Background: Ubiquitin-mediated proteolysis is the principal mechanism for regulating protein half-lives in cells.
Results: Cdc34 promotes ubiquitin chain assembly onto protein substrates and contains an essential acidic loop near the active site.
Conclusion: The Cdc34 acidic loop promotes both protein-protein interactions and catalysis of ubiquitin chain formation.
Significance: These results uncover specific biochemical activities for the Cdc34 acidic loop.
Keywords: Kinetics, Ubiquitin-conjugating Enzyme (Ubc), Ubiquitin Ligase, Ubiquitination, Yeast Genetics
Abstract
Together with ubiquitin ligases (E3), ubiquitin-conjugating enzymes (E2) are charged with the essential task of synthesizing ubiquitin chains onto protein substrates. Some 75% of the known E2s in the human proteome contain unique insertions in their primary sequences, yet it is largely unclear what effect these insertions impart on the ubiquitination reaction. Cdc34 is an important E2 with prominent roles in cell cycle regulation and signal transduction. The amino acid sequence of Cdc34 contains an insertion distal to the active site that is absent in most other E2s, yet this acidic loop (named for its four invariably conserved acidic residues) is critical for Cdc34 function both in vitro and in vivo. Here we have investigated how the acidic loop in human Cdc34 promotes ubiquitination, identifying two key molecular events during which the acidic loop exerts its influence. First, the acidic loop promotes the interaction between Cdc34 and its ubiquitin ligase partner, SCF. Second, two glutamic acid residues located on the distal side of the loop collaborate with an invariably conserved histidine on the proximal side of the loop to suppress the pKa of an ionizing species on ubiquitin or Cdc34 which greatly contributes to Cdc34 catalysis. These results demonstrate that insertions can guide E2s to their physiologically relevant ubiquitin ligases as well as provide essential modalities that promote catalysis.
Introduction
Ubiquitin-mediated proteolysis is the principal mechanism in eukaryotic cells for regulating the half-lives of proteins as well as ridding cells of damaged or misfolded proteins (1–5). The process involves the attachment of polyubiquitin chains onto protein substrates, which localizes them to the 26S proteasome for degradation (6, 7). This pathway is encoded by some 1000 genes in mammalian cells and influences various biological functions such as apoptosis, NFκB signaling, circadian rhythms, perception of metabolites, and cell division. Ubiquitin-mediated proteolysis is critical for cellular homeostasis, as the misregulation of molecular events controlled by the ubiquitin pathway has been linked to multiple forms of cancer and neurodegenerative disorders (8–11).
Ubiquitin chains are synthesized by a large family of enzymes named ubiquitin ligases (E3)3 (12). E3s act as a scaffold recruiting the protein substrate as well as a charged ubiquitin-conjugating enzyme (E2) that has been preloaded with ubiquitin by E1 enzyme. E2s share a conserved fold and bind to E3s using a common molecular surface, and most E3s contain a domain or subunit (named RING) that recognizes E2s. Mammalian cells contain some 40 different E2s and at least 600 unique E3s, with nearly half of all E3s belonging to the Cullin-RING ligase (CRL) family. The impact of the CRLs on human cells is impressive, with some 20% of all proteasome-dependent degradation controlled by these enzymes (13). The ubiquitin-conjugating enzyme Cdc34 serves as the principal E2 enzyme for the majority if not all of the CRLs (14–16). Although ubiquitin contains seven lysine residues that may covalently connect adjacent protomers in a ubiquitin chain, Cdc34 catalyzes the synthesis of chains that are predominantly linked through lysine 48, which promotes the recognition of the ubiquitinated substrates by the 26S proteasome (17).
Although all E2s share a common catalytic domain, Cdc34 contains two molecular add-ons including an acidic tail appended to the C terminus of the catalytic domain (18–20) as well as a 12-residue insertion near the active site, commonly referred to as the acidic loop (21). These two molecular features are conserved in all Cdc34 orthologs, and their combined presence is unique to Cdc34.
Much has been discovered about how the Cdc34 acidic tail contributes to Cdc34 function. Soon after the discovery of Cdc34, it was determined that the acidic tail promotes Cdc34 interaction with the SCF ubiquitin ligase, the archetypal CRL (22, 23). This important result demonstrates that an E2 can participate in bidentate interactions with an E3 and also hinted that the presence of the acidic tail may alter Cdc34-SCF interactions in a manner that benefits the specific needs for that system. For instance, the kinetics for the ubiquitination of Cdc34-SCF substrates are atypically fast (24), and the acidic tail promotes this by enabling the rapid association of Cdc34 and SCF (14).
Much less is known about the molecular mechanism of how the acidic loop participates in Cdc34 catalysis; however evidence from several laboratories has established the importance of the acidic loop for Cdc34 function both in vitro and in vivo. One striking feature of the loop is some four highly conserved acidic residues, with two aspartic acid residues on the N-terminal side of the loop and two glutamic acid residues on the C-terminal side (Fig. 1A). The acidic loop is necessary for proper Cdc34 function in the cell, and a mutant Cdc34 allele with mutations in three acidic loop residues is unable to support growth in the absence of the wild-type allele in Saccharomyces cerevisiae (25). In 2005, Petroski and Deshaies (17) conducted a biochemical characterization of the acidic loop in yeast Cdc34, and mutation of the acidic residues to alanine resulted in a substantial reduction of Cdc34 activity. In 2007, Pan and colleagues (26) were the first to show that the mutation of acidic residues in the human Cdc34 acidic loop also leads to a loss of Cdc34 activity in vitro. More recently, molecular dynamics simulations predicted changes in the conformations of the yeast Cdc34 acidic loop in response to the phosphorylation of two active site serine residues previously identified as potential sites of casein kinase 2 phosphorylation (27, 28). Finally, members of the ubiquitin-conjugating enzyme family Ube2G2 (called Ubc7 in yeast) also contain an acidic loop with four conserved acidic residues, and their mutation also leads to the loss of human Ube2G2 activity in vitro (29).
FIGURE 1.

Cdc34 and Ube2G2 family members contain a conserved acidic loop that is distal to the active site. A, multiple sequence alignment of the acidic loop region from Cdc34 orthologs and Ube2G2 (also referred to as Ubc7) from yeast. The active site cysteine has been marked by a red asterisk, the acidic loop histidine residue by a green asterisk, and the four highly conserved acidic loop residues by black asterisks. B–E, structural superposition of four apoE2 structures in the RCSB database containing coordinates for the acidic loop. Some nine structures for either apoCdc34 or Ube2G2 were available as of September, 2013 (PDB codes 1PZV, 2AWF, 3H8K, 3FSH, 2KLY, 2UCZ, 3RZ3, 2OB4, 2CYX, 4LAD, and 2LXP). The structures containing ordered loops (3H8K, gray; 2UCZ, yellow; 3FSH, orange; and 2CYX, red) were superimposed in PyMOL and rendered as ribbon diagrams. Notice that the acidic loop conformations all significantly differ from each other.
The ubiquitination reaction involves numerous steps in which the acidic loop may potentially exert its influence. These steps include: the E1-catalyzed formation of the thioester bond between the C terminus of ubiquitin (commonly referred to as donor ubiquitin) and the Cdc34 active site cysteine residue (30), the formation of a non-covalent interface between Cdc34 and donor ubiquitin that promotes Cdc34 function (31), the deprotonation of the side-chain primary amine on lysine residues either on protein substrate or the distal ubiquitin on a substrate-modified ubiquitin chain (commonly referred to as acceptor ubiquitin), the binding of acceptor ubiquitin to Cdc34, and the association and dissociation of Cdc34 with E3s such as SCF. We and others have recently developed a quantitative kinetics platform for studying Cdc34 activity (14, 24, 32, 33), and its application allows for the careful dissection of the multistep ubiquitination reaction into its individual components. Here we demonstrate that the Cdc34 acidic loop modulates the affinity between Cdc34 and SCF and also plays a role in the deprotonation of an important ionizable species on Cdc34 or ubiquitin.
EXPERIMENTAL PROCEDURES
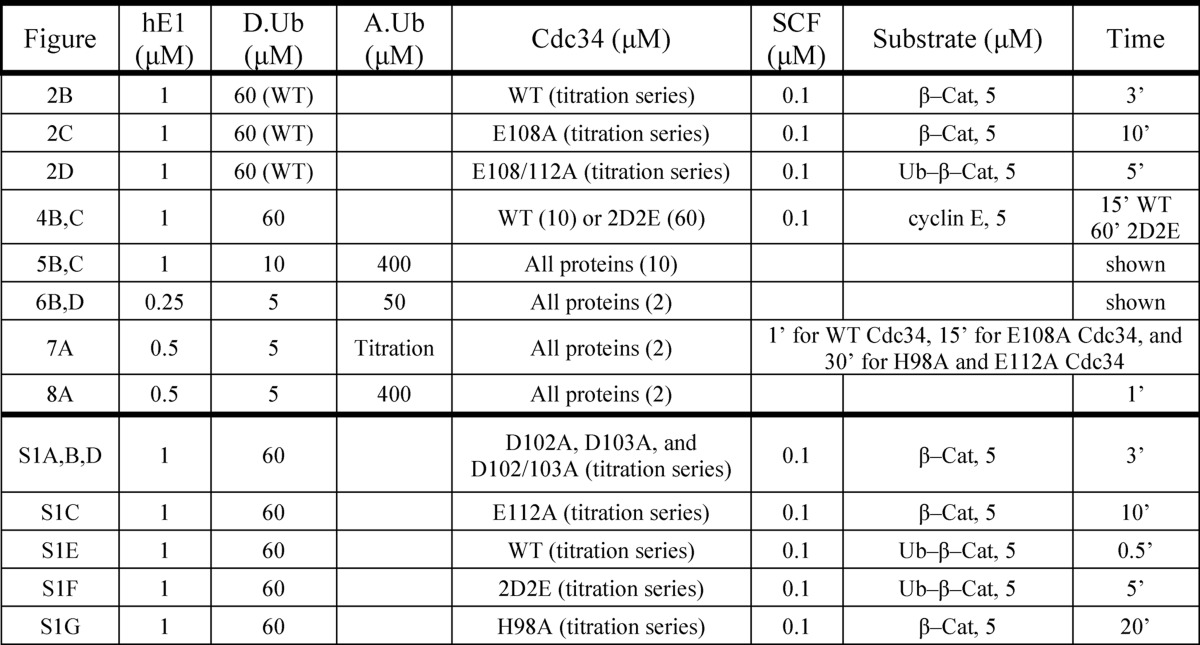
Expression and Purification of Recombinant Proteins
All proteins were expressed recombinantly in either Escherichia coli or Hi5 insect cells and purified using standard procedures (supplemental Table S1). The final buffer condition for all proteins prior to drop-freezing in liquid nitrogen for long-term storage was 30 mm Tris-Cl (pH 7.5), 100 mm NaCl, 1 mm DTT, and 10% glycerol. The Rbx1-Cul1 complex was expressed using a previously described “Split-n-Coexpress” protocol where the Cul1 protein is expressed as two fragments, referred to as the NTD (N-terminal domain) and the CTD (C-terminal domain) (34). This system allows higher expression of these complexes in E. coli. All ubiquitination reactions containing SCF utilized neddylated Cul1-Rbx1; the detailed purification strategy of this complex has been described previously (33). Production and purification of the Ub-β-catenin peptide substrate has also been described previously (33). All in vitro experiments described in this article were performed with human proteins. Mammalian wild-type ubiquitin was used for all relevant assays (Boston Biochem).
Multi-turnover Ubiquitination Assays
Ubiquitination assays were performed using the β-catenin peptide, Ub-β-catenin peptide, or cyclin E peptide as described previously (14, 33). Briefly, 50 μm peptide was labeled with 5 kilo units of cAMP-dependent protein kinase (New England Biolabs) in the presence of [γ32P]ATP for 1 h at 30 °C. All ubiquitination experiments were performed at room temperature in the following buffer: 30 mm Tris-Cl (pH 7.5), 100 mm NaCl, 5 mm MgCl2, 2 mm DTT, and 2 mm ATP. Multi-turnover reactions (Fig. 2 and supplemental Fig. S1) were initiated by the addition of labeled substrate and quenched with an equal volume of 2× reducing SDS-PAGE buffer. All samples were resolved by SDS-PAGE followed by autoradiography and quantitation using ImageQuant software (GE Healthcare). All reported values are the average of at least two experiments. Further details on the reaction conditions (including protein concentrations and incubation periods) may be found in the figure legends (Figs. 2 and 4) and in Table 1. Note that the 1 μm E1 concentration was shown to be sufficient to fully convert all Cdc34 proteins to the Cdc34∼ubiquitin thioester form, even when the Cdc34 concentration was as high as 60 μm (data not shown).
FIGURE 2.
Acidic loop residues Glu-108 and Glu-112 in human Cdc34 function in SCF binding. A, schematic for the assembly and order of addition of the multi-turnover ubiquitination reaction. Reactions were initiated by the addition of either 32P-labeled β-Cat peptide or Ub-β-catenin peptide (β-Cat-(Ub)1) substrate followed by quenching and SDS-PAGE. B, increasing amounts of WT Cdc34 were titrated into ubiquitination reactions followed by phosphorimaging and quantitation of products and reactants. Note that each lane corresponds to a single reaction and quantity of Cdc34. The initial velocities of the ubiquitination reaction were plotted against WT Cdc34 concentration and used to estimate the Km and kcat of WT Cdc34 for SCF using nonlinear regression (GraphPad Prism software). C, same as B with E108A Cdc34. D, same as B except with E108A/E112A Cdc34 and Ub-β-catenin peptide. The asterisk corresponds to a contaminant of the substrate preparation. E, graph showing the ratio of Km values for all mutant Cdc34 proteins used in this investigation divided by the Km for WT Cdc34. Note that all experiments were done in duplicate, and the error bars represent the standard error of measurement. Reaction conditions are summarized in Table 1.
FIGURE 4.
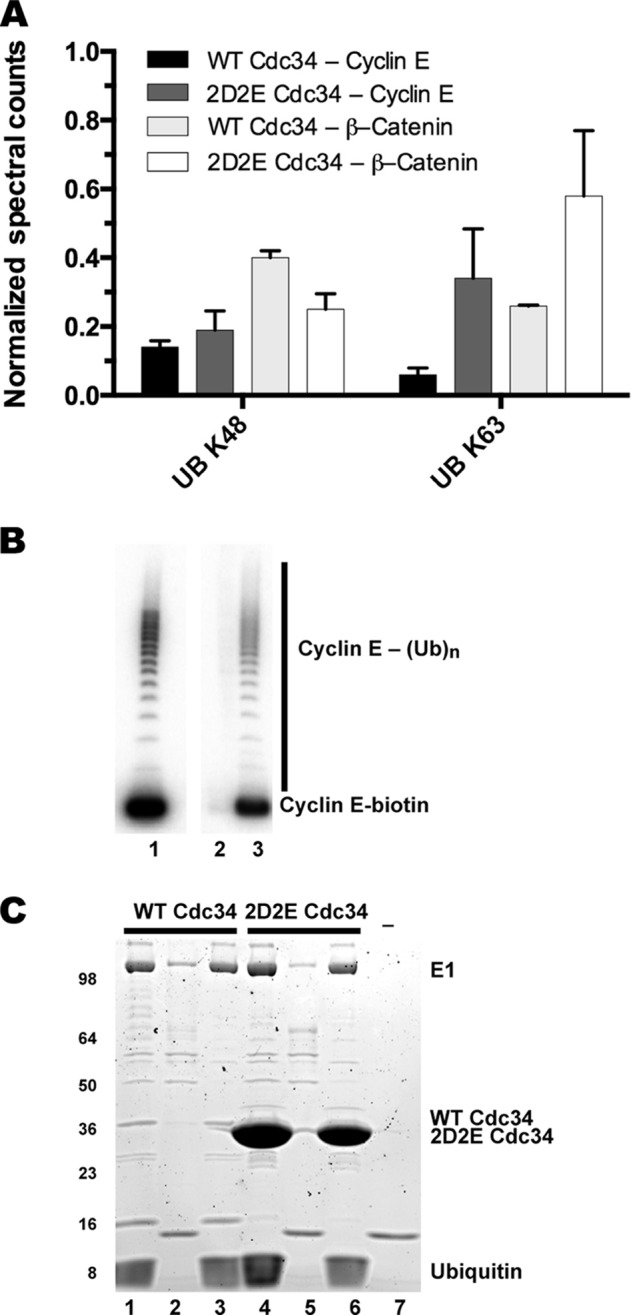
Both WT and 2D2E Cdc34 synthesize Lys-48-linked (K48) ubiquitin chains. A, SCF-dependent ubiquitination reactions were initiated in the presence of either WT or 2D2E Cdc34 and either β-catenin or cyclin E peptide substrates. Ubiquitin chain linkages were determined for each sample by counting ubiquitin-derived peptides containing a Gly-Gly signature using mass spectrometry. The total number of peptides observed for either Lys-48 or Lys-63 linkages were then normalized by the total spectral counts matched to the E1 enzyme. Error bars represent the standard deviation of triplicate measurements for β-catenin reactions and 6× replicates for cyclin E reactions (see “Experimental Procedures”). B, autoradiogram of a ubiquitination reaction containing WT Cdc34 and 32P-labeled cyclin E peptide substrate containing a C-terminal biotin before and after purification (see “Experimental Procedures”). Lane 1 shows product formation for the reaction prior to purification. Lane 2 shows the contents from the first wash step after incubation of the reaction components with magnetized streptavidin beads. Lane 3 shows bead-bound substrates and products after the addition of 2× SDS-PAGE loading buffer. Notice that both 32P-labeled cyclin E substrates and products are efficiently captured by the beads even after five washings. C, same as B except reactions with either WT or 2D2E Cdc34 and unlabeled cyclin E peptide were processed followed by Coomassie staining of the gel. Lanes 1 and 4 correspond to the ubiquitination reactions prior to bead purification (WT and 2D2E Cdc34, respectively); lanes 2 and 5 correspond to substrates and products after bead purification; and lanes 3 and 6 correspond to the contents of the first wash step. Lane 7 is from a mock reaction containing only beads. Notice that nearly all of the 2D2E Cdc34 is efficiently removed by the purification process.
TABLE 1.
Reaction conditions for ubiquitination assays used in this study
hE1, human E1 enzyme; D.Ub, donor ubiquitin; A.Ub, acceptor ubiquitin.
The initial velocities of the multi-turnover SCF-dependent ubiquitination reactions containing either β-catenin peptide or Ub-β-catenin (Fig. 2 and supplemental Fig. S1) were calculated as follows. The amount of substrate and product were quantified and used to calculate the percentage of substrates that were modified by one or more ubiquitins. This is equal to the sum of the intensities of all bands above the substrate band divided by the total intensity in the lane (each lane represents individual ubiquitination reactions containing a defined quantity of Cdc34 protein from the titration series). This percentage was then multiplied by the ratio of the total concentrations of substrate and enzyme (in this case, SCF). The resulting value was then divided by the time of incubation yielding the reaction rate in units of min−1.
FRET Binding Assay
Cdc34 labeling reactions were performed using Lumio Green (Invitrogen) as described previously (33). Purified WT or 2D2E Cdc34 (where 2D2E is a quadruple mutant in which the acidic residues Asp-102/Asp-103/Glu-108/Glu-112 have been mutated to alanine), with the amino acid sequence CCPGCCHHHHHH appended to the C terminus, was incubated at 30 μm with 40 μm Lumio Green in a buffer containing 20 mm Tris-Cl (pH 7.5), 100 mm NaCl, 2 mm Tris(2-carboxyethyl)phosphine hydrochloride (TCEP, Thermo Scientific, San Jose, CA), 1 mm EDTA, and 5% glycerol at 22 °C for 2 h. Rbx1-Cul1-CFP was purified as described previously (33).
Equilibrium fluorescence measurements were collected on a FluoroLog-3 spectrofluorimeter (Jobin Yvon). Binding reactions containing the FRET donor Rbx1-Cul1-CFP (at a concentration below Kd) and various concentrations of Cdc34 proteins were incubated for 20 min at 22 °C in a buffer containing 20 mm Tris-Cl (pH 7.5), 50 or 100 mm NaCl, 0.5 mm DTT, and 5% glycerol. Samples were excited at 430 nm, and the emission spectra were acquired from 450 to 570 nm.
Ubiquitin Chain Linkage Analysis
β-Catenin ubiquitination reactions were carried out in standard reaction buffer (see above). 10 μm WT or 60 μm 2D2E Cdc34 was charged in the presence of 1 μm E1 and 60 μm ubiquitin for 2 min prior to the addition of 0.1 μm SCFTrCP followed by an additional 2-min incubation period. Reactions were initiated by adding either 5 μm β-catenin (WT Cdc34) or Ub-β-catenin peptide substrate (2D2E Cdc34) and incubation for 3 or 30 min, respectively, prior to the addition of 10 mm DTT, which terminated the reactions. Samples containing unlabeled substrates were drop-frozen in liquid nitrogen prior to mass spectrometry. Cyclin E ubiquitination reactions were carried out under similar conditions, except 10 μm WT Cdc34 or 50 μm 2D2E Cdc34 was included with 0.1 μm SCFFbxw7 and 5 μm cyclin E substrate containing a biotin label at the C-terminal end of the peptide. Reactions were terminated in 10 mm DTT followed by a 60-min incubation with magnetized streptavidin beads (Dynabeads, Invitrogen) on ice with occasional mixing. Beads were then washed five times with PBST (PBS with 0.1% Tween 20) and drop-frozen in liquid nitrogen for storage prior to mass spectrometry. Verification of the purification strategy is shown in Fig. 4, B and C.
Ubiquitination reactions containing either β-catenin or cyclin E peptide substrates were resuspended in 5% acetonitrile (ACN) in 50 mm ammonium bicarbonate (pH 7.8) and digested with 0.5 μg of trypsin overnight at room temperature. The resulting peptides were desalted with in-house-prepared C18 stage tips using a previously described protocol and dried in a vacuum centrifuge (35). Samples were resuspended in 5% formic acid, 5% ACN and analyzed in triplicate by LC-MS/MS using a Q-Exactive mass spectrometer (Thermo Scientific) with the following conditions. Peptides were first separated by reverse-phase chromatography using a fused silica microcapillary column (100 μm ID, 22 cm) packed with C18 reverse-phase resin (Magic C18AQ, 3-μm particles, 200-Å pore size; Michrom Bioresources, Auburn, CA) using an in-line nanoflow EASY-nLC 1000 UHPLC (Thermo Scientific). Peptides were eluted over a 45-min period in a 10–30% ACN gradient followed by a 4-min 30–45% ACN gradient and then a 1-min 45–98% gradient, with a final 10-min isocratic step at 98% ACN for a total run time of 60 min at a flow rate of 500 nL min−1. MS/MS data were collected in a data-dependent fashion using a Top10 method with a full MS mass range from 400 to 1800 m/z, resolution 70,000, and an automatic gain control target of 1 e6. MS2 scans were triggered when an ion intensity threshold of 2.5 e2 was reached. A dynamic exclusion time of 8 s was used, and the peptide match setting was enabled. Singly charged ions and unassigned charge states were excluded. The resultant RAW files were converted into mzXML format using the ReAdW program. The SEQUEST search algorithm (version 28) was used to search MS/MS spectra against a concatenated target-decoy database composed of forward and reversed sequences from the reviewed UniProt human FASTA database and appended mutant Cdc34 sequences. The search parameters used were as follows: 50 ppm precursor ion tolerance and 0.01 Da fragment ion tolerance; up to two missed cleavages allowed; and dynamic modifications of 15.99491 Da on methionine (oxidation) and 114.04293 Da on lysine. Peptide matches were filtered to a false discovery rate of 1% using the linear discriminant analysis (36).
Di-ubiquitin Synthesis Assays
The Cdc34 di-ubiquitin synthesis assays (thioester chase and steady state) were adapted from previously established protocols (31, 37, 38). Note that it is necessary to suppress product formation during the thioester bond formation stage between Cdc34 and ubiquitin (Figs. 5A and 6A). Here 32P-labeled K48R ubiquitin is chosen as the donor ubiquitin, as Cdc34 is highly selective for lysine 48 during ubiquitin chain synthesis. Similarly, the unlabeled acceptor ubiquitin should not be available as a donor (this is especially relevant during the steady-state di-ubiquitin synthesis assay, where E1 enzyme remains active during the product formation step and could thus charge hydrolyzed apoCdc34 with unlabeled ubiquitin). Therefore, the acceptor ubiquitin contains the wild-type 76 residues with an aspartate residue appended to the C terminus (often referred to as Asp-77 ubiquitin in other articles), blocking its ability to form thioesters with Cdc34.
FIGURE 5.
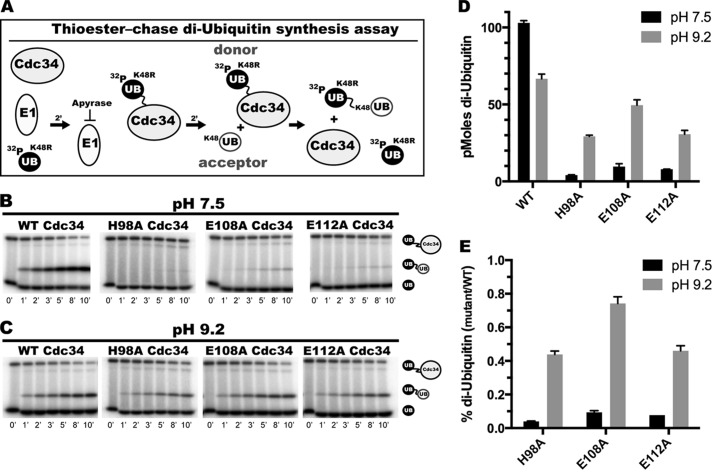
Human Cdc34 residues His-98, Glu-108, and Glu-112 participate in the deprotonation of an ionizable group on Cdc34 or ubiquitin. A, schematic describing the thioester-chase di-ubiquitin synthesis assay. 32P-labeled K48R ubiquitin was incubated in the presence of Cdc34 and E1 to form covalent Cdc34∼ubiquitin complexes followed by treatment with apyrase. Unlabeled acceptor ubiquitin was added to initiate the reaction. B, di-ubiquitin synthesis for WT, H98A, E108A, and E112A Cdc34 as a function of time at pH 7.5. Phosphorimages show the quantities of 32P-labeled K48R ubiquitin, di-ubiquitin, and Cdc34∼ubiquitin as separated by nonreducing SDS-PAGE. C, same as in B except reactions were incubated at pH 9.2. D, bar graph showing the final amounts of di-ubiquitin product (pmol) for the reactions in B and C. E, bar graph showing the percentages of di-ubiquitin products for H98A, E108A, and E112A Cdc34 as compared with WT Cdc34 at either pH 7.5 or 9.2.
FIGURE 6.
Mutations in conserved residues in the acidic loop cause moderate to severe defects in SCF-independent Cdc34 activity. A, schematic describing the steady-state di-ubiquitin synthesis assay. 32P-labeled K48R ubiquitin was incubated in the presence of Cdc34 and E1 to form covalent Cdc34∼ubiquitin complexes followed by the introduction of unlabeled acceptor ubiquitin, which initiates the reaction. B, di-ubiquitin synthesis for WT, E108A, E112A, H98A, H98N, H98Q, H98A/E108A, and H98A/E112A Cdc34 as a function of time (min) and pH 7.5. The phosphorimage shows the quantities of 32P-labeled K48R ubiquitin and di-ubiquitin as separated by reducing SDS-PAGE. C and D, the rates of di-ubiquitin synthesis for Cdc34 proteins in B were estimated by plotting the concentration of di-ubiquitin (normalized by Cdc34 concentration; see “Experimental Procedures”) against time followed by linear regression analysis. Rates are summarized in Table 3. E, the rates of all Cdc34 mutant proteins were calculated as a percentage of the rate of product formation for WT Cdc34. Each data point in C and D represents the mean of duplicate measurements, and the magnitudes of the error bars are equal S.E. For reaction conditions, see Table 1.
Thioester-Chase Di-ubiquitin Synthesis Assay
Cdc34 was charged with 32P-labeled K48R ubiquitin in standard reaction buffer containing Tris-Cl at pH 7.5 or 9.2. Charging efficiency and equivalency were confirmed at both pH values. Reactions were treated with 1 unit of apyrase (Sigma) for 1 min to inactivate E1 enzyme. The zero time point represents the state of the reaction immediately prior to the addition of acceptor ubiquitin. Note that there are two potential products of this reaction: the hydrolysis of the Cdc34∼ubiquitin thioester bond or the formation of di-ubiquitin product. Samples taken at the indicated time points were quenched in nonreducing SDS-PAGE sample buffer containing 5 mm N-ethylmaleimide (which has been shown to better preserve the Cdc34∼ubiquitin thioester bond during electrophoresis).
Steady-state Di-ubiquitin Synthesis Assay
Details on the steady-state di-ubiquitin synthesis assay are provided in the Fig. 6A figure legend. Reactions were quenched in reducing SDS-PAGE loading buffer at the indicated time points. The initial velocities of the di-ubiquitin synthesis reactions were calculated as follows. The amount of substrate (32P-labeled K48R ubiquitin) and di-ubiquitin product was quantified and used to calculate the percentage of product. This is equal to the intensity of the di-ubiquitin band divided by the total intensity in the lane. This percentage was then multiplied by the ratio of the total concentrations of substrate and Cdc34. These values were plotted on a graph as a function of time followed by linear regression (GraphPad Prism software). The resulting slope is equal to the initial velocity of the reaction (see Table 3). A similar procedure was used to calculate the initial velocities for determining the Km of acceptor ubiquitin for Cdc34 (Fig. 7) and the pH dependence of Cdc34 activity (Fig. 8), except a single time point was used rather than time courses.
TABLE 3.
Estimates for the rates of ubiquitin transfer in steady-state di-ubiquitin synthesis assays
| Cdc34 | SCF | pH | Acceptor Ub | kobs |
|---|---|---|---|---|
| μm | h−1 | |||
| Wild-type | — | 7.5 | 50 | 1.1 ± 0.04 |
| D103A | — | 7.5 | 50 | 0.3 ± 0.01 |
| E108A | — | 7.5 | 50 | 0.13 ± 0.01 |
| E112A | — | 7.5 | 50 | 0.05 ± 0.003 |
| H98A | — | 7.5 | 50 | 0.05 ± 0.004 |
| H98N | — | 7.5 | 50 | 0.06 ± 0.004 |
| H98Q | — | 7.5 | 50 | 0.04 ± 0.002 |
| H98A/E108A | — | 7.5 | 50 | 0.04 ± 0.005 |
| H98A/E112A | — | 7.5 | 50 | NDa |
a Not determined. All reactions were performed in the absence of SCF (−).
FIGURE 7.
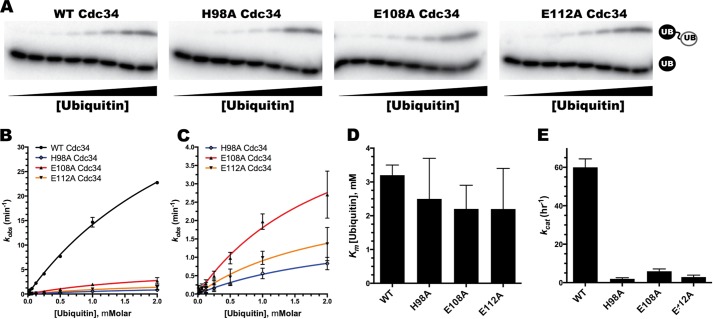
The mutation of Cdc34 residues His-98, Glu-108, or Glu-112 does not disrupt acceptor ubiquitin binding to Cdc34. A, di-ubiquitin synthesis assays (Fig. 6A) were used to determine the initial velocities of reactions containing increasing concentrations of cold acceptor ubiquitin. Note that each lane represents a single ubiquitination assay at pH 7.5 containing increasing concentrations of acceptor ubiquitin (the rightmost lane in each gel shows substrate and product for reactions containing 2 mm ubiquitin). B and C, graphs showing the rate of di-ubiquitin formation (kobs) as a function of the acceptor ubiquitin concentration. Initial velocities (kobs) were calculated by determining the percentage of di-ubiquitin product multiplied by the ratio of the substrate and Cdc34 concentrations divided by the reaction time (see Table 1 for reaction conditions and incubation periods for the reactions). The Km (D) and kcat (E) values of acceptor ubiquitin for Cdc34 proteins were determined by nonlinear curve fitting to the Michaelis-Menten equation (GraphPad Prism software). All experiments were done in duplicate, and error bars represent the standard error of measurement.
FIGURE 8.

The mutation of acidic loop residues His-98 or E112A Cdc34 causes an increase in the pKa for an ionizable group on Cdc34 or ubiquitin. A, di-ubiquitin synthesis assays were performed to estimate the initial velocities of Cdc34 ubiquitination reactions as a function of pH (a titration series between pH 5.8 and 10.7 was accomplished using bis-Tris propane for the buffer). B, the data were fit to either a monophasic sigmoidal equation (WT Cdc34) or a biphasic sigmoidal equation corresponding to a doubly ionizing system (H98A and E112A Cdc34) (49). The apparent pKa values are shown in parentheses. For reaction conditions, see Table 1. All experiments were done in duplicate, and error bars represent S.E.
Yeast Cdc34 Expression and Growth Assays
A previously described plasmid shuffle approach was used to replace the chromosomal yeast CDC34 gene with a plasmid containing various Cdc34 alleles with mutations in the acidic loop region (39). Briefly, we began with a haploid yeast strain in which the essential function of the WT CDC34 gene was temporarily provided by a plasmid containing both the WT CDC34 open reading frame controlled by the CDC34 promoter and the URA3 gene (strain RJD 3255; MATa cdc34Δ::kanRhis3Δ1 leu2Δ0 ura3Δ0::[RDB1967], where RDB1967 is pRS316 with the WT CDC34 gene). This strain was then transformed with various pRS315-based constructs (empty vector, WT CDC34, H100A, E109A, or E113A CDC34) followed by selection of transformants on growth media lacking leucine (−LEU). These strains were used to inoculate liquid YPD (yeast/peptone/dextrose) cultures grown at 30 °C with shaking. Once exponential growth had been verified, each strain was diluted in YPD to an A600 of 0.5 followed by the formation of a 3-fold dilution series in PBS. The yeast were then replica-plated onto either SD medium with 5-fluoroorotic acid or SD −LEU plates and incubated at 30 °C for 2 days.
Western Blots
Some six A600 units of mid-log phase yeast cultures were harvested by centrifugation and resuspended in 1 ml of ice-cold stop buffer (50 mm Tris-Cl (pH 7.5), 50 mm NaF, and 0.02% NaN3). Cells were pelleted and resuspended in 100 μl of 1× SDS-PAGE sample loading buffer. An equal volume of 0.5-mm-diameter glass beads (Sigma) was added, and samples were quickly vortexed followed by incubation at 95 °C for 3 min. The samples were allowed to cool for 1 min and then harshly vortexed for 2 min. Samples were again heated at 95 °C for 1 min and centrifuged at maximum speed in a tabletop centrifuge for 2 min. Yeast extracts were separated by SDS-PAGE and transferred onto a PVDF membrane. After blocking for 1 h with 5% nonfat milk, the membrane was probed with a 1:1000 anti-V5 monoclonal antibody (Sigma, clone V5-10). Actin was also probed for as a loading control (EMD Millipore). Protein levels were detected with HRP-conjugated secondary antibodies (Sigma) using WesternBright ECL (BioExpress) and imaged using a Bio-Rad ChemiDoc.
RESULTS
Two Conserved Acidic Residues in the Human Cdc34 Acidic Loop Promote the Interaction of Cdc34 with Ubiquitin Ligase SCF
Many enzymes contain flexible loops near the active site that participate in catalysis. These loops can adopt “open” or disordered structures in the absence of substrate or cofactors but become ordered during catalysis. A comprehensive examination of the available structures of Cdc34 and Ube2G2 in the absence of their E3 partners suggests that the acidic loop can adopt several conformations. As of September 2013, the Protein Data Bank contained some nine x-ray or NMR structures of apoCdc34 or Ube2G2 orthologs. Residues encompassing the acidic loop were disordered in five of the structures (including the sole NMR structure (40)); however coordinates for the loop were included in four of the Ube2G2 structures. The superposition of all four structures shows that whereas the E2 fold is highly conserved, the loop conformations are all distinct from each other (Fig. 1, B–E) and, in at least some cases, may be influenced by crystal packing forces. We also reasoned that the conformation of the acidic loop may be controlled by the presence of donor ubiquitin that is bound to the active site. However, the acidic loop remained disordered in a recently solved x-ray structure of a Cdc34∼ubiquitin complex (41).
Recently, Papaleo et al. (42) used molecular dynamics simulations to model conformations of the Cdc34 acidic loop in the presence of E3. Interestingly, their results predict that several acidic loop residues may form intermolecular interactions with the Rbx1 subunit of the E3. Furthermore, Ube2G2 was recently co-crystallized with the RING subunit of its cognate E3 gp78 (43). Although the majority of the acidic loop was still disordered, the position of the distal glutamic acid residue was resolved, and the side-chain carboxylate forms an electrostatic interaction with E3. Thus, we hypothesized that the acidic loop may promote E2 interaction with E3.
To test our hypothesis, we used Michaelis-Menten kinetics to assess the affinities between human Cdc34 proteins containing mutations in the acidic loop and the SCF ubiquitin ligase. Specifically, we constructed a suite of acidic loop mutants in which key conserved residues were mutated to alanine. To control for potential aberrations to the structures, each Cdc34 mutant protein used here was demonstrated capable of forming thioester bonds with ubiquitin at levels comparable with the wild type (data not shown).
It is well established that the Michaelis constant (Km) of a substrate for enzyme is similar to the equilibrium dissociation constant (Kd) when the rate of substrate dissociation from enzyme, koff, is substantially faster than the rate of catalysis, kcat (in the case of E2-E3 interactions, Cdc34∼ubiquitin is considered the “substrate” and SCF is the “enzyme”). In fact, we previously showed that Cdc34 dissociation from E3 (∼100 s−1) is much faster than the maximum rate of ubiquitin transfer (2–6 s−1) (14, 24). Therefore, the affinity between Cdc34 and SCF can be estimated by measuring the initial velocities of ubiquitination reactions containing increasing concentrations of Cdc34 and solving for the Michaelis-Menten parameters, Km and kcat (Fig. 2A).
Cdc34 proteins containing mutations at either Glu-108 or Glu-112 in the acidic loop showed decreased affinities for SCF, whereas Cdc34 mutants at the other two conserved positions (Asp-102 or Asp-103) were comparable with WT Cdc34. Ubiquitination reactions containing 32P-labeled β-catenin peptide substrate and SCF were initiated in the presence of increasing concentrations of WT Cdc34 (Fig. 2B). Nonlinear curve fitting resulted in a Km of 460 nm, consistent with previously published results under similar conditions. Results for both D102A and D103A Cdc34 were comparable with the WT (supplemental Fig. S1, A and B); however the Km of E108A Cdc34 for SCF was 4.7 μm, corresponding to a 10-fold decrease in affinity (Fig. 2, C and E, and Table 2). E112A Cdc34 also showed a decrease in affinity (Fig. 2E and supplemental Fig. S1C).
TABLE 2.
Estimates of Km and kcat for WT or mutant Cdc34 proteins and SCF

The combination of the E108A and E112A Cdc34 single mutants into an E108A/E112A double mutant led to an even greater decrease in Cdc34 affinity for SCF, whereas a combination of the D102A and D103A mutations still resulted in WT-like affinity for SCF. Because all four acidic residues in the Cdc34 loop are highly conserved in both Cdc34 and Ube2G2 orthologs, we were surprised that the mutation of Asp-102 or Asp-103 to alanine had no effect on the Km of Cdc34 for SCF, whereas mutation of either Glu-108 or Glu-112 to alanine did. We reasoned that a D102A/D103A Cdc34 double mutant might result in a higher Km; however this was not the case (Fig. 2E and supplemental Fig. S1D). On the other hand, E108A/E112A Cdc34 was completely inactive in the presence of β-catenin peptide substrate, even when present at concentrations as high as 100 μm. It was shown previously that a SCF substrate modified with one ubiquitin (Ub-β-catenin) is significantly more reactive than unmodified substrate, and product formation was observable for E108A/E112A Cdc34 in the presence of Ub-β-catenin peptide. The Km of WT Cdc34 for SCF was 170 nm when using this substrate (supplemental Fig. S1E); however the Km for E108A/E112A Cdc34 was 17 μm (Fig. 2, D and E), 2 orders of magnitude higher than for WT Cdc34. The mutation of all four acidic residues to alanine (referred to herein as 2D2E Cdc34) had a similar Km for SCF as E108A/E112A Cdc34 (Fig. 2E and supplemental Fig. S1F).
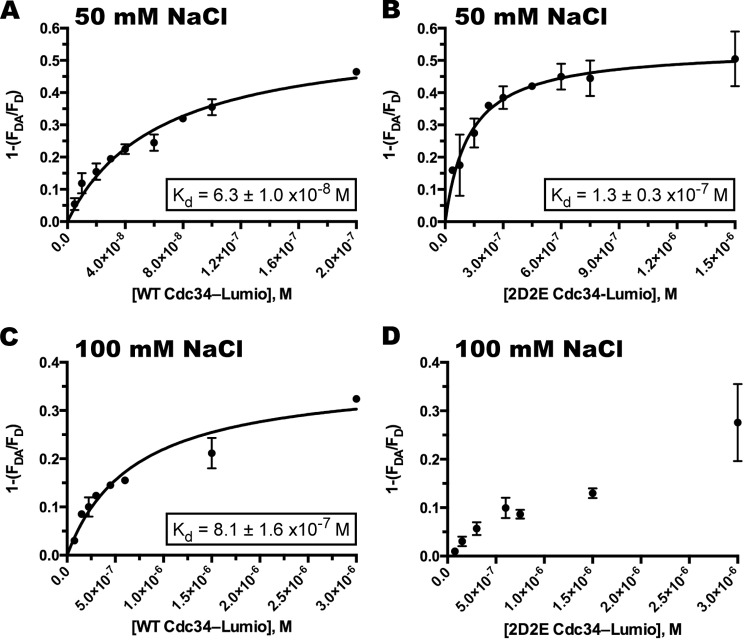
Although it was noted earlier that the kinetic rate constants for the interaction of Cdc34 and SCF suggest that the Km values are reasonable estimates of Cdc34 affinity for SCF, the reactions used in this study are highly complex, and thus direct binding experiments were carried out to confirm these findings. Using a FRET-based binding assay, the affinities of either wild-type or 2D2E Cdc34 were estimated for SCF. Surprisingly, the mutation of all four conserved acidic residues in the loop to alanine resulted in only a modest 2-fold increase in the Kd as compared with the wild type (Fig. 3, A and B). Note that the affinity of Cdc34 for SCF is highly dependent on the concentration of salt in the reaction (14), and the relatively low ionic strength of the binding buffer may have suppressed the effect of the acidic loop on SCF binding. Thus, the binding experiments were repeated at a higher ionic strength. Although the affinity of WT Cdc34 for SCF decreased ∼10-fold as expected (Fig. 3C), the titration of fluorescently labeled 2D2E Cdc34 resulted in significantly less FRET signal as compared with WT (Fig. 3D). In fact, despite the addition of high concentrations of labeled 2D2E Cdc34, we were unable to achieve saturation of SCF with 2D2E Cdc34. Thus, these results confirm that Cdc34 residues Glu-108 and Glu-112 appear to be important determinants in Cdc34 affinity for SCF, and their combined mutation profoundly affected the ability of Cdc34 to bind to SCF, whereas residues Asp-102 and Asp-103 do not appear to have similar roles.
FIGURE 3.
2D2E Cdc34 has decreased affinity for SCF as compared with wild-type Cdc34, especially at higher ionic strength. A, FRET efficiency (defined as 1 minus the ratio of the fluorescence for the donor-acceptor pair and the fluorescence of donor only (33)) was plotted versus the concentration of labeled wild-type Cdc34. The Kd of Cdc34 for SCF was estimated by fitting the data to a one-site binding model using nonlinear regression (GraphPad Prism software). B, same as A except with fluorescently labeled 2D2E Cdc34. C and D, same as A and B, respectively, except the reaction buffer contained 100 mm NaCl. All measurements were done at least in duplicate.
2D2E Cdc34 Maintains Lys-48 Chain Specificity
Cdc34 synthesizes ubiquitin chains onto its protein substrates in which isopeptide bond formation between adjacent ubiquitin protomers through lysine 48 is greatly preferred over the six other lysine residues on ubiquitin; this is referred to as Lys-48 specificity. Because the acidic loop is located adjacent to the enzyme active site, we reasoned that it may participate in the ability of Cdc34 to generate Lys-48-specific chains. To determine if this was the case, we used mass spectrometry to assess the lysine linkages of the ubiquitin chains from products generated in SCF-dependent ubiquitination reactions using two different peptide substrates. As expected, Lys-48-linked chains were the predominant chain linkages detected from reactions containing β-catenin peptide and WT Cdc34, and a similar frequency of Lys-48 linkages was observed for reactions containing 2D2E Cdc34 as well (Fig. 4A). Interestingly, ubiquitin chains from reactions containing WT Cdc34 also contained Lys-63 linkages, and reactions containing 2D2E Cdc34 showed an even greater frequency of Lys-63 linkages as compared with the Lys-48 ones. Similar results were observed when using cyclin E peptide as substrate. We reasoned that Lys-63-linked ubiquitin chains may have resulted from off-target reactions such as Cdc34 auto-ubiquitination. To address this likelihood, a protocol to purify the SCF-dependent substrates from the other reaction components was developed (Fig. 4, B and C). Indeed, the purification of a biotinylated cyclin E peptide substrate eliminated the signal for Lys-63 linkages from reactions containing either WT or 2D2E Cdc34 and resulted in the detection of only Lys-48-linked ubiquitin chains (data not shown). Thus, mutation of the conserved acidic residues in the Cdc34 acidic loop did not affect the lysine specificity of ubiquitin chains assembled by Cdc34 onto SCF-bound substrates.
Human Acidic Loop Residues His-98, Glu-108, and Glu-112 Assist in the Deprotonation of an Ionizable Group on Cdc34 or Ubiquitin
Our results presented thus far, and in combination with previous investigations into the molecular function of the Cdc34 acidic loop (17, 26), firmly establish that two of the conserved acidic loop residues (Glu-108 and Glu-112 in human Cdc34) play an important catalytic role during the ubiquitination reaction. For instance, notice that the kcat values for E108A, E112A, and E108A/E112A Cdc34 were substantially reduced as compared with WT Cdc34 even though Cdc34 was saturating SCF (Table 2).
In a seminal article on Ubc9, the E2 responsible for transferring SUMO to protein targets (SUMO is highly similar in sequence to ubiquitin), Chris Lima and Ali Yunus (44) revealed how Ubc9 deprotonates the lysine on protein substrates for SUMO. (Note that this is a required step during both SUMOylation and ubiquitination. As the typical pKa for the primary amine on lysine side chains is 10–11, the spontaneous dissociation of a proton is rare at physiological pH in the absence of an enzyme.) Specifically, Ubc9 contains several residues, including an aspartic acid residue (Asp-127 in human Ubc9), that catalyze SUMOylation by suppressing the pKa of the lysyl primary amine. Although this residue is conserved in both human Ubc9 as well as many ubiquitin-conjugating enzymes, it is not conserved in Cdc34. We therefore wondered whether Glu-108 and/or Glu-112 in the Cdc34 acidic loop may function similarly during the ubiquitination reaction.
If acidic loop mutant Cdc34 proteins are defective in lysyl deprotonation (or more generally in the deprotonation of any ionizable species on ubiquitin or Cdc34 that is required during the ubiquitination reaction), increasing the reaction pH should at least partially bypass the necessity for enzyme-catalyzed deprotonation. Because the prolonged exposure of E1 enzyme to high pH is known to reduce its activity (31), thioester-chase di-ubiquitin synthesis protocol was employed (Fig. 5A). First, Cdc34 and 32P-labeled K48R ubiquitin (donor ubiquitin) were incubated with E1 at either pH 7.5 or 9.2, forming the Cdc34∼ubiquitin complex. E1 activity was then quenched by the addition of apyrase, and the reactions were initiated by the addition of free, unlabeled acceptor ubiquitin.
The amounts of di-ubiquitin formation were first assessed for the WT, E108A, and E112A Cdc34 proteins at pH 7.5. Product formation was negligible for both E108A and E112A Cdc34 as compared with the WT (Fig. 5B and supplemental Fig. S2, A–C). In fact, nearly all of the Cdc34∼ubiquitin thioester bonds partitioned to hydrolysis instead of di-ubiquitin formation after E1 quenching. Importantly, these defects were not particular to the assay conditions (such as the elimination of E1 activity), as similar trends were observed when using a previously established steady-state di-ubiquitin synthesis assay (Fig. 6, A–C and E, and Table 3) (37, 38). The pH of the reaction buffer was then increased to 9.2, and the reactions were repeated. Remarkably, the amount of di-ubiquitin produced by either E108A or E112A Cdc34 was substantially restored to WT levels (Fig. 5, C–E, and supplemental Fig. S2, D–F) even though the rate of hydrolysis of the Cdc34∼ubiquitin bond was increased in the absence of ubiquitin acceptor at pH 9.2 (data not shown).
We next considered whether there might be additional residues in proximity to either Glu-108 or Glu-112 in the acidic loop that also participate in a deprotonation event during ubiquitination. Specifically, both Cdc34 and Ube2G2 orthologs contain an invariably conserved histidine residue (His-98 in human Cdc34 (Fig. 1A)) that is in close proximity to the active site, and its substitution to alanine in Ube2G2 results in nearly complete loss of E2 activity (29). Therefore, H98A Cdc34 was produced and assayed using the same thioester-chase di-ubiquitin synthesis assay as described above (Fig. 5A). Although product formation for H98A Cdc34 was barely detectable at pH 7.5 (Fig. 5B and supplemental Fig. S2, A–C), increasing the pH to 9.2 led to similar increases in H98A Cdc34 activity as observed for both Glu-108 and Glu-112 Cdc34 (Fig. 5, C–E, and supplemental Fig. S2, D–F).
Although these results indicate that Cdc34 residues His-98, Glu-108, and Glu-112 are involved in the deprotonation of an important group on Cdc34 or ubiquitin, including potentially lysine 48 on the acceptor ubiquitin, it is unclear how they may function at the molecular level. For instance, note that histidine residues can participate in catalysis in several ways, such as in the direct transfer of a proton from substrate to the imidazole ring, in the formation of a chemical environment that suppresses the pKa of an ionizable group on substrate, or in the stabilization of either the enzyme active site or the enzyme-substrate complex through hydrogen bonds. To help distinguish between these possibilities, both H98N and H98Q Cdc34 were produced, and their activities were compared with H98A Cdc34. This comparison is based on the notion that if His-98 forms hydrogen bonds with the acceptor ubiquitin, these more conservative mutations may still be capable of forming hydrogen bonds to substrate and may result in less severe reduction in Cdc34 activity as compared with H98A. Interestingly, the activities of both H98N and H98Q Cdc34 proteins were comparable with H98A Cdc34 (Fig. 6, B, D, and E), a result that is more consistent with the first two hypotheses.
To more generally explore whether Cdc34 residues His-98, Glu-108, and Glu-112 may function in the binding of acceptor ubiquitin, the affinities of WT, H98A, E108A, and E112A Cdc34 for acceptor ubiquitin were estimated using Michaelis-Menten kinetics (Fig. 7). The Km of ubiquitin for WT Cdc34 was 3.1 ± 0.3 mm, and the values for H98A, E108A, and E112A Cdc34 were also similar (2.5 ± 1.2, 2.2 ± 0.8, and 2.2 ± 1.2 mm, respectively). On the other hand, the kcat for WT Cdc34 was 59 ± 4.4 h−1, whereas the kcat values for H98A, E108A, and E112A Cdc34 were all at least 1 order of magnitude lower (1.9 ± 0.6, 5.9 ± 1.2, and 2.9 ± 1.0 mm, respectively). Taken together, these results are most consistent with a role of the acidic loop residues in assisting in the deprotonation of an important group on ubiquitin or Cdc34 and not in substrate binding.
Considering how His-98, Glu-108, and Glu-112 may participate in deprotonation, it is possible that they function synergistically. For instance, recall the catalytic triad mechanism for serine proteases, in which these enzymes have conserved active sites containing both a histidine and an adjacent aspartic acid residue. The hallmark feature of serine proteases is that the histidine and aspartic acid residues function only when both are present, as demonstrated by the substitution of either residue to alanine resulting in reductions in enzyme activity comparable to when both residues are substituted in combination (45). Therefore, both H98A/E108A and H98A/E112A Cdc34 proteins were generated and their activities were compared with the single mutants (Fig. 6, B, D, and E). The rate of di-ubiquitin formation for H98A/E108A Cdc34 was nearly identical to H98A Cdc34; however the activity of H98A/E112A Cdc34 was significantly lower (in fact, product formation was barely detectable over the time course). Finally, if His-98 and Glu-108 function together by forming an ion pair, the disruption of an electrostatic interaction by the H98A mutation would be predicted to have an effect on the Km of H98A Cdc34 for SCF similar to that of the E108A mutation (recall that the Km of E108A Cdc34 for SCF was increased relative to the wild type), and this was shown to be the case (supplemental Fig. S1G). Thus, these results suggest that His-98 and Glu-108 may function together during catalysis; however Glu-112 may have an independent role.
When considering the mechanism of the enzyme-catalyzed deprotonation of substrate and/or enzyme, it is often illuminating to measure enzyme activity as a function of pH. For example, in a study by Yunus and Lima (44), a single ionization event was deduced from the pH dependence of Ubc9-catalyzed SUMOylation reactions, corresponding to the primary amine of the lysine on protein substrate (that is, the lysine that becomes conjugated to SUMO). This was useful because it allowed for the estimation of the substrate lysine pKa, and thus the effects on lysine deprotonation for mutations of potentially important active site residues could be assessed directly. For example, mutation of Asp-127 to alanine in Ubc9 resulted in a substantial increase in the pKa of the substrate lysine. We performed a similar analysis on WT Cdc34, also demonstrating that Cdc34 activity varied with pH in a manner consistent with a singly titratable species with an apparent pKa of 6.4 ± 0.1 (Fig. 8). Similar to Ubc9, the mutation of Glu-112 to alanine also resulted in a large increase in the apparent pKa value. However, note that the pH profile for E112A Cdc34 was also more complex and suggested the presence of at least one additional ionizing species. For instance, although the majority of E112A Cdc34 activity titrated with an apparent pKa of 8.7 ± 0.1, there was also a small but reproducible amount of activity occurring with an apparent pKa of 6.4 ± 0.1. In addition, the same doubly ionizing behavior was also observed for H98A Cdc34. Therefore, these results potentially uncover the participation of at least two ionizable groups during Cdc34 catalysis.
The Expression of H100A Cdc34 in S. cerevisiae Results in a Slow Growth Phenotype
It is not uncommon for mutations that result in severe defects in Cdc34 activity in vitro to show little to no effect on cell viability when expressed in the absence of WT Cdc34 in vivo. To test for the effects of Cdc34 acidic loop mutations in living cells, alanine mutations were introduced in the yeast CDC34 gene at structurally equivalent positions to His-98, Glu-108, or Glu-112 in human Cdc34 (His-100, Glu-109, and Glu-113, respectively), and their impact was assessed on the growth of haploid yeast cells. All three mutant Cdc34 proteins were capable of sustaining viability; however a significant defect in the growth of a haploid strain carrying H100A Cdc34 was observed (Fig. 9A). Note that this was not caused by the expression level of Cdc34 proteins, as H100A Cdc34 levels were comparable to WT (Fig. 9B), although the expression of E113A Cdc34 was increased, which may explain why this mutant shows a normal phenotype in the assay. Furthermore, the reduction in growth rate was not due to a dominant-negative effect, as yeast expressing both WT and H100A Cdc34 grew at the same rate as yeast containing only WT Cdc34 protein (Fig. 9C).
FIGURE 9.
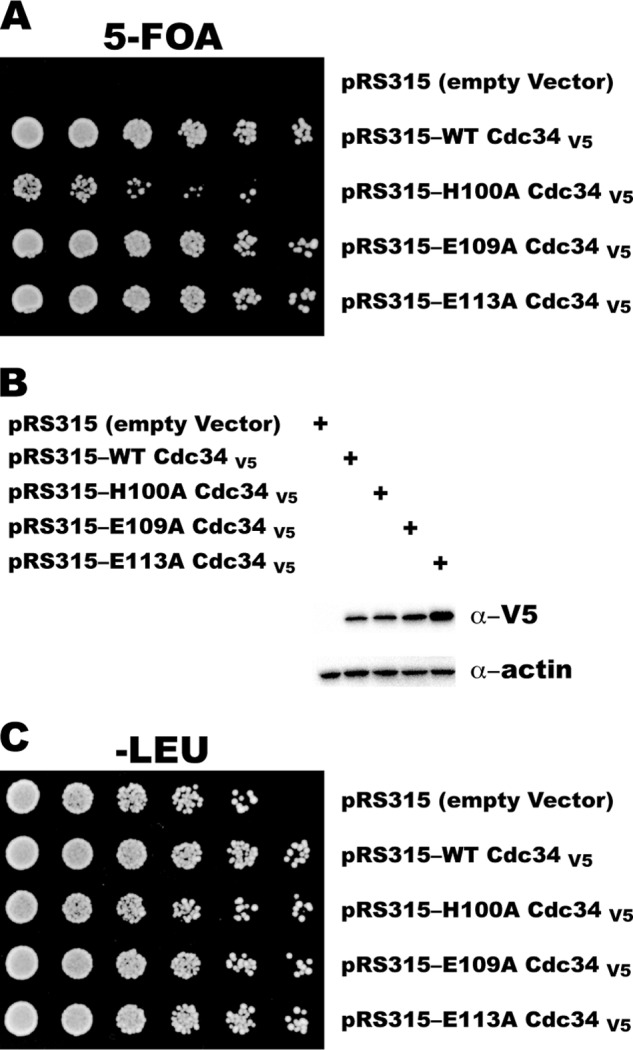
The replacement of WT Cdc34 with H100A Cdc34 in yeast results in a slow growth phenotype. A, haploid yeast strains lacking the chromosomal CDC34 gene were transformed with both pRS316 expressing WT Cdc34 and pRS315 expressing either WT or mutant Cdc34 with a C-terminal V5 epitope. Strains were grown in liquid media, serially diluted 3-fold, and plated onto SD medium with 5-fluoroorotic acid (5-FOA). B, Western blot showing the expression levels of the V5-tagged Cdc34 proteins. C, same as A, except the yeast strains were plated onto SD −LEU medium.
DISCUSSION
We conducted a thorough biochemical characterization of the human Cdc34 acidic loop with the goal of obtaining molecular insight into how conserved residues in the loop exert their influence during the ubiquitination reaction. Our investigation implied two roles for the loop: promoting the interaction between Cdc34 and SCF and the deprotonation of a key ionizable species on either Cdc34 or acceptor ubiquitin, possibly the primary amine on the lysine 48 side chain from the acceptor ubiquitin.
Interpreting the kcat and Km of Cdc34 for SCF and the Implications for Cdc34-SCF Binding and Catalysis
The kcat values reported here for the multi-turnover reactions are complex and could be affected by multiple microscopic rate constants, including the rates of substrate and/or product association and dissociation from SCF as well as the rate of ubiquitin transfer to substrate. Fortunately, these rate constants have been estimated by the quench-flow technique and enable the interpretation of the kinetic constants reported here (24). For instance, the rates of substrate and product dissociation (∼20 min−1) were substantially faster than the rate of the first ubiquitin transfer to β-catenin peptide, and thus the association and dissociation of substrates and products during the multi-turnover reactions in this study were unlikely to be rate-limiting for this event. Secondly, the rate of ubiquitin transfer to a β-catenin peptide that has been modified by one or more ubiquitins is 2 orders of magnitude faster than the rate of the first ubiquitin transfer to unmodified β-catenin (24), and thus the first ubiquitin transfer to substrate is the rate-limiting step for the multi-turnover ubiquitination reaction when β-catenin peptide is the substrate.
These observations have at least two relevant consequences. First, the kcat for the β-catenin peptide ubiquitination reaction estimates the rate of transfer of the first ubiquitin to substrate, and the obtained results here indicate a modest but significant defect for the acidic loop mutants in this step (especially E112A and H98A Cdc34 (Table 2)). Secondly, the value for kcat when using the Ub-β-catenin peptide as substrate and WT Cdc34 was undoubtedly limited by the rates of substrate and/or product dissociation from SCF; however this was less likely the case for the acidic loop mutants given their substantially reduced rates of ubiquitin transfer. Therefore, the differences in kcat for WT and either E108/112A or 2D2E Cdc34 (which were based on reactions containing Ub-β-catenin) greatly underestimate the actual differences in the rates of ubiquitin transfer (Table 2).
Previous studies on the interaction of Cdc34 and SCF (14), as well as the results presented here, indicate that the Km of Cdc34 for SCF from ubiquitination reactions is a reasonable estimate of their affinity. In fact, during the process of confirming this using a FRET-based binding assay, it was observed that an acidic loop Cdc34 mutant (2D2E Cdc34) had only a modestly reduced affinity for SCF in low salt conditions as compared with the wild type; however a greater effect on binding was observed when the ionic strength of the reaction buffer was increased. This curious result can be reconciled with recent work by Brenda Schulman and colleagues (46), where it was demonstrated that the interaction between Cdc34 and SCF is almost completely controlled by the acidic “tail” on Cdc34 at lower ionic strength. Thus, the influence of the acidic “loop” is only apparent at higher salt concentrations.
These results indicate that Cdc34 utilizes three points of contact during binding, including the Cdc34 catalytic domain (which binds to Rbx1, the RING subunit for SCF ligases), the acidic tail (which binds to the basic canyon on the Cul1 subunit surface), and the acidic loop. Why does Cdc34 form tridentate interactions with SCF, considering that there are E2s, such as the UbcH5 family, that have a minimal catalytic domain that is nevertheless sufficient to bind to E3s? One potential reason is that molecular add-ons can impart unique abilities to the E2. For example, the presence of the Cdc34 acidic tail does not simply increase the affinity for E3, it enables Cdc34 to associate with SCF with very rapid kinetics (14). Secondly, the existence of loops and tails can regulate E2 activity. For instance, multiple sites of phosphorylation in the Cdc34 acidic tail have been found that may serve as a mechanism for regulating Cdc34 activity throughout the cell cycle (47, 48). Finally, the presence of the acidic loop can promote allostery (see below).
The Deprotonation of a Key Group on Cdc34 or Ubiquitin Is Promoted by the Conserved Cdc34 Acidic Loop Residues His-98, Glu-108, and Glu-112
The kinetic results reported here, in combination with previous investigations (17, 26), firmly establish a molecular role for the Cdc34 acidic loop in catalysis. The molecular mechanism of action appears to involve promoting the deprotonation of an ionizable species on Cdc34 or ubiquitin, as the activities of several Cdc34 mutants were substantially rescued by increasing the pH of the ubiquitination reaction. It was reasoned that His-98, Glu-108, and/or Glu-112 may somehow function together, and whereas H98A/E108A Cdc34 was no more defective than either of the two single mutants (hinting that His-98 and Glu-108 may physically contact each other in the active site), this was not the case for H98A/E112A Cdc34.
It is tempting to speculate that the identity of the deprotonated species may be the primary amine on lysine 48 on acceptor ubiquitin, because the pH titrations involving the acidic loop mutants resulted in large increases in the apparent pKa values (Fig. 8) and were similar to results from a related and higher resolution investigation on Ubc9. Indeed, the mechanism of lysine activation is well established for Ubc9, the E2 responsible for SUMO conjugation (44). Human Ubc9 contains a conserved aspartic acid residue (Asp-127) that along with other key residues forms a microenvironment that suppresses the pKa of the lysyl primary amine on protein substrate. Notably, yeast Ubc9 contains a serine residue at a structurally equivalent position. However, the mutation of Asp-127 to serine in human Ubc9 was still competent in lysine activation, demonstrating that serine can function in a capacity similar to the aspartic acid residue. Like yeast Ubc9, human Cdc34 contains a serine residue (Ser-138) that occupies a structural position equivalent to Asp-127 in human Ubc9, hinting that this residue may function in lysine activation as well. In support of this suggestion, a human S138A Cdc34 mutant was highly defective in in vitro ubiquitin reactions (26); however the equivalent mutation in yeast Cdc34 complemented a cdc34 null mutation with no apparent defects in growth (21). Thus, it remains possible that Cdc34 promotes lysine activation by a distinct mechanism.
If our contention regarding lysine 48 on acceptor ubiquitin is correct, how then do Cdc34 residues His-98, Glu-108, and Glu-112 promote lysine deprotonation on ubiquitin? We acknowledge that the results presented here are insufficiently detailed to unambiguously assign a mechanism of action for these residues, and although it may be the case that they are directly involved in lysine activation (or more generally in the deprotonation of a group on Cdc34 or ubiquitin), it is also possible that their mutation affects deprotonation indirectly (see below). If they are directly involved in lysine activation, they may function by forming a microenvironment that suppresses the pKa of the lysyl primary amine, similar to the mechanism of lysine activation for Ubc9. Alternatively, His-98 may act as a general base and accept a proton directly from the lysine. Combining these models, His-98 may participate in acid-base chemistry, whereas Glu-112 complements this by suppressing the lysyl pKa on acceptor ubiquitin.
One caveat to these interpretations is the absence of structural evidence, and the mutation of the acidic loop residues in Cdc34 to alanine may have somehow indirectly perturbed the active site. For example, consider that Cdc34 and donor ubiquitin are known to form a non-covalent interface (in addition to the covalent thioester bond) that is important for catalysis (31). The superposition of a molecular model of the Cdc34∼ubiquitin structure onto the Ube2G2-RING x-ray structure (43) showed that the C terminus of ubiquitin was in close proximity to the distal end of the acidic loop (data not shown). Thus, if the mutation of the acidic loop residues changes the conformation of the loop, this could disrupt the position of the donor ubiquitin on Cdc34. In fact, a mutation that disrupts the Cdc34∼ubiquitin interface is partially rescued by increasing the pH (31). Regardless of the detailed mechanism of action, these results shed new light on the molecular function of the Cdc34 acidic loop and likely will aid in the interpretation of an atomic structure when it becomes available.
Understanding the Cdc34 Acidic Loop: Where Do We Go Next?
The presence of SCF stimulates Cdc34 activity by at least 1 order of magnitude (17), although it remains unclear how this allosteric activation is achieved. It may be that some of the acidic loop residues make direct contact with the SCF structure, which optimizes the conformation of the acidic loop for Cdc34 activity. In fact, preliminary investigations suggested that the acidic loop mutants used in this study were not significantly activated while in the presence of SCF (data not shown), and a comparison of a Cdc34∼ubiquitin model with the Ube2G2-RING structure suggested that the acidic loop may contact the C-terminal end of ubiquitin (see above). Thus, the acidic loop may activate Cdc34 by optimizing the position of donor ubiquitin on the surface of Cdc34.
Finally, note that the first two acidic residues in the loop (Asp-102 and Asp-103 in human Cdc34) appeared to have little or no contribution to the ubiquitination reaction, as their mutation to alanine resulted in mild defects at best. The strict conservation of these residues in both Cdc34 and Ube2G2 family members suggests their importance, and further investigation into their roles will be necessary to uncover how they contribute to Cdc34 function.
Acknowledgments
We thank Casey Hall and Shirley Shen (University of Nevada, Las Vegas, Genomics Center) for DNA sequencing, care of critical instruments, and expert advice. We also thank Anjanabha Saha for kindly providing the Ub-β-catenin peptide and both Nathan Pierce and Chris Lima for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants 5P20RR016464-11 from NCRR and 8P20GM103440-11 from NIGMS (to A. Z. and G. K.). This work was also supported by the Ellison Medical Foundation and the Sidney Kimmel Cancer Research Foundation (both to E. J. B.).

This article contains supplemental Figs. S1 and S2 and Table S1.
- E3
- ubiquitin ligase
- E1
- ubiquitin-activating enzyme
- E2
- ubiquitin-conjugating enzyme
- SCF
- Skp1-Cullin-Fbox ligase
- CRL
- Cullin-RING ligase
- Ub-β-catenin
- β-catenin peptide modified by a single ubiquitin
- ACN
- acetonitrile
- β-Cat
- β-catenin
- Ubc
- ubiquitin-conjugating enzyme.
REFERENCES
- 1. Dye B. T., Schulman B. A. (2007) Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu. Rev. Biophy. Biomol. Struct. 36, 131–150 [DOI] [PubMed] [Google Scholar]
- 2. Hershko A., Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 3. Pickart C. M. (2001) Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70, 503–533 [DOI] [PubMed] [Google Scholar]
- 4. Pickart C. M. (2004) Back to the future with ubiquitin. Cell 116, 181–190 [DOI] [PubMed] [Google Scholar]
- 5. Pickart C. M., Eddins M. J. (2004) Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta 1695, 55–72 [DOI] [PubMed] [Google Scholar]
- 6. Chau V., Tobias J. W., Bachmair A., Marriott D., Ecker D. J., Gonda D. K., Varshavsky A. (1989) A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243, 1576–1583 [DOI] [PubMed] [Google Scholar]
- 7. Thrower J. S., Hoffman L., Rechsteiner M., Pickart C. M. (2000) Recognition of the polyubiquitin proteolytic signal. EMBO J. 19, 94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cardozo T., Pagano M. (2007) Wrenches in the works: drug discovery targeting the SCF ubiquitin ligase and APC/C complexes. BMC Biochem. 8, Suppl. 1, S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deshaies R. J. (2009) Drug discovery: fresh target for cancer therapy. Nature 458, 709–710 [DOI] [PubMed] [Google Scholar]
- 10. Frescas D., Pagano M. (2008) Deregulated proteolysis by the F-box proteins SKP2 and β-TrCP: tipping the scales of cancer. Nat. Rev. Cancer 8, 438–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nakayama K. I., Nakayama K. (2006) Ubiquitin ligases: cell-cycle control and cancer. Nat. Rev. Cancer 6, 369–381 [DOI] [PubMed] [Google Scholar]
- 12. Deshaies R. J., Joazeiro C. A. (2009) RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 [DOI] [PubMed] [Google Scholar]
- 13. Soucy T. A., Smith P. G., Milhollen M. A., Berger A. J., Gavin J. M., Adhikari S., Brownell J. E., Burke K. E., Cardin D. P., Critchley S., Cullis C. A., Doucette A., Garnsey J. J., Gaulin J. L., Gershman R. E., Lublinsky A. R., McDonald A., Mizutani H., Narayanan U., Olhava E. J., Peluso S., Rezaei M., Sintchak M. D., Talreja T., Thomas M. P., Traore T., Vyskocil S., Weatherhead G. S., Yu J., Zhang J., Dick L. R., Claiborne C. F., Rolfe M., Bolen J. B., Langston S. P. (2009) An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736 [DOI] [PubMed] [Google Scholar]
- 14. Kleiger G., Saha A., Lewis S., Kuhlman B., Deshaies R. J. (2009) Rapid E2-E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell 139, 957–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Petroski M. D., Deshaies R. J. (2005) Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20 [DOI] [PubMed] [Google Scholar]
- 16. Schwob E., Böhm T., Mendenhall M. D., Nasmyth K. (1994) The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell 79, 233–244 [DOI] [PubMed] [Google Scholar]
- 17. Petroski M. D., Deshaies R. J. (2005) Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell 123, 1107–1120 [DOI] [PubMed] [Google Scholar]
- 18. Goebl M. G., Yochem J., Jentsch S., McGrath J. P., Varshavsky A., Byers B. (1988) The yeast cell cycle gene CDC34 encodes a ubiquitin-conjugating enzyme. Science 241, 1331–1335 [DOI] [PubMed] [Google Scholar]
- 19. Kolman C. J., Toth J., Gonda D. K. (1992) Identification of a portable determinant of cell cycle function within the carboxyl-terminal domain of the yeast CDC34 (UBC3) ubiquitin-conjugating (E2) enzyme. EMBO J. 11, 3081–3090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Silver E. T., Gwozd T. J., Ptak C., Goebl M., Ellison M. J. (1992) A chimeric ubiquitin-conjugating enzyme that combines the cell cycle properties of CDC34 (UBC3) and the DNA repair properties of RAD6 (UBC2): implications for the structure, function, and evolution of the E2s. EMBO J. 11, 3091–3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu Y., Mathias N., Steussy C. N., Goebl M. G. (1995) Intragenic suppression among CDC34 (UBC3) mutations defines a class of ubiquitin-conjugating catalytic domains. Mol. Cell. Biol. 15, 5635–5644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mathias N., Steussy C. N., Goebl M. G. (1998) An essential domain within Cdc34p is required for binding to a complex containing Cdc4p and Cdc53p in Saccharomyces cerevisiae. J. Biol. Chem. 273, 4040–4045 [DOI] [PubMed] [Google Scholar]
- 23. Wu K., Chen A., Tan P., Pan Z. Q. (2002) The Nedd8-conjugated ROC1-CUL1 core ubiquitin ligase utilizes Nedd8 charged surface residues for efficient polyubiquitin chain assembly catalyzed by Cdc34. J. Biol. Chem. 277, 516–527 [DOI] [PubMed] [Google Scholar]
- 24. Pierce N. W., Kleiger G., Shan S. O., Deshaies R. J. (2009) Detection of sequential polyubiquitylation on a millisecond timescale. Nature 462, 615–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pitluk Z. W., McDonough M., Sangan P., Gonda D. K. (1995) Novel CDC34 (UBC3) ubiquitin-conjugating enzyme mutants obtained by charge-to-alanine scanning mutagenesis. Mol. Cell. Biol. 15, 1210–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gazdoiu S., Yamoah K., Wu K., Pan Z. Q. (2007) Human Cdc34 employs distinct sites to coordinate attachment of ubiquitin to a substrate and assembly of polyubiquitin chains. Mol. Cell. Biol. 27, 7041–7052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Coccetti P., Tripodi F., Tedeschi G., Nonnis S., Marin O., Fantinato S., Cirulli C., Vanoni M., Alberghina L. (2008) The CK2 phosphorylation of catalytic domain of Cdc34 modulates its activity at the G1 to S transition in Saccharomyces cerevisiae. Cell Cycle 7, 1391–1401 [DOI] [PubMed] [Google Scholar]
- 28. Papaleo E., Ranzani V., Tripodi F., Vitriolo A., Cirulli C., Fantucci P., Alberghina L., Vanoni M., De Gioia L., Coccetti P. (2011) An acidic loop and cognate phosphorylation sites define a molecular switch that modulates ubiquitin charging activity in Cdc34-like enzymes. PLoS Comput. Biol. 7, e1002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li W., Tu D., Brunger A. T., Ye Y. (2007) A ubiquitin ligase transfers preformed polyubiquitin chains from a conjugating enzyme to a substrate. Nature 446, 333–337 [DOI] [PubMed] [Google Scholar]
- 30. Huang D. T., Walden H., Duda D., Schulman B. A. (2004) Ubiquitin-like protein activation. Oncogene 23, 1958–1971 [DOI] [PubMed] [Google Scholar]
- 31. Saha A., Lewis S., Kleiger G., Kuhlman B., Deshaies R. J. (2011) Essential role for ubiquitin-ubiquitin-conjugating enzyme interaction in ubiquitin discharge from Cdc34 to substrate. Mol. Cell 42, 75–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kleiger G., Hao B., Mohl D. A., Deshaies R. J. (2009) The acidic tail of the Cdc34 ubiquitin-conjugating enzyme functions in both binding to and catalysis with ubiquitin ligase SCFCdc4. J. Biol. Chem. 284, 36012–36023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Saha A., Deshaies R. J. (2008) Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol. Cell 32, 21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li T., Pavletich N. P., Schulman B. A., Zheng N. (2005) High-level expression and purification of recombinant SCF ubiquitin ligases. Methods Enzymol. 398, 125–142 [DOI] [PubMed] [Google Scholar]
- 35. Rappsilber J., Ishihama Y., Mann M. (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 36. Huttlin E. L., Jedrychowski M. P., Elias J. E., Goswami T., Rad R., Beausoleil S. A., Villén J., Haas W., Sowa M. E., Gygi S. P. (2010) A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hofmann R. M., Pickart C. M. (1999) Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96, 645–653 [DOI] [PubMed] [Google Scholar]
- 38. Hofmann R. M., Pickart C. M. (2001) In vitro assembly and recognition of Lys-63 polyubiquitin chains. J. Biol. Chem. 276, 27936–27943 [DOI] [PubMed] [Google Scholar]
- 39. Petroski M. D., Kleiger G., Deshaies R. J. (2006) Evaluation of a diffusion-driven mechanism for substrate ubiquitination by the SCF-Cdc34 ubiquitin ligase complex. Mol. Cell 24, 523–534 [DOI] [PubMed] [Google Scholar]
- 40. Ju T., Bocik W., Majumdar A., Tolman J. R. Solution structure and dynamics of human ubiquitin conjugating enzyme Ube2g2. Proteins 78, 1291–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huang H., Ceccarelli D. F., Orlicky S., St-Cyr D., Ziemba A., Garg P., Plamondon S., Auer M., Sidhu S., Marinier A., Kleiger G., Tyers M., Sicheri F. (2013) E2 enzyme inhibition by stabilization of a low affinity interface with ubiquitin. Nat. Chem. Biol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Papaleo E., Casiraghi N., Arrigoni A., Vanoni M., Coccetti P., De Gioia L. (2012) Loop 7 of E2 enzymes: an ancestral conserved functional motif involved in the E2-mediated steps of the ubiquitination cascade. PLoS One 7, e40786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Das R., Liang Y. H., Mariano J., Li J., Huang T., King A., Tarasov S. G., Weissman A. M., Ji X., Byrd R. A. (2013) Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine. EMBO J. 32, 2504–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yunus A. A., Lima C. D. (2006) Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nat. Struct. Mol. Biol. 13, 491–499 [DOI] [PubMed] [Google Scholar]
- 45. Carter P., Wells J. A. (1988) Dissecting the catalytic triad of a serine protease. Nature 332, 564–568 [DOI] [PubMed] [Google Scholar]
- 46. Duda D. M., Olszewski J. L., Tron A. E., Hammel M., Lambert L. J., Waddell M. B., Mittag T., DeCaprio J. A., Schulman B. A. (2012) Structure of a glomulin-RBX1-CUL1 complex: inhibition of a RING E3 ligase through masking of its E2-binding surface. Mol. Cell 47, 371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Block K., Appikonda S., Lin H. R., Bloom J., Pagano M., Yew P. R. (2005) The acidic tail domain of human Cdc34 is required for p27Kip1 ubiquitination and complementation of a cdc34 temperature sensitive yeast strain. Cell Cycle 4, 1421–1427 [DOI] [PubMed] [Google Scholar]
- 48. Sadowski M., Mawson A., Baker R., Sarcevic B. (2007) Cdc34 C-terminal tail phosphorylation regulates Skp1/Cullin/F-box (SCF)-mediated ubiquitination and cell cycle progression. Biochem. J. 405, 569–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fersht A. (1985) Enzyme Structure and Mechanism, 2nd Ed., p. 157, W. H. Freeman, New York [Google Scholar]