Background: Human and malaria orotate phosphoribosyltransferases (OPRTs) in the de novo pyrimidine biosynthesis pathway are characterized by ribocation transition states with fully dissociated leaving orotate groups.
Results: Transition state analogues with varied ribocation substitutions, different linkers, and distinct leaving groups exhibited nanomolar binding affinities for the OPRT enzymes.
Conclusion: Components crucial for inhibitor binding to the OPRT active sites have been identified, and powerful inhibitors were characterized.
Significance: Transition state analogues of OPRTs provide new insights into the nature of potential antimalarials and anticancer agents.
Keywords: Biosynthesis, Enzyme Catalysis, Malaria, Plasmodium, Pyrimidine, Inhibitor Design, N-Ribosyltransferases, Orotidine 5′-Monophosphate
Abstract
The survival and proliferation of Plasmodium falciparum parasites and human cancer cells require de novo pyrimidine synthesis to supply RNA and DNA precursors. Orotate phosphoribosyltransferase (OPRT) is an indispensible component in this metabolic pathway and is a target for antimalarials and antitumor drugs. P. falciparum (Pf) and Homo sapiens (Hs) OPRTs are characterized by highly dissociative transition states with ribocation character. On the basis of the geometrical and electrostatic features of the PfOPRT and HsOPRT transition states, analogues were designed, synthesized, and tested as inhibitors. Iminoribitol mimics of the ribocation transition state in linkage to pyrimidine mimics using methylene or ethylene linkers gave dissociation constants (Kd) as low as 80 nm. Inhibitors with pyrrolidine groups as ribocation mimics displayed slightly weaker binding affinities for OPRTs. Interestingly, p-nitrophenyl riboside 5′-phosphate bound to OPRTs with Kd values near 40 nm. Analogues designed with a C5-pyrimidine carbon–carbon bond to ribocation mimics gave Kd values in the range of 80–500 nm. Acyclic inhibitors with achiral serinol groups as the ribocation mimics also displayed nanomolar inhibition against OPRTs. In comparison with the nucleoside derivatives, inhibition constants of their corresponding 5′-phosphorylated transition state analogues are largely unchanged, an unusual property for a nucleotide-binding site. In silico docking of the best inhibitor into the HsOPRT active site supported an extensive hydrogen bond network associated with the tight binding affinity. These OPRT transition state analogues identify crucial components of potent inhibitors targeting OPRT enzymes. Despite their tight binding to the targets, the inhibitors did not kill cultured P. falciparum.
Introduction
Malaria kills nearly 1 million people each year (1). Plasmodium falciparum accounts for >95% of the fatal cases. Due to the lack of an effective vaccine and emerging resistance of P. falciparum parasites to current first-line drugs, novel chemotherapeutic agents are in demand to combat drug-resistant strains. P. falciparum parasites require robust supplies of nucleic acid precursors for growth. Because P. falciparum parasites do not encode enzymes for pyrimidine salvage, synthesis of pyrimidine nucleotides in parasites relies on the de novo pathway (2, 3). Therefore, pyrimidine nucleotide synthesis is a target for novel antimalarial drugs.
In human cancer cells, de novo pyrimidine synthesis plays a similar role in providing DNA and RNA precursors for maintaining rapid proliferation (4). Inhibitors targeting components within the de novo pyrimidine synthesis have validated this pathway, as they arrest the growth of rapidly dividing cells by restricting the pyrimidine supply (5–7). In addition, inhibitors acting on this pathway have been successfully applied to the treatment of malignant neoplastic and autoimmune diseases (8–12).
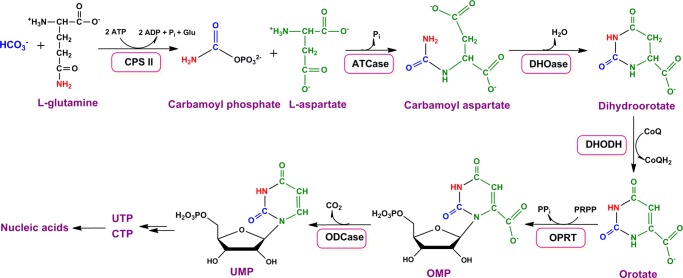
Orotate phosphoribosyltransferase (OPRT)2 is the fifth enzyme in the pyrimidine de novo synthesis pathway and catalyzes the formation of orotidine 5′-monophosphate (OMP), which is subsequently converted to UMP by OMP decarboxylase (Fig. 1). In most prokaryotes and lower eukaryotes, OPRT exists as a monofunctional protein, but OPRT from eukaryotes is encoded by a bifunctional protein fused to OMP decarboxylase and named UMP synthase (3, 13–15).
FIGURE 1.
De novo pyrimidine synthesis. CPS II, carbamoyl phosphate synthetase II; ATCase, aspartate carbamoyltransferase; DHOase, dihydroorotase; DHODH, dihydroorotate dehydrogenase; ODCase, OMP decarboxylase; PRPP, 5′-phospho-d-ribosyl-1-pyrophosphate.
Previous OPRT inhibition studies reported that pyrazofurin, a C-riboside antibiotic, inhibits P. falciparum OPRT (PfOPRT) activity and shows antimalarial activities with submicromolar IC50 values (16). Similarly, 5′-substituted orotate analogues bind to PfOPRT and block the growth of parasites with IC50 values at micromolar levels (16–18). Substituted pyridines have also shown submicromolar inhibition against the OPRT from Mycobacterium tuberculosis (19). In addition, 5-selenated uridine derivatives exhibit submicromolar inhibition against PfOPRT and Homo sapiens OPRT (HsOPRT) (20).
Transition state analogues bind more tightly to their cognate enzymes than substrates (21–24). Such tight binding affinity is usually combined with high specificity, and transition state analogues hold promise for the development of drugs. In practice, transition state analogues with binding constants as low as 10−14 m have been achieved (25–27). Moreover, transition state analogues for other N-ribosyltransferases have been designed on the basis of experimentally solved transition state structures to give picomolar inhibitors. Some of these are currently in clinical trials for the treatment of leukemia and gout (28–33).
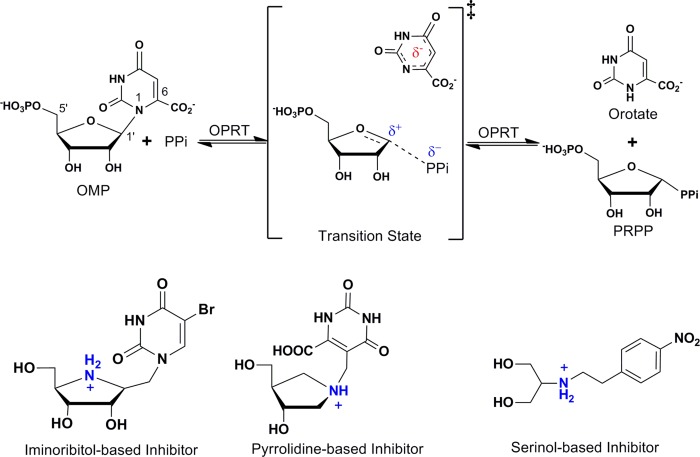
Enzymatic transition state structure reflects the instantaneous geometrical and electrostatic characteristics of reactants at the transition state and provides the basis for the design of transition state analogues. The combination of experimental kinetic isotope effect measurements and computational chemistry permits access to information about transition state structure. Experimental kinetic isotope effects for yeast OPRT support an SN1 transition state with partially dissociative orotate and ribooxacarbenium ion character (34). A proposed transition state analogue amidrazone displays tight binding affinity for the yeast enzyme (35). PfOPRT and HsOPRT transition state structures are characterized by well developed ribocations, fully dissociated dianionic orotate, and weak participation of the pyrophosphate nucleophiles (Fig. 2) (36, 37). On the basis of mechanistic studies, leaving group activation has been established to be the major catalytic force in OPRT catalysis (38).
FIGURE 2.
OPRT-catalyzed OMP pyrophosphorolysis, ribocation transition state, and transition state mimics. OPRT transition states are characterized by fully dissociated dianionic orotate, an oxacarbenium ion, and weak nucleophile participation. Examples of transition state analogues designed with iminoribitol or pyrrolidine groups as mimics of the oxacarbenium ion and methylene or ethylene linkers to resemble extended C1–N1′ bond distance are shown at the bottom. PRPP, 5′-phospho-d-ribosyl-1-pyrophosphate.
Here, we designed and synthesized analogues that resemble the geometric and electrostatic features of the PfOPRT and HsOPRT transition states and tested them as inhibitors. Inhibition studies showed that transition state analogues with several distinct chemical scaffolds display dissociation constants (Kd) at nanomolar levels. Specifically, cyclic iminoribitol, hydroxypyrrolidine, and acyclic serinol groups were exploited as ribocation mimics. At physiological pH, the nitrogen atoms in these groups are protonated, mimicking the ribocationic characteristic of the transition states. A methylene or ethylene linker between the ribocation mimic and orotate derivative resembles the extended N-ribosidic bond distance characteristic of OPRT transition states. Orotate derivatives were explored to approximate the electrostatic potential at the transition state and to interact with amino acids interacting with the pyrimidine leaving group. In silico docking of the 40 nm inhibitor into the OPRT active site revealed an extensive interaction network surrounding the bound inhibitor. The binding model suggests how the transition state analogues capture the binding energy. These analogues reveal essential chemical features for favorable binding energy at the OPRT active sites.
EXPERIMENTAL PROCEDURES
Reagents
PfOPRT and HsOPRT were expressed in Escherichia coli cells and purified as described previously (37). Specifically, a stop codon was introduced at Leu-246 of human UMP synthase for expression of the HsOPRT domain alone. All other reagents were purchased from readily available commercial sources and used without further purification.
Inhibitors
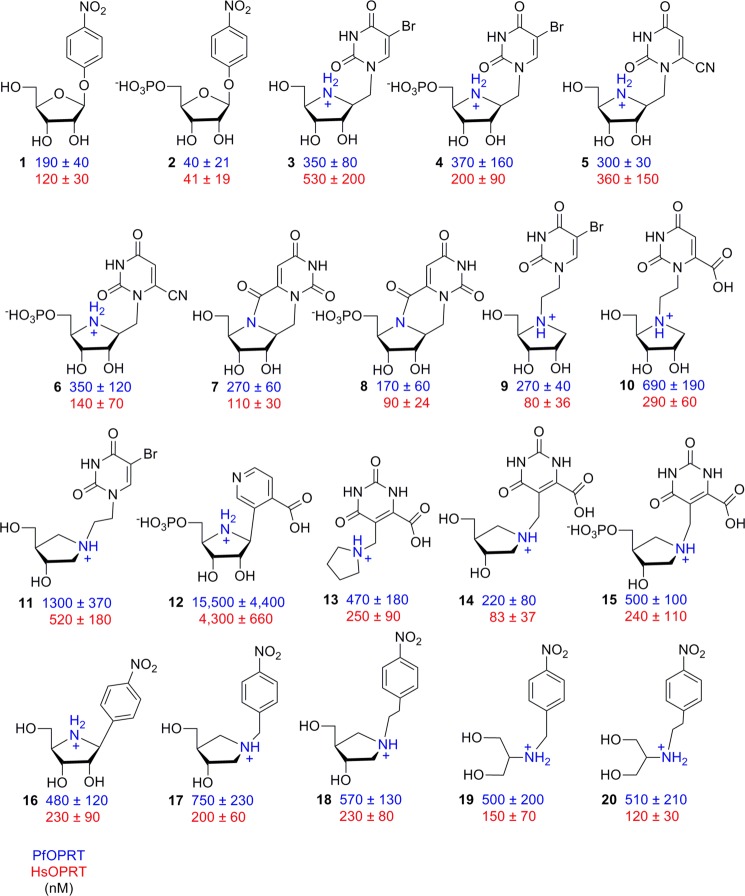
Chemical synthesis of the inhibitors (Fig. 3) was achieved by modifications of published methods. Full details of synthetic steps and chemical characterization of the inhibitor products is described in the supplemental data. The synthetic chemistry approach incorporated and extended the methods published previously to achieve the chemical synthesis of transition state analogues for N-ribosyltransferases (39–44). Generally, p-nitrophenyl riboside inhibitors (1 and 2) were synthesized according to a previous report (45). Inhibitor 16 was synthesized using methods previously described (46). The iminoribitol-, pyrrolidine-, and serinol-based inhibitors were synthesized by an extension of published methods for the synthesis of immucillin-H, 4′-deaza-1′-aza-2′-deoxy-1′-(9-methylene)-immucillin-H, and serinol-N-(9-methylene)-immucillin-H, respectively (see supplemental data) (40, 47–49). The identities of all inhibitors were confirmed by high-resolution mass spectrometry. Their purity was established by 1H and 13C NMR spectrometry.
FIGURE 3.
Inhibition constants of transition state analogues for PfOPRT (blue) and HsOPRT (red). The values are inhibition constants (Ki) treated as competitive against OMP.
Inhibition Assay
Inhibition constants (Ki, equivalent to Kd for competitive inhibitors) were measured at 25 °C by adding purified OPRT enzymes (25 nm for PfOPRT and 100 nm for HsOPRT) to reaction mixtures with 50 mm Tris-HCl (pH 8.0), 5 mm MgCl2, 1 mm PPi, 0.5 mm OMP, and inhibitor at varied concentrations. In all cases, the inhibitor concentration was at least 10 times the enzyme concentration. Reaction rates were monitored through the absorbance increase at 295 nm to give an extinction coefficient of 3.67 mm−1 cm−1 (20). Slow-onset inhibition patterns were not observed for the inhibitors described here in direct competition assays or in enzyme inhibitor preincubation studies. The Ki values were determined by fitting the initial reaction rate and inhibitor concentrations to the expression for competitive inhibition: (V′0/V0) = (Km + [S])/(Km + [S] + (Km[I]/Ki)), where V′0 is the initial reaction rate in the presence of inhibitor, V0 is the initial reaction rate in the absence of inhibitor, [S] is the substrate concentration, and [I] is the inhibitor concentration. The Km values of OMP were determined to be 3.7 μm for PfOPRT and 1.5 μm for HsOPRT under these assay conditions.
In Silico Docking of Inhibitor into the OPRT Active Site
Inhibitors 2 (p-nitrophenyl β-d-ribose 5′-phosphate), 16, and 17 were virtually docked into the HsOPRT active site (Protein Data Bank entry 2WNS, chains A and B) using the SwissDock web server (50, 51).
Activity of OPRT Inhibitors against Cultured P. falciparum
Cell growth inhibition of the compounds against P. falciparum D2 (chloroquine- and mefloquine-resistant) was conducted in cultured human erythrocytes as described previously (52). Stock solutions of each test compound were made in sterile water and diluted into the culture medium. Ten point dilutions were used to test the dose response at concentrations ranging from 200 to 0.04 μm. BCX4945, an inhibitor of human and Plasmodium purine nucleoside phosphorylases, was used as a control.
RESULTS AND DISCUSSION
Nitrophenyl Riboside Inhibitors
p-Nitrophenyl β-d-ribose 5′-phosphate is a mechanistic tool to distinguish relative forces for leaving group activation and for ribocation formation (45). Enzymes achieving catalysis through formation of a ribocation show the p-nitrophenyl to be an excellent leaving group and therefore give rapid rates of chemical reaction. In contrast, if enzymatic contacts to the pyrimidine leaving group dominate catalytic forces, no significant catalysis is expected, as the p-nitrophenyl group does not have the requisite hydrogen bond acceptor and donor sites required for leaving group activation of a pyrimidine group.
Substrate specificity studies indicate that PfOPRT and HsOPRT do not recognize nitrophenyl ribosides as substrates. The result establishes leaving group activation as a major catalytic force in the OPRT-catalyzed reaction, providing that p-nitrophenyl β-d-ribose 5′-phosphate can be shown to bind to the enzyme.
Despite its lack of substrate activity, p-nitrophenyl β-d-ribose 5′-phosphate bound to both PfOPRT and HsOPRT with Ki values near 40 nm (Fig. 3, 2). The role of the 5′-phosphate in inhibitor binding was evaluated by comparing inhibition of p-nitrophenyl β-d-ribose with that of p-nitrophenyl β-d-ribose 5′-phosphate. In vitro inhibition assays showed that p-nitrophenyl β-d-ribose (Fig. 3, 1) has Ki values of 190 and 120 nm for PfOPRT and HsOPRT, respectively. Thus, the 5′-phosphate causes only 3–4-fold increased binding affinities, indicating a relatively weak role of the phosphate region for inhibitor binding. Compared with the OMP substrate, p-nitrophenyl β-d-ribose 5′-phosphate bound more tightly to OPRT with Km/Ki values of 93 and 38 for PfOPRT and HsOPRT, respectively.
Methylene-bridged Iminoribitol Inhibitors
Transition state analogues with iminoribitol as a ribocation mimic and a methylene bridge to a 5-bromouracil analogue showed inhibitory activities against OPRT at nanomolar concentrations (Fig. 3, 3 and 4). The nucleoside analogue 3 displayed a modest preference for PfOPRT with Ki values of 350 and 530 nm for PfOPRT and HsOPRT, respectively. We tested the effect of the 5′-phosphate in the conversion of 3 to 4 to determine whether the nucleotide effect was the same with different pyrimidine analogue substituents. We found, at most, only a 2.5-fold increased affinity in response to the 5′-phosphate. This was similar to the response with 1 and 2 and confirms that 5′-phosphorylation has an unusually weak effect on inhibitor binding for PfOPRT and HsOPRT transition state analogues.
Substitution of the 5-bromouracil group by a 6-cyanouracil (5) gave Ki values similar to those obtained with 3. Conversion of 5 to its 5′-phosphate (6) gave a 3-fold increased affinity for HsOPRT but had little effect on PfOPRT (Fig. 3). An internal carboxamide between the pyrimidine ring and the iminoribitol nitrogen (7) and its 5′-phosphate (8) gave stereochemically locked inhibitors with Ki values in the range of 90–270 nm (Fig. 3).
It is significant that even without the 5′-phosphate group, 3, 5, and 7 are nanomolar inhibitors. The 5′-phosphate group increased the binding of 1 by a factor of 3–4 but had a more dramatic effect on substrate activity, where the presence of the 5′-phosphate decreased the Km for OMP relative to orotidine by factors of 26 and 57 for PfOPRT and HsOPRT, respectively. However, conversions of 5 and 7 to their 5′-phosphorylated forms had even more modest effects on binding affinities. The modest effects of binding affinity as a result of 5′-phosphorylation is an advantage in targeting PfOPRT and HsOPRT because drug delivery of phosphate monoesters is challenging, but nucleoside analogues are usually found to be cell-permeable as a result of membrane transporters (43).
Ethylene-bridged Iminoribitol Inhibitors
Transition state analogues with iminoribitol mimics of the ribocation and ethylene bridges at N4′ to 5-bromouracil (9) or orotate (10) showed Ki values of 80–690 nm, a range similar to that for methylene-bridged iminoribitol inhibitors (Fig. 3). Among these iminoribitol inhibitors, the interaction of 9 with HsOPRT was the tightest one at 80 nm. By comparison, 10 with PfOPRT displayed the lowest affinity with a Ki of 690 nm. We tested a geometry in which an ethylene bridge to N1′ of a hydroxypyrrolidine might better capture the position of the ribocation (11). The 5-bromouracil substituent in this construct gave dissociation constants of 1300 and 520 nm for PfOPRT and HsOPRT, respectively (Fig. 3). Comparison of the three 5-bromouracil analogues with three distinct ribocation analogues (3, 9, and 11) was revealing because they support similar Ki values with different ribocation chemical scaffolds. The result supports the original hypothesis that the OPRTs gain most of their transition state interaction energy from leaving group interactions. Even the p-nitrophenyl riboside (1), without a ribocation mimic, showed similar binding affinity, suggesting a preference for the p-nitrophenyl group, and we further explored that interaction (see below).
The strong dependence of affinity for a pyrimidine analogue led us to explore other motifs. The carboxypyridine linked to 5′-phosphorylated iminoribitol (12) can be compared with 4, 6, and 8 to establish the interaction energy of the carboxypyridine group. 4, 6, and 8 bound better than 12 by at least an order of magnitude; hence, the carboxypyridine does not provide a favored interaction.
Pyrrolidine-based C-Linked Orotate Analogues
Linking the 5-position of an orotate analogue to pyrrolidine mimics of the ribocation theoretically places the pyrimidine keto and carboxyl groups in appropriate positions for hydrogen bonding in the catalytic site and the cation in an appropriate geometry as a ribocation mimic (13, 14, and 15). A simple unsubstituted pyrrolidine cation (13) gave Ki values of 250–470 nm. Incorporation of two stereospecific hydroxyl groups was intended to capture some of the ribocation binding energy in 14 and gave improved Ki values of 83–220 nm. Because 5′-phosphorylation increased binding energy in other constructs, phosphorylation of the primary hydroxyl group was accomplished but, in this geometry, decreased the affinity (15). The decreased affinities for 13–15 may be due to the lack of a 2′-hydroxyl group, which promotes binding in this series of compounds. At the OPRT catalytic sites, the 2′-hydroxyl group of 5′-phospho-d-ribosyl-1-pyrophosphate is involved in interactions with an Mg2+ ion and forms hydrogen bonds with the side chain of a conserved aspartate and with an ordered water molecule (13). The interactions associated with the 2′-hydroxyl group are important for binding and also for catalysis and ribocation stabilization. However, a previous inhibition study of yeast OPRT with amidrazone analogues indicated only a modest energetic contribution to binding from the 2′- and 3′-hydroxyl groups, suggesting that binding energy contributed by the oxacarbenium ion and orotate mimics dominates in this series of compounds (34). Nonetheless, for pyrrolidine inhibitors, the decreased binding affinities relative to some of the related iminoribitol-based inhibitors imply the importance of the ribosyl hydroxyl groups for inhibitor binding. The similar nanomolar binding affinities for the methylene- and ethylene-bridged inhibitors described above reveal that these linkers could be exploited to resemble the extended N-glycosidic bond distance at the OPRT transition states.
Considering inhibitors with nanomolar Ki values despite large variation in the chemical structures, all share common features of ribocation mimics, orotate mimics, or p-nitrophenyl leaving groups. The relatively tight binding of ribocation mimics such as 14 with geometry distinct from that of the ribocation transition state suggested additional exploration of related structures. Finally, the favorable Ki values for 1 and 2, where the p-nitrophenyl group is presumed to be a major contributor to binding energy, led us to examine a variety of ribocation mimics with a constant being the p-nitrophenyl group to replace the pyrimidine.
Ribocation Mimics Bearing p-Nitrophenyl Leaving Groups
Iminoribitol directly linked to a p-nitrophenyl group (16) was a 2-fold weaker inhibitor than the p-nitrophenyl riboside (1). To test if this is due to insufficient distance between the ribocation and the nitrophenyl group, 17 and 18 with methylene and ethylene bridges between the groups were tested. There was no significant improvement. Serinol-based inhibitors with p-nitrophenyl groups (19 and 20) were also tested for inhibitory activities against PfOPRT and HsOPRT (Fig. 3). Inhibitors 19 and 20 displayed similar nanomolar binding affinities for OPRT enzymes, supporting the incorporation of both linkers for inhibitor design and that the p-nitrophenyl leaving groups capture pyrimidine group interactions. These acyclic inhibitors provide a new strategy for OPRT-targeted drug development because 19 and 20 are achiral molecules.
Inhibitor Docking into the HsOPRT Active Site
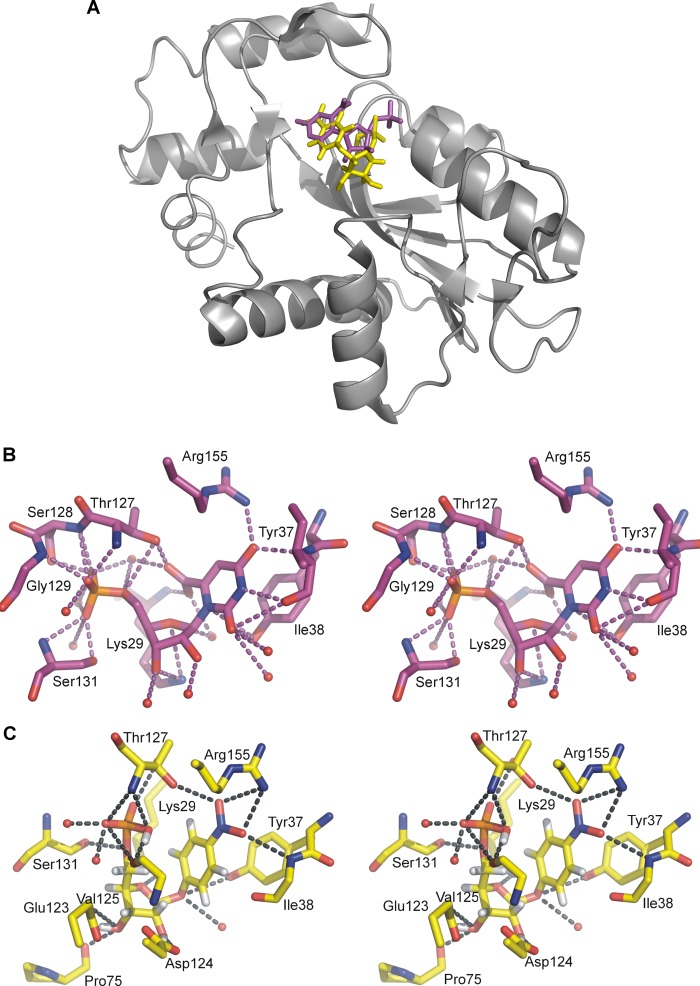
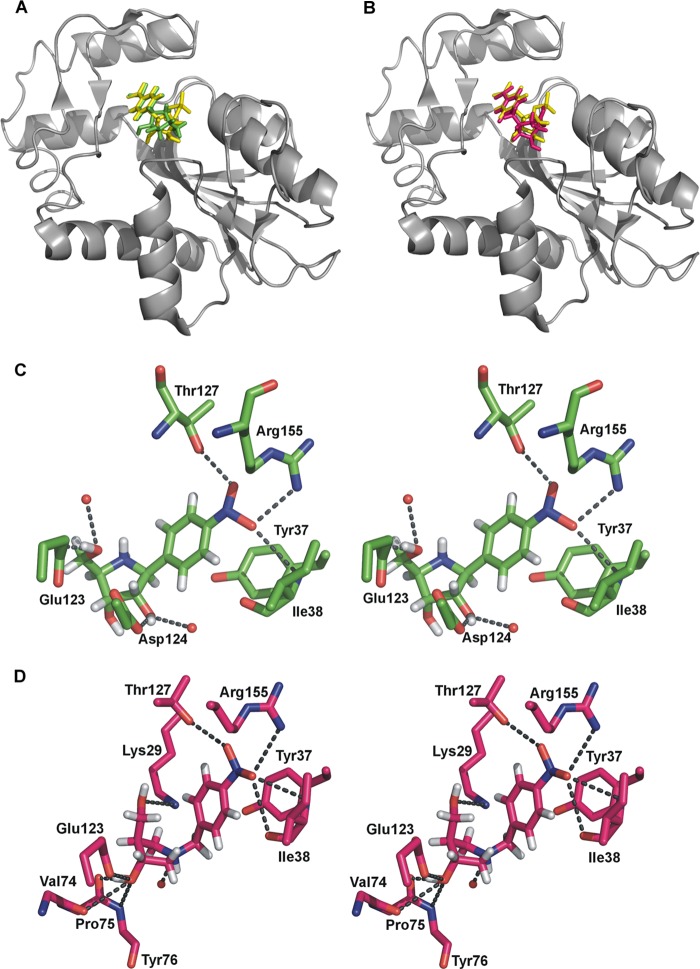
To predict the interactions between OPRT active site residues and the transition state analogue, inhibitor 2 with a Ki of 40 nm was docked into the HsOPRT active site using the SwissDock web server and the previously solved HsOPRT crystal structure in complex with OMP (Protein Data Bank entry 2WNS, chains A and B). The binding modes of the inhibitor were predicted by sampling different dihedral angle space to optimize binding interactions in the enzyme. The virtual docking result indicated that eight conformers of inhibitor 2 could be docked at the HsOPRT active site with the predicted binding free energy varying from −10.44 to −7.73 kcal/mol. On the basis of the predicted most favorable binding free energy of −10.44 kcal/mol, inhibitor 2 has a calculated binding affinity of 22 nm for HsOPRT, in good agreement with the experimental Ki of 41 nm. Compared with the OMP geometry at the HsOPRT active site, 2 exhibits a more restrained conformation (Fig. 4A). Moreover, due to the extended O-glycosidic bond, the ribosyl moiety of 2 adopts a geometry with 2′- and 3′-hydroxyl groups in hydrogen bonds to active site residues, whereas the p-nitrophenyl leaving group of 2 is predicted to be at a position similar to the orotate leaving group.
FIGURE 4.
Inhibitor 2 docking into the HsOPRT active site. A, overall structure of HsOPRT in complex with OMP (purple) or docked inhibitor 2 (yellow). B, stereoview of the HsOPRT active site with residues and water molecules in hydrogen bonds to OMP. C, stereoview of the HsOPRT active site with residues and water molecules in hydrogen bonds to docked inhibitor 2.
Specifically, both the orotate and p-nitrophenyl leaving groups form hydrogen bonds with the side chain of Arg-155 and the main chain of Ile-38 (Fig. 4, B and C). The side chain of Thr-127 is in a hydrogen bond with the 6-carboxyl group of orotate from OMP, but it is predicted to interact with the 4-nitro group upon binding to inhibitor 2. In both structures, the side chain of Tyr-37 is parallel to the leaving group and is predicted to form a π-stacking interaction.
The Tyr-37 hydroxyl group is near N1 of OMP. With inhibitor 2, the hydroxyl of Tyr-37 is in hydrogen bond distance of the O-glycoside bond and a water molecule. Significant differences between bound OMP and 2 occur at the ribosyl group, where O4′ of 2 is closer to the side chain of Lys-29 compared with O4′ OMP. In addition, the 2′- and 3′-hydroxyl groups of 2 form multiple interactions with the side chains of Glu-123 and Asp-124 and the main chain of Pro-75, interactions that are not observed for bound OMP. The 5′-phosphate groups in both structures are involved in multiple hydrogen bond interactions including several water molecules.
On the basis of the docked structure of HsOPRT with inhibitor 2, the inhibitor conformation captures binding energy through an extensive interaction network with surrounding residues and water molecules. New interactions relative to the active site with OMP include interactions with the 2′- and 3′-hydroxyl groups. These interactions collectively contribute to binding of 2 to the OPRT enzyme and may explain the observed nanomolar binding affinity.
The extended O-glycoside bond of 2 resembles the known OPRT transition state geometry (37). The docking result indicates that by mimicking the OPRT transition state geometry, inhibitor 2 captures properly positioned hydrogen bond donors and acceptors in the ribocation mimics and leaving group. When combined with the optimized bond distance found in the transition state, improved inhibitor binding affinities are observed. Exhaustive attempts to crystallize the OPRTs described here were not successful, and additional structure-based studies would be desirable to characterize these ribocation mimics of the transition state.
To further explore components of 2 that are important for tight binding, 16 and 17 were docked into HsOPRT to give predicted binding free energy varying from −8.89 to −6.12 kcal/mol. The predicted binding free energies for 16 and 17 are less than for 2 and are consistent with the observed weaker binding of 16 and 17. The p-nitrophenyl groups of 2, 16, and 17 exhibit similar docked conformations, whereas their ribocation mimics are predicted to be in different conformations (Fig. 5, A and B). The p-nitrophenyl groups are predicted to be in hydrogen bonds with the side chains of Arg-155 and Thr-127 and the main chain of Ile-38 and in parallel to the side chain of Tyr-37 (Fig. 5, C and D).
FIGURE 5.
Docking of 16 and 17 into the HsOPRT active site. A, overall structure of HsOPRT in complex with docked inhibitor 2 (yellow) or inhibitor 16 (green). B, overall structure of HsOPRT in complex with docked inhibitor 2 (yellow) or inhibitor 17 (pink). C, stereoview of the HsOPRT active site with residues and water molecules in hydrogen bonds to inhibitor 16. D, stereoview of the HsOPRT active site with residues and water molecules in hydrogen bonds to docked inhibitor 17.
Docking results place the ribocation mimics of 16 and 17 to form weaker hydrogen bond interactions with surrounding residues and water molecules in comparison with 2. Specifically, the 2′- and 5′-hydroxyl groups of 16 are in hydrogen bonds with the side chains of Asp-124 and Glu-123, respectively (Fig. 5, C and D). Unlike O4′ of 2 interacting with the side chain of Lys-29, N4′ of 16 is cationic and forms no interaction with surrounding residues. The lack of interactions to the ribocation mimics of 16 may result from the short distance between the ribocation mimic and the p-nitrophenyl group. Inhibitor 2, with its extended O-glycosidic bond, enables better positioning of ribocation mimics for extensive interactions with active site residues while maintaining p-nitrophenyl stabilization. The methylene linker of 17 permits the 3′-hydroxyl to form multiple hydrogen bonds interactions with the side chain of Glu-123 and the main chains of Val-74, Pro-75, and Tyr-76 (Fig. 5, C and D), but the lack of a 2′-hydroxyl group and the interactions with N1′ could explain its weaker binding affinity relative to 2.
Inhibitors Do Not Affect the Growth of P. falciparum in Culture
Compounds 1-3, 5-7, 9-14, and 17-20 were tested against the growth of P. falciparum parasites cultured in human erythrocytes. No growth inhibition was found at concentrations up to 100 μm. Thus, the compounds do not gain access to the OPRT compartment in cultured parasites, do not block pyrimidine synthesis, or are by-passed by pyrimidine sources from erythrocytes in this complex culture medium, although it contains no added pyrimidine source. The last option is considered unlikely, as 5-fluoroorotate inhibits P. falciparum growth under these conditions (18). As antimalarials must penetrate the erythrocyte and parasite plasma membranes to contact the target OPRT enzyme, the lack of cell growth inhibition could be due to permeability barriers. More detailed metabolic studies will be needed to distinguish these mechanisms. With several of the inhibitors demonstrating dissociation constants below 200 nm, the inhibitor concentration would be expected to cause >99% inhibition of OPRT if freely available to the enzyme. In preliminary studies, growth of several cultured mammalian cell lines was unaffected by these inhibitors up to 10 μm.
In conclusion, transition state analogues of PfOPRT and HsOPRT were generated to resemble the geometrical and electrostatic characteristics of the transition states solved earlier for these enzymes. Analogues characterized with varied ribocation substitutions, different linkers, and distinct leaving group mimics displayed nanomolar binding affinities for OPRT enzymes. Comparative analysis of the binding constants of these analogues revealed crucial components for the inhibitor binding at OPRT active sites. Protonated iminoribitol, pyrrolidine, and serinol groups as ribocation mimics promoted inhibitor binding. Methylene, ethylene, and O-ribosidic linkers resembling extended N-ribosidic bonds, a characteristic of the transition state, gave rise to increased binding affinity. Both orotate derivatives and p-nitrophenyl are good leaving groups to capture leaving group interaction forces. The binding affinity of these analogues is predicted to arise from an extensive interaction network at the OPRT active site. These transition state analogues provide new insights into the design of potential antimalarials and anticancer agents. However, problems of permeability, target access, and species specificity will need to be addressed in the context of these nanomolar inhibitors.
This work was supported, in whole or part, by National Institutes of Health Grants AI049512 and GM041916 and Training Grant GM007288. This work was also supported by New Zealand Foundation for Research Science and Technology Contract C08X0701.

This article contains supplemental data, Schemes 1–5, and additional references.
- OPRT
- orotate phosphoribosyltransferase
- OMP
- orotidine 5′-monophosphate
- PfOPRT
- P. falciparum OPRT
- HsOPRT
- H. sapiens OPRT.
REFERENCES
- 1. Guerin P. J., Olliaro P., Nosten F., Druilhe P., Laxminarayan R., Binka F., Kilama W. L., Ford N., White N. J. (2002) Malaria: current status of control, diagnosis, treatment, and a proposed agenda for research and development. Lancet Infect. Dis. 2, 564–573 [DOI] [PubMed] [Google Scholar]
- 2. Gero A. M., O'Sullivan W. J. (1990) Purines and pyrimidines in malarial parasites. Blood Cells 16, 467–484 [PubMed] [Google Scholar]
- 3. Jones M. E. (1980) Pyrimidine nucleotide biosynthesis in animals: genes, enzymes, and regulation of UMP biosynthesis. Annu. Rev. Biochem. 49, 253–279 [DOI] [PubMed] [Google Scholar]
- 4. Quéméneur L., Beloeil L., Michallet M. C., Angelov G., Tomkowiak M., Revillard J. P., Marvel J. (2004) Restriction of de novo nucleotide biosynthesis interferes with clonal expansion and differentiation into effector and memory CD8 T cells. J. Immunol. 173, 4945–4952 [DOI] [PubMed] [Google Scholar]
- 5. Krungkrai J., Krungkrai S. R., Phakanont K. (1992) Antimalarial activity of orotate analogs that inhibit dihydroorotase and dihydroorotate dehydrogenase. Biochem. Pharmacol. 43, 1295–1301 [DOI] [PubMed] [Google Scholar]
- 6. Phillips M. A., Gujjar R., Malmquist N. A., White J., El Mazouni F., Baldwin J., Rathod P. K. (2008) Triazolopyrimidine-based dihydroorotate dehydrogenase inhibitors with potent and selective activity against the malaria parasite Plasmodium falciparum. J. Med. Chem. 51, 3649–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Seymour K. K., Lyons S. D., Phillips L., Rieckmann K. H., Christopherson R. I. (1994) Cytotoxic effects of inhibitors of de novo pyrimidine biosynthesis upon Plasmodium falciparum. Biochemistry 33, 5268–5274 [DOI] [PubMed] [Google Scholar]
- 8. Allison A. C. (2000) Immunosuppressive drugs: the first 50 years and a glance forward. Immunopharmacology 47, 63–83 [DOI] [PubMed] [Google Scholar]
- 9. Christopherson R. I., Lyons S. D., Wilson P. K. (2002) Inhibitors of de novo nucleotide biosynthesis as drugs. Acc. Chem. Res. 35, 961–971 [DOI] [PubMed] [Google Scholar]
- 10. Chu E., Callender M. A., Farrell M. P., Schmitz J. C. (2003) Thymidylate synthase inhibitors as anticancer agents: from bench to bedside. Cancer Chemother. Pharmacol. 52, S80–S89 [DOI] [PubMed] [Google Scholar]
- 11. Herrmann M. L., Schleyerbach R., Kirschbaum B. J. (2000) Leflunomide: an immunomodulatory drug for the treatment of rheumatoid arthritis and other autoimmune diseases. Immunopharmacology 47, 273–289 [DOI] [PubMed] [Google Scholar]
- 12. Weber G. (2001) Ordered biochemical program of gene expression in cancer cells. Biochemistry 66, 1164–1173 [DOI] [PubMed] [Google Scholar]
- 13. González-Segura L., Witte J. F., McClard R. W., Hurley T. D. (2007) Ternary complex formation and induced asymmetry in orotate phosphoribosyltransferase. Biochemistry 46, 14075–14086 [DOI] [PubMed] [Google Scholar]
- 14. Suchi M., Mizuno H., Kawai Y., Tsuboi T., Sumi S., Okajima K., Hodgson M. E., Ogawa H., Wada Y. (1997) Molecular cloning of the human UMP synthase gene and characterization of point mutations in two hereditary orotic aciduria families. Am. J. Hum. Genet. 60, 525–539 [PMC free article] [PubMed] [Google Scholar]
- 15. Yablonski M. J., Pasek D. A., Han B. D., Jones M. E., Traut T. W. (1996) Intrinsic activity and stability of bifunctional human UMP synthase and its two separate catalytic domains, orotate phosphoribosyltransferase and orotidine-5′-phosphate decarboxylase. J. Biol. Chem. 271, 10704–10708 [DOI] [PubMed] [Google Scholar]
- 16. Queen S. A., Jagt D. L., Reyes P. (1990) In vitro susceptibilities of Plasmodium falciparum to compounds which inhibit nucleotide metabolism. Antimicrob. Agents Chemother. 34, 1393–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Krungkrai S. R., Aoki S., Palacpac N. M., Sato D., Mitamura T., Krungkrai J., Horii T. (2004) Human malaria parasite orotate phosphoribosyltransferase: functional expression, characterization of kinetic reaction mechanism and inhibition profile. Mol. Biochem. Parasitol. 134, 245–255 [DOI] [PubMed] [Google Scholar]
- 18. Rathod P. K., Khatri A., Hubbert T., Milhous W. K. (1989) Selective activity of 5-fluoroorotic acid against Plasmodium falciparum in vitro. Antimicrob. Agents Chemother. 33, 1090–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Breda A., Machado P., Rosado L. A., Souto A. A., Santos D. S., Basso L. A. (2012) Pyrimidin-2(1H)-ones based inhibitors of Mycobacterium tuberculosis orotate phosphoribosyltransferase. Eur. J. Med. Chem. 54, 113–122 [DOI] [PubMed] [Google Scholar]
- 20. Abdo M., Zhang Y., Schramm V. L., Knapp S. (2010) Electrophilic aromatic selenylation: new OPRT inhibitors. Org. Lett. 12, 2982–2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schramm V. L. (2003) Enzymatic transition state poise and transition state analogues. Acc. Chem. Res. 36, 588–596 [DOI] [PubMed] [Google Scholar]
- 22. Schramm V. L. (2005) Enzymatic transition states: thermodynamics, dynamics and analogue design. Arch. Biochem. Biophys. 433, 13–26 [DOI] [PubMed] [Google Scholar]
- 23. Schramm V. L. (2007) Enzymatic transition state theory and transition state analogue design. J. Biol. Chem. 282, 28297–28300 [DOI] [PubMed] [Google Scholar]
- 24. Wolfenden R., Snider M. J. (2001) The depth of chemical time and the power of enzymes as catalysts. Acc. Chem. Res. 34, 938–945 [DOI] [PubMed] [Google Scholar]
- 25. Schramm V. L. (2003) Transition state analogues for enzymes of nucleic acid metabolism. Nucleic Acids Res. Suppl. 3, 107–108 [DOI] [PubMed] [Google Scholar]
- 26. Singh V., Evans G. B., Lenz D. H., Mason J. M., Clinch K., Mee S., Painter G. F., Tyler P. C., Furneaux R. H., Lee J. E., Howell P. L., Schramm V. L. (2005) Femtomolar transition state analogue inhibitors of 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Escherichia coli. J. Biol. Chem. 280, 18265–18273 [DOI] [PubMed] [Google Scholar]
- 27. Wolfenden R. (1969) Transition state analogues for enzyme catalysis. Nature 223, 704–705 [DOI] [PubMed] [Google Scholar]
- 28. Bantia S., Parker C., Upshaw R., Cunningham A., Kotian P., Kilpatrick J. M., Morris P., Chand P., Babu Y. S. (2010) Potent orally bioavailable purine nucleoside phosphorylase inhibitor BCX-4208 induces apoptosis in B- and T-lymphocytes–a novel treatment approach for autoimmune diseases, organ transplantation and hematologic malignancies. Int. Immunopharmacol. 10, 784–790 [DOI] [PubMed] [Google Scholar]
- 29. Kicska G. A., Tyler P. C., Evans G. B., Furneaux R. H., Kim K., Schramm V. L. (2002) Transition state analogue inhibitors of purine nucleoside phosphorylase from Plasmodium falciparum. J. Biol. Chem. 277, 3219–3225 [DOI] [PubMed] [Google Scholar]
- 30. Kicska G. A., Tyler P. C., Evans G. B., Furneaux R. H., Schramm V. L., Kim K. (2002) Purine-less death in Plasmodium falciparum induced by immucillin-H, a transition state analogue of purine nucleoside phosphorylase. J. Biol. Chem. 277, 3226–3231 [DOI] [PubMed] [Google Scholar]
- 31. Lewandowicz A., Schramm V. L. (2004) Transition state analysis for human and Plasmodium falciparum purine nucleoside phosphorylases. Biochemistry 43, 1458–1468 [DOI] [PubMed] [Google Scholar]
- 32. Lewandowicz A., Shi W., Evans G. B., Tyler P. C., Furneaux R. H., Basso L. A., Santos D. S., Almo S. C., Schramm V. L. (2003) Over-the-barrier transition state analogues and crystal structure with Mycobacterium tuberculosis purine nucleoside phosphorylase. Biochemistry 42, 6057–6066 [DOI] [PubMed] [Google Scholar]
- 33. Lewandowicz A., Tyler P. C., Evans G. B., Furneaux R. H., Schramm V. L. (2003) Achieving the ultimate physiological goal in transition state analogue inhibitors for purine nucleoside phosphorylase. J. Biol. Chem. 278, 31465–31468 [DOI] [PubMed] [Google Scholar]
- 34. Goitein R. K., Chelsky D., Parsons S. M. (1978) Primary 14C and α secondary 3H substrate kinetic isotope effects for some phosphoribosyltransferases. J. Biol. Chem. 253, 2963–2971 [PubMed] [Google Scholar]
- 35. Witte J. F., Bray K. E., Thornburg C. K., McClard R. W. (2006) 'Irreversible' slow-onset inhibition of orotate phosphoribosyltransferase by an amidrazone phosphate transition-state mimic. Bioorg. Med. Chem. Lett. 16, 6112–6115 [DOI] [PubMed] [Google Scholar]
- 36. Zhang Y., Schramm V. L. (2010) Pyrophosphate interactions at the transition states of Plasmodium falciparum and human orotate phosphoribosyltransferases. J. Am. Chem. Soc. 132, 8787–8794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang Y., Luo M., Schramm V. L. (2009) Transition states of Plasmodium falciparum and human orotate phosphoribosyltransferases. J. Am. Chem. Soc. 131, 4685–4694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang Y., Deng H., Schramm V. L. (2010) Leaving group activation and pyrophosphate ionic state at the catalytic site of Plasmodium falciparum orotate phosphoribosyltransferase. J. Am. Chem. Soc. 132, 17023–17031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Clinch K., Evans G. B., Fleet G. W., Furneaux R. H., Johnson S. W., Lenz D. H., Mee S. P., Rands P. R., Schramm V. L., Taylor Ringia E. A., Tyler P. C. (2006) Syntheses and bio-activities of the l-enantiomers of two potent transition state analogue inhibitors of purine nucleoside phosphorylases. Org. Biomol. Chem. 4, 1131–1139 [DOI] [PubMed] [Google Scholar]
- 40. Clinch K., Evans G. B., Fröhlich R. F., Furneaux R. H., Kelly P. M., Legentil L., Murkin A. S., Li L., Schramm V. L., Tyler P. C., Woolhouse A. D. (2009) Third-generation immucillins: syntheses and bioactivities of acyclic immucillin inhibitors of human purine nucleoside phosphorylase. J. Med. Chem. 52, 1126–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Clinch K., Evans G. B., Furneaux R. H., Lenz D. H., Mason J. M., Mee S. P., Tyler P. C., Wilcox S. J. (2007) A practical synthesis of (3R,4R)-N-tert-butoxycarbonyl-4-hydroxymethylpyrrolidin-3-ol. Org. Biomol. Chem. 5, 2800–2802 [DOI] [PubMed] [Google Scholar]
- 42. Evans G. B., Furneaux R. H., Hausler H., Larsen J. S., Tyler P. C. (2004) Imino-C-nucleoside synthesis: heteroaryl lithium carbanion additions to a carbohydrate cyclic imine and nitrone. J. Org. Chem. 69, 2217–2220 [DOI] [PubMed] [Google Scholar]
- 43. Huang M., Wang Y., Gu J., Yang J., Noel K., Mitchell B. S., Schramm V. L., Graves L. M. (2008) Determinants of sensitivity of human T-cell leukemia CCRF-CEM cells to immucillin-H. Leuk. Res. 32, 1268–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Longshaw A. I., Adanitsch F., Gutierrez J. A., Evans G. B., Tyler P. C., Schramm V. L. (2010) Design and synthesis of potent “sulfur-free” transition state analogue inhibitors of 5′-methylthioadenosine nucleosidase and 5′-methylthioadenosine phosphorylase, J. Med. Chem. 53, 6730–6746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mazzella L. J., Parkin D. W., Tyler P. C., Furneaux R. H., Schramm V. L. (1996) Mechanistic diagnoses of N-ribohydrolases and purine nucleoside phosphorylase. J. Am. Chem. Soc. 118, 2111–2112 [Google Scholar]
- 46. Parkin D. W., Limberg G., Tyler P. C., Furneaux R. H., Chen X. Y., Schramm V. L. (1997) Isozyme-specific transition state inhibitors for the trypanosomal nucleoside hydrolases. Biochemistry 36, 3528–3534 [DOI] [PubMed] [Google Scholar]
- 47. Evans G. B., Furneaux R. H., Lewandowicz A., Schramm V. L., Tyler P. C. (2003) Synthesis of second-generation transition state analogues of human purine nucleoside phosphorylase. J. Med. Chem. 46, 5271–5276 [DOI] [PubMed] [Google Scholar]
- 48. Furneaux R. H., Schramm V. L., Tyler P. C. (1999) Transition state analogue inhibitors of protozoan nucleoside hydrolases. Bioorg. Med. Chem. 7, 2599–2606 [DOI] [PubMed] [Google Scholar]
- 49. Furneaux R. H., Limberg G., Tyler P. C., Schramm V. L. (1997) Synthesis of transition state inhibitors for N-riboside hydrolases and transferases. Tetrahedron 53, 2915–2930 [Google Scholar]
- 50. Grosdidier A., Zoete V., Michielin O. (2011) Fast docking using the CHARMM force field with EADock DSS. J. Comput. Chem. 32, 2149–2159 [DOI] [PubMed] [Google Scholar]
- 51. Grosdidier A., Zoete V., Michielin O. (2011) SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 39, W270–W277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cassera M. B., Hazleton K. Z., Riegelhaupt P. M., Merino E. F., Luo M., Akabas M. H., Schramm V. L. (2008) Erythrocytic adenosine monophosphate as an alternative purine source in Plasmodium falciparum. J. Biol. Chem. 283, 32889–32899 [DOI] [PMC free article] [PubMed] [Google Scholar]