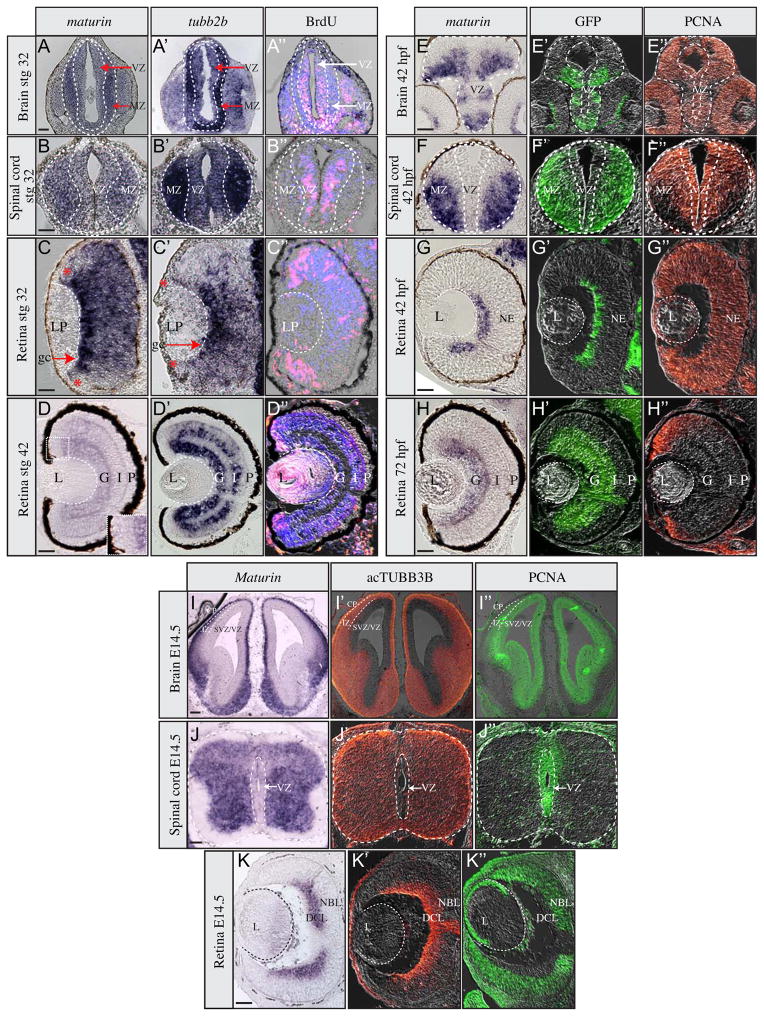
Whole mount in situ hybridization indicated that frog maturin transcripts were expressed in developing neural tissues (Fig. 1). To confirm this observation and determine if maturin is similarly expressed in other vertebrates, in situ hybridization was used to detect maturin expression in brain, spinal cord and retinal sections. Markers that label mitotic and differentiated regions were also used to compare the maturin expression patterns of actively proliferating and post-mitotic regions of the nervous system.
Regions of neural differentiation can be identified by the very early expression of neural-specific type II β tubulin (tubb2b) (Chitnis et al., 1995; Oschwald et al., 1991). Similar to tubb2b, maturin transcripts were detected in the dividing (BrdU-positive) cells of the frog ventricular zone, but were more intensely expressed in post-mitotic (BrdU-negative) neurons of the stage 32 spinal cord and brain (Fig. 2A–B″). Similar results were observed in the retina. However, maturin was expressed more broadly than and preceded tubb2b in later differentiating regions (Fig. 2C–C″). Retinal ganglion cells (RGCs) are the first neurons of the Xenopus retina to exit the cell cycle and differentiate (Holt et al., 1988). Maturin expression was detected most intensely throughout the presumptive RGC layer before tubb2b expression (compare Fig. 2C to 2C′). Similarly, maturin expression precedes that of tubb2b throughout the central retina, but was significantly reduced or undetectable in the most peripheral region, where retinal progenitors continue to divide (Fig. 2C and 2C″). In stage 42 tadpoles, when central retinal differentiation is complete, maturin expression is significantly reduced (relative to earlier stages and tubb2b; Fig. 2D–D′). Maturin transcripts were not detected in the proliferating cells of the ciliary marginal zone, but were detected adjacent to the CMZ, where maturin expression overlaps that of tubb2b (Fig. 2D–D′, inset). Maturin transcripts were not detected in the lens at any stage tested (Fig. 2C and D and not shown).
Fig. 2.
Maturin expression during neural differentiation. Expression of maturin (A–K), differentiation (A′–K′) and proliferation markers (A″–K″) in frog (A–D″), zebrafish (E–H″) and mouse (I–K″) neural tissues. Expression of maturin (A–K) and tubb2b (A′–D′) was determined using in situ hybridization. Differentiating neurons were identified by GFP immunolabeling to detect elav3l:GFP transgene expression in zebrafish (green in E′–H′) and by acTUBB3b (orange in I′–K′) immunolabeling in mouse. Proliferating neuroblasts were identified by either BrdU in Xenopus (pink in A″–D″) or PCNA immunolabeling in zebrafish and mouse (orange in E″– H″ and green in I″– K″). Nuclei were stained blue with DAPI (A″–D″). Inset in panel D is magnified view of region encompassing the dorsal CMZ. MZ, marginal zone; VZ, ventricular zone; LP, lens placode; gc, ganglion cells; *, retinal periphery; L, lens; G, ganglion cell layer; I, inner plexiform layer; P, photoreceptor layer; NE, neuroepithelium; CP, cortical plate; IZ, intermediate zone; DCL, differentiated cell layer; NBL, neuroblastic layer. Scale bars, 50 μm.
Zebrafish maturin expression in the developing retina, brain and spinal cord was similar to that of GFP driven from the elav3l promoter/enhancer (elav3l:GFP), which marks early born, differentiating neurons (Park et al., 2000). Similar to frog, zebrafish maturin expression was absent or reduced in proliferating (PCNA-positive) cells of the ventricular zones (Fig. 2E–F″). In the early retina, maturin transcripts were detected in elav3l:GFP-positive presumptive ganglion cells (Fig. 2G–G″). Maturin was not detected in the retina before the onset of ganglion cell differentiation (not shown). At later stages, similar to elav3l:GFP, maturin was expressed in differentiated ganglion cells and the IPL (Fig. 2H and H′), yet was absent from proliferating cells of the ciliary marginal zone (Fig. 2H and H″). elav3l:GFP expression appeared to preceded maturin expression in the fish INL (Fig. 2H and H′). As in frog, fish maturin was not detected in the lens at any developmental stage tested (Fig. 2G, H and not shown).
As in fish and frog, mouse Maturin transcripts were detected in differentiating cells of the embryonic brain, spinal cord and retina (Fig. 2I–K″). Mouse Maturin transcription was most intense in differentiating (acTUBB3B-positive) cells and was undetectable or reduced in proliferating (PCNA-positive) cells (Fig. 2I–K″).
In summary, maturin transcripts are expressed prior to and in differentiating neurons and are reduced or absent in regions of the developing nervous system that are actively proliferating. Furthermore, this pattern of maturin expression appears to be evolutionarily conserved as similar patterns are observed in fish, frog and mice.
Maturin knockdown results in neural plate expansion
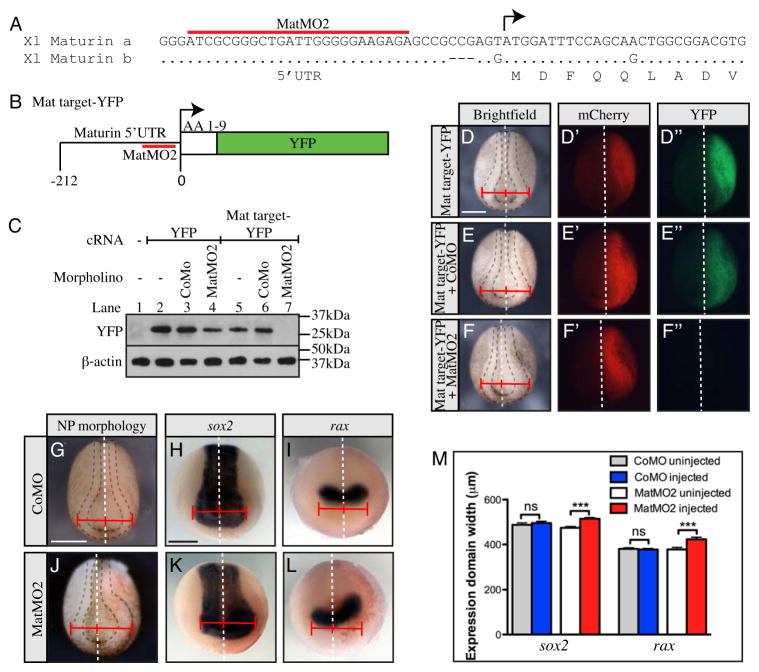
Two non-overlapping antisense morpholino oligonucleotides (MatMO1 and MatMO2) were generated to inhibit maturin translation (Figs. 3A, S1A). A reporter construct (Mat target-YFP), generated by fusing the morpholino target sites with the open reading frame of YFP, was used to determine if the morpholinos could inhibit protein translation (Figs. 3B, S1B). Embryos were injected with capped RNA (cRNA) coding for YFP or the Mat target-YFP fusion alone, or with morpholinos. Western blot analysis was performed on whole embryo extracts to detect expression of YFP or fusion protein (Fig. 3C). YFP protein was detected when YFP cRNA was injected alone, or with control or Maturin morpholinos (Fig. 3C; lanes 2–4 and not shown). Similarly, Mat target-YFP protein was detected when injected alone or with control morpholino (Fig. 3C; lanes 5 and 6). In contrast, the Mat target-YFP fusion protein was undetectable when coinjected with MatMO2 (Fig. 3C; lane 7). To confirm target knockdown in living embryos, fluorescence was analyzed in neurula embryos that had been injected in one blastomere at the two-cell stage with cRNA coding for Mat target-YFP, mCherry RFP (as a tracer) and either control or Maturin morpholinos (Fig. 3D–F″, S1C–F″). Both mCherry and YFP fluorescence were detected when injected alone (Fig. 3D–D″; 100% of embryos mCherry and YFP positive, n=40) or in combination with control morpholino (Fig. 3E–E″; 100% of embryos mCherry and YFP positive; n=50). However, only mCherry was detected with coinjection of Maturin morpholino (Fig. 3F–F″; 100% of embryos mCherry positive; 0% of embryos YFP positive; n=44). These results demonstrate the Maturin morpholinos can inhibit the translation of transcripts containing the maturin target sequence in vivo.
Fig. 3.
Maturin knockdown results in neural plate expansion. (A–F″) Design and test of Maturin morpholino activity. (A) Sequence alignment of a and b homeologs of X. laevis maturin showing relative position of the Maturin morpholino MatMO2 (red overline). (B) Schematic of the Mat target-YFP reporter construct used to test morpholino activity. (C) Western blots were used to detect the expression of YFP and β-actin (loading control) in extracts prepared from embryos injected with the indicated morpholino, and cRNA coding for YFP or Mat target-YFP. (D–F″) Brightfield (D, E and F), mCherry fluorescent (D′, E′ and F′) and YFP fluorescent (D″, E″ and F″) images of stage 15 embryos unilaterally injected with cRNA for mCherry and Mat target-YFP alone (D – D″), with CoMO (E–E″), or MatMO2 (F–F″). (G–M) Neural plate expansion following Maturin knockdown. The extent of neural plate expansion was determined by comparing the distance between the embryonic midline (white dashed line) and outer edge of the neural ridge on the control (uninjected) and injected side of embryos injected with CoMO (G–I) or MatMO2 (J–L). In situ hybridization for sox2 (H, K) and rax (I, L) was used to more precisely quantitate the extent of neural plate expansion (M). Graph shows the size of the sox2 and rax expression domains in the uninjected and injected side of either CoMO or MatMO2 embryos. Error bars show the s.e.m. Asterisks indicate P-values calculated using a one-way ANOVA analysis (ns P>0.05; *** P<0.0001). Right side (viewer’s perspective) of all embryos is the injected side. Scale bars, 400μm.
Coincident with the loss of YFP fluorescence, we also observed an expansion of the neural plate on the Maturin morpholino-injected side of embryos (compare Fig. 3D and E with F). Both MatMO1 and MatMO2 expanded the neural plate (Fig. 3 and S1). However, due to the greater frequency with which it could generate the expanded neural plate phenotype (MatMO1; 67%, n=58 vs. MatMO2; 80%, n=45), MatMO2 was used for all subsequent experiments. To confirm neural plate expansion, in situ hybridization was used to detect changes in the expression of sox2 (neural plate) and rax (eye field) (Andreazzoli et al., 2003; Mathers et al., 1997; Mizuseki et al., 1998). Unilateral injection of the control morpholino did not alter neural plate size on the injected side (Fig. 3G; 0%, n=65), and the expression patterns of sox2 and rax remained bilaterally symmetric (Fig. 3H, I, M; sox2, 487.4 ± 9.1 μm uninjected side vs. 495.4 ± 8.1 μm injected side, n=65; and rax, 380 ± 5.1 μm uninjected side vs. 377.2 ± 4.8 μm injected side, n=58; ns P>0.05). In contrast, the neural plate (Fig. 3J) and expression domains of both sox2 and rax (Fig. 3K and L), were enlarged with Maturin knockdown (Fig. 3M; sox2, 474.1 ± 5.5 uninjected side vs. 514.6 ± 5.2 μm injected side, n=74; and rax, 377.8 ± 8.8 uninjected side vs. 423.5 ± 9.0 μm injected side, n=63; *** P<0.0001). These results, and the observation that two, non-overlapping Maturin morpholinos generate the same phenotype, strongly suggest that Maturin knockdown results in neural plate expansion.
Maturin knockdown expands the neural plate by increasing cell number
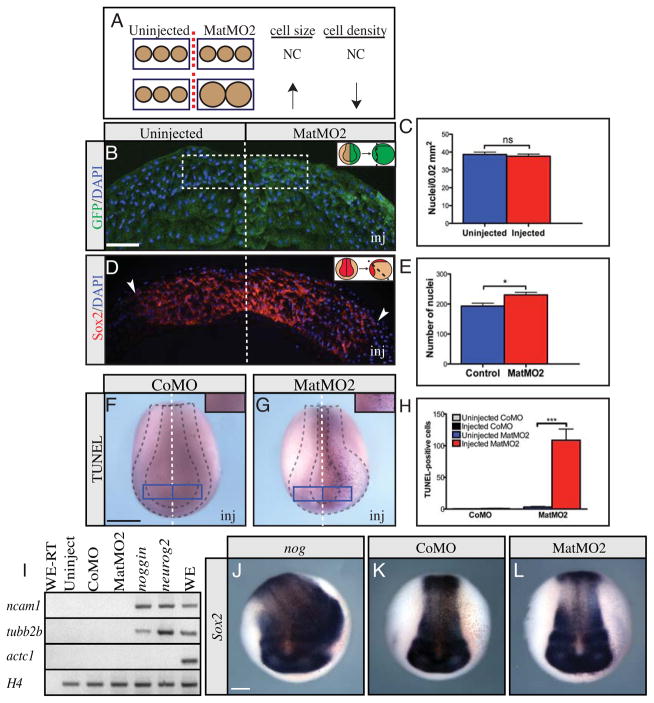
Neural plate enlargement could result from either larger cells, or an increase in the number of cells. If due to an increase in cell size, the number of cell nuclei per unit area (nuclear density) should be reduced in morpholino injected embryos (Fig. 4A). Nuclear density was compared on both sides of the embryonic midline in embryos unilaterally injected with Maturin morpholino. For comparisons, we selected bilaterally symmetric regions of equal size within the areas of the anterior neural plate showing the greatest change in neural plate size (Fig. 4B). Counts of DAPI-labeled nuclei showed that the average nuclear density on the uninjected and Maturin morpholino-injected sides of embryos were similar (Fig. 4C; uninjected side 39 ± 1 cells/0.02 mm2, n=2,520 cells vs. MatMO2-injected side 38 ± 1 cells/0.02 mm2, n=2,583 cells, P=0.5812). This result suggests Maturin knockdown did not alter cell size and was therefore not likely to be responsible for neural plate expansion.
Fig. 4.
Maturin knockdown increases neural cell number without reducing cell death or inducing neural cell fate. (A–C) Neural plate cell density in MatMO2-injected embryos. (A) Diagram illustrating the predicted effect on cell density if a change in cell size is responsible for neural plate expansion. (B) The number of nuclei (Blue - DAPI) in a 100 μm by 200 μm region on either side of the midline of embryos unilaterally injected with MatMO2 and GFP cRNA (green) was determined. Inset shows how the embryos were positioned for sectioning. (C) Average number of nuclei observed in 0.02 mm2 area. (D, E) Neural plate cell number in MatMO2-injected embryos. (D) Neural plate sections were stained for sox2 expression (red) and DAPI (blue). Dashed line indicates embryonic midline, and the lateral extent of sox2 expression is indicated by the arrowheads. Inset shows how the embryos were positioned for sectioning. (E) Total number of cells (nuclei) within the sox2 expression domain. (F–H) TUNEL staining was used to detect cell death in CoMO- (F) and MatMO2-injected embryos (G). Insets in F and G show the location of the 0.138 mm2 area on the control and injected side of each embryo scored to generate results (H). (I) RT-PCR for ncam1, tubb2b, actc1 (actin, alpha cardiac muscle 1) and H4 on animal cap explants from embryos injected into both blastomere at the 2-cell stage with CoMO, MatMO2, noggin or neurog2. Actc1 was used to confirm neural induction resulting from nog and neurog2 injection was direct (not via mesoderm induction) (Hemmati-Brivanlou and Melton, 1994). H4 was used as a loading control. Controls included RNA isolated from whole embryos and processed without (WE-RT) and with (WE) reverse transcriptase, as well as uninjected animal cap explants. Similar results were obtained from two independent experiments and when explants were cultured to the equivalent of stg 22. (J–L) Sox2 in situ hybridization on embryos injected into one ventral blastomere at the 8-cell stage with noggin (J), CoMO (K) or MatMO2 (L). The injected side is on the right (viewer’s perspective). In the graphs the error bars show the s.e.m. P-values obtained using Student’s t-test: ns, not significant; *P=0.0101; ***P<0.0001. Scale bars, (B)=100 μm; (F and J)=400 μm.
To determine if Maturin knockdown increased cell number within the expanded neural plate region, sections of neurula embryos were costained for sox2 by in situ hybridization to identify the boundary of the neural plate, and with DAPI to identify cell nuclei. Sox2 expression was clearly expanded in sections, consistent with the sox2 expansion observed in whole mount embryos (Fig. 3K and 4D). Importantly, the average number of DAPI-labeled cells within the sox2 expressing domain was significantly higher on the Maturin morpholino-injected side (Fig. 4E; uninjected side 193 ± 10 cells/section, n=5,649 cells vs. MatMO2-injected side 230 ± 9 cells/section, n=6,702 cells, P=0.0101). Together, these results indicate that Maturin knockdown expands the neural plate by increasing cell number and not by altering cell size.
The increase in neural plate cell number following Maturin knockdown does not result from a reduction in cell death or a change in cell fate
Neural plate enlargement following Maturin knockdown could result from a reduction in cell death during embryonic development, ultimately leading to a larger pool of neural progenitors and a larger neural plate. To determine if Maturin knockdown inhibited cell death, embryos were stained using the Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) method. The number of TUNEL-positive cells on the control and morpholino-injected sides of embryos were compared (Fig. 4F–H). The control morpholino did not significantly alter the number of TUNEL-positive cells (Fig. 4F and H; uninjected side 0 ± 0 vs. CoMO-injected side 1 ± 1; n=15 embryos). In contrast, Maturin knockdown dramatically increased the number of TUNEL-positive cells (Fig. 4G and H; uninjected side 3 ± 1 cells vs. MatMO2-injected side 109 ± 19; n=16 embryos). These results are consistent with other reports in which an increase in apoptotic cell death was observed following excessive proliferation in the Xenopus nervous system (Klisch et al., 2006; Zuber et al., 1999). We conclude that neural plate expansion following Maturin knockdown, did not result from a reduction in cell death.
Neural plate enlargement in response to Maturin knockdown could result from a change in cell fate. Conversion of cells from a non-neural to neural lineage would result in an increase in the number of neural plate cells and a larger neural plate. To determine if Maturin loss of function was sufficient to induce neural tissue, we injected Maturin morpholinos into both blastomeres of two-cell staged embryos, collected animal poles prior to gastrulation (stage 8.5) and cultured the explants to the equivalent of neurula stages. In contrast to the neural inducers noggin (nog) and neurog2, Maturin morpholinos did not induce the expression of the neural progenitor markers neural cell adhesion molecule 1 (ncam1) or tubb2b, suggesting Maturin knockdown is not sufficient to induce neural tissue (Fig. 4I).
Although unable to induce neural markers in isolated primitive ectoderm, it is still possible that morpholino-mediated knockdown of Maturin could result in neural expansion in vivo. Therefore we injected Maturin morpholinos into ventral blastomeres at the 8-cell stage, cultured the embryos to neurula stages and used in situ hybridization to detect sox2 expression. Noggin was sufficient to induce ectopic neural tissue in every injected embryo, resulting in expansion of the sox2 expression domain (Fig. 4J, 100%, n=82). In contrast, control and Maturin morpholinos neither expanded the neural plate nor induced ectopic sox2 expression in ventrally injected embryos (Fig. 4K, L; CoMO, 0%, n=56; MatMO2, 0%, n=40).
These results indicate that Maturin knockdown is not sufficient to induce neural markers in vitro or in vivo, and is therefore unlikely to drive neural plate expansion by conversion of early progenitors from a non-neural to neural lineage.
Maturin knockdown expands the neural plate by inhibiting neuronal differentiation
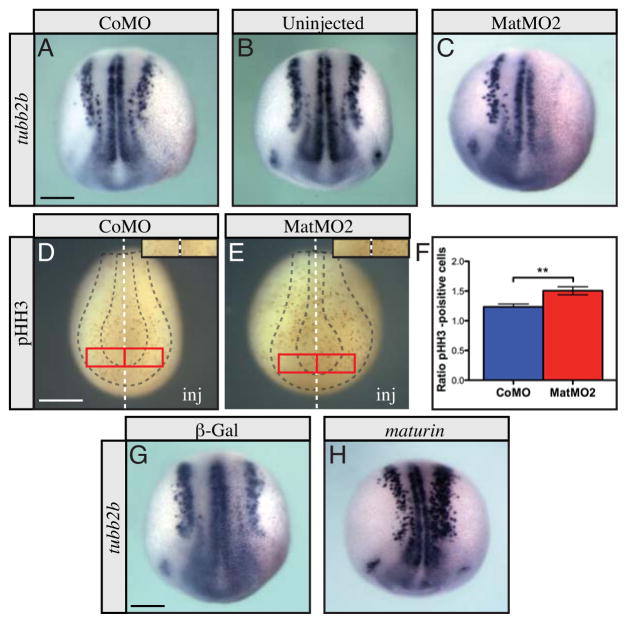
An increase in neural plate cell number could result from an accumulation of neural progenitor cells resulting from a failure to differentiate. To test this possibility, we used whole mount in situ hybridization for tubb2b, to determine the effect of Maturin knockdown on primary neurogenesis. In embryos injected unilaterally with control morpholino, tubb2b expression remained bilaterally symmetric in the vast majority of embryos, with only a small minority showing asymmetric expression (Fig. 5A; 3%, n=105). The frequency of asymmetry was similar to that observed in uninjected embryos, suggesting an infrequent, yet detectable variation in tubb2b expression during normal development (Fig. 5B; 3%, n=91). In contrast, tubb2b expression was dramatically reduced or completely lost on the injected side of every Maturin morpholino-injected embryo (Fig. 5C; 100%; n=290). Similar results were observed with MatMO1 (Fig. S1G). These results suggest that Maturin knockdown blocks the differentiation of the primary neurons.
Fig. 5.
Maturin knockdown inhibits differentiation of neural progenitors. (A–C) Whole mount in situ hybridization for tubb2b following Maturin knockdown. tubb2b expression in CoMO injected (A), uninjected (B), or MatMO2 injected embryos (C). A similar effect on tubb2b expression was also observed with MatMO1 (Fig. S1G). (D–F) Proliferation in Morpholino-injected embryos. Proliferating cells were labeled using pHH3 whole mount immunostaining in CoMO (D) and MatMO2 injected embryos (E). Insets in D and E show the 0.138 mm2 area on the control and injected side of each embryo in which pHH3-positive cells were counted to generate the results (F). (G–I) Whole mount in situ hybridization for tubb2b after maturin overexpression. tubb2b expression in β-Gal (G) and maturin cRNA (H) injected embryos. In all panels of this figure, the injected side is to the right (viewer’s perspective). The s.e.m. is shown. P-values obtained using Student’s t-test: **P=0.0023. Scale bars, 400μm.
Inhibition of differentiation could result from an increase in the number of proliferating neural progenitor cells. To determine if Maturin knockdown also increased proliferation in the neural plate, mitotic cells were labeled using phosphohistone H3 wholemount immunostaining. The ratio of mitotic cells on the injected vs. uninjected side of control and Maturin morpholino injected embryos was compared (Fig. 5D–F). Since neural plate expansion was most dramatic anteriorly, we selected this region of the neural plate to score. Control morpholinos only slightly increased the number of mitotic cells detected (injected/uninjected = 1.234 ± 0.047; n=1,916 pHH3+ cells on injected side versus 1,684 cells on uninjected side of 116 embryos; Fig. 5D and F). However, the ratio of mitotic cells between the injected and uninjected side was consistently higher in Maturin morpholino injected embryos (injected/uninjected = 1.505 ± 0.068; n=2,249 pHH3+ cells on the injected side versus 1,759 cells on uninjected side of 148 embryos; Fig. 5E and F). From these results we conclude that Maturin knockdown not only inhibited neuronal differentiation, but also increased the number of proliferating cells in the neural plate.
The above results suggest that Maturin function is required for primary neurogenesis. If so, excessive Maturin might be expected to promote the differentiation of the primary neurons. To test this possibility, we overexpressed Maturin unilaterally by cRNA injection, and stained embryos for tubb2b expression. Injection of βgal cRNA, had no effect on primary neurogenesis (Fig. 5G). In contrast, maturin cRNA resulted in an increase in the number of tubb2b-positive cells only on the injected side of embryos (Fig. 5H; 43%; n=172 embryos). The increase in tubb2b expression was restricted to the neural plate, as ectopic tubb2b expression was not detected in more lateral or ventral regions. We conclude, that Maturin is sufficient to induce the differentiation of primary neurons in the context of the neural plate.
Our results indicate that Maturin function is required for the differentiation of the primary neurons. When misexpressed, Maturin can induce excessive neural differentiation. Conversely, blocking Maturin function inhibits neural differentiation, increases the number of proliferating neural progenitors and ultimately results in an enlarged neural plate.
Maturin transcription is induced by the proneural transcription factors Neurog2, Neurod1 and Ebf3
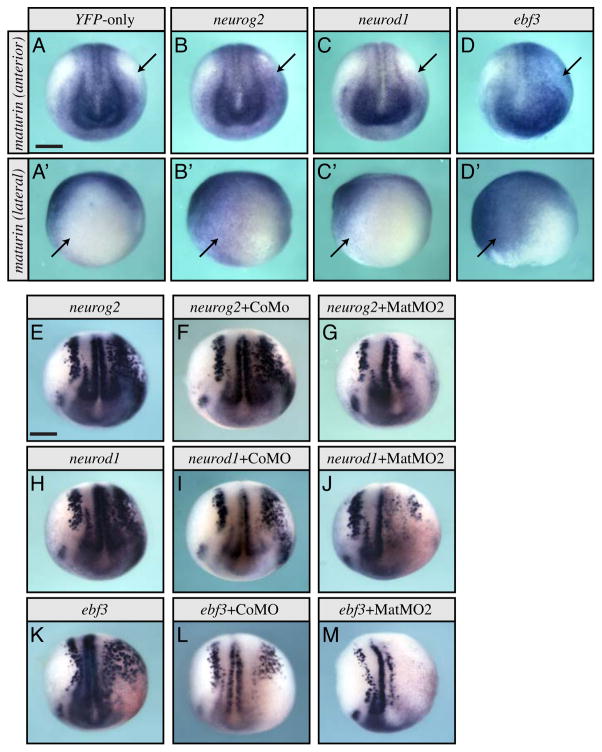
The above results suggest Maturin is required for normal primary neurogenesis. Differentiation of the primary neurons is driven by transcription factors of the proneural pathway (Henningfeld et al., 2007). To determine if maturin transcription could be induced by the proneural transcription factors, we injected cRNA coding for neurog2, neurod1 or ebf3 into one blastomere at the 2-cell stage and used whole mount in situ hybridization to determine the effect on maturin expression. No change in maturin expression was detected when YFP was expressed alone (Fig. 6A–A′; 0%, n=27). In contrast, expression of neurog2, neurod1, or ebf3 all resulted in unilateral, ectopic transcription of maturin (Fig. 6B–D′; neurog2, 54%, n=35; neurod1 1, 39%, n=36; ebf3, 86%; n=35). These results suggested to us that Maturin may function downstream of the proneural transcription factors.
Fig. 6.
Maturin transcription is induced by, and Maturin function is required for the activity of, the proneural transcription factors. (A–D′) Anterior (A–D) and lateral (A′–D′) views of maturin expression in embryos misexpressing YFP-only (A, A′), neurog2 (B, B′), neurod1 (C, C′), and ebf3 (D, D′), respectively. Arrows show areas normally lacking maturin (A, A′), express ectopic maturin when proneural transcription factors are misexpressed (B–D′). (E–M) One blastomere of 2-cell staged embryos were injected with the indicated proneural transcription factor alone (E, H and K), with control (F, I and L) or Maturin (G, J and M) morpholinos. The effects on tubb2b expression were detected by whole mount in situ hybridization at stage 15. Right side (viewer’s perspective) is the injected side. Scale bar, 400μm.
Maturin function is required for the activity of the proneural transcription factors Neurog2, Neurod1 and Ebf3
When overexpressed, proneural genes induce ectopic primary neurons as determined by expression of the neuronal marker tubb2b (Lee et al., 1995; Ma et al., 1996; Pozzoli et al., 2001). To determine if Maturin might function within the proneural pathway, we coinjected cRNA coding for the proneural transcription factors neurog2, neurod1 or ebf3, with either CoMO or MatMO2. Misexpression of neurog2, neurod1, or ebf3 by cRNA injection resulted in unilateral, ectopic expression of tubb2b as previously reported (Fig. 6E, H and K; neurog2, 99%, n=86; neurod1, 100%, n=67; ebf3, 100%; n=30) (Lee et al., 1995; Ma et al., 1996; Pozzoli et al., 2001). Coinjection of control morpholino did not alter the ability of the proneural transcription factors to induce ectopic tubb2b expression (Fig. 6F, I and L; neurog2 + CoMO, 100%, n=75; neurod1 + CoMO, 100%, n=61; ebf3 + CoMO, 97%; n=32). In contrast, Maturin knockdown reduced or completely blocked ectopic tubb2b expression in 92%, 89% and 100% of embryos injected with neurog2, neurod1 and ebf3, respectively (neurog2 + MatMO2, n=66; neurod1 + MatMO2, n=59; ebf3 + MatMO2, n=15; Fig. 6G, J and M). These results demonstrate that maturin can be transcriptionally regulated by all three proneural transcription factors, is required for the activity of Neurog2, Neurod1 and Ebf3 in vivo and therefore most likely functions downstream of these proneural pathway transcription factors during primary neurogenesis.
Maturin and Pak3 are both required for, and synergize to promote neural differentiation
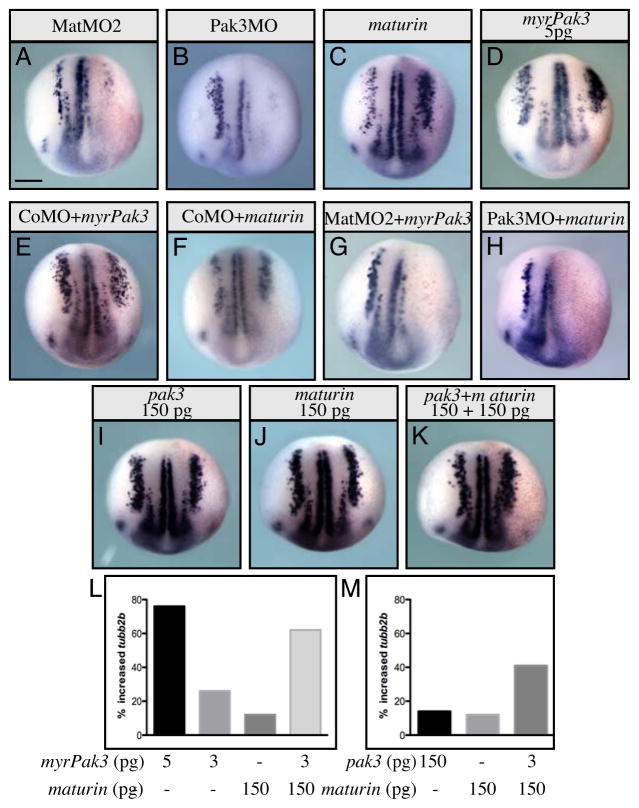
The maturin expression pattern, loss-of-function and overexpression phenotypes indicate Maturin regulates the differentiation of the primary neurons. The p21-activated kinase 3 (Pak3) regulates the cell cycle exit and differentiation of primary neurons (Souopgui et al., 2002). Pak3 is expressed in differentiating neural cells, and like Maturin, is required downstream of the proneural transcription factors. Similar to Maturin knockdown, Pak3 knockdown (with Pak3MO) also inhibits primary neurogenesis and increases cell proliferation resulting in neural plate expansion (compare Fig. 7A and 7B; MatMO2, 100%, n=290 and Pak3MO, 81%, n=84). Misexpression of Pak3 by cRNA injection alters neither tubb2b expression nor embryonic development (Souopgui et al., 2002). However, when localized to the cell membrane by artificial myristoylation, myrPak3 is made constitutively active and at very low doses (5 pg) induces additional tubb2b expression in the neural plate in a manner similar to maturin misexpression (compare Fig. 7C and 7D; maturin, 62%, n=149 and myrPak3, 74%, n=38 and see (Hing et al., 1999; Souopgui et al., 2002)).
Fig. 7.
Maturin and Pak3 synergy. (A–K) Embryos were injected unilaterally into one blastomere at the 2-cell stage with the indicated morpholino(s) and/or cRNA(s). At stage 15 in situ hybridization was used to determine the effect on tubb2b expression. (L, M) Graphs illustrate the percent of embryos with additional tubb2b-positive cells in the lateral stripe, when injected with sub-maximal amounts of maturin, myrPak3, pak3 or the indicated combination of cRNAs. Scale bars, 400μm. Injected side in all embryos is right side (viewer’s perspective).
To determine if Maturin function was required before, after, or with Pak3, we blocked the function of endogenous Maturin or Pak3 with MatMO2 or Pak3MO, and asked if primary neurogenesis could be rescued by coinjection with myrPak3 and maturin, respectively. Control morpholino did not block the ability of either myrPak3 or Maturin to induce tubb2b expression (Fig. 7E and 7F; CoMO + myrPak3, 82%, n=38 and CoMO + maturin, 62%, n=40). Interestingly, Maturin loss-of-function could not be rescued by constitutively active myrPak3 (Fig. 7G; 0%, n=20). Similarly, Maturin could not significantly rescue Pak3 loss-of-function (Fig. 7H; 15%, n=27). Together, these results demonstrate that both Maturin and Pak3 are required for primary neurogenesis. To determine if they might function synergistically, we coinjected sub-maximal amounts of maturin and myrPak3 cRNAs. Reducing the amount of maturin (400 to 150 pg) or myrPak3 (5 to 3 pg) cRNA, lowered the per cent of embryos with an increase in tubb2b expression from 62% to 12% and from 74% to 26%, respectively (Fig. 7L). Interestingly, when coexpressed at sub-maximal levels, Maturin and myrPak3 induced tubb2b expression synergistically (Fig. 7L). To test this idea further, we injected maturin with non-myristoylated pak3 cRNA (pak3). In contrast to constitutively active myrPak3 (74% at 5 pg), concentrations as high as 150 pg of pak3 only weakly (14%) induced tubb2b expression (Fig. 7I and 7M). Surprisingly, Maturin even functioned synergistically with non-myristoylated Pak3 (Fig. 7I–K and M). Together, these results indicate that functional Maturin and Pak3 are both required for primary neurogenesis. Furthermore, Maturin can function synergistically not only with constitutively active myrPak3, but also non-myristoylated Pak3 to activate neural differentiation.
Discussion
We report the identification of Maturin, a novel protein required for differentiation during primary neurogenesis. Our conclusions are supported by the observations that: (1) maturin transcripts are enriched in the neural plate at the time of primary neurogenesis and in other neural progenitors as they differentiate; (2) Maturin knockdown inhibits, while overexpression induces neuronal differentiation in the neural plate; (3) maturin expression is induced by the proneural transcription factors Neurog2, Neurod1 and Ebf3; (4) Maturin is required for the proneural transcription factors to induce neurogenesis; (5) Maturin and Pak3 are both required for primary neurogenesis; and (6) Maturin functions synergistically with Pak3 to promote neural differentiation.
Evolutionary conservation of Maturin
A striking feature of Maturin is its high degree of sequence conservation, particularly the 29 residue Maturin Motif which is invariant among vertebrates tested (Fig. 1). Due to its perfect sequence conservation, it is logical to assume that the Maturin Motif is critical for the protein’s function. Structure-function analysis of the Maturin Motif (and other conserved regions) will help define the importance of these regions during primary neurogenesis and for Pak3 synergy.
Primary neurogenesis in Xenopus laevis has proven to be an invaluable tool for discovering conserved genes and genetic interactions required for neurogenesis (Henningfeld et al., 2007). In frog, maturin has a dynamic, yet consistent, developmental expression pattern - low in proliferating neural progenitors, high during differentiation, then reduced again following differentiation (Fig. 1 and 2). In the embryonic zebrafish and mouse nervous system, a remarkably similar expression pattern is also observed (Fig. 2 and R. I. Martinez-De Luna and M. E. Zuber, unpublished results). Conservation of both the primary amino acid sequence and expression patterns of zebrafish, frog and mouse maturins does not of course demonstrate an identical function for Maturin in all three species. However, it does strongly suggests Maturin may have an important role, not only in Xenopus primary neurogenesis, but also during neural differentiation in other vertebrate species.
The nature of Maturin/Pak3 synergy
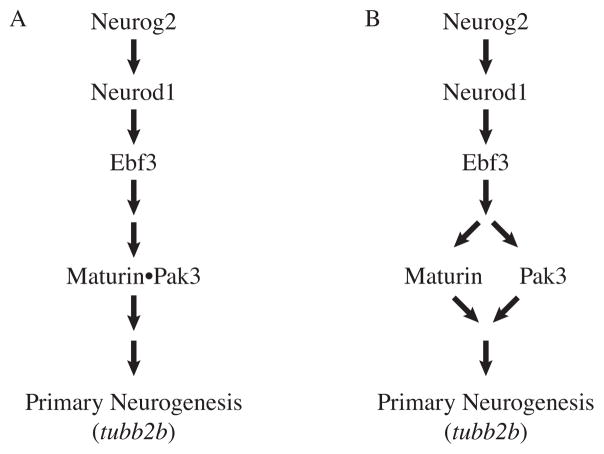
Maturin and Pak3 share many similarities. Both are required for primary neurogenesis. Morpholino knockdown of either Maturin or Pak3 inhibits neural differentiation, increases cell proliferation and results in neural plate expansion. Neurog2, Neurod1 and Ebf3 can all induce both maturin and pak3 transcription (this work and (Souopgui et al., 2002)). Both Maturin and Pak3 loss-of-function also block the ability of the proneural transcription factors Neurog2, Neurod1 and Ebf3 to induce ectopic neural differentiation. Both Maturin and Pak3 are sufficient for primary neurogenesis. Maturin and Pak3 (myrPak3) gain-of-function by cRNA injection promotes neural differentiation (this work and (Souopgui et al., 2002)). Despite the fact they can both induce primary neurogenesis, Maturin and Pak3 appear to be interdependent. MyrPak3 and Maturin cannot rescue the Maturin and Pak3 knockdown phenotypes, respectively. Furthermore, Maturin functions synergistically not only with constitutively active myrPak3, but also Pak3. Together, these results are most consistent with a role for Maturin in the proneural pathway, downstream of the proneural transcription factors where both Maturin and Pak3 functions are required during primary neurogenesis (Fig. 8).
Fig. 8.
Proposed gene network illustrating the most likely position of Maturin in the proneural pathway. Maturin and Pak3 function together downstream of the proneural transcription factors to promote differentiation of the primary neurons. Our results are consistent with models in which Maturin and Pak3 form a complex (A), or function independently (B). In both models, Maturin and Pak3 are both required for normal primary neurogenesis.
Pak3 kinase activity is required for myrPak3 to induce differentiation, and 14 of the 131 Maturin residues are serines or threonines (Fig. 1 and (Souopgui et al., 2002)). Although only eight of these residues are conserved and none lie in a Pak consensus phosphorylation site, it will be important to determine if a physical association of these proteins is required for synergy, Pak3 kinase activity, and/or possible Maturin phosphorylation (Fig. 1 and (Gururaj et al., 2005; King et al., 2001; Rennefahrt et al., 2007)).
Known activators of Pak protein kinase activity are Rac1 and Cdc42 GTPases, and Paks are recruited to the plasma membrane via their interactions with adaptor proteins (Arias-Romero and Chernoff, 2008; Bokoch, 2003; Kreis and Barnier, 2009). For example, human Pak1 is recruited to active tyrosine kinase receptors in the membrane by the SH3 domain of the adaptor protein Nck and its membrane localization results in kinase activation (Galisteo et al., 1996; Lu and Mayer, 1999). The increase in neural differentiation observed after misexpression of Maturin with non-myristoylated Pak3 may suggest a role for Maturin in recruiting or targeting Pak3 to the plasma membrane. However, Maturin may be part of a protein complex that recruits Pak3 to the membrane for kinase activation and subsequent activation of downstream differentiation signals. In this model, a direct interaction of Maturin and Pak3 would be predicted (Fig. 8A)
It is important to note, that we cannot discount alternative hypotheses. Unlike pak3 (and the proneural transcription factors), maturin expression is not only detected in the differentiating primary neurons, but also throughout the neural plate before and during primary neurogenesis (Fig. 1 and (Chalmers et al., 2002; Lee et al., 1995; Ma et al., 1996; Pozzoli et al., 2001; Souopgui et al., 2002)). Maturin and Pak3 may function in independent pathways, both of which are required for primary neurogenesis (Fig. 8B). In addition, Maturin may also have a role in regulating neural progenitor proliferation prior to neural differentiation. This possibility is consistent with our observation that Maturin knockdown results in an expansion of the sox2 and rax expression domains in the anterior neural plate, where neural differentiation has not yet started (Fig. 3K and L).
Primary neurogenesis in the Xenopus neural plate has served as an important model for identifying the molecules and mechanisms responsible for neural differentiation in other tissues and species. The expression of Maturin in differentiating retina, brain and spinal cord neurons in fish, frog, and mice, implies a role not only in primary neurogenesis, but also in the differentiation of cells in other regions of the nervous system. Maturin is an evolutionarily conserved, novel protein with no identifiable functional domains. Understanding the mechanism(s) by which Maturin regulates neurogenesis will identify it as a new molecular component of know signaling systems, or as part of a new, previously unidentified signaling system required for differentiation of the primary neurons.
Supplementary Material
Acknowledgments
We would like thank the following for providing plasmids: Monica Vetter (pCS2+.Ebf3), Sally Moody (pBSSK+.Sox2, pBSKS+.tubb2b, pCS2+.neurog2, and pCS2+MT.NeuroD), Tomas Pieler (pCS2+.myrPak3 and pBK-CMV.Pak3). We also thank Heather Nihart, Matthew Mellini, Jeffrey Amack and Andrea Viczian for technical assistance, Nancy Papalopulu for the detailed fish gelatin embedding and sectioning protocol, Jeffrey Amack and Katharine Lewis for providing wild type and elav3l:GFP transgenic zebrafish embryos, and Andrea Viczian and Francesca Pignoni for suggestions on the manuscript. Research reported in this publication was supported by the National Eye Institute of the National Institutes of Health under award numbers R01EY017964 and R01EY015748 (MEZ), a Research to Prevent Blindness unrestricted grant to the Upstate Medical University Department of Ophthalmology, and the Lions Club of Central New York.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agathocleous M, Harris WA. From progenitors to differentiated cells in the vertebrate retina. Annu Rev Cell Dev Biol. 2009;25:45–69. doi: 10.1146/annurev.cellbio.042308.113259. [DOI] [PubMed] [Google Scholar]
- Andreazzoli M, Gestri G, Cremisi F, Casarosa S, Dawid IB, Barsacchi G. Xrx1 controls proliferation and neurogenesis in Xenopus anterior neural plate. Development. 2003;130:5143–5154. doi: 10.1242/dev.00665. [DOI] [PubMed] [Google Scholar]
- Arias-Romero LE, Chernoff J. A tale of two Paks. Biol Cell. 2008;100:97–108. doi: 10.1042/BC20070109. [DOI] [PubMed] [Google Scholar]
- Bokoch GM. Biology of the p21-activated kinases. Annu Rev Biochem. 2003;72:743–781. doi: 10.1146/annurev.biochem.72.121801.161742. [DOI] [PubMed] [Google Scholar]
- Chalmers AD, Strauss B, Papalopulu N. Oriented cell divisions asymmetrically segregate aPKC and generate cell fate diversity in the early Xenopus embryo. Development. 2003;130:2657–2668. doi: 10.1242/dev.00490. [DOI] [PubMed] [Google Scholar]
- Chalmers AD, Welchman D, Papalopulu N. Intrinsic differences between the superficial and deep layers of the Xenopus ectoderm control primary neuronal differentiation. Dev Cell. 2002;2:171–182. doi: 10.1016/s1534-5807(02)00113-2. [DOI] [PubMed] [Google Scholar]
- Chitnis A, Henrique D, Lewis J, Ish-Horowicz D, Kintner C. Primary neurogenesis in Xenopus embryos regulated by a homologue of the Drosophila neurogenic gene Delta. Nature. 1995;375:761–766. doi: 10.1038/375761a0. [DOI] [PubMed] [Google Scholar]
- Dehay C, Kennedy H. Cell-cycle control and cortical development. Nat Rev Neurosci. 2007;8:438–450. doi: 10.1038/nrn2097. [DOI] [PubMed] [Google Scholar]
- Donovan SL, Dyer MA. Regulation of proliferation during central nervous system development. Semin Cell Dev Biol. 2005;16:407–421. doi: 10.1016/j.semcdb.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Dyer MA, Cepko CL. Regulating proliferation during retinal development. Nat Rev Neurosci. 2001;2:333–342. doi: 10.1038/35072555. [DOI] [PubMed] [Google Scholar]
- Galisteo ML, Chernoff J, Su YC, Skolnik EY, Schlessinger J. The adaptor protein Nck links receptor tyrosine kinases with the serine-threonine kinase Pak1. J Biol Chem. 1996;271:20997–21000. doi: 10.1074/jbc.271.35.20997. [DOI] [PubMed] [Google Scholar]
- Good PJ, Richter K, Dawid IB. The sequence of a nervous system-specific, class II beta-tubulin gene from Xenopus laevis. Nucleic Acids Res. 1989;17:8000. doi: 10.1093/nar/17.19.8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemot F. Cell fate specification in the mammalian telencephalon. Prog Neurobiol. 2007;83:37–52. doi: 10.1016/j.pneurobio.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Gururaj AE, Rayala SK, Kumar R. p21-activated kinase signaling in breast cancer. Breast Cancer Res. 2005;7:5–12. doi: 10.1186/bcr961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenstein V. Early neurogenesis in Xenopus: the spatio-temporal pattern of proliferation and cell lineages in the embryonic spinal cord. Neuron. 1989;3:399–411. doi: 10.1016/0896-6273(89)90200-6. [DOI] [PubMed] [Google Scholar]
- Hemmati-Brivanlou A, Melton DA. Inhibition of activin receptor signaling promotes neuralization in Xenopus. Cell. 1994;77:273–281. doi: 10.1016/0092-8674(94)90319-0. [DOI] [PubMed] [Google Scholar]
- Henningfeld KA, Locker M, Perron M. Xenopus primary neurogenesis and retinogenesis. Functional Development and Embryology. 2007;1:26–36. [Google Scholar]
- Hensey C, Gautier J. Programmed cell death during Xenopus development: a spatio-temporal analysis. Dev Biol. 1998;203:36–48. doi: 10.1006/dbio.1998.9028. [DOI] [PubMed] [Google Scholar]
- Hindley C, Philpott A. Co-ordination of cell cycle and differentiation in the developing nervous system. Biochem J. 2012;444:375–382. doi: 10.1042/BJ20112040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hing H, Xiao J, Harden N, Lim L, Zipursky SL. Pak functions downstream of Dock to regulate photoreceptor axon guidance in Drosophila. Cell. 1999;97:853–863. doi: 10.1016/s0092-8674(00)80798-9. [DOI] [PubMed] [Google Scholar]
- Holt CE, Bertsch TW, Ellis HM, Harris WA. Cellular determination in the Xenopus retina is independent of lineage and birth date. Neuron. 1988;1:15–26. doi: 10.1016/0896-6273(88)90205-x. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- King AJ, Wireman RS, Hamilton M, Marshall MS. Phosphorylation site specificity of the Pak-mediated regulation of Raf-1 and cooperativity with Src. FEBS Lett. 2001;497:6–14. doi: 10.1016/s0014-5793(01)02425-5. [DOI] [PubMed] [Google Scholar]
- Klisch TJ, Souopgui J, Juergens K, Rust B, Pieler T, Henningfeld KA. Mxi1 is essential for neurogenesis in Xenopus and acts by bridging the pan-neural and proneural genes. Dev Biol. 2006;292:470–485. doi: 10.1016/j.ydbio.2005.12.037. [DOI] [PubMed] [Google Scholar]
- Kreis P, Barnier JV. PAK signalling in neuronal physiology. Cell Signal. 2009;21:384–393. doi: 10.1016/j.cellsig.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Lee JE, Hollenberg SM, Snider L, Turner DL, Lipnick N, Weintraub H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- Lu W, Mayer BJ. Mechanism of activation of Pak1 kinase by membrane localization. Oncogene. 1999;18:797–806. doi: 10.1038/sj.onc.1202361. [DOI] [PubMed] [Google Scholar]
- Ma Q, Kintner C, Anderson DJ. Identification of neurogenin, a vertebrate neuronal determination gene. Cell. 1996;87:43–52. doi: 10.1016/s0092-8674(00)81321-5. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathers PH, Grinberg A, Mahon KA, Jamrich M. The Rx homeobox gene is essential for vertebrate eye development. Nature. 1997;387:603–607. doi: 10.1038/42475. [DOI] [PubMed] [Google Scholar]
- Mizuseki K, Kishi M, Matsui M, Nakanishi S, Sasai Y. Xenopus Zic-related-1 and Sox-2, two factors induced by chordin, have distinct activities in the initiation of neural induction. Development. 1998;125:579–587. doi: 10.1242/dev.125.4.579. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal Table of Xenopus Laevis (Daudin): A Systematical & Chronological Survey of the Development from the Fertilized Egg till the End of. the Fertilized Egg Till the End of Metamorp) Garland Science; New York: 1994. [Google Scholar]
- Oschwald R, Richter K, Grunz H. Localization of a nervous system-specific class II beta-tubulin gene in Xenopus laevis embryos by whole-mount in situ hybridization. Int J Dev Biol. 1991;35:399–405. [PubMed] [Google Scholar]
- Pagni M, Ioannidis V, Cerutti L, Zahn-Zabal M, Jongeneel CV, Hau J, Martin O, Kuznetsov D, Falquet L. MyHits: improvements to an interactive resource for analyzing protein sequences. Nucleic Acids Res. 2007;35:W433–W437. doi: 10.1093/nar/gkm352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HC, Kim CH, Bae YK, Yeo SY, Kim SH, Hong SK, Shin J, Yoo KW, Hibi M, Hirano T, Miki N, Chitnis AB, Huh TL. Analysis of upstream elements in the HuC promoter leads to the establishment of transgenic zebrafish with fluorescent neurons. Dev Biol. 2000;227:279–293. doi: 10.1006/dbio.2000.9898. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8(10):785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Pozzoli O, Bosetti A, Croci L, Consalez GG, Vetter ML. Xebf3 is a regulator of neuronal differentiation during primary neurogenesis in Xenopus. Dev Biol. 2001;233:495–512. doi: 10.1006/dbio.2001.0230. [DOI] [PubMed] [Google Scholar]
- Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regad T, Roth M, Bredenkamp N, Illing N, Papalopulu N. The neural progenitor-specifying activity of FoxG1 is antagonistically regulated by CKI and FGF. Nat Cell Biol. 2007;9:531–540. doi: 10.1038/ncb1573. [DOI] [PubMed] [Google Scholar]
- Rennefahrt UE, Deacon SW, Parker SA, Devarajan K, Beeser A, Chernoff J, Knapp S, Turk BE, Peterson JR. Specificity profiling of Pak kinases allows identification of novel phosphorylation sites. J Biol Chem. 2007;282:15667–15678. doi: 10.1074/jbc.M700253200. [DOI] [PubMed] [Google Scholar]
- Saka Y, Smith JC. Spatial and temporal patterns of cell division during early Xenopus embryogenesis. Dev Biol. 2001;229:307–318. doi: 10.1006/dbio.2000.0101. [DOI] [PubMed] [Google Scholar]
- Salomoni P, Calegari F. Cell cycle control of mammalian neural stem cells: putting a speed limit on G1. Trends Cell Biol. 2010;20:233–243. doi: 10.1016/j.tcb.2010.01.006. [DOI] [PubMed] [Google Scholar]
- Sigrist CJ, Cerutti L, de Castro E, Langendijk-Genevaux PS, Bulliard V, Bairoch A, Hulo N. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 2010;38:D161–D166. doi: 10.1093/nar/gkp885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM. Early development of Xenopus laevis: a laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y: 2000. [Google Scholar]
- Souopgui J, Solter M, Pieler T. XPak3 promotes cell cycle withdrawal during primary neurogenesis in Xenopus laevis. EMBO J. 2002;21:6429–6439. doi: 10.1093/emboj/cdf644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viczian AS, Bang AG, Harris WA, Zuber ME. Expression of Xenopus laevis Lhx2 during eye development and evidence for divergent expression among vertebrates. Dev Dyn. 2006;235:1133–1141. doi: 10.1002/dvdy.20708. [DOI] [PubMed] [Google Scholar]
- Viczian AS, Solessio EC, Lyou Y, Zuber ME. Generation of functional eyes from pluripotent cells. PLoS Biol. 2009;7:e1000174. doi: 10.1371/journal.pbio.1000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viczian AS, Vignali R, Zuber ME, Barsacchi G, Harris WA. XOtx5b and XOtx2 regulate photoreceptor and bipolar fates in the Xenopus retina. Development. 2003;130:1281–1294. doi: 10.1242/dev.00343. [DOI] [PubMed] [Google Scholar]
- Zuber ME, Gestri G, Viczian AS, Barsacchi G, Harris WA. Specification of the vertebrate eye by a network of eye field transcription factors. Development. 2003;130:5155–5167. doi: 10.1242/dev.00723. [DOI] [PubMed] [Google Scholar]
- Zuber ME, Perron M, Philpott A, Bang A, Harris WA. Giant eyes in Xenopus laevis by overexpression of XOptx2. Cell. 1999;98:341–352. doi: 10.1016/s0092-8674(00)81963-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.