Background: Cellular ligands of the activating natural killer (NK) cell receptor NKp30 are poorly characterized.
Results: The identified domain of the cellular ligand BCL2-associated athanogene 6 (BAG-6) is essential and sufficient to bind NKp30 and inhibits NK cell function.
Conclusion: The BAG-6 domain from amino acid 686 to 936 is an important element of BAG-6-dependent tumor immune escape.
Significance: This study gives the first molecular insights into BAG-6-mediated inhibition of NKp30-dependent NK cell cytotoxicity.
Keywords: Immunosuppression, Innate Immunity, Natural Killer (NK) Cell, Tumor Immunology, Tumor Marker, BAG-6, BAT3, NKp30, Natural Killer Cell Receptors (NCR), Tumor Immune Escape
Abstract
Immunosurveillance of tumor cells depends on NKp30, a major activating receptor of human natural killer (NK) cells. The human BCL2-associated athanogene 6 (BAG-6, also known as BAT3; 1126 amino acids) is a cellular ligand of NKp30. To date, little is known about the molecular details of this receptor ligand system. Within the current study, we have located the binding site of NKp30 to a sequence stretch of 250 amino acids in the C-terminal region of BAG-6 (BAG-6686–936). BAG-6686–936 forms a noncovalent dimer of 57–59 kDa, which is sufficient for high affinity interaction with NKp30 (KD < 100 nm). As our most important finding, BAG-6686–936 inhibits NKp30-dependent signaling, interferon-γ release, and degranulation of NK cells in the presence of malignantly transformed target cells. Based on these data, we show for the first time that BAG-6686–936 comprises a subdomain of BAG-6, which is sufficient for receptor docking and inhibition of NKp30-dependent NK cell cytotoxicity as part of a tumor immune escape mechanism. These molecular insights provide an access point to restore tumor immunosurveillance by NK cells and to increase the efficacy of cellular therapies.
Introduction
NK2 cells eliminate tumor cells by the polarized release of cytotoxic granules containing perforin and granzymes or by binding of the tumor necrosis factor family members FAS/CD95, TRAIL receptor (TRAILR), and tumor necrosis factor receptor (TNFR) on tumor cells to their cognate ligands FAS-L, TRAIL, and TNF, respectively, on NK cells (1, 2). The mechanisms leading to NK cell activation are described by the principles of “missing self” and “induced self,” which imply that target cells with low or absent expression of major histocompatibility complex (MHC) class I or other non-MHC class I inhibitory ligands (missing self) and/or stress-induced expression of ligands for activating NK receptors (induced self) are preferentially eliminated by NK cells (3). Thus, a balance of activating and inhibitory signals determines whether NK responses are initiated or not. The major activating receptors involved in recognition and killing of malignantly transformed cells include the natural cytotoxicity receptors (NCRs) NKp30, NKp44, and NKp46, whose few ligands identified so far remain poorly characterized on the molecular level, and NKG2D, recognizing several related ligands such as the MHC class I polypeptide-related sequence A and B (MICA, MICB) and the UL16-binding proteins (ULBPs) (4–7). The importance of the NCRs for NK cell activation is underscored by the fact that expression of an insufficient amount of NCRs results in resistance of leukemia cells to NK cell cytotoxicity in patients with acute myeloid leukemia (8–10). Moreover, blocking of these receptors resulted in significantly decreased killing of virus-infected and malignantly transformed cells (6). Recently, the BCL2-associated athanogene 6 (BAG-6, also known as BAT3) was identified as a cellular ligand of NKp30 (11, 12). The domain organization and three-dimensional structure of BAG-6 are mostly unknown except for an N-terminal ubiquitin-like domain (Protein Data Bank (PDB) IDs: 4EEW and 4DWF). Additionally, a domain of unknown function (DUF3538) and a C-terminal BAG domain were identified (see Fig. 1A). BAG-6 is present in various tissues, on the plasma membrane of immune cells and tumor cells as well as on exosomes. In addition, BAG-6 is expressed as a soluble protein upon cellular stress (11–14). Therefore, BAG-6 represents an important tumor antigen and might be part of a tumor immune escape strategy reminiscent of the NKG2D ligands ULBP2 and ULBP3 (14, 15).
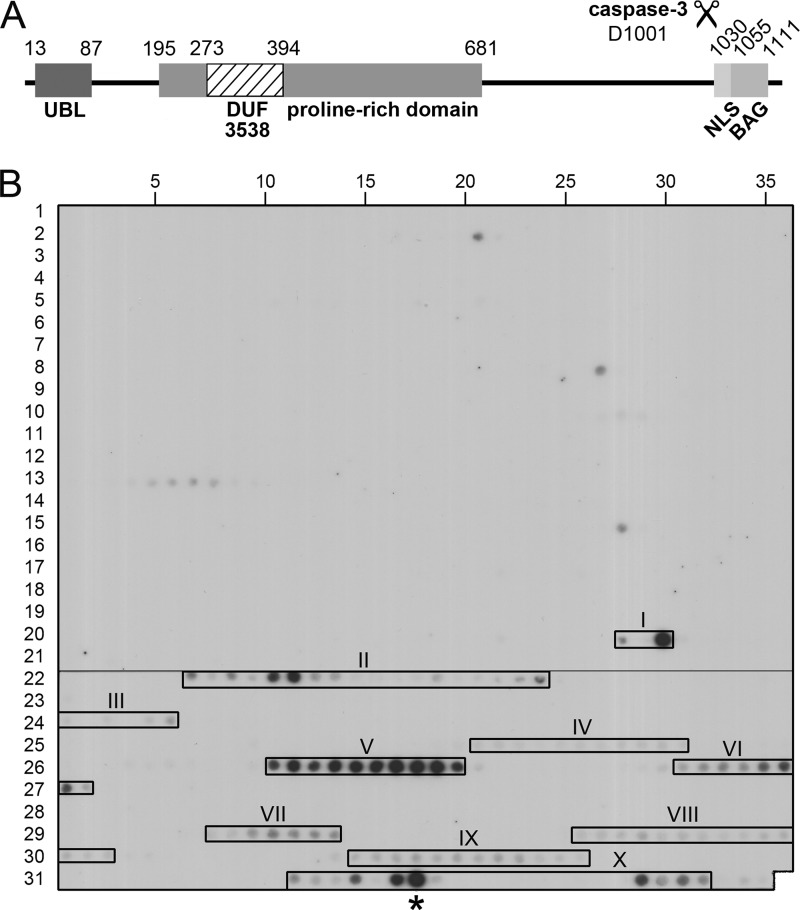
FIGURE 1.
NKp30 binding to peptide arrays of BAG-6. A, domain organization of human BAG-6 (P46379-1). The domain borders are given by numbers corresponding to the amino acid positions. The caspase-3 cleavage site at position 1001 is indicated. UBL, ubiquitin-like domain; DUF, domain of unknown function; NLS, nuclear localization signal; BAG, Bcl-2-associated athanogene domain. B, overlapping peptide arrays (18-amino acid-long peptides, offset by one amino acid) of human BAG-6 were probed with soluble NKp30-IgG1-Fc fusion proteins. Bound NKp30 was detected with a human Fc-specific HRP-conjugated antibody and visualized by chemiluminescence imaging. The identified interaction hot spots (I–X) are marked with boxes. *, nonspecific spot (compare supplemental Fig. S1).
Recent data suggest that BAG-6 on the plasma membrane of immature DCs triggers NK cell killing, whereas mature DCs escape from killing by up-regulation of MHC class I expression (12). By killing of immature DCs (cells that are associated with the induction of tolerogenic responses), activated NK cells might select a more immunogenic subset of DCs during a protective immune response (16). However, it remains puzzling whether up-regulation of BAG-6 at the plasma membrane of immature dendritic cells is a consequence of NK cell cytotoxicity via a different NK cell receptor-ligand system or a prerequisite for NKp30-BAG-6-dependent killing of immature dendritic cells.
The molecular details of the interaction between NKp30 and BAG-6 are largely unknown. However, detailed knowledge about this receptor-ligand system is required to understand NK cell function and for the development of NK cell-based therapies. Therefore, within the current study, we investigated the interaction of NKp30 and BAG-6 in immunosurveillance of tumor cells.
EXPERIMENTAL PROCEDURES
Antibodies
The following antibodies were used: mouse monoclonal anti-polyhistidine tag HRP-conjugate and anti-human HRP-conjugate (both Sigma-Aldrich), anti-Strep-mAb classic HRP (IBA), anti-NKp30 clone p30-15 (kindly provided by Carsten Watzl), goat anti-mouse IgG1 APC conjugate (Life Technologies), and anti-CD4 APC conjugate (eBioscience).
Cells
Human embryonic kidney cells (293T) and natural killer lymphoma cells (NK-92) were purchased from Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ). Murine pro-B cells (Ba/F3 cells) transduced with B7-H6 (Ba/F3-B7-H6) or the empty vector (Ba/F3-GFP) were provided by C. Watzel (Leibniz Research Centre for Working Environment and Human Factors (IfADo), Dortmund, Germany). Generation of A5-GFP reporter cell lines was described before (17). Spodoptera frugiperda Sf9 and Trichoplusia ni High Five cells were purchased from Life Technologies.
Flow Cytometry of Cells
NK-92 (0.5–1 × 106 cells) were blocked with 2% FCS (v/v) and 5% BSA (w/v) in PBS prior to incubation with specific antibodies or recombinant protein for 1 h at 4 °C. Following detection with secondary fluorophore-conjugated antibodies for 1 h at 4 °C, a minimum of 20,000 cells were analyzed on a FACSCanto II instrument (BD Biosciences).
Protein Production and Purification
NKp30-IgG1-Fc fusion proteins were produced as described previously (17). BAG-6686–936 (with a C-terminal hexahistidine tag) was heterologously expressed in Escherichia coli BL21 cells as a soluble cytosolic protein and in High Five insect cells as a secreted protein (with a C-terminal Strep-tag II).
Fragments of the BAG-6 gene were amplified from human cDNA clone IRAUp969B1019D (isoform 2 (P46379-2); Source BioScience) by PCR using gene-specific primers and were cloned into the pET21a expression vector. Transformed E. coli BL21 cells were grown to an A600 of 0.6, and protein expression was induced with 0.3 mm isopropyl-β-d-thiogalactoside for 4 h. Bacteria were harvested by centrifugation and disrupted by sonication. Cell debris was removed by centrifugation following purification of BAG-6 protein from the supernatant by immobilized metal ion affinity chromatography (IMAC) on nickel-nitrilotriacetic acid-agarose beads (Qiagen) and subsequent size exclusion chromatography (SEC) in 100 mm HEPES buffer supplemented with 25 mm NaCl (pH 7.5) on a Superdex 200 column (GE Healthcare). To produce BAG-6 variants in insect cells, BAG-6 genes were cloned into transfer vector pFastBac1 with an N-terminal secretion sequence from gp67 and a C-terminal Strep-tag II. Transformed E. coli DH10Bac YFP (kind provided by Imre Berger, Grenoble, France) integrate target genes from the transfer vector into bacmid DNA by Tn7 transposition. Sf9 cells were transfected with bacmid DNA to produce recombinant baculovirus within 72 h at 27 °C. Initial viral supernatant (V0) was used to amplify viral particles in Sf9 cells in shaking flasks for a further 72 h at 27 °C (V1). Afterward, BAG-6 variants were produced in High Five cells (7 × 105 cells/ml) using V1 (1:100 (v/v)). After removal of viral particles by ultracentrifugation (2 h, 100,000 × g), harvested supernatants were sterile filtered and purified using StrepTactin Sepharose (IBA) and subsequent SEC in 100 mm Tris/HCl (pH 8) supplemented with 150 mm NaCl and 1 mm EDTA.
Size Exclusion Chromatography-Multiangle Laser Light Scattering (SEC-MALS)
Size exclusion chromatography coupled with multiangle light scattering (SEC-MALS) was performed using a TSK-GEL G3000SWXL column (15 ml, Tosoh Bioscience), a light-scattering detector (miniDAWN TREOS) and a refractometer (Optilab rEX) from Wyatt Technology, and a UV detector, an HPLC, pump, and degasser from Jasco. The system was equilibrated with 2 column volumes of SEC buffer (25 mm sodium Pi, 25 mm NaCl (pH 7.0), filtered through 0.1-μm pore size VVLP filters (Millipore)) following a recirculation through the system for at least 1 day at 0.1 ml/min to improve the base line. Per measurement, 150–300 μg of protein in 100 μl were injected and analyzed at a flow rate of 0.5 ml/min at 4 °C. The obtained signals were processed with the ASTRA software package version 5.3.4.13 (Wyatt Technology) to calculate the molecular mass using the refractive index (RI) signal for concentration determination taking a differential index of refraction of 0.185 ml/g.
Analytical Ultracentrifugation
Sedimentation velocity experiments were conducted with 300 μl of protein samples at a protein concentration of 0.3–0.6 mg/ml in 100 mm Tris/HCl (pH 8) supplemented with 150 mm NaCl and 1 mm EDTA or 25 mm sodium phosphate buffer (pH 8) with an Optima XL-A centrifuge (Beckman Coulter Instruments). Absorbance data were acquired at a wavelength of 280 nm, at rotor speeds of 40,000 rpm, using a 0.003-cm step scan with three averages, and at a temperature of 20 °C. Data were analyzed with the sedfit program (18). The protein partial-specific volume, the buffer density, and viscosity were calculated using the SEDNTERP program, kindly provided by Dr. J. Philo.
Peptide Arrays
Peptide arrays of human BAG-6 (P46379-1; peptides of 18 amino acids offset by one amino acid) were synthesized by Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry at activated PEG spacers on cellulose membranes by automated parallel peptide synthesis on a MultiPep RS instrument (Intavis) and used for binding experiments as described previously (19, 20). After saturation of unspecific binding sites, peptide membranes were probed with soluble human NKp30-IgG1-Fc fusion proteins (17). Bound IgG1-Fc fusion proteins were detected with a human IgG1-Fc-specific HRP-conjugated antibody (Sigma-Aldrich) and visualized by chemiluminescence imaging.
CD Spectroscopy
The CD spectra of BAG-6 (7 μl; 2 mg/ml) in 25 mm phosphate buffer (pH 8.0) were recorded by using a CaF2 cuvette with a path length of 50 μm. The spectra were collected on a J-720 CD spectrometer (Jasco) with a resolution of 0.1 nm; 10 scans per spectra were taken. The sample holder was temperature-controlled by an external water bath.
Immunoprecipitation
Tagged BAG-6686–936 proteins were mixed with IgG1-Fc fusion proteins for 1 h and incubated with 20 μl of magnetic beads covalently coated with polyhistidine tag-specific mouse monoclonal antibodies (GenScript) or StrepTactin-coated magnetic beads (IBA) for 1 h at 4 °C. As negative control, an IgG1-Fc fusion protein of the human interferon receptor subunit 2 (IFNAR2-IgG1-Fc) was used (17). Bead-associated proteins were eluted with SDS sample buffer by boiling for 5 min (His-tagged BAG-6686–936) or with 2 mm biotin (Strep-tag II-tagged BAG-6686–936) and analyzed by SDS-PAGE and Western blotting.
ELISAs
96-well ELISA plates (Greiner Bio-One) were coated with recombinant BAG-6686–936 protein (1 μg/100 μl/well) in 100 mm sodium carbonate buffer (pH 9.6), blocked with 5% (w/v) bovine serum albumin (BSA) in PBS, and incubated with graded amounts of IgG1-Fc fusion proteins (0–2 μm). The amount of bound IgG1-Fc fusion proteins was quantified after immunodetection (anti-human IgG-Fc-HRP) and visualization with 3,3′,5,5′-tetramethylbenzidine substrate in a microtiter plate reader by measuring the absorbance at 450 and 650 nm (reference). KD values were determined after fitting the curves to a 1:1 Langmuir binding model using the Prism 5 software (GraphPad).
Signaling Reporter Assays
ELISA plates (96-well round-bottom plates, Corning Costar) were coated with recombinant proteins (1 μg/200 μl/well) in PBS for 16 h at 4 °C, washed once with PBS, and incubated with A5-GFP effector cells transduced with NKp30 or empty lentiviral vectors (17). After a 16-h incubation at 37 °C, GFP-positive A5 cells were quantified by flow cytometry. For competition experiments, A5-GFP effector cells were co-incubated with Ba/F3-B7-H6 target cells. A5-GFP cells were preincubated with recombinant proteins (20 μg/well) or NKp30-specific antibodies (p30-15, 1 μg/well), and Ba/F3-B7-H6 cells were preincubated with NKp30-IgG1-Fc fusion proteins (10 μg/well) for 1 h at 37 °C. Then A5-GFP effector cells and Ba/F3-B7-H6 target cells were mixed at an effector:target ratio of 1:1 and incubated for 16 h. As positive control, A5 cells were incubated with 0.05 μg/ml phorbol 12-myristate 13-acetate and 0.75 μg/ml ionomycin (Life Technologies). For analysis, cells were stained with a CD4-specific antibody, and GFP expression of CD4-A5 cells was determined on a flow cytometer.
Cytokine Production and Degranulation Assays
NK-92 cells (1 × 105) were incubated with recombinant proteins (10–20 μg) or anti-NKp30 antibodies (p30-15, 10 μg/ml) for 1 h at 37 °C. As control, Ba/F3-B7-H6 cells were preincubated with NKp30-IgG1-Fc or IFNAR2-IgG1-Fc (10 μg). NK and target cells (Ba/F3-B7-H6, Ba/F3-GFP) were mixed at an effector:target ratio of 1:1, and cells were incubated for 1 h at 37 °C in the presence of anti-human CD107a pacific blue-conjugated antibodies (Miltenyi Biotech). As positive control, NK cells were incubated with 2.5 μg/ml phorbol 12-myristate 13-acetate and 0.75 μg/ml ionomycin. Cells were treated with monensin (6 μg/ml) and brefeldin A (10 μg/ml; Sigma-Aldrich) and incubated for 4 h at 37 °C. Cells were harvested, fixed with 4% (w/v) paraformaldehyde, and permeabilized with 0.2% (w/v) saponin. Afterward, permeabilized cells were stained for intracellular INF-γ using anti-human IFN-γ APC-coupled antibodies. IFN-γ and CD107a expression was quantified by flow cytometry.
RESULTS
NKp30 Interacts with the C-terminal Part of BAG-6
To determine the binding interface of NKp30 and BAG-6, we probed overlapping peptide arrays (peptides of 18 amino acids in length, offset by one amino acid) covering the entire amino acid sequence of human BAG-6 (Fig. 1A) with soluble NKp30-IgG1-Fc fusion proteins. After detection with a human Fc-specific antibody, we identified 10 clustered interaction hot spots (I–X) for NKp30 within the C-terminal half of BAG-6 (Fig. 1B and supplemental S1 and supplemental Table S1). The reactivity of the hot spots ranged from weak (III, IV, VIII, IX) over moderate (VI, VII) to strong (I, II, V, X). For reference, only one spot within region X was recognized on the peptide arrays by the human Fc-specific antibody alone (supplemental Fig. S1), demonstrating high specificity of the binding assay for the other regions. The strongest specific interaction hot spots (I, II, V) were localized in the sequence stretch from amino acids 686 to 936 (BAG-6686–936, nomenclature according to isoform 2 of BAG-6; spots 692–925, compare supplemental Table S1).
Expression and Purification of BAG-6686–936
The BAG-6 fragment, which contained the strongest specific interaction hot spots (I, II, V) for NKp30 (BAG-6686–936), was cloned with a C-terminal hexahistidine tag or Strep-tag II. These constructs were expressed as soluble cytosolic proteins in E. coli BL21 bacteria (hexahistidine tag) and High Five insect cells (Strep-tag II) after secretion into the culture supernatant. Both proteins were purified to homogeneity by IMAC or by StrepTactin-Sepharose as demonstrated by SDS-PAGE and Western blotting (Fig. 2, A and B). Per liter of culture, roughly 1.5 and 3 mg of pure protein were obtained from E. coli and insect cells, respectively.
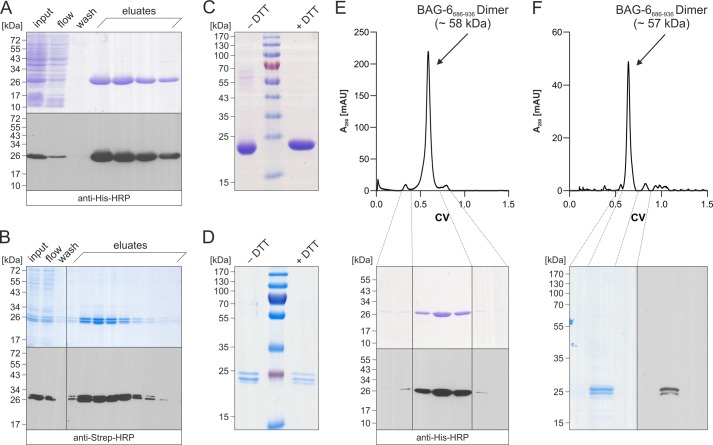
FIGURE 2.
Expression and purification of BAG-6686–936. A, soluble hexahistidine-tagged BAG-6686–936 was purified from BL21 E. coli bacteria by IMAC after isopropyl-β-d-thiogalactoside induction and sonication. Representative fractions of the purification were analyzed by Coomassie Blue-stained reducing SDS-PAGE and by Western blot with a mouse monoclonal polyhistidine tag-specific HRP-conjugated antibody prior to chemiluminescence imaging. B, after heterologous expression, secreted Strep-tag II-tagged BAG-6686–936 was purified from High Five insect cell culture supernatant via StrepTactin. Representative fractions of the purification were analyzed by Coomassie Blue-stained reducing SDS-PAGE and Western blotting with a mouse monoclonal Strep-tag-specific HRP-conjugated antibody prior to chemiluminescence imaging. C and D, aliquots of the concentrated elution fractions from IMAC (C) and StrepTactin (D) purification were analyzed by Coomassie Blue-stained nonreducing (− DTT) and reducing (+ DTT) SDS-PAGE. E and F, top, IMAC-purified (E) and StrepTactin-purified BAG-6686–936 (F) were subjected to size exclusion chromatography on a Superdex 200 column to remove minor contaminations. E and F, bottom, representative aliquots of the peak fractions were analyzed by Coomassie Blue-stained SDS-PAGE and Western blotting with a mouse monoclonal anti-polyhistidine tag- (E) or Strep-tag-specific HRP-conjugated antibody (F). CV, column volume.
The BAG-6686–936 protein appeared as a monomer of roughly 26 kDa (as predicted from its primary sequence, BAG-6686–936 (E. coli) 28.6 kDa; BAG-6686–936 (High Five cells) 29.3 kDa) in SDS-PAGE and corresponding Western blot analyses. Reducing and nonreducing SDS-PAGE gels demonstrate the absence of covalently linked oligomers (Fig. 2, C and D). Notably, BAG-6686–936 from insect cells appears as a double band in SDS-PAGE and corresponding Western blot analyses. The reason for these two species is unknown. However, phosphorylation as well as N- and O-linked glycosylation were excluded because they were not detected by phosphotyrosine-specific antibodies and their electrophoretic mobility remained unchanged in enzymatic deglycosylation assays. Interestingly, BAG-6686–936 appeared as a monodisperse peak, which corresponds to a dimer of 57–58 kDa in size exclusion chromatography on a Superdex 200 column (Fig. 2, E and F). The peak annotation and protein purity were validated by Coomassie Blue-stained SDS-PAGE and Western blotting.
BAG-6686–936 Forms a Noncovalent Dimer with a Predominantly α-Helical Conformation
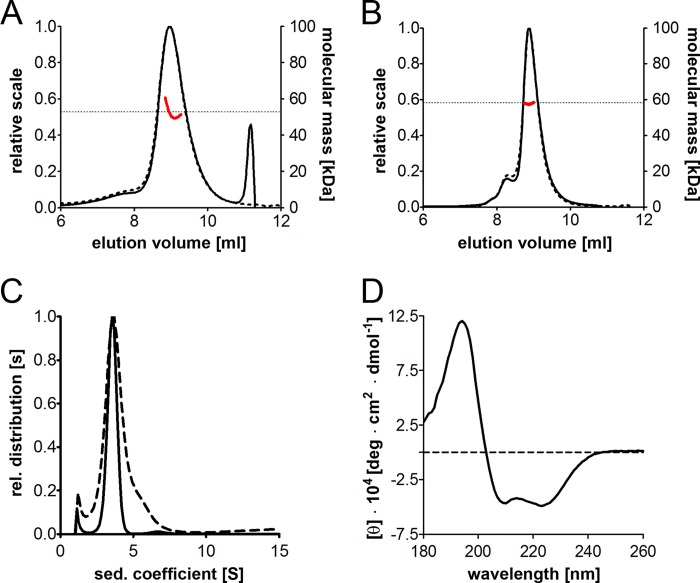
To determine the oligomeric state of BAG-6686–936, we performed SEC-MALS measurements. In these experiments, the predominant protein species had a molecular mass of 57 kDa corresponding to a dimer of BAG-6686–936 (Fig. 3, A and B). To analyze the oligomerization state of BAG-6686–936 in solution, we performed sedimentation velocity measurements. We found that the BAG-6686–936 proteins migrate at a sedimentation coefficient of 3.5 s as a single peak corresponding to more than 80% of the material loaded (Fig. 3C), confirming BAG-6686–936 dimerization. Based on the SEC-MALS and sedimentation velocity measurements, the molecular mass of the BAG-6686–936 dimer is 57–59 kDa, which is in agreement with its theoretical mass (BAG-6686–936 (E. coli) 57.2 kDa; BAG-6686–936 (High Five cells) 58.6 kDa) calculated from primary sequences. To assess the folding and secondary structure of the BAG-6686–936 dimers, we employed CD spectroscopy. The far UV-CD spectrum between 178 and 260 nm (at 22 °C) indicates a predominantly α-helical conformation (roughly 80%, Fig. 3D). These findings are in accordance with secondary structure prediction performed with the Phyre2 web interface (21). Here, the α-helix content of BAG-6686–936 is 70% with a high confidence score, whereas annotations for the rest of BAG-6 (except for the ubiquitin-like domain) are poor.
FIGURE 3.
Oligomeric state and secondary structure of BAG-6686–936. A and B, SEC-MALS measurements of IMAC-purified E. coli (A) and StrepTactin-purified High Five BAG-6686–936 (B). The UV absorbance (dashed line) and the differential refractive index (solid line) were analyzed, and the resulting molecular weight affiliations (red) are indicated. C, sedimentation velocity measurements of E. coli BAG-6686–936 (dashed line) and High Five BAG-6686–936 (solid line). The main protein species has a sedimentation coefficient of 3.5 s. rel., relative. D, CD spectroscopy of BAG-6 proteins. The far UV-CD spectrum between 178 and 260 nm (at 22 °C) of E. coli BAG-6686–936 shows a mainly α-helical folding. deg, degree.
NKp30 Interacts Specifically with BAG-6686–936
To analyze the interaction of BAG-6686–936 with NKp30 on a molecular level, we initially performed co-immunoprecipitation experiments. Therefore, recombinant BAG-6686–936 from E. coli or High Five insect cells was mixed with a 15-fold molar excess of recombinant NKp30-IgG1-Fc fusion proteins. Protein complexes were recovered on magnetic beads coupled to polyhistidine tag-specific antibodies or StrepTactin and analyzed by SDS-PAGE and Western blotting (Fig. 4, A and B). Based on our results, NKp30 binds specifically to both BAG-6686–936 proteins, demonstrating that the BAG-6686–936 domain is essential and sufficient for binding to NKp30. In contrast, IgG1-Fc fusion proteins of the interferon receptor subunit IFNAR2 (IFNAR2-IgG1-Fc), used as a negative control, showed no binding to BAG-6686–936, confirming specificity of the assay.
FIGURE 4.
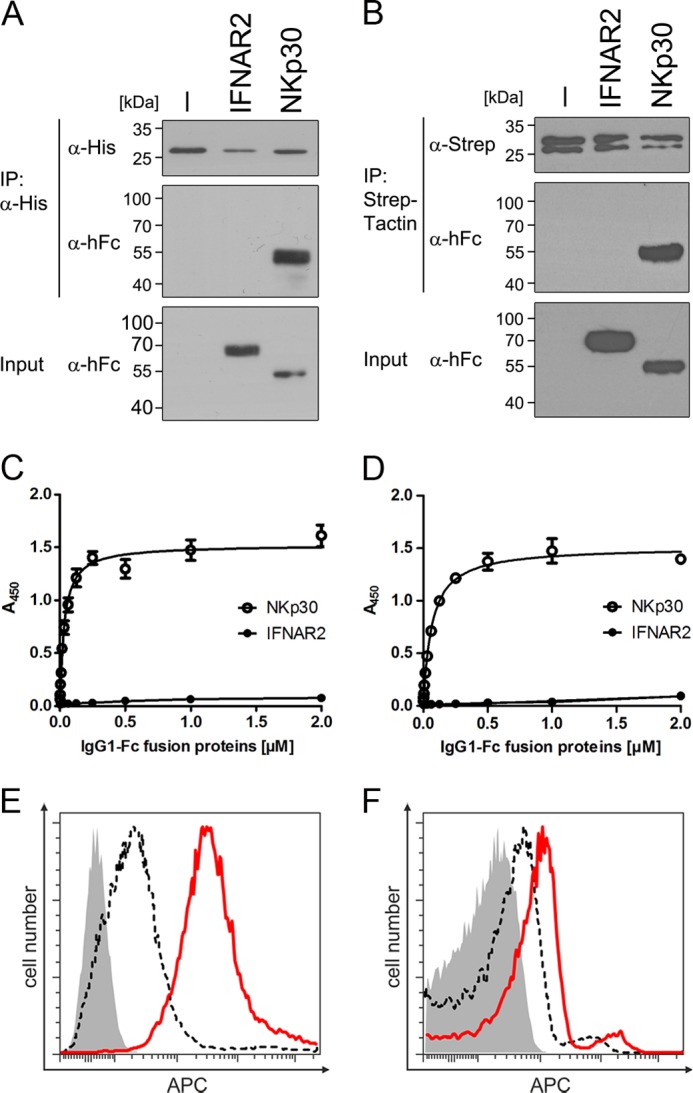
BAG-6686–936 binds to NKp30 in vitro and on NK cells. Co-immunoprecipitation (IP) of NKp30-IgG1-Fc and BAG-6686–936. A and B, E. coli BAG-6686–936 immobilized on beads via polyhistidine tag-specific antibody (A) and High Five insect cell BAG-6686–936 immobilized on StrepTactin-coupled beads (B) were tested for IgG1-Fc fusion protein binding and analyzed by SDS-PAGE and Western blot with anti-human Fc (α-hFc) antibodies. C and D, graded amounts of IgG1-Fc fusion proteins were probed for binding to E. coli BAG-6686–936 (C) and to High Five insect cell BAG-6686–936 (D), and KD values were calculated after fitting of the curves to a 1:1 Langmuir binding model. E and F, binding of E. coli (E) and High Five (F) BAG-6686–936 to NK-92 cells was analyzed via flow cytometry using mouse monoclonal polyhistidine tag-specific or Strep-tag II-specific antibodies prior to detection with APC-conjugated anti-mouse secondary antibodies. Data are representative of at least three independent experiments. Error bars represent mean values ± S.E.
For binding affinity measurements of NKp30 and BAG-6686–936, we developed an ELISA-based assay. In brief, recombinant BAG-6686–936 proteins were immobilized on an ELISA plate and probed with graded amounts of NKp30-IgG1-Fc or IFNAR2-IgG1-Fc fusion proteins (negative control). NKp30 bound specifically to BAG-6 in a concentration-dependent manner, whereas IFNAR2 binding was close to the detection limit of the assay (Fig. 4, C and D). After fitting of the binding curves to a 1:1 Langmuir binding model, equilibrium binding constants (KD) of 32 ± 4 and 64 ± 3 nm were calculated for NKp30 binding to BAG-6686–936 produced in E. coli and insect cells, respectively.
Following detailed biochemical characterization of the interaction between NKp30 and BAG-6686–936, we investigated whether the BAG-6686–936 fragment binds to NKp30 in the plasma membrane of living NK cells. Therefore, NK-92 cells were incubated with purified BAG-6686–936 and analyzed by flow cytometry after detection with tag-specific antibodies. In accordance with the results from ELISA measurements, BAG-6686–936 bound specifically to NKp30 in the plasma membrane of NK cells (Fig. 4, E and F).
Soluble BAG-6686–936 Impairs NKp30-dependent Signaling, Degranulation, and Cytokine Production of NK Cells
BAG-6686–936 interacts specifically with NKp30. Therefore, we investigated whether BAG-6686–936 can induce NKp30 signaling in NKp30-transduced A5-GFP reporter cells, which couple NKp30-CD3ζ signaling to proportional expression of GFP (17) (Fig. 5A). Soluble BAG-6686–936 did not induce NKp30-dependent signaling, presumably because it lacks the ability to cross-link NKp30. Therefore, we tried to activate NKp30 with BAG-6686–936 immobilized on a solid support, mimicking the situation as it is found on exosomes. Surprisingly, immobilized BAG-6686–936 also did not induce NKp30-CD3ζ signaling, as indicated by the absence of GFP expression (Fig. 5B). In contrast, the immobilized ectodomain of the NKp30 activating ligand B7-H6 resulted in robust NKp30-CD3ζ signaling, indicating the appropriateness of the experimental setup. Results from these experiments suggested that the BAG-6686–936 domain, and thus BAG-6, is an inhibitory ligand of NKp30. To test this hypothesis, NKp30-transduced A5-GFP reporter cells were stimulated with Ba/F3 cells stably transduced with B7-H6 in the absence and presence of soluble BAG-6686–936 (Fig. 5C). As expected, in the absence of BAG-6686–936,strong NKp30-CD3ζ signaling was observed. Strikingly, BAG-6686–936 purified from insect cells led to suppression of NKp30-CD3ζ signaling similar to the efficiency of NKp30-specific monoclonal antibodies, whereas BAG-6686–936 purified from E. coli showed no inhibitory potential. Notably, NKp30-IgG1-Fc fusion proteins could drastically inhibit NKp30-CD3ζ signaling as well. For reference, IFNAR2-IgG1-Fc fusion proteins showed no inhibitory potential, demonstrating specificity of the assay.
FIGURE 5.
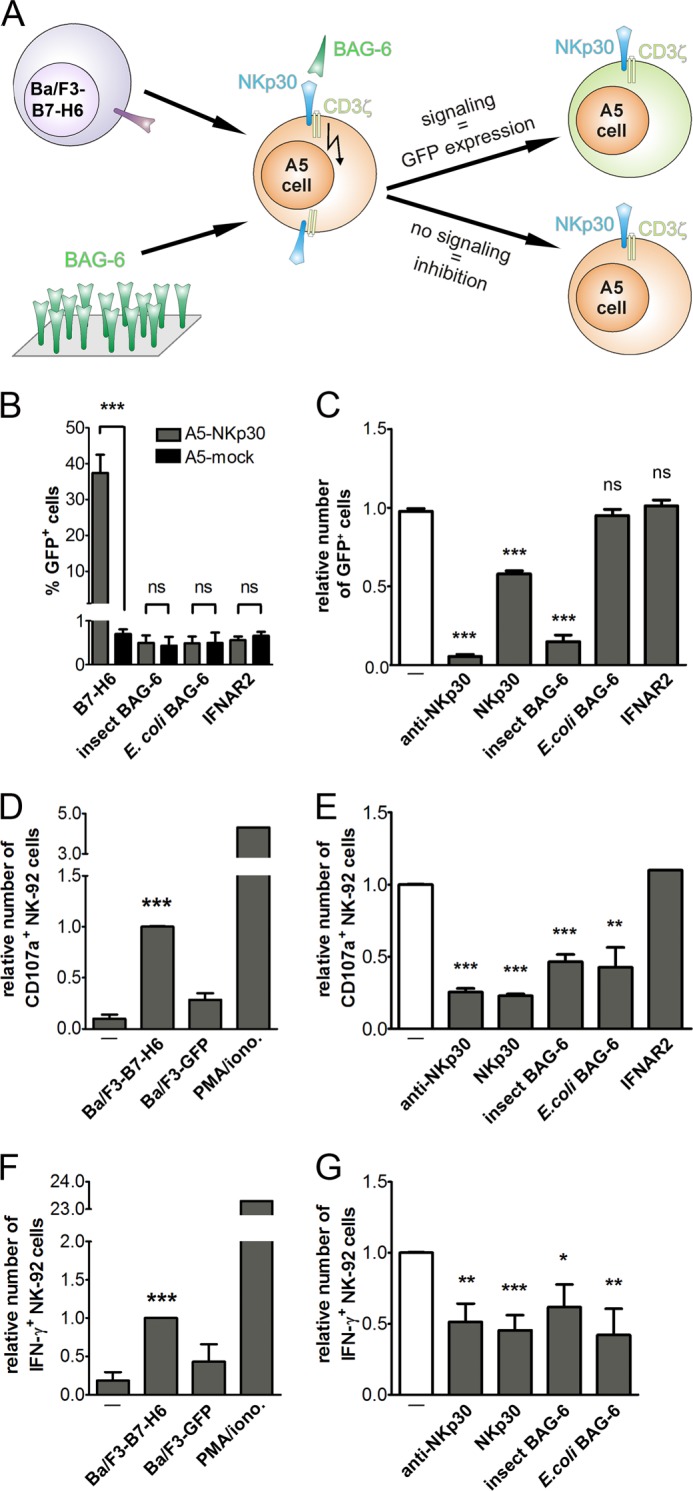
BAG-6686–936 inhibits NK cell activation and cytotoxicity. A, scheme of the experimental setup using NKp30-transduced A5-GFP reporter cells for detection of NKp30-dependent CD3ζ-signaling based on induced GFP expression. B, NKp30- or mock-transduced A5-GFP reporter cells were cultivated in wells with immobilized proteins and analyzed for the number of GFP positive cells. C, NKp30-transduced A5-GFP reporter cells were co-cultivated with Ba/F3 cells transduced with the NKp30 ligand B7-H6 (Ba/F3-B7-H6). GFP expression was analyzed in the absence or presence of recombinant BAG-6686–936, NKp30-IgG1-Fc, IFNAR2-IgG1-Fc, or an anti-NKp30 antibody. D, CD107a surface expression, a marker for NK cell cytotoxicity, was measured by flow cytometry after co-incubation of NK-92 cells with Ba/F3-B7-H6 cells or mock-transduced Ba/F3 cells (Ba/F3-GFP). For reference, cells were treated with phorbol 12-myristate 13-acetate (PMA) and ionomycin (iono.). E, BAG-6686–936 and control proteins were probed for inhibition of Ba/F3-B7-H6 cell-induced CD107a surface expression by flow cytometry. F, in parallel, NK-92 cells were permeabilized and analyzed for intracellular IFN-γ expression after co-incubation with Ba/F3-B7-H6 cells and Ba/F3-GFP cells or treatment with phorbol 12-myristate 13-acetate and ionomycin. G, IFN-γ expression of NK-92 cells, stimulated with Ba/F3-B7-H6 cells, was analyzed by flow cytometry in the presence of recombinant proteins. Data points represent mean values ± S.E. obtained from least five individual experiments performed; *, p < 0.01; **, p < 0.001; ***, p < 0.0001 by Student's t test. ns, not significant.
Next, we evaluated the influence of BAG-6686–936 on NK cell-mediated activation and cytotoxicity by analyzing the expression of CD107a, a marker for degranulation of NK cells, and IFN-γ production. In the presence of Ba/F3 cells expressing B7-H6 (Ba/F3-B7-H6, effector:target ratio 1:1), NK-92 cells show high CD107a surface expression (Fig. 5D), whereas only a few NK cells show CD107a expression upon co-cultivation with mock-transduced Ba/F3 cells (Ba/F3-GFP). Strikingly, Ba/F3-B7-H6-induced degranulation of NK-92 cells was impaired in the presence of BAG-6686–936 from insect cells and E. coli (Fig. 5E). For reference, reduced CD107a expression was also achieved by anti-NKp30 antibodies and co-incubation of Ba/F3 cells with NKp30-IgG1-Fc. Moreover, Ba/F3-B7-H6 cells could induce IFN-γ expression in NK-92 cells, as analyzed by intracellular staining and analyses by flow cytometry (Fig. 5F). In accordance with our results from degranulation assays, BAG-6686–936 led to significantly reduced production of IFN-γ by NK-92 stimulated with Ba/F3-B7-H6 cells. For reference, NKp30-IgG1-Fc and anti-NKp30 antibodies showed comparable inhibition of IFN-γ production (Fig. 5G). In addition, inhibition of NK cell cytotoxicity by BAG-6686–936 was confirmed in killing experiments with K562 target cells (supplemental Materials and Methods and Fig. S2). Notably, inhibition was comparable with an NKp30 blocking antibody.
In summary, BAG-6686–936 comprises a subdomain of BAG-6, which is essential and sufficient for receptor docking and inhibition of NKp30-dependent NK cell cytotoxicity. Interestingly, recombinant BAG-6686–936 derived from E. coli and High Five insect cells displayed equal biochemical and biophysical properties with respect to its binding affinity to NKp30, folding, and oligomerization. However, although BAG-6686–936 derived from E. coli has the ability to suppress degranulation and IFN-γ production of NK cells, it failed to inhibit NKp30-CD3ζ chain-dependent signaling. In contrast, High Five insect cell-derived BAG-6686–936 is a strong inhibitor of NKp30. The precise reason for this phenomenon is unknown. Possible reasons might be subtle changes in protein folding, post-translational modifications other than glycosylation or phosphorylation, or a certain degree of disturbing LPS contamination in E. coli-derived BAG-6686–936 (22).
DISCUSSION
The human BAG-6 (also known as BAT3; 1126 amino acids) is a multifunctional protein as indicated by its diverse cellular localization. As a nuclear protein, BAG-6 is involved in p53-dependent DNA repair (23) and in modulation of histone methylation, thus regulating gene expression (24). Further, BAG-6 is cleaved by caspase-3 at position 1001 (DEQD), and the resulting C-terminal fragment of 131 amino acids mediates ricin-induced apoptotic morphological changes (25). BAG-6 also promotes stress-induced apoptosis by stabilizing the apoptosis-inducing factor AiF (26). In addition, BAG-6 binds to heat shock protein 70 (Hsp70) and acts as co-chaperone (27). BAG-6 links the targeting and ubiquitination pathway as it plays a role in translocation of tail-anchored proteins into the endoplasmic reticulum membrane (28, 29). Full-length BAG-6, but not a C-terminally truncated form of BAG-6, binds to TGF-β receptors, modulates TGF-β signaling, and enhances TGF-β1-induced type I collagen expression in mouse mesangial cells (30). The fact that homozygous BAG-6 knock-out in mice led to embryonic or perinatal lethality, associated with pronounced developmental defects in several tissues, indicates that BAG-6 is critical for normal mammalian development (31). BAG-6 is up-regulated in colorectal tumor tissue (13). In addition, BAG-6 is released by heat-shocked macrophages, and treatment of macrophages with purified BAG-6 led to reduced nitric oxide after INF-γ stimulation as well as IL-1β and IL-12p70 production upon LPS treatment.
Only very recently, BAG-6 on tumor cells and immature DCs was proposed to be a cellular ligand of the major activating NK cell receptor NKp30 (11, 12). However, the molecular details of the NKp30/BAG-6 receptor-ligand system are poorly characterized.
Within the current study, we have localized the binding site of NKp30 on BAG-6 to a sequence stretch of 250 amino acids in the C-terminal half of BAG-6 (BAG-6686–936). These data are in accordance with a previous study, which identified a C-terminal fragment of BAG-6 (BAG-6-CT, amino acids 555-(Δ1055–1111)-1032) by a yeast two-hybrid screen that interacts with the extracellular domain of NKp30 (11). The identified cDNA fragment of BAG-6 was characterized by a deletion of the conserved BAG domain (amino acids 1055–1111), which is responsible for binding of BAG-6 to Hsp70 (27, 32). Both variants, either including or lacking the BAG domain, are expressed in tumor tissues, cell lines, and monocyte-derived dendritic cells (11, 13, 33).
BAG-6 contains a lengthy proline-rich stretch (amino acids 195–681), making it an attractive molecular scaffold that could mediate multiple protein-protein interactions in several cellular compartments. By contrast to the previous study (11), the identified NKp30 binding fragment of BAG-6 (BAG-6686–936) does not contain sequences from this proline-rich stretch.
Soluble recombinant BAG-6686–936 formed stable noncovalent dimers and bound with nanomolar affinity to NKp30, suggesting that BAG-6686–936 forms a separate domain fold within the BAG-6 protein. Therefore, the current study provides the first information on the structural organization of the sequence stretch of BAG-6 in between the N-terminal ubiquitin-like domain (amino acids 1–87) and a C-terminal BAG domain (amino acids 1055–1111). Notably, ab initio modeling of BAG-6 did not reveal significant results.
Strikingly, BAG-6686–936 inhibited NKp30-dependent signaling in a reporter cell assay and in NK cells (inhibition of IFN-γ release, degranulation, and cytotoxicity). Based on these data, we show for the first time that soluble BAG-6 is an inhibitory ligand of NKp30 and might thus be part of a potent tumor immune escape mechanism. Further, BAG-6 is the first inhibitory ligand of NKp30 that acts via binding to the ectodomain of NKp30, mechanistically different from cytomegalovirus pp65, which acts via dissociation of the NKp30-CD3ζ chain complex (34).
Taken together, these data suggest that soluble, and most likely also exosomal BAG-6 acts as a potent tumor immunoevasin reminiscent of exosomal ULBP3 and soluble ULBP2, which lead to down-modulation of NKG2D (15).
Acknowledgments
We thank Carsten Watzl (Leibniz Research Centre for Working Environment and Human Factors (IfADo), Dortmund, Germany) for critical review of the manuscript and helpful discussions and Elke Pogge von Strandmann (Medical School, University of Cologne, Germany) for helpful discussions and technical advice. The Georg-Speyer-Haus is funded jointly by the German Federal Ministry of Health (BMG) and the Ministry of Higher Education, Research and the Arts of the State of Hessen (HMWK).
This work was supported by institutional funds of the Georg-Speyer-Haus and by grants from the Schleicher Stiftung and the LOEWE Center for Cell and Gene Therapy Frankfurt funded by Hessisches Ministerium für Wissenschaft und Kunst (HMWK) (funding reference number: III L 4-518/17.004 (2010)).

This article contains supplemental Table 1 and Figs. S1 and S2.
- NK
- natural killer
- NCR
- natural cytotoxicity receptor
- MICA, MICB
- MHC class I polypeptide-related sequence A and B
- ULBPs
- UL16-binding proteins
- BAG-6
- BCL2-associated athanogene 6
- DC
- dendritic cell
- APC
- allophycocyanin
- IMAC
- immobilized metal ion affinity chromatography
- SEC
- size exclusion chromatography
- SEC-MALS
- size exclusion chromatography-multiangle laser light scattering.
REFERENCES
- 1. Farag S. S., Caligiuri M. A. (2006) Human natural killer cell development and biology. Blood Rev. 20, 123–137 [DOI] [PubMed] [Google Scholar]
- 2. Moretta L., Bottino C., Pende D., Castriconi R., Mingari M. C., Moretta A. (2006) Surface NK receptors and their ligands on tumor cells. Semin. Immunol. 18, 151–158 [DOI] [PubMed] [Google Scholar]
- 3. Funke J., Dürr R., Dietrich U., Koch J. (2011) Natural killer cells in HIV-1 infection: a double-edged sword. AIDS Rev. 13, 67–76 [PubMed] [Google Scholar]
- 4. Cerwenka A., Lanier L. L. (2003) NKG2D ligands: unconventional MHC class I-like molecules exploited by viruses and cancer. Tissue Antigens 61, 335–343 [DOI] [PubMed] [Google Scholar]
- 5. Lanier L. L. (2005) NK cell recognition. Annu. Rev. Immunol. 23, 225–274 [DOI] [PubMed] [Google Scholar]
- 6. Moretta A., Bottino C., Vitale M., Pende D., Cantoni C., Mingari M. C., Biassoni R., Moretta L. (2001) Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annu. Rev. Immunol. 19, 197–223 [DOI] [PubMed] [Google Scholar]
- 7. Vivier E., Tomasello E., Baratin M., Walzer T., Ugolini S. (2008) Functions of natural killer cells. Nat. Immunol. 9, 503–510 [DOI] [PubMed] [Google Scholar]
- 8. Arnon T. I., Achdout H., Lieberman N., Gazit R., Gonen-Gross T., Katz G., Bar-Ilan A., Bloushtain N., Lev M., Joseph A., Kedar E., Porgador A., Mandelboim O. (2004) The mechanisms controlling the recognition of tumor- and virus-infected cells by NKp46. Blood 103, 664–672 [DOI] [PubMed] [Google Scholar]
- 9. Costello R. T., Sivori S., Marcenaro E., Lafage-Pochitaloff M., Mozziconacci M. J., Reviron D., Gastaut J. A., Pende D., Olive D., Moretta A. (2002) Defective expression and function of natural killer cell-triggering receptors in patients with acute myeloid leukemia. Blood 99, 3661–3667 [DOI] [PubMed] [Google Scholar]
- 10. Fauriat C., Just-Landi S., Mallet F., Arnoulet C., Sainty D., Olive D., Costello R. T. (2007) Deficient expression of NCR in NK cells from acute myeloid leukemia: Evolution during leukemia treatment and impact of leukemia cells in NCRdull phenotype induction. Blood 109, 323–330 [DOI] [PubMed] [Google Scholar]
- 11. Pogge von Strandmann E., Simhadri V. R., von Tresckow B., Sasse S., Reiners K. S., Hansen H. P., Rothe A., Böll B., Simhadri V. L., Borchmann P., McKinnon P. J., Hallek M., Engert A. (2007) Human leukocyte antigen-B-associated transcript 3 is released from tumor cells and engages the NKp30 receptor on natural killer cells. Immunity 27, 965–974 [DOI] [PubMed] [Google Scholar]
- 12. Simhadri V. R., Reiners K. S., Hansen H. P., Topolar D., Simhadri V. L., Nohroudi K., Kufer T. A., Engert A., Pogge von Strandmann E. (2008) Dendritic cells release HLA-B-associated transcript-3 positive exosomes to regulate natural killer function. PLoS One 3, e3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu W., Song W., Li S., Ouyang S., Fok K. L., Diao R., Miao S., Chan H. C., Wang L. (2012) Regulation of apoptosis by Bat3-enhanced YWK-II/APLP2 protein stability. J. Cell Sci. 125, 4219–4229 [DOI] [PubMed] [Google Scholar]
- 14. Reiners K. S., Topolar D., Henke A., Simhadri V. R., Kessler J., Sauer M., Bessler M., Hansen H. P., Tawadros S., Herling M., Krönke M., Hallek M., Pogge von Strandmann E. (2013) Soluble ligands for NK cell receptors promote evasion of chronic lymphocytic leukemia cells from NK cell anti-tumor activity. Blood 121, 3658–3665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fernández-Messina L., Ashiru O., Boutet P., Agüera-González S., Skepper J. N., Reyburn H. T., Valés-Gómez M. (2010) Differential mechanisms of shedding of the glycosylphosphatidylinositol (GPI)-anchored NKG2D ligands. J. Biol. Chem. 285, 8543–8551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Koch J., Steinle A., Watzl C., Mandelboim O. (2013) Activating natural cytotoxicity receptors of natural killer cells in cancer and infection. Trends Immunol. 34, 182–191 [DOI] [PubMed] [Google Scholar]
- 17. Hartmann J., Tran T. V., Kaudeer J., Oberle K., Herrmann J., Quagliano I., Abel T., Cohnen A., Gatterdam V., Jacobs A., Wollscheid B., Tampé R., Watzl C., Diefenbach A., Koch J. (2012) The stalk domain and the glycosylation status of the activating natural killer cell receptor NKp30 are important for ligand binding. J. Biol. Chem. 287, 31527–31539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schuck P. (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hilpert K., Winkler D. F., Hancock R. E. (2007) Peptide arrays on cellulose support: SPOT synthesis, a time and cost efficient method for synthesis of large numbers of peptides in a parallel and addressable fashion. Nat. Protoc. 2, 1333–1349 [DOI] [PubMed] [Google Scholar]
- 20. Plewnia G., Schulze K., Hunte C., Tampé R., Koch J. (2007) Modulation of the antigenic peptide transporter TAP by recombinant antibodies binding to the last five residues of TAP1. J. Mol. Biol. 369, 95–107 [DOI] [PubMed] [Google Scholar]
- 21. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 22. Hammond T., Lee S., Watson M. W., Flexman J. P., Cheng W., Fernandez S., Price P. (2010) Toll-like receptor (TLR) expression on CD4+ and CD8+ T-cells in patients chronically infected with hepatitis C virus. Cell. Immunol. 264, 150–155 [DOI] [PubMed] [Google Scholar]
- 23. Sasaki T., Gan E. C., Wakeham A., Kornbluth S., Mak T. W., Okada H. (2007) HLA-B-associated transcript 3 (Bat3)/Scythe is essential for p300-mediated acetylation of p53. Genes Dev. 21, 848–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nguyen P., Bar-Sela G., Sun L., Bisht K. S., Cui H., Kohn E., Feinberg A. P., Gius D. (2008) BAT3 and SET1A form a complex with CTCFL/BORIS to modulate H3K4 histone dimethylation and gene expression. Mol. Cell. Biol. 28, 6720–6729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu Y. H., Shih S. F., Lin J. Y. (2004) Ricin triggers apoptotic morphological changes through caspase-3 cleavage of BAT3. J. Biol. Chem. 279, 19264–19275 [DOI] [PubMed] [Google Scholar]
- 26. Desmots F., Russell H. R., Michel D., McKinnon P. J. (2008) Scythe regulates apoptosis-inducing factor stability during endoplasmic reticulum stress-induced apoptosis. J. Biol. Chem. 283, 3264–3271 [DOI] [PubMed] [Google Scholar]
- 27. Corduan A., Lecomte S., Martin C., Michel D., Desmots F. (2009) Sequential interplay between BAG6 and HSP70 upon heat shock. Cell. Mol. Life Sci. 66, 1998–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Leznicki P., Clancy A., Schwappach B., High S. (2010) Bat3 promotes the membrane integration of tail-anchored proteins. J. Cell Sci. 123, 2170–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mariappan M., Li X., Stefanovic S., Sharma A., Mateja A., Keenan R. J., Hegde R. S. (2010) A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature 466, 1120–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kwak J. H., Kim S. I., Kim J. K., Choi M. E. (2008) BAT3 interacts with transforming growth factor-β (TGF-β) receptors and enhances TGF-β1-induced type I collagen expression in mesangial cells. J. Biol. Chem. 283, 19816–19825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Desmots F., Russell H. R., Lee Y., Boyd K., McKinnon P. J. (2005) The Reaper-binding protein Scythe modulates apoptosis and proliferation during mammalian development. Mol. Cell. Biol. 25, 10329–10337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Thress K., Song J., Morimoto R. I., Kornbluth S. (2001) Reversible inhibition of Hsp70 chaperone function by Scythe and Reaper. EMBO J. 20, 1033–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tsukahara T., Kawaguchi S., Torigoe T., Kimura S., Murase M., Ichimiya S., Wada T., Kaya M., Nagoya S., Ishii T., Tatezaki S., Yamashita T., Sato N. (2008) Prognostic impact and immunogenicity of a novel osteosarcoma antigen, papillomavirus binding factor, in patients with osteosarcoma. Cancer Sci. 99, 368–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Arnon T. I., Achdout H., Levi O., Markel G., Saleh N., Katz G., Gazit R., Gonen-Gross T., Hanna J., Nahari E., Porgador A., Honigman A., Plachter B., Mevorach D., Wolf D. G., Mandelboim O. (2005) Inhibition of the NKp30 activating receptor by pp65 of human cytomegalovirus. Nat. Immunol. 6, 515–523 [DOI] [PubMed] [Google Scholar]