Background: Stability of Far1, an essential enzyme for plasmalogen synthesis, is regulated by the cellular plasmalogen level.
Results: Expression of a mutant Far1 harboring the mutation in its C-terminal membrane-flanking region increased plasmalogen synthesis because of the inhibition of its degradation.
Conclusion: Elevation of plasmalogen synthesis is achieved by expression of Far1.
Significance: Far1 is a rate-limiting enzyme for plasmalogen synthesis.
Keywords: Lipid Ether, Peroxisomes, Plasmalogen, Protein Stability, Protein Targeting, Ether Lipid
Abstract
Peroxisomal fatty acyl-CoA reductase 1 (Far1) is essential for supplying fatty alcohols required for ether bond formation in ether glycerophospholipid synthesis. The stability of Far1 is regulated by a mechanism that is dependent on cellular plasmalogen levels. However, the membrane topology of Far1 and how Far1 is targeted to membranes remain largely unknown. Here, Far1 is shown to be a peroxisomal tail-anchored protein. The hydrophobic C terminus of Far1 binds to Pex19p, a cytosolic receptor harboring a C-terminal CAAX motif, which is responsible for the targeting of Far1 to peroxisomes. Far1, but not Far2, was preferentially degraded in response to the cellular level of plasmalogens. Experiments in which regions of Far1 or Far2 were replaced with the corresponding region of the other protein showed that the region flanking the transmembrane domain of Far1 is required for plasmalogen-dependent modulation of Far1 stability. Expression of Far1 increased plasmalogen synthesis in wild-type Chinese hamster ovary cells, strongly suggesting that Far1 is a rate-limiting enzyme for plasmalogen synthesis.
Introduction
Peroxisomes are single membrane-bounded organelles that participate in several metabolic pathways, including plasmalogen synthesis, β-oxidation of very long chain fatty acids, β-oxidation of branched fatty acids, and H2O2 metabolism (1, 2). These peroxisomal functions are essential for human development. Indeed, several diseases are caused by defects in peroxisomal activity, either the impaired function of a peroxisomal enzyme or defects in peroxisome biogenesis (3). The physiological significance of plasmalogens is demonstrated by the human peroxisomal disorders such as Zellweger syndrome and rhizomelic chondrodysplasia punctata (4). Plasmalogens are enriched in the central nervous system (5), and the metabolic stability of plasmalogens varies between different regions of the brain. For instance, plasmalogens are relatively stable in myelin, whereas there is a dynamic pool of plasmalogens in gray matter that is rapidly metabolized and has a half-life of less than an hour (6). The levels of plasmalogens are reduced in several diseases, including sporadic Alzheimer disease and Pelizaeus-Merzbacher disease (7–9). Therefore, it is important to elucidate the molecular mechanisms that regulate the synthesis and degradation of plasmalogens.
Plasmalogens are synthesized via a seven-step reaction pathway that starts with the conversion of dihydroxyacetonephosphate (DHAP)2 to 1-acyl-DHAP by peroxisomal matrix dihydroxyacetonephosphateacyltransferase. Then, the formation of an ether bond is catalyzed by another peroxisomal matrix protein, alkyl-dihydroxyacetonephosphate synthase (ADAPS), which replaces the acyl chain of 1-acyl-DHAP with a long-chain fatty alcohol. The peroxisomal enzyme fatty acyl-CoA reductase 1 (Far1) is essential for the formation of fatty alcohols (10). We demonstrated previously that the cellular level of plasmalogens modulates the stability of Far1 (11). In this study, we investigated the biogenesis and topology of Far1 and characterized the region of Far1 that is required for its plasmalogen-dependent degradation.
EXPERIMENTAL PROCEDURES
Biochemicals
Restriction enzymes and DNA-modifying enzymes were purchased from Nippon Gene (Tokyo, Japan) and Takara (Kyoto, Japan). Fetal bovine serum, DMEM, and Ham's F-12 medium were from Invitrogen. Anti-human Far2 antibody was raised in rabbits by injection of DNA coding for amino acid residues at positions 155–253 of human Far2 using genomic antibody technology (Strategic Diagnostics, Newark, DE). Rabbit antibodies against human Far1 (11), rat 70-kDa peroxisomal integral membrane protein (PMP70) (12), Pex3p (13), Pex19p (14), Pex14p (15), peroxisomal targeting signal type 1 (PTS1) (16), GFP (MBL, Nagoya, Japan), and malate dehydrogenase (17, 18); the goat antibody against lactate dehydrogenase (Rockland, Gilbertsville, PA); and mouse antibodies against actin (Millipore, Billerica, MA), GFP (Santa Cruz Biotechnology, Inc.), FLAG (catalog no. M2, Sigma), and influenza virus HA (catalog no. 16B12, Covance) were used. Plasmenylethanolamine purified from bovine brain was purchased from Doosan Serdary Research Laboratories (Kyungki-Do, South Korea). [14C]palmitate and [14C]palmitoyl-CoA were purchased from Moravek Biochemicals Inc. (Brea, CA). The solvent for TLC and N-ethylmaleimide (NEM) were purchased from Nakarai Tesque (Kyoto, Japan). StealthTM siRNA (Invitrogen) was used to knock down PEX19 in HeLa cells and FAR2 in MCF7 cells. The target sequences of the siRNA are as follows: human PEX19, 5′-GCCAGTGGTGAACAGTGTCTGATCA-3′; human FAR2-61, 5′-GGGAAAGGGTTTCTTCGGGCCATAA-3′; human FAR2-62, 5′-GACCTTTCAGGAGGCCAAATGCTAA-3′; and human FAR2-63, 5′-GAGCATCCAGCACGCTCAAAGTTTA-3′.
Cell Culture
MCF7 cells were purchased from RIKEN BRC Cell Bank (Tsukuba, Japan). MCF7 and HeLa cells were cultured in DMEM supplemented with 10% fetal bovine serum in 5% CO2 and 95% air. CHO-K1 and ZPEG251 cells (19) were cultured in Ham's F-12 medium supplemented with 10% fetal bovine serum in 5% CO2 and 95% air (19, 20). CHO-K1 cells stably expressing FLAG-FAR1, FLAG-FAR1490FAR2, and ZPEG251 cells stably expressing FLAG-FAR1-HA2 were isolated by selection with ZeocinTM (250 μg/ml). Plasmalogen levels in ZPEG251 cells were restored by adding 10 μm 1-O-hexadecylglycerol (HG) to the culture media every 24 h for the indicated period (19). Plasmalogen levels in CHO-K1 cells were increased by adding bovine brain plasmenylethanolamine (37.5 μg/ml) or ethanolamine (2 μm).
Lipid Analysis
Plasmalogen biosynthesis was assessed by labeling cells with [14C]palmitate for 5 h. Then, cells were subjected to alkaline methanolysis to obtain 1-alkeyl-glycerophosphoethanolamine (GPE), as described previously (11). Plasmalogen was converted to 2-acyl-GPE using trichloroacetic acid (19). Phospholipids, 1-alkeyl-GPE, and 2-acyl-GPE were resolved on TLC plates (silica gel 60, Merck KgaA) using a chloroform/methanol/acetic acid solution (v/v/v, 65/25/10). In vitro Far enzyme activity was determined using [14C]palmitoyl-CoA as described (11).
Construction of FAR1
To construct pcDNAZeo3.1FLAG-FAR1-HA2, pcDNAZeo3.1FLAG-FAR1 (11) was digested with NheI and BglII. FAR1-HA2 (11) was digested with BglII and ApaI. The fragments were ligated between the NheI and ApaI sites of pcDNA3.1/Zeo.
FARΔC Mutants
C-terminal truncation mutants of FAR1 were generated by PCR using NheI-FLAG-Far-Fw (5′-gcggctagcccgccatggattacaaggatgacgacgataagggcggcgtttcaatcccagaa-3′) as the forward primer and one of the following reverse primers: FAR-507stop-ApaI-Rv (5′-cgcgggccctcagaagtatgacaaaaacttgtaaca-3′), FAR-490stop-ApaI-Rv (5′-ccgctcgagggccctcatcttgccatttgtgatcttgc-3′), or FAR-467stop-ApaI-Rv (5′-ccgctcgagggccctcaacgtatattccgcaacttgttc-3′). The PCR products were digested with NheI and ApaI and were ligated between the NheI and ApaI sites of pcDNA3.1/Zeo. The resulting clones were FLAG-Far1507, FLAG-Far1490, and FLAG-Far1467.
Enhanced GFP (EGFP)-FAR1 Variants
DNA fragments encoding a C-terminal fragment of Far1 were amplified by PCR using FAR-515-ApaI-Rv (11) as the reverse primer and one of the following forward primers: BglII-451-Fw (5′-ctcagatctggcctccctgcagccaga-3′), BglII-469-Fw (5′-cgggagatctggttttaatactatccttgt-3′), or BglII-490-Fw (5′-cgggagatctatctggtactttgtggttag-3′). The PCR products were digested with BglII and ApaI and were ligated between the BglII and ApaI sites of pEGFP-C1 (Clontech). Because the BglII site was used to construct EGFP-Far1490–515, the asparagine residue at position 491 of Far1 was changed to serine. EGFP-Far1451–507 and EGFP-Far1469–507 were similarly constructed using FAR-507stop-ApaI-Rv as the reverse primer and BglII-451-Fw or BglII-469-Fw as the forward primer, respectively. To generate EGFP-Far1508–515, a DNA fragment encoding amino acids 508–515 of Far1 was fused to EGFP using the CMV.Fw (5′-cgcaaatgggcggtaggcgtg-3′) and EGFP-C8-ApaI-Rv (5′-cgcgggccctcagtatctcatagtgctggatgctcgcttgtacagctcgtccatgccgag-3′) primers. The PCR product was digested with NheI and ApaI and was ligated between the NheI and ApaI sites of pEGFP-C1.
Construction of FAR2 and Chimeric FAR1 and FAR2 Constructs
FLAG-FAR2 was constructed by PCR using FLJ10462 (Toyobo, Tokyo, Japan) as the template and the EcoRI-FLAG-Far2 (5′-gccgaattcgccaccatggattacaaggatgacgacgataagtccacaattgcagct-3′) and Far2XhoI (5′-ggcctcgagttaaactttgagcgtgc-3′) primers. The PCR product was digested with EcoRI and XhoI and was ligated between the EcoRI and XhoI sites of pcDNA3.1/Zeo. To generate FAR2, a KpnI site was introduced at the 5′ end of FAR2 by PCR using the Kpn-Far2-Fw (5′-gccggtaccgccaccatgtccacaattgca-3′) and Far2XhoI primers and FLAG-FAR2 as the template. The product was digested with KpnI and XhoI and was ligated between the KpnI and XhoI sites of pcDNA3.1/Zeo.
FLAG-FAR2-HA2
To generate FLAG-FAR2-HA2, a SpeI site was introduced at the 3′ end of FLAG-FAR2 by PCR using the FL-Far2–5′NotI Fw (5′-gccgcggccgccaccatggattacaaggat-3′) and FL-Far2 1545 SpeI Rv (5′- ctagactagtaactttgagcgtgct-3′) primers and FLAG-FAR2 as the template. The product was digested with NotI and SpeI and was ligated between the NotI and NheI sites of pUcD2HygPEX16-HA2 (21).
FLAG-FAR1466FAR2
To generate FLAG-FAR1466FAR2, a DNA fragment encoding the 50 amino acids at the C terminus of Far2 was amplified by PCR using the FL-Far1/466-Far2/467.Fw (5′-cctgcagccagaaaacatctgaacaagttgcggaatattcactacctctttaatac-3′) and BGH.Rv (5′-agaaggcacagtcgagg-3′) primers and FLAG-FAR2 as the template. This product was digested with PstI and XhoI, and pcDNA3.1/FLAG-FAR1 was digested with NheI and PstI. The fragments were ligated between the NheI and XhoI sites of pcDNA3.1/Zeo.
FLAG-FAR1490FAR2
To generate FLAG-FAR1490FAR2, a DNA fragment encoding the 25 amino acids at the C terminus of Far2 was amplified by PCR using the FL-Far1/490-Far2/491.Rv (5′-cagaagcttacaatgaagaaccagacatttcttgccatttgtgatc-3′) and CMV.Fw primers and FLAG-FAR1 as the template. This product was digested with PstI and HindIII, and FLAG-FAR1 was digested with NheI and PstI. These fragments were ligated between the NheI and HindIII sites of FLAG-FAR2.
FLAG-FAR1507FAR2
A DNA fragment encoding the eight amino acids at the C terminus of Far2 was amplified by PCR using the CMV.Fw and Far1/507-Far2/508.Rv (5′-tagggccctaaactttgagcgtgctggatgctctgaagtatgacaaaaa-3′) primers. The product was digested with PstI and ApaI and was ligated between the PstI and ApaI sites of pcDNA3.1/Zeo/FLAG-FAR1.
FLAG-FAR1FAR2491/507
To generate FLAG-FAR1FAR2491–507, a DNA fragment encoding amino acids 491–507 of Far2 and the eight amino acids at the C terminus of Far1 was amplified by PCR using the FL-Far2/491/507/Far1.Rv (5′-gggggccctcagtatctcatcgtgctggatgctctaaagtaggagag-3′) and CMV.Fw primers and FL-FAR1490FAR2 as the template. The product was digested with BspEI and ApaI, and FLAG-FAR1 was digested with NheI and BspEI. These fragments were ligated between the NheI and ApaI sites of pcDNA3.1/Zeo.
FLAG-FAR2FAR1491/507
In amino acids 491–507, only the residues at positions 492, 494, 496, and 499 differ between Far1 and Far2. These four amino acids in Far2 were mutated to the corresponding residues of Far1. A DNA fragment encoding amino acids 491–507 of Far1 was amplified by PCR using the Far2Far1/491/507 (5′-caagatctcagatggctcggaatatctggtacttcgtggtaagcctgtgttataaattcctc-3′) and BGH.Rv primers and pcDNA3.1/Zeo/FL-FAR2 as the template. In a second PCR, this fragment and CMV.Fw were used as the primers and pcDNA3.1/Zeo/FLAG-FAR2 was used as the template. The product was digested with BamHI and XhoI, and pcDNA3.1/Zeo/FLAG-FAR2 was digested with EcoRI and BamHI. These fragments were ligated between the EcoRI and XhoI sites of pcDNA3.1/Zeo.
FLAG-FAR2FAR1355/507
To generate FLAG-FAR1355/507, a DNA fragment encoding amino acids 350–354 of Far2 and 161 amino acids of FLAG-Far1507Far2 was amplified by PCR using the Far2/354Far1/355.Fw (5′-tcacagtactggaatgcggtaagccataaggcccca-3′) and BGH.Rv primers and FLAG-FAR1507FAR2 as the template. The product was digested with ScaI and ApaI, and pcDNA3.1/Zeo/FLAG-FAR2 was digested with EcoRI and ScaI. These fragments were ligated between the EcoRI and ApaI sites of pcDNA3.1/Zeo.
FLAG-FAR2FAR1286/507
To generate FLAG-FAR286/507, a DNA fragment encoding 230 amino acids of FLAG-Far1507Far2 was amplified by PCR using the NheFar1286.Fw (5′-catgaggctagcggcagcctgg-3′) and BGH.Rv primers and FLAG-FAR1507FAR2 as the template. The product was digested with NheI and ApaI, and pcDNA3.1/Zeo/FLAG-FAR2 was digested with EcoRI and NheI. These fragments were ligated between the EcoRI and ApaI sites of pcDNA3.1/Zeo. In FLAG-Far2Far1286/507, an arginine residue was introduced between the methionine residue at position 285 of Far1 and the leucine residue at position 286 of Far2 because of the creation of a NheI site.
FLAG-FAR2FAR1466/515
To generate FLAG-FAR2FAR1466/515, a DNA fragment encoding the C-terminal 50 amino acid sequence of Far1 was amplified by PCR using a set of primers, Far2/465Far1/466.Fw (5′-gggatcccaaaagcaaagcaacgcttaaaaaggctccgaaatatacgttatggttttaatactatcc-3′) and BGH.Rv (5′-agaaggcacagtcgagg-3′), and FLAG-FAR1 as a template. The product was digested with BamHI and ApaI, and pcDNA3.1/FLAG-FAR2 was digested with EcoRI and BamHI. The fragments were ligated between the EcoRI and ApaI sites of pcDNA3.1/Zeo.
FLAG-FAR2FAR1491/515
The three amino acids from the C terminus of FLAG-FAR2FAR1491/507 was mutated to the corresponding residues of Far1. A DNA fragment encoding amino acids of FLAG-FAR2FAR1491/515 was amplified by PCR using the primers Far1507Rv Xho (5′-cctacgagtcagtatctcatcgtgctggatgctcggaag-3′) and CMV.Fw and pcDNA3.1/Zeo/FLAG-FAR2Far1491/507 as a template. The product was digested with EcoRI and XhoI and ligated between the EcoRI and XhoI sites of pcDNA3.1/Zeo.
Morphological Analysis
Immunofluorescence of FLAG-tagged Far1 and its derivatives was performed as described previously (22). Differential membrane permeabilization using digitonin was performed as described previously (23).
Coimmunoprecipitation Assay
EGFP-Far1 variants or Far1 mutants lacking the C terminus were expressed with HA2-PEX19 in pex19 ZP119 cells (24) for 14 h. The cells were lysed with ice-cold PBS containing 0.2% digitonin and protease inhibitors for 15 min on ice and then further solubilized at 4 °C for 15 min. After centrifugation, cell lysates were subjected to immunoprecipitation using an anti-GFP antibody or anti-FLAG M2-conjugated agarose.
Biochemical Analysis
Subcellular fractionation and carbonate extraction were performed as described previously (22). For proteinase K treatment, postnuclear supernatant (PNS) was prepared in the absence of protease inhibitors, and 20 μg of PNS was digested with proteinase K (1 μg) on ice for 30 min in a 200-μl reaction volume. Tryptic digestion was performed likewise.
In vitro transcription/translation reactions in a rabbit reticulocyte-lysate protein-synthesizing system were performed using the quick-coupled transcription/translation system (TNT T7, Promega) according to the instructions of the manufacturer.
RESULTS
Far1 Is a Peroxisomal Integral Membrane Protein
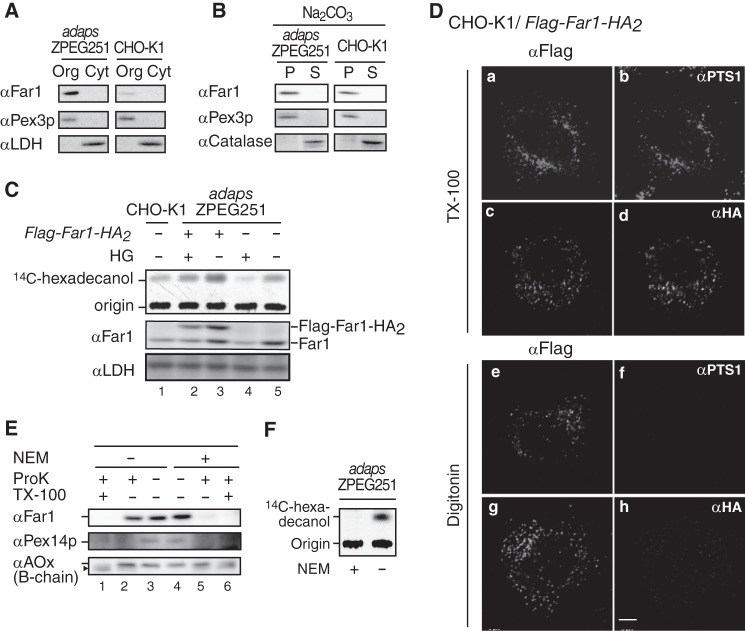
We demonstrated previously that endogenous Far1 is localized to peroxisomes in ZPEG251 cells, in which plasmalogen synthesis is defective (11). In this study, we used sodium carbonate extraction to analyze the integrity of Far1 in ZPEG251 and CHO-K1 cells (Fig. 1B). Subcellular fractionation demonstrated that Far1 was exclusively recovered in the membrane fraction (Fig. 1A). Far1 was not extracted by sodium carbonate treatment and was recovered in the membrane fractions of ZPEG251 and CHO-K1 cells. Pex3p, a peroxisomal membrane peroxin, was similarly present in the membrane pellet, whereas catalase, a peroxisomal matrix protein, was recovered in the soluble fraction (Fig. 1B). These results indicate that Far1 is an integral peroxisomal membrane protein.
FIGURE 1.
Far1 is a tail-anchored type II peroxisomal membrane protein. A, organelle (Org) and cytosolic (Cyt) fractions prepared from ZPEG251, an ADAPS-defective CHO mutant cell line (adapsZPEG251), and CHO-K1 cells were assessed by Western blotting with antibodies against Far1, the peroxisomal membrane protein Pex3p, and lactate dehydrogenase. B, organelle fractions of ZPEG251 and CHO-K1 cells were treated with sodium carbonate, separated into membrane (P) and soluble (S) fractions, and subjected to immunoblotting with antibodies as indicated on the left. C, CHO-K1 cells (lane 1), ZPEG251 cells (lanes 4 and 5), and ZPEG251 cells stably expressing FLAG-Far1-HA2 (lanes 2 and 3) were cultured for 2 days in the presence (+) or absence (−) of HG. PNSs were prepared from the cells, and Far1 activity was assessed using [14C]palmitoyl-CoA as a substrate (top panel). Expression levels of endogenous Far1, FLAG-Far1-HA2, and lactate dehydrogenase were assessed by immunoblotting using antibodies against Far1 and lactate dehydrogenase (center and bottom panels, respectively). Origin, the spots where the extracted lipids were placed. D, transmembrane topology of FLAG-Far1-HA2. CHO-K1 cells transfected with FLAG-Far1-HA2 were fixed, treated with 1% TX-100 (a–d) or 25 μg/ml digitonin (e–h), and subjected to dual labeling with antibodies against FLAG (a and e) and PTS1 (b and f) or FLAG (c and g) and HA (d and h). Scale bar = 5 μm. E, PNS was prepared from ZPEG251 cells in the presence (+) or absence (−) of 1 mm NEM and subjected to proteinase K (ProK) (1 μg) digestion. The protease sensitivities of Far1, Pex14p, and acyl-CoA oxidase (αAOx) were assessed by immunoblotting with their corresponding antibodies. Note that Far1 was effectively degraded by proteinase K in the presence of NEM. The arrowhead indicates a cleavage product of AOx (42). F, PNS was prepared as described in E and assessed for Far activity with [14C]palmitoyl-CoA as a substrate in the presence (+) or absence (−) of NEM.
We next analyzed the membrane topology of Far1 in CHO-K1 cells by expressing FLAG-Far1-HA2, which encodes both N- and C-terminally epitope-tagged Far1. The accumulation of hexadecanol was evident when FLAG-Far1-HA2 was expressed in ZPEG251 cells (Fig. 1C, lanes 3 and 5), and this accumulation was reduced when plasmalogen levels were restored by the addition of HG (lane 2). This implies that FLAG-Far1-HA2 is functional. CHO-KI cells expressing FLAG-Far1-HA2 were permeabilized with Triton X-100 and colabeled with anti-FLAG and anti-HA antibodies or anti-FLAG and anti-PTS1 antibodies. Colocalization indicated that FLAG-Far1-HA2 was localized in peroxisomes (Fig. 1D, a–d). When CHO-K1 cells expressing FLAG-Far-HA2 were permeabilized with 25 μg/ml digitonin under conditions in which plasma membranes are selectively permeabilized and intraperoxisomal proteins are inaccessible to exogenous antibodies (25, 26), labeling with an anti-FLAG antibody was punctate (Fig. 1D, e and g), whereas labeling with anti-HA and anti-PTS1 antibodies generated no signal (h and f). FLAG-Far1-HA2 showed the same membrane topology in ZPEG251 and CHO-K1 cells (data not shown).
We further assessed the topology of endogenous Far1. When a PNS fraction prepared from ZPEG251 cells was treated with proteinase K, Far1 was resistant to protease treatment, whereas it was completely digested in the presence of NEM (Fig. 1E, lanes 2 and 5). NEM treatment inactivated Far1 (Fig. 1F), as demonstrated previously (27). Interestingly, Far1 migrated slightly slower in NEM-treated cells than in untreated cells (Fig. 1E, lanes 3 and 4). We speculate that this indicates that a catalytic domain of Far1 is tightly packed and that the structure of this domain is modified by NEM treatment so that it becomes accessible to proteases. Taken together, these results suggest that the C terminus of Far1 is exposed to the peroxisome matrix, whereas a large catalytic domain in its N terminus is located outside of peroxisomes.
Far1 contains 515 amino acids. The NADPH-binding domain corresponds to amino acids 15–285. Hydropathy analysis according to a Doolittle and Kyte plot predicted that the transmembrane segment of Far1 is located in its C terminus region (amino acids 466–483). To assess the topogenesis of Far1, we examined whether several deletion mutants of Far1 localize to peroxisomes (Fig. 2A). FLAG-Far1507, which lacks the final eight amino acids at the C terminus, was resistant to carbonate extraction (supplemental Fig. S1) and localized to peroxisomes, as assessed by colocalization with the peroxisomal membrane protein Pex14p (Fig. 2B, c and d). By contrast, FLAG-Far1490 and FLAG-Far1467 were localized in the mitochondrion and cytosol, respectively (Fig. 2B). These results suggest that the C terminus of Far1 is important for its peroxisomal localization. We further assessed the minimum region required for the peroxisomal targeting of Far1 using EGFP as a reporter protein. The 65 and 47 amino acids at the C terminus of Far1 were attached to the C terminus of EGFP to generate EGFP-Far1451–515 andEGFP-Far1469–515, respectively. These fusion proteins were localized to peroxisomes, suggesting that Far1 associates with peroxisomes via its C-terminal region (Fig. 3B). Further deletion of the predicted hydrophobic region of Far1 abrogated its peroxisomal localization, as demonstrated by analysis of the EGFP-Far1490–515 and EGFP-Far1508–515 fusion proteins. By contrast, EGFP-Far1451–507 was localized to peroxisomes in a manner resistant to carbonate extraction (Fig. 3, B and C), consistent with the results that indicated that the final eight amino acids at the C terminus of Far1 are not essential for its peroxisomal localization (Fig. 2). EGFP-Far1469–507 contains the predicted transmembrane domain and 22 amino acids of the luminal domain of Far1. This protein less efficiently bound to Pex19p and localized to peroxisomes and mitochondria, suggesting that amino acids 451–468 are required for the efficient binding to Pex19p. These results indicate that amino acids 451–507 in the C-terminal region of Far1 are sufficient for the targeting and integration of Far1 into peroxisomal membranes, which strongly implies that Far1 is a peroxisomal tail-anchored protein.
FIGURE 2.
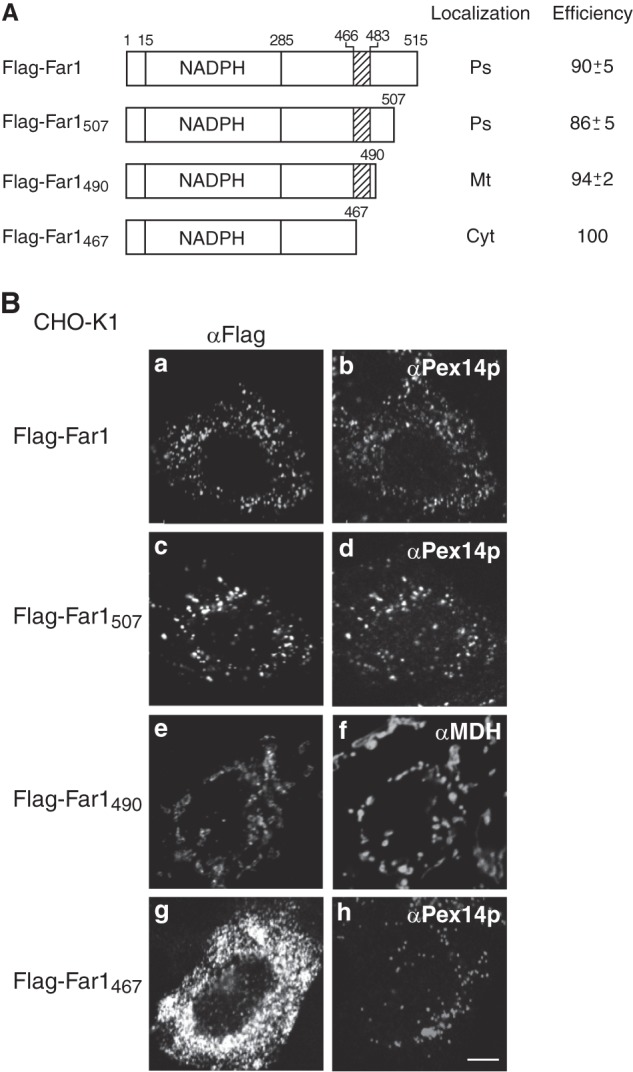
C-terminal portion of Far1 is important for its peroxisome localization. A, Far1 deletion mutant constructs. NADPH indicates the predicted NADPH-binding domain of Far1, and the hatched box indicates the predicted transmembrane domain of Far1. The intracellular localizations of these constructs are indicated in the center column. Ps, peroxisomes; Mt, mitochondria; Cyt, cytosol. The numbers in the right column show the percentages of cells showing the organelles targeted by the respective Far1 truncated mutants. Data were collected by counting more than 100 cells expressing Far1 variants from three independent experiments and are represented by the mean ± S.D. B, intracellular localizations of these Far1 mutants were examined in CHO-K1 cells. Cells were stained with antibodies against FLAG (a, c, e, and g), Pex14p (b, d, and h), and malate dehydrogenase (f). Scale bar = 5 μm.
FIGURE 3.
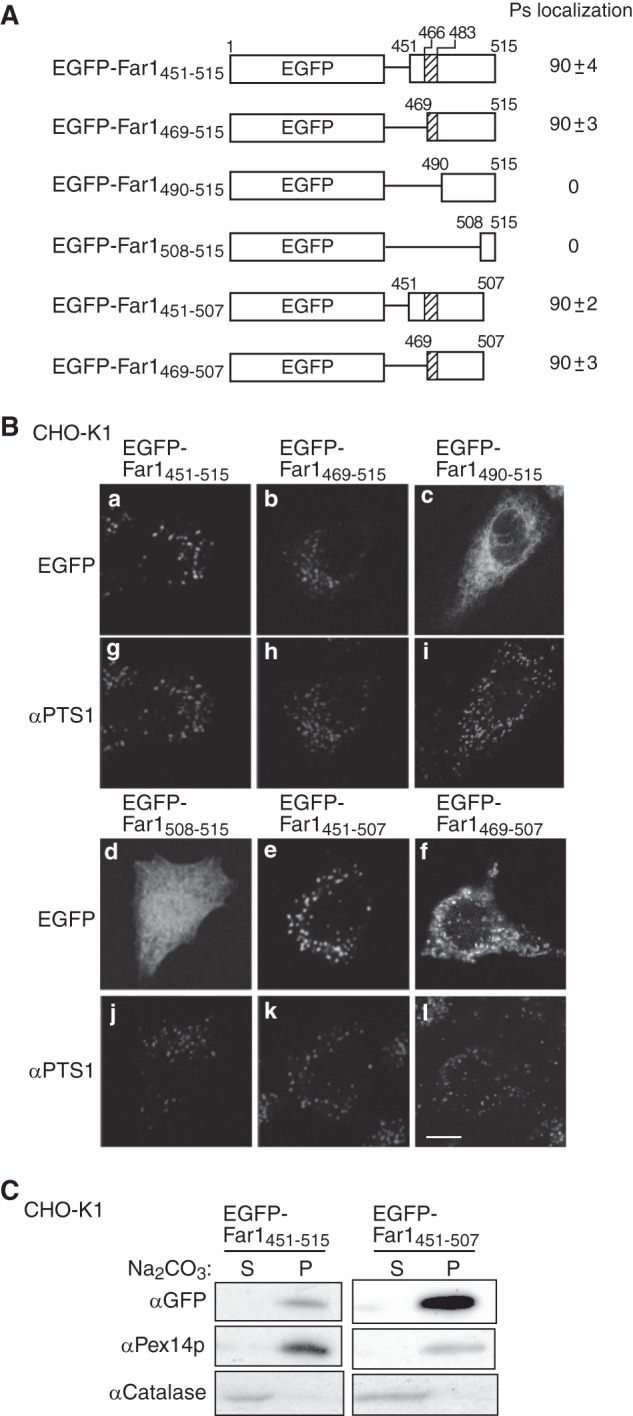
The C-terminal portion of Far1 is sufficient for its peroxisome localization. A, schematic representation of the EGFP fusion proteins used to search for the minimum region of Far1 that is required for the localization of this protein to peroxisomes. The peroxisomal targeting activity of each fusion protein is represented by percentage. Data were collected by counting more than 100 cells expressing EGFP-fused Far1 mutants from three independent experiments and are represented by the mean ± S.D. Note that EGFP-Far1469–507 was localized to both peroxisomes and mitochondria. B, each construct was expressed in CHO-K1 cells for 14 h and detected by monitoring GFP fluorescence (a–f) and labeling with an anti-PTS1 antibody (g–l). Scale bar = 5 μm. C, the membrane integrities of two peroxisome-localizing fusion proteins expressed in CHO-K1 cells was assessed by carbonate extraction as described in Fig. 1B. P and S are membrane pellet and soluble fractions, respectively. Pex14p was used as a positive control for a peroxisomal integral membrane protein.
Far1 Is Localized to Peroxisomes in a Pex19p-dependent Manner
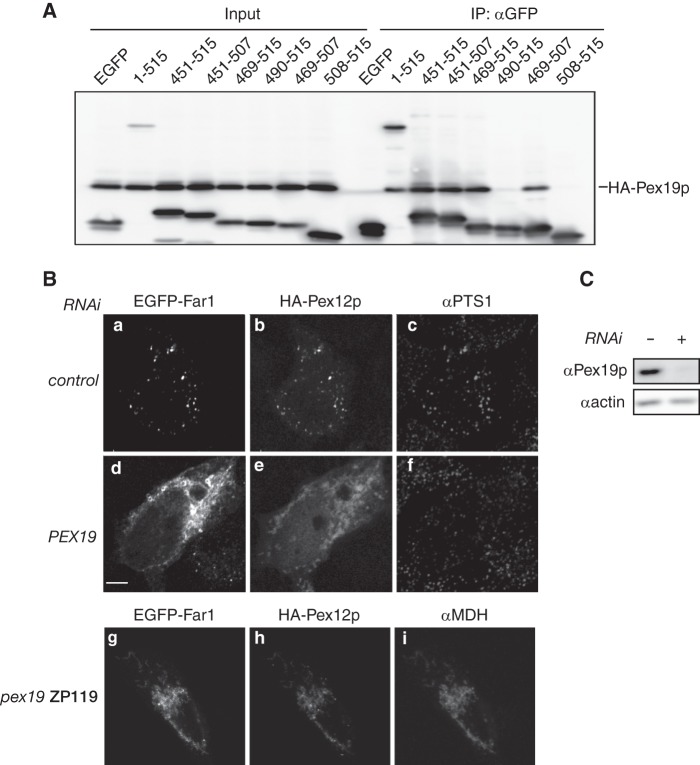
Mammalian Pex26p is a peroxisomal tail-anchored protein, and Pex19p is required for the correct targeting of this protein (28). To assess whether Far1 is localized to peroxisomes in a Pex19p-dependent manner, we first analyzed the interaction between Pex19p and EGFP-tagged Far1 proteins. HA-Pex19p and various EGFP-Far1 fusion proteins were expressed in pex19 ZP119 cells, and these fusion proteins were immunoprecipitated using an anti-GFP antibody. Pex19p was coimmunoprecipitated with EGFP-tagged Far1 fusion proteins that localized to peroxisomes, strongly suggesting that Far1 is transported to peroxisomes in a Pex19p-dependent manner (Fig. 4A). We further verified the requirement of Pex19p for the peroxisomal localization of Far1 by reducing Pex19p expression in HeLa cells and by using the PEX19-deficient CHO cell line pex19 ZP119 (24). Expression of Pex19p was efficiently reduced in HeLa cells by transfection of dsRNA against PEX19 (Fig. 4C). When HA-Pex12p and EGFP-Far1 were coexpressed in HeLa cells, both proteins colocalized with labeling of an anti-PTS1 antibody, indicating that they localized to peroxisomes (Fig. 4B, a–c). By contrast, HA-Pex12p had a mitochondrion-like localization in HeLa cells treated with dsRNA against PEX19, indicating that expression of Pex19p was efficiently reduced in these cells. EGFP-Far1 was also not targeted to peroxisomes in these cells, despite peroxisomes being recognized with an anti-PTS1 antibody (Fig. 4B, d–f). EGFP-Far1 and HA-Pex12p were localized in mitochondria in pex19 ZP119 cells (Fig. 4B, g–i). Taken together, these results strongly suggest that Far1 is targeted to peroxisomes in a Pex19p-dependent manner.
FIGURE 4.
Far1 is transported to peroxisomes in a Pex19p-dependent manner. A, EGFP fusion proteins containing full-length Far1 or a portion of the C terminus were coexpressed with HA-Pex19p in pex19 ZP119 cells for 14 h and subjected to immunoprecipitation with an anti-GFP antibody. EGFP-tagged Far1 proteins and HA-Pex19p were detected using antibodies against GFP and HA in the same membrane. Input, 2.5% of the total cell lysate used in each immunoprecipitation (IP). Note that HA-Pex19p bound all EGFP-tagged Far1 proteins that localized to peroxisomes in CHO-K1 cells (see Fig. 3B). B, HeLa cells were treated without (top panel) or with a dsRNA against PEX19 (center panel) for 72 h. EGFP-Far1 and HA-Pex12p (43) were coexpressed for the final 12 h of this treatment. EGFP-Far1 and HA-Pex12p were also coexpressed in pex19 ZP119 cells (bottom panel). The intracellular localizations of EGFP-Far1, HA-Pex12p, PTS1 proteins, and malate dehydrogenase were assessed by monitoring GFP fluorescence and labeling with antibodies against HA, PTS1, and malate dehydrogenase. Note that the localization of EGFP-Far1 and HA-Pex12p to peroxisomes was impaired when PEX19 expression was reduced, despite peroxisomes remaining intact in these cells as indicated by the punctate labeling of the anti-PTS1 antibody. Scale bar = 5 μm. C, depletion of Pex19p expression was confirmed by immunoblotting. Actin was used as a loading control.
Far1 Is Specifically Degraded in Response to Cellular Plasmalogen Levels
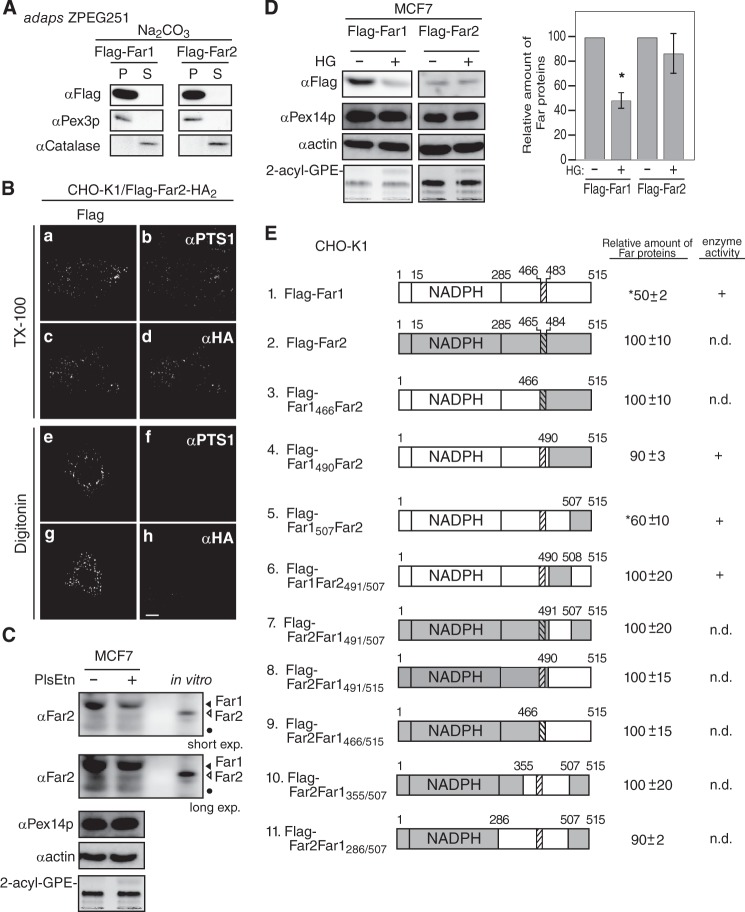
We demonstrated previously that the stability of Far1 is regulated in response to plasmalogen levels (11). The amino acid sequence of human Far2 has 59% identity and 78% similarity to that of human Far1. Far1 and Far2 both localize to peroxisomes and catalyze fatty alcohol formation. The substrate specificities of the proteins slightly differ because Far2 only catalyzes the formation of saturated C16 and C18 fatty acids (10). We investigated whether the stability of Far2 is also regulated in a plasmalogen-dependent manner. To this end, FLAG-Far2 and FLAG-Far2-HA2 were expressed in CHO-K1 cells. Both proteins localized to peroxisomes, as assessed by colocalization with PTS1-containing proteins (Fig. 5B). Furthermore, the N terminus and C terminus of Far2 were located on the cytosolic side and the peroxisomal side of the peroxisomal membrane, respectively, as judged by the accessibility of the anti-FLAG antibody under conditions in which the plasma membrane was selectively permeabilized (Fig. 5B). The localization of FLAG-Far2 to peroxisomes in ZPEG251 cells was not affected by carbonate treatment (Fig. 5A). Together, these results strongly suggest that Far2 is also a peroxisomal tail-anchored protein. However, FLAG-Far2 was not degraded following the restoration of plasmalogen levels in ZPEG251 cells, whereas FLAG-Far1 was efficiently degraded under these conditions (Fig. 5F). This indicates that Far1, but not Far2, is degraded when plasmalogen levels reach a certain threshold.
FIGURE 5.
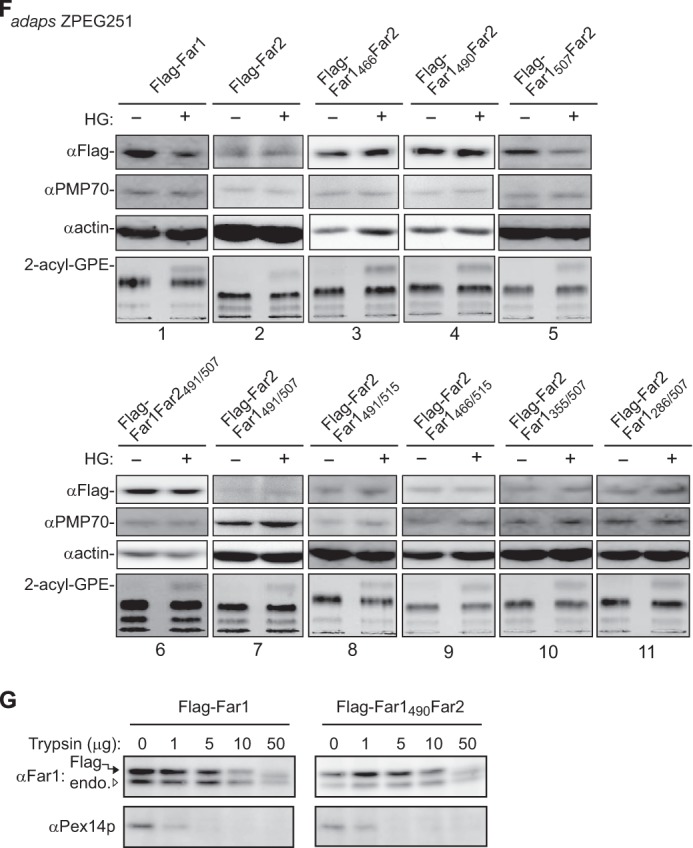
The transmembrane-flanking region of Far1 is required for its plasmalogen-dependent degradation. A, FLAG-tagged Far1 and Far2 were expressed in ZPEG251 cells, and their membrane insertion was assessed by carbonate extraction as described in Fig. 1B. P and S are membrane pellet and soluble fractions, respectively. B, the membrane topology of FLAG-Far2-HA2 in CHO-K1 cells was assessed by differential membrane permeabilization with digitonin as described in Fig. 1D. Scale bar = 5 μm. C, MCF7 cells were cultured for 2 days in the presence (+) or absence (−) of plasmenylethanolamine (PlsEtn). The expression levels of endogenous Far1 (solid arrowhead) and Far2 (open arrowhead), Pex14p, and actin were assessed by immunoblotting with antibodies to Far2, Pex14p, and actin, respectively. Note that anti-Far2 antibody recognized both Far1 and Far2. An in vitro-translated Far2 was loaded in the same SDS-PAGE. The same blot for Far1 and Far2 at a longer exposure is also shown (second panel). The plasmalogen level was detected as 2-acyl-GPE and is shown in the fifth panel. Dots indicates a nonspecific band. D, FLAG-Far1 and FLAG-Far2 were transfected to MCF7 cells. Cells were divided into two dishes and cultured for 2 days in the presence (+) or absence (−) of HG. Proteins were detected by Western blotting with the antibodies indicated on the left. FLAG-tagged Far proteins were quantified by a LAS-4000 mini lumino image analyzer. Relative expression levels of FLAG-Far1 and FLAG-Far2 are shown, where the levels of respective Far proteins in untreated MCF7 cells were designated as 100. Scale bars represent the mean ± S.D. of three experiments. *, p < 0.05; Student's t test versus untreated MCF7 cells. E, schematic of Far1 and Far2 chimeric fusion proteins. Plasmalogen-dependent degradation and enzyme activity of these fusion proteins are summarized on the right. n.d., not done. The relative expression levels of Far1-Far2 chimera proteins were determined, where the expression levels of respective Far1-Far2 chimera proteins in plasmalogen-deficient ZPEG251 cells were designated as 100. Numbers represent the mean ± S.D. of three experiments. *, p < 0.05; Student's t test versus untreated ZPEG251. F, ZPEG251 cells were transfected with FLAG-Far1, FLAG-Far2, or the chimeric fusion proteins and then cultured in the presence (+) or absence (−) of HG for 48 h. The levels of each of the fusion proteins, PMP70, and actin were assessed by labeling with their corresponding antibodies. The plasmalogen level is shown in the bottom panel. G, PNS prepared from ZPEG251 cells expressing FLAG-Far1 or FLAG-Far1490Far2 was digested with trypsin. The digestion was terminated by the addition of trichloroacetic acid, and the protease sensitivities of FLAG-Far1 and FLAG-Far1490Far2 were assessed by immunoblotting with antibodies against Far1 and Pex14p. Solid and open arrowheads indicate FLAG-tagged and endogenous Far proteins, respectively.
We next assessed whether endogenous Far2 is degraded in response to the cellular level of plasmalogens. To this end, we screened several cell lines and found that Far2 was expressed in MCF7 cells but not in HeLa cells (supplemental Fig. S2). Our anti-Far2 antibody recognized both Far1 and Far2 with apparently distinct migration in SDS-PAGE, which was confirmed by the immunoblotting using in vitro translation products (supplemental Fig. S2). MCF7 cells were defective in plasmalogen synthesis (Fig. 5C), consistent with a previous study (29). In MCF7 cells, the degradation of Far1 was evident upon elevating plasmalogen levels by supplementing with plasmenylethanolamine, whereas the expression level of Far2 was not altered significantly (Fig. 5C). When FLAG-tagged Far2 was expressed in MCF7, FLAG-Far2 was relatively more stable than FLAG-Far1 upon elevating the plasmalogen level by supplementing with HG (Fig. 5D). Taken together, these results suggest that Far1 stability is more dynamically regulated than Far2 in MCF7 cells.
To elucidate the molecular mechanism underlying plasmalogen-dependent degradation of Far1, we took advantage of the similarities in the domain structures of Far1 and Far2 and constructed several chimeric proteins (Fig. 5E). In FLAG-Far1466Far2 and FLAG-Far1490Far2, the 49 and 25 amino acids at the C terminus of Far1 were replaced with the corresponding region of Far2, respectively. Both chimeric proteins were efficiently targeted to peroxisomes (supplemental Fig. S3). Neither chimeric protein was degraded in ZPEG251 cells when plasmalogen levels were restored. This suggests that the C-terminal region of Far1 that is exposed to the peroxisome matrix is required for plasmalogen-dependent degradation of Far1 (Fig. 5F).
Next, the C terminus region of Far1 was divided into two parts, and these fragments were replaced with the corresponding fragments of Far2 (Fig. 5E). Substitution of the final eight amino acids of Far1 with those of Far2 did not abrogate plasmalogen-dependent degradation of Far1. By contrast, degradation of FLAG-Far1Far2491/507, in which 17 amino acids in the transmembrane-flanking region of Far1 were replaced with the corresponding residues of Far2, was not stimulated in ZPEG251 cells when plasmalogen levels were restored. This indicates that these 17 amino acids in the transmembrane-flanking region of Far1 are necessary for plasmalogen-dependent degradation of Far1.
Finally, we investigated whether these 17 amino acids in the transmembrane-flanking region of Far1 are sufficient for the plasmalogen-dependent degradation of this protein. To this end, the transmembrane-flanking region of Far2 was replaced with the corresponding region of Far1 to generate FLAG-Far2Far1491/507. This protein was efficiently targeted to peroxisomes. However, its expression level was not reduced when plasmalogen levels were restored in ZPEG251 cells. This indicates that amino acids 491–507 in the transmembrane-flanking region of Far1 are necessary but not sufficient for the degradation of Far1 in response to plasmalogen levels. In addition, FLAG-Far2Far1491/515 and FLAG-Far2Far1466/515 were not degraded, suggesting that the C-terminal 8 amino acids of Far1 do not influence its plasmalogen-dependent degradation. Moreover, FLAG-Far2Far1355/507 and FLAG-Far2Far1285/507 were not degraded in a plasmalogen-dependent manner. Together, these data indicate that almost the entire sequence of Far1 is needed for its plasmalogen-dependent degradation.
These results led us examine whether replacement of the transmembrane-flanking region of Far1 induces a conformational change in the catalytic domain of the protein that abrogates plasmalogen-dependent degradation of mutant Far1 proteins. To address this issue, PNS fractions prepared from ZPEG251 cells expressing FLAG-Far1 and FLAG-Far1490Far2 were treated with trypsin (Fig. 5G). FLAG-Far1 was largely resistant to trypsin digestion and was partially digested upon incubation with a large amount of trypsin. Pex14p was efficiently digested in the presence of a small amount of trypsin. FLAG-Far1490Far2 was similarly resistant to trypsin digestion, which indicates that replacement of the transmembrane-flanking region of Far1 does not induce a large conformational change in the catalytic domain of Far1 mutant proteins. Taken together, these results suggest that plasmalogen-dependent regulation of Far1 stability is largely mediated by its transmembrane-flanking region and that the catalytic domain may play an ancillary role.
Finally, we asked whether elevation of Far1 expression increases plasmalogen synthesis. To this end, we established cell lines stably expressing FLAG-Far1 and FLAG-Far1490Far2, and labeled these cells with [14C]palmitate to assess plasmalogen synthesis. As expected, plasmalogen synthesis was higher in CHO-K1 cells expressing FLAG-Far1490Far2 and FLAG-Far1 than in the parental cell line (Fig. 6 and supplemental Fig. S4).
FIGURE 6.
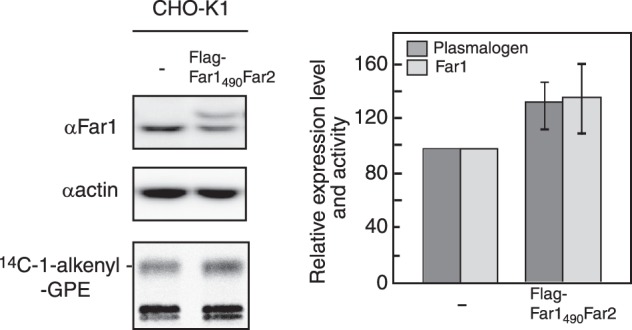
Expression of FLAG-Far1490Far2 increased plasmalogen synthesis. Plasmalogen biosynthesis was examined in CHO-K1 cells stably expressing FLAG-Far1490Far2 by labeling with [14C]palmitate in the presence of 2 μm ethanolamine for 5 h. Levels of endogenous Far1, FLAG-Far1490Far2, and actin were assessed by immunoblotting using antibodies against Far1 and actin. Band intensities were measured with a LAS-4000 mini lumino image analyzer. FLAG-Far1490Far2 migrated slightly slower than endogenous Far1. Relative expression levels of Far1 or Far1 plus FLAG-Far1490Far2 (light gray bars) and plasmalogen biosynthesis (dark gray bars) are shown, where levels in control CHO-K1 cells were designated as 100. Scale bars represent the mean ± S.D. of three experiments.
DISCUSSION
Far1 Is a Peroxisomal Tail-anchored Protein
In this study, we demonstrated that Far1 is a peroxisomal tail-anchored protein and that a region within the C terminus of Far1 is sufficient for its localization to peroxisomes. Experiments in which the plasma membrane was selectively permeabilized revealed that the catalytic domain in the N-terminal region of Far1 is located on the outer surface of peroxisomes. Protease digestion experiments in which purified peroxisomes were treated with trypsin at 37 °C indicated the same (30). However, the catalytic domain of Far1 is relatively resistant to protease digestion because most peroxisomal membrane proteins are digested by treatment with trypsin at 0 °C for 5 min (31). Indeed, the catalytic domain of Far1 was largely resistant to proteinase K digestion in the absence of NEM (Fig. 1), which indicates that the catalytic domain of Far1 is tightly packed or is surrounded by a lipid bilayer. Hydropathy plots did not show any significant hydrophobic region in the catalytic domain of Far1, and so we conclude that the tightly packed conformation of the Far1 catalytic domain model is the more likely of these two possibilities. Cysteine residues seem to be involved in this tight packing because Far1 was sensitive to proteinase K in the presence of NEM. Far1 has six Cys residues, and the Cys residue at position 500, which is conserved in several species, including human, rat, and mouse, is located on the peroxisomal matrix side. This suggests that at least one Cys residue in the catalytic domain of Far1 remains accessible to NEM modification. Notably, Far activity is abolished by sulfhydryl reagents, and the active site of the enzyme was suggested to contain an essential thiol residue (27).
The mammalian proteins Pex26p, Far1, mitochondrial fission protein 1 (Fis1), and mitochondrial fission factor are peroxisomal tail-anchored proteins, and Fis1 and mitochondrial fission factor localize to both mitochondria and peroxisomes (28, 32–34). Of these proteins, the peroxisomal localizations of Pex26p and Fis1 are mediated by Pex19p. Pex26p has two neighboring Pex19p-binding sites in its transmembrane domain and luminal C terminus. The luminal Pex19p-binding site is essential for correct targeting of full-length Pex26p to peroxisomes (28). By contrast, the Pex19p-binding site of Fis1 is located in the region of the C terminus that contains the transmembrane domain (33). Coimmunoprecipitation of Far1 with Pex19p revealed that the Pex19p-binding site of Far1 overlaps with its transmembrane domain. Consistent with our experimental findings, a potential Pex19p-binding site of Far1 is predicted in its transmembrane domain by mPTS Predictor (35). Positively charged residues in the transmembrane domain that flanks the C terminus were suggested recently to be important for Pex19p-mediated peroxisome localization of tail-anchored proteins (36). Far1 has five basic amino acids in its C-terminal luminal region. Interestingly, FLAG-Far1490, which has arginine residues at positions 485 and 490, interacted with Pex19p but was mislocalized to mitochondria in a carbonate-resistant manner (supplemental Fig. S1). This suggests that the luminal region of Far1 might participate in the integration of Far1 into peroxisomal membranes.
In this study, we showed that the N terminus and C terminus of Far2 are oriented toward the cytosolic and luminal faces of peroxisomes, respectively (Fig. 5B). The C-terminal region of Far2 is sufficient for its peroxisomal localization, as demonstrated by experiments with FLAG-Far1466Far2. In this construct, the C-terminal region of Far2, including the transmembrane domain, was fused to a Far1 mutant lacking the region necessary for peroxisomal localization. Together with the results of these experiments, the similarities in the domain structures of Far1 and Far2 strongly suggest that Far2 is also a peroxisomal C-tail-anchored protein.
Regulation of Far1 Expression Levels
Two Far isozymes, Far1 and Far2, localize to peroxisomes and synthesize long-chain fatty alcohols by reducing fatty acyl-CoA with slightly different substrate specificities (10). Interestingly, we showed that the stability of Far1, but not Far2, is regulated in response to the level of plasmalogens in CHO-K1 cells and that the transmembrane-flanking region of Far1 is necessary for this plasmalogen-dependent degradation. Together with the topological analysis of Far1, these observations suggest that peroxisomal membrane or matrix proteins participate in the regulation of Far1 stability. However, we suspect that it is more likely that a peroxisomal membrane protein is involved because Far1 was efficiently degraded when plasmalogen levels were restored in mutant cells in which the import of peroxisomal matrix proteins is defective (11). We showed that the catalytic domain of Far1 is required for plasmalogen-dependent regulation of this protein. However, the conformational feature of Far1 appears to be less important for plasmalogen-dependent degradation of this protein because FLAG-Far1 and FLAG-Far1490Far2 were similarly resistant to trypsin digestion (Fig. 5G), whereas FLAG-Far1490Far2 was not degraded in a plasmalogen-dependent manner. Together, these results suggest that the transmembrane-flanking region of Far1 plays a major role in its degradation.
On the basis of our finding that Far1 activity is regulated by modulation of its stability in response to cellular plasmalogen levels, we suggested previously that Far1 is a rate-limiting enzyme in plasmalogen synthesis (11). Dihydroxyacetonephosphateacyltransferase is essential for plasmalogen synthesis but appears not to be the rate-limiting enzyme on the basis of the observation of the plasmalogen synthesis in the plasmalogen-deficient CHO mutant NRel-4 cells expressing severalfold of dihydroxyacetonephosphateacyltransferase that are restored in the plasmalogen levels up to that in wild-type CHO-K1 (37). In this study, plasmalogen synthesis was indeed increased when Far1 expression was elevated (Fig. 6 and supplemental Fig. S4). We attempted to isolate a cell line expressing a higher level of FLAG-Far1 or FLAG-Far1490Far2. However, the expressed proteins were not localized only to peroxisomes but rather mislocalized to other organelles, including mitochondria (data not shown), most likely because of the overexpression of these proteins. Therefore, we could not dramatically increase the plasmalogen level in our cell lines.
It remains to be elucidated whether the reduced level of plasmalogens causes the pathologies of Zellweger syndrome, rhizomelic chondrodysplasia punctata, and sporadic Alzheimer disease. Nevertheless, various approaches have been used in attempts to increase cellular plasmalogen levels. Dietary plasmalogen increases plasmalogen levels only in plasma without cleaving the vinyl ether bond at the sn-1 position of plasmalogen (38). 1-O-alkylglycerol was successfully applied to restore plasmalogen levels in plasmalogen-deficient cells (19, 39, 40) and the peripheral tissues of PEX7 knockout mice (41). However, such applications appear unable to efficiently increase plasmalogen levels in nervous tissues such as the sciatic nerve, cerebrum, and cerebellum (41). Therefore, modulation of Far1 activity may be a more efficient means of increasing plasmalogen levels in nervous tissues.
Acknowledgments
We thank Y. Nanri and S. Okuno for technical assistance, M. Nishi and K. Shimizu for preparing the figures, and the other members of our laboratory for discussions.
This work was supported in part by a Science and Technology Agency of Japan CREST grant (to Y. F.); by grants-in-aid for scientific research (to M. H. and Y. F.); by the global COE program and grants for excellent graduate schools from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; and by grants from the Japan Foundation for Applied Enzymology (to Y. F.) and the Takeda Science Foundation (to M. H. and Y. F.).

This article contains supplemental Figs. S1–S4.
- DHAP
- dihydroxyacetonephosphate
- ADAPS
- alkyl-dihydroxyacetonephosphate synthase
- NEM
- N-ethylmaleimide
- HG
- 1-O-hexadecylglycerol
- EGFP
- enhanced GFP
- PNS
- postnuclear supernatant.
REFERENCES
- 1. Wanders R. J., Waterham H. R. (2006) Peroxisomal disorders. The single peroxisomal enzyme deficiencies. Biochim. Biophys. Acta 1763, 1707–1720 [DOI] [PubMed] [Google Scholar]
- 2. van den Bosch H., Schutgens R. B., Wanders R. J., Tager J. M. (1992) Biochemistry of peroxisomes. Annu. Rev. Biochem. 61, 157–197 [DOI] [PubMed] [Google Scholar]
- 3. Weller S., Gould S. J., Valle D. (2003) Peroxisome biogenesis disorders. Annu. Rev. Genomics Hum. Genet. 4, 165–211 [DOI] [PubMed] [Google Scholar]
- 4. Steinberg S. J., Dodt G., Raymond G. V., Braverman N. E., Moser A. B., Moser H. W. (2006) Peroxisome biogenesis disorders. Biochim. Biophys. Acta 1763, 1733–1748 [DOI] [PubMed] [Google Scholar]
- 5. Nagan N., Zoeller R. A. (2001) Plasmalogens. Biosynthesis and functions. Prog. Lipid Res. 40, 199–229 [DOI] [PubMed] [Google Scholar]
- 6. Rosenberger T. A., Oki J., Purdon A. D., Rapoport S. I., Murphy E. J. (2002) Rapid synthesis and turnover of brain microsomal ether phospholipids in the adult rat. J. Lipid Res. 43, 59–68 [PubMed] [Google Scholar]
- 7. Ginsberg L., Rafique S., Xuereb J. H., Rapoport S. I., Gershfeld N. L. (1995) Disease and anatomic specificity of ethanolamine plasmalogen deficiency in Alzheimer's disease brain. Brain Res. 698, 223–226 [DOI] [PubMed] [Google Scholar]
- 8. Han X., Holtzman D. M., McKeel D. W., Jr. (2001) Plasmalogen deficiency in early Alzheimer's disease subjects and in animal models. Molecular characterization using electrospray ionization mass spectrometry. J. Neurochem. 77, 1168–1180 [DOI] [PubMed] [Google Scholar]
- 9. Wood P. L., Smith T., Pelzer L., Goodenowe D. B. (2011) Targeted metabolomic analyses of cellular models of Pelizaeus-Merzbacher disease reveal plasmalogen and myo-inositol solute carrier dysfunction. Lipids Health Dis. 10, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cheng J. B., Russell D. W. (2004) Mammalian wax biosynthesis. I. Identification of two fatty acyl-Coenzyme A reductases with different substrate specificities and tissue distributions. J. Biol. Chem. 279, 37789–37797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Honsho M., Asaoku S., Fujiki Y. (2010) Posttranslational regulation of fatty acyl-CoA reductase 1, Far1, controls ether glycerophospholipid synthesis. J. Biol. Chem. 285, 8537–8542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tsukamoto T., Yokota S., Fujiki Y. (1990) Isolation and characterization of Chinese hamster ovary cell mutants defective in assembly of peroxisomes. J. Cell Biol. 110, 651–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ghaedi K., Tamura S., Okumoto K., Matsuzono Y., Fujiki Y. (2000) The peroxin Pex3p initiates membrane assembly in peroxisome biogenesis. Mol. Biol. Cell 11, 2085–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Matsuzono Y., Kinoshita N., Tamura S., Shimozawa N., Hamasaki M., Ghaedi K., Wanders R. J., Suzuki Y., Kondo N., Fujiki Y. (1999) Human PEX19. cDNA cloning by functional complementation, mutation analysis in a patient with Zellweger syndrome and potential role in peroxisomal membrane assembly. Proc. Natl. Acad. Sci. U.S.A. 96, 2116–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mukai S., Ghaedi K., Fujiki Y. (2002) Intracellular localization, function, and dysfunction of the peroxisome-targeting signal type 2 receptor, Pex7p, in mammalian cells. J. Biol. Chem. 277, 9548–9561 [DOI] [PubMed] [Google Scholar]
- 16. Otera H., Okumoto K., Tateishi K., Ikoma Y., Matsuda E., Nishimura M., Tsukamoto T., Osumi T., Ohashi K., Higuchi O., Fujiki Y. (1998) Peroxisome targeting signal type 1 (PTS1) receptor is involved in import of both PTS1 and PTS2. Studies with PEX5-defective CHO cell mutants. Mol. Cell. Biol. 18, 388–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Miyata N., Fujiki Y. (2005) Shuttling mechanism of peroxisome targeting signal type 1 receptor, Pex5. ATP-independent import and ATP-dependent export. Mol. Cell. Biol. 25, 10822–10832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kawajiri K., Harano T., Omura T. (1977) Biogenesis of the mitochondrial matrix enzyme, glutamate dehydrogenase, in rat liver cells. I. Subcellular localization, biosynthesis, and intracellular translocation of glutamate dehydrogenase. J. Biochem. 82, 1403–1416 [DOI] [PubMed] [Google Scholar]
- 19. Honsho M., Yagita Y., Kinoshita N., Fujiki Y. (2008) Isolation and characterization of mutant animal cell line defective in alkyl-dihydroxyacetonephosphate synthase: Localization and transport of plasmalogens to post-Golgi compartments. Biochim. Biophys. Acta 1783, 1857–1865 [DOI] [PubMed] [Google Scholar]
- 20. Okumoto K., Bogaki A., Tateishi K., Tsukamoto T., Osumi T., Shimozawa N., Suzuki Y., Orii T., Fujiki Y. (1997) Isolation and characterization of peroxisome-deficient Chinese hamster ovary cell mutants representing human complementation group III. Exp. Cell Res. 233, 11–20 [DOI] [PubMed] [Google Scholar]
- 21. Honsho M., Tamura S., Shimozawa N., Suzuki Y., Kondo N., Fujiki Y. (1998) Mutation in PEX16 is causal in the peroxisome-deficient Zellweger syndrome of complementation group D. Am. J. Hum. Genet. 63, 1622–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Honsho M., Hiroshige T., Fujiki Y. (2002) The membrane biogenesis peroxin Pex16p. Topogenesis and functional roles in peroxisomal membrane assembly. J. Biol. Chem. 277, 44513–44524 [DOI] [PubMed] [Google Scholar]
- 23. Honsho M., Fujiki Y. (2001) Topogenesis of peroxisomal membrane protein requires a short, positively charged intervening-loop sequence and flanking hydrophobic segments: Study using human membrane protein PMP34. J. Biol. Chem. 276, 9375–9382 [DOI] [PubMed] [Google Scholar]
- 24. Kinoshita N., Ghaedi K., Shimozawa N., Wanders R. J., Matsuzono Y., Imanaka T., Okumoto K., Suzuki Y., Kondo N., Fujiki Y. (1998) Newly identified Chinese hamster ovary cell mutants are defective in biogenesis of peroxisomal membrane vesicles (peroxisomal ghosts), representing a novel complementation group in mammals. J. Biol. Chem. 273, 24122–24130 [DOI] [PubMed] [Google Scholar]
- 25. Motley A., Hettema E., Distel B., Tabak H. (1994) Differential protein import deficiencies in human peroxisome assembly disorders. J. Cell Biol. 125, 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Okumoto K., Fujiki Y. (1997) PEX12 encodes an integral membrane protein of peroxisomes. Nat. Genet. 17, 265–266 [DOI] [PubMed] [Google Scholar]
- 27. Bishop J. E., Hajra A. K. (1981) Mechanism and specificity of formation of long chain alcohols by developing rat brain. J. Biol. Chem. 256, 9542–9550 [PubMed] [Google Scholar]
- 28. Halbach A., Landgraf C., Lorenzen S., Rosenkranz K., Volkmer-Engert R., Erdmann R., Rottensteiner H. (2006) Targeting of the tail-anchored peroxisomal membrane proteins PEX26 and PEX15 occurs through C-terminal PEX19-binding sites. J. Cell Sci. 119, 2508–2517 [DOI] [PubMed] [Google Scholar]
- 29. Welsh C. J., Robinson M., Warne T. R., Pierce J.H., Yeh G.C., Phang J.M. (1994) Accumulation of fatty alcohol in MCF-7 breast cancer cells. Arch. Biochem. Biophys. 315, 41–47 [DOI] [PubMed] [Google Scholar]
- 30. Burdett K., Larkins L. K., Das A. K., Hajra A. K. (1991) Peroxisomal localization of acyl-coenzyme A reductase (long chain alcohol forming) in guinea pig intestine mucosal cells. J. Biol. Chem. 266, 12201–12206 [PubMed] [Google Scholar]
- 31. Fujiki Y., Rachubinski R. A., Lazarow P. B. (1984) Synthesis of a major integral membrane polypeptide of rat liver peroxisomes on free polysomes. Proc. Natl. Acad. Sci. U.S.A. 81, 7127–7131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kobayashi S., Tanaka A., Fujiki Y. (2007) Fis1, DLP1, and Pex11p coordinately regulate peroxisome morphogenesis. Exp. Cell Res. 313, 1675–1686 [DOI] [PubMed] [Google Scholar]
- 33. Delille H. K., Schrader M. (2008) Targeting of hFis1 to peroxisomes is mediated by Pex19p. J. Biol. Chem. 283, 31107–31115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gandre-Babbe S., van der Bliek A. M. (2008) The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell 19, 2402–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Halbach A., Lorenzen S., Landgraf C., Volkmer-Engert R., Erdmann R., Rottensteiner H. (2005) Function of the PEX19-binding site of human adrenoleukodystrophy protein as targeting motif in man and yeast. PMP targeting is evolutionarily conserved. J. Biol. Chem. 280, 21176–21182 [DOI] [PubMed] [Google Scholar]
- 36. Yagita Y., Hiromasa T., Fujiki Y. (2013) Tail-anchored PEX26 targets peroxisomes via a PEX19-dependent and TRC40-independent class I pathway. J. Cell Biol. 200, 651–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu D., Nagan N., Just W. W., Rodemer C., Thai T. P., Zoeller R. A. (2005) Role of dihydroxyacetonephosphate acyltransferase in the biosynthesis of plasmalogens and nonether glycerolipids. J. Lipid Res. 46, 727–735 [DOI] [PubMed] [Google Scholar]
- 38. Nishimukai M., Wakisaka T., Hara H. (2003) Ingestion of plasmalogen markedly increased plasmalogen levels of blood plasma in rats. Lipids 38, 1227–1235 [DOI] [PubMed] [Google Scholar]
- 39. Nagan N., Hajra A. K., Das A. K., Moser H. W., Moser A., Lazarow P., Purdue P. E., Zoeller R. A. (1997) A fibroblast cell line defective in alkyl-dihydroxyacetone phosphate synthase. A novel defect in plasmalogen biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 94, 4475–4480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nagan N., Hajra A. K., Larkins L. K., Lazarow P., Purdue P. E., Rizzo W. B., Zoeller R. A. (1998) Isolation of a Chinese hamster fibroblast variant defective in dihydroxyacetonephosphate acyltransferase activity and plasmalogen biosynthesis. Use of a novel two-step selection protocol. Biochem. J. 332, 273–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brites P., Ferreira A. S., da Silva T. F., Sousa V. F., Malheiro A. R., Duran M., Waterham H. R., Baes M., Wanders R. J. (2011) Alkyl-glycerol rescues plasmalogen levels and pathology of ether-phospholipid deficient mice. PLoS ONE 6, e28539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Miura S., Kasuya-Arai I., Mori H., Miyazawa S., Osumi T., Hashimoto T., Fujiki Y. (1992) Carboxyl-terminal consensus Ser-Lys-Leu-related tripeptide of peroxisomal proteins functions in vitro as a minimal peroxisome-targeting signal. J. Biol. Chem. 267, 14405–14411 [PubMed] [Google Scholar]
- 43. Matsuzaki T., Fujiki Y. (2008) The peroxisomal membrane-protein import receptor Pex3p is directly transported to peroxisomes by a novel Pex19p- and Pex16p-dependent pathway. J. Cell Biol. 183, 1275–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]