Background: The S1A serine proteases function in many biological processes and contain a conserved disulfide bond near the active site.
Results: This disulfide in the S1A mast cell protease, βII-tryptase, exists in oxidized and reduced states in the enzyme, which influences the specificity and efficiency of the enzyme.
Conclusion: βII-Tryptase is regulated by an allosteric disulfide bond.
Significance: Other S1A proteases are likely similarly regulated.
Keywords: Allosteric Regulation, Disulfide, Mast Cell, Redox Regulation, Serine Protease
Abstract
The S1A serine proteases function in many key biological processes such as development, immunity, and blood coagulation. S1A proteases contain a highly conserved disulfide bond (Cys191–Cys220 in chymotrypsin numbering) that links two β-loop structures that define the rim of the active site pocket. Mast cell βII-tryptase is a S1A protease that is associated with pathological inflammation. In this study, we have found that the conserved disulfide bond (Cys220–Cys248 in βII-tryptase) exists in oxidized and reduced states in the enzyme stored and secreted by mast cells. The disulfide bond has a standard redox potential of −301 mV and is stoichiometrically reduced by the inflammatory mediator, thioredoxin, with a rate constant of 350 m−1 s−1. The oxidized and reduced enzymes have different substrate specificity and catalytic efficiency for hydrolysis of both small and macromolecular substrates. These observations indicate that βII-tryptase activity is post-translationally regulated by an allosteric disulfide bond. It is likely that other S1A serine proteases are similarly regulated.
Introduction
Over 3% of human genes encode for proteolytic enzymes, and approximately one-quarter of these are serine proteases (1). The name stems from the nucleophilic serine in the active site of the enzyme, which attacks the carbonyl moiety of the substrate peptide bond. The serine proteases function in most biological processes, including development, fertilization, immunity, blood coagulation, digestion, and apoptosis. The majority of serine proteases have a trypsin fold, which is defined by a two β-barrel architecture, and trypsin-like substrate specificity, which is a preference for Arg or Lys side chains at the P1 position in the substrate (2). One class of the trypsin-like serine proteases are the mast cell tryptases.
Activated mast cells undergo degranulation, releasing tryptases, proteoglycans, and histamine into the extracellular space, promoting inflammation. Tryptases are one of the most abundant proteins stored in human mast cells (∼90% of the total protein in secretory granules), which contribute to inflammatory processes in an incompletely understood manner (3).
Among the identified forms of human tryptase, the β form is by far the most abundant in mast cells (3). The polymorphic TPSAB1 and TPSAB2 gene loci on chromosome 16p13.3 encode four tryptase isoforms: α and three nearly identical forms of β (βI, βII, and βIII). The α-tryptase isoform is inactive and is genetically absent in many human subjects (4), whereas the β-tryptases are proteolytically active, with βII predominant in the lung and βI abundant in the skin (5, 6). Little is known about the βIII form. Because the individual β-tryptase isoforms can be difficult to differentiate in vivo, they are often referred to generically as β-tryptase. β-Tryptase is involved in many acute and chronic inflammatory processes. This includes promotion of tissue remodeling, fibrosis, airway hyperreactivity, and bronchoconstriction (reviewed in Ref. 7). Increased levels of β-tryptase are commonly found in the bronchoalveolar lavage fluid of patients with allergic rhinitis (8) and atopic asthma (9). Increased levels of β-tryptase are also found in the joints of patients with inflammatory arthritis (10), whereas tryptase knock-out mice have significantly reduced joint disease during induction of inflammatory arthritis models (11).
Despite the growing correlation between β-tryptase and inflammatory responses, the actual biological targets of tryptase are relatively unknown. β-Tryptase has been proposed to cleave fibronectin, fibrinogen, protease-activated receptor 2, and kininogen and is also thought to activate matrix metalloproteinase-3 and pro-urokinase by discrete proteolysis (reviewed in Ref. 3). Also, no endogenous inhibitors of β-tryptase have been identified as yet. This poses puzzling questions as to how β-tryptase activity is regulated in vivo.
We have observed that the βII-tryptase Cys220–Cys248 disulfide bond exists in oxidized and reduced states in the enzyme stored and secreted by mast cells. The disulfide bond is reduced by the inflammatory mediator, thioredoxin. Importantly, the oxidized and reduced isoforms have different specificity and catalytic efficiency for hydrolysis of both small peptide and macromolecular protein substrates. These observations indicate that βII-tryptase activity is allosterically regulated by a redox active disulfide bond. The disulfide bond is highly conserved in S1A serine proteases, which suggests that other enzymes in this class will be similarly regulated.
EXPERIMENTAL PROCEDURES
Collection of Secreted Human β-Tryptase
The human leukemia mast cell line HMC-1 was kindly provided by Dr. J. H. Butterfield (Mayo Clinic, Rochester, MN). Cells were thawed from liquid nitrogen and grown in flasks containing Iscove's modified Dulbecco's medium supplemented with 10% FBS, 2 mm l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 0.01% α-thioglycerol. The cells were split and resuspended at 1 × 106 cells/ml into Iscove's modified Dulbecco's medium supplemented with 2 mm l-glutamine, 20 units/ml penicillin, 20 μg/ml streptomycin, and no serum. The cells were placed at 37 °C for 24 h before collection of the conditioned medium was collected by centrifugation and concentrated using Amicon Ultra 10-kDa cut-off centrifugal filters. For experiments where media supernatant and cell lysates were collected, α-thioglycerol and FBS were excluded from the media for 24 h prior to harvesting.
Collection of Murine Bone Marrow Mast Cells
The femurs and tibias of both wild type C57BL/6 and mouse mast cell protease 6 (mMCP-6−/−)2 null C57BL/6 mice (12) were harvested and placed in sterile RPMI 1640 medium supplemented with penicillin and streptomycin, after humane sacrifice of the mice. After removing extraneous tissue from the bone, the ends of the bones were removed with a scalpel, and 5 ml of enriched medium (RPMI 1640 medium supplemented with 10% FBS, 2 mm l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, 0.1 mm minimal essential medium nonessential amino acid solution, and 50 μm 2-mercaptoethanol) was injected into the core of the bone using a 25-gauge needle, expelling the bone marrow stem cells. The cells were cultured 50% enriched medium and 50% Wehi-3b conditioned medium supplemented with 50 units/ml of recombinant mouse IL-3. Media was changed weekly until >4 weeks post-harvest, when the presence of mouse bone marrow mast cells was confirmed by preparing cytospins and toluidine blue staining.
Labeling of Unpaired Cysteine Thiols in βII-Tryptase
HMC-1 conditioned medium, HMC-1 cell lysate, or bone marrow mast cell lysate was incubated with Me2SO or 2 mm PEG-maleimide (TMM(PEG)12; Thermo Scientific) for 1 h. A sample of the reaction was resolved on SDS-PAGE using NuPAGE Novex 4–12% Bis-Tris gel (Invitrogen) with MOPS running buffer under nonreducing conditions, protein-transferred to PVDF membrane (Millipore), and blotted with anti-mMCP-6 (R&D Systems; AF3736) and rabbit anti-goat peroxidase antibodies (Dako). The blots were visualized using chemiluminescence (NEN, PerkinElmer Life Sciences).
Recombinant βII-tryptase (Promega) was produced in Pichia pastoris cells and purified from the conditioned medium. βII-Tryptase (1 μm) was incubated without or with thioredoxin (0.5-1 μm) and then labeled with PEG-maleimide for 1 h at room temperature. Human thioredoxin-1 was produced as previously described (13, 14). Just before use, the active site disulfide bond of the oxidoreductase was reduced using tris-(2-carboxyethyl)phosphine-agarose (Thermo Scientific) for 30 min at room temperature before desalting the thioredoxin into buffer using a Zeba spin desalting column (Thermo Scientific). The βII-tryptase was resolved on SDS-PAGE, transferred to PVDF membrane, and blotted with an anti-tryptase antibody (anti-tryptase clone AA1, Dako; or anti-tryptase, clone G3, Millipore) and secondary goat anti-mouse peroxidase antibody.
Identification of the Redox Active Disulfide Bond in βII-Tryptase Using Mass Spectrometry
βII-Tryptase (1 μm) was incubated without or with reduced thioredoxin (0.5 μm) for 1 h at 30 °C in phosphate-buffered saline. We employed mass spectrometry with differential cysteine labeling to identify the redox active disulfide bond in the enzyme (15). Briefly, any free thiols in the samples were labeled with iodoacetamide before SDS-PAGE. Gels were stained with SYPRO Ruby (Invitrogen), and bands corresponding to tryptase were excised, destained, dried, and incubated with 100 mm DTT and then washed and incubated with 50 mm methyl methanethiolsulfonate (Sigma-Aldrich). The slices were washed and dried before digestion of proteins with 20 ng/μl of trypsin (Promega) or 12 ng/μl of chymotrypsin (Roche Applied Science) in 25 mm NH4CO2 overnight at 25 °C. Peptides were eluted from the slices with 5% formic acid, 50% acetonitrile and separated by liquid chromatography on an Ultimate 3000 HPLC (Dionex, Amsterdam, The Netherlands). Samples (2.5 μl) were concentrated and desalted onto a micro C18 precolumn (500 μm × 2 mm; Michrom Bioresources, Auburn, CA) with H2O:CH3CN (98:2, 0.05% trifluoroacetic acid) at 15 μl/min. After a 4-min wash, the precolumn was switched (Valco 10 port valve; Dionex) into line with a fritless nano column (75 μm × ∼10 cm) containing C18 medium (5 μ, 200 Å Magic; Michrom) manufactured as described (16). Peptides were eluted using a linear gradient of H2O:CH3CN (98:2, 0.1% formic acid) to H2O:CH3CN (64:36, 0.1% formic acid) at 250 nl/min over 30 min. High voltage (2000 V) was applied to low volume tee (Upchurch Scientific), and the column tip was positioned ∼0.5 cm from the heated capillary (280 °C) of an Orbitrap Velos (Thermo Electron, Bremen, Germany) mass spectrometer. Positive ions were generated by electrospray, and the Orbitrap operated in data-dependent acquisition mode.
A survey scan m/z 350–1750 was acquired in the Orbitrap (resolution = 30,000 at m/z 400, with an accumulation target value of 1,000,000 ions) with lockmass enabled. Up to the 10 most abundant ions (>5,000 counts) with charge states of >+2 were sequentially isolated and fragmented within the linear ion trap using collisionally induced dissociation with an activation q of 0.25 and an activation time of 30 ms at a target value of 30,000 ions. The m/z ratios selected for MS/MS were dynamically excluded for 30 s. MS data were searched using Mascot (V2.2; Matrix Science) against the nonredundant database from the National Center for Biotechnology Information. Search parameters were: precursor tolerance, 6 ppm; and product ion tolerances, ±0.6 Da. Methionine oxidation, Cys-methyldisulfide, and Cys-carboxyamidomethyl were selected as variable modifications with full tryptic and chymotryptic cleavage of up to five missed cleavages. To determine the extent of alkylated cysteine residues in tryptase, the relative ion abundance of peptides containing Cys-carboxyamidomethyl and Cys-methyldisulfide was used. To calculate ion abundance of peptides, extracted ion chromatograms were generated using the XCalibur Qual Browser software (v2.1.0; Thermo Scientific). The area was calculated using the automated peak detection function built into the software. The ratio of carboxyamidomethyl to methyldisulfide (the methyl methanethiolsulfonate adduct) labeling represents the fraction of the cysteine in the population that is in the reduced state.
Zymography
Zymography was performed using gelatin 10% Tris-glycine gels according to the manufacturer (Invitrogen).
Chemical Properties of the βII-Tryptase Cys220–Cys248 Disulfide Bond
Recombinant βII-tryptase (1 μm) was incubated in argon-flushed phosphate-buffered saline containing 0.1 mm EDTA, 0.5 mm DTTox (oxidized DTT or 4,5-dihydroxy-1,2-dithiane; Sigma) and various concentrations of DTTred (reduced DTT; dithiothreitol; Sigma) for 18 h at room temperature to allow equilibrium to be reached. Microcentrifuge tubes were flushed with argon prior to sealing to prevent oxidation during the incubation period. Reactions were quenched with 2 mm 3-(N-maleimido-propionyl) biocytin (MPB; Invitrogen) for 60 min at 22 °C and unreacted MPB quenched with glutathione (4 mm) for 60 min at 22 °C. Samples were resolved on SDS-PAGE as described above, protein-transferred to PVDF membrane, and blotted with 1:10,000 dilution of streptavidin-peroxidase to detect the MPB label. Band intensity was quantified using ImageJ version 1.43u software. The fraction of reduced Cys220–Cys248 disulfide bond was measured from the intensity of labeling with MPB with reference to labeling of fully reduced disulfide bond. The results were expressed as the ratio of reduced to oxidized protein and fitted to Equation 1,
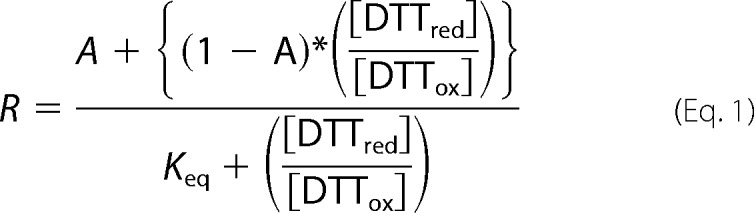 |
where R is the fraction of reduced protein at equilibrium, A is the fraction of reduced βII-tryptase in the native recombinant protein, and Keq is the equilibrium constant. The standard redox potential (E0′) of the Cys220–Cys248 bond was calculated using the Nernst equation (Equation 2),
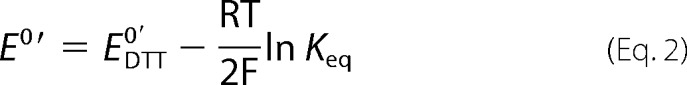 |
using a value of −307 mV for the redox potential of the DTT disulfide bond (17).
The rate of reduction by thioredoxin and rate of air oxidation of the Cys220–Cys248 disulfide were measured by incubating 1 μm recombinant βII-tryptase and 10 or 1 μm thioredoxin at 37 °C in argon-flushed phosphate-buffered saline containing 0.1 mm EDTA. Aliquots were removed at specified time points, and free thiols were immediately labeled with 200 μm MPB for 1 h before quenching unreacted MPB with 400 μm glutathione. For the rate of reduction of the β-tryptase disulfide bond, the data were fit to one-phase exponential association using Prism (GraphPad). For the rate of oxidation of β-tryptase, the data were fit to one-phase exponential decay using Prism (GraphPad).
Hydrolysis of a Tripeptide Chromogenic Substrate by Oxidized and Reduced βII-Tryptase
βII-Tryptase was incubated in the presence or absence of 2 μm thioredoxin in one of four different buffer conditions for 1 h at 37 °C: pH 7.4, ionic strength 50 (50 mm HEPES, 30 mm NaCl, 10% glycerol, 1 mg/ml PEG 6000); pH 7.4, ionic strength 150 (50 mm HEPES, 130 mm NaCl, 10% glycerol, 1 mg/ml PEG 6000); pH 6.2, ionic strength 50 (50 mm MES, 30 mm NaCl, 10% glycerol, 1 mg/ml PEG 6000); and pH 6.2, ionic strength 150 (50 mm MES, 130 mm NaCl, 10% glycerol, 1 mg/ml PEG 6000). After diluting into corresponding buffer with substrate, the initial rate of hydrolysis of pyro-Glu-Pro-Arg-p-nitroanilide (Hyphen Biomed or Sigma-Aldrich) by oxidized or reduced βII-tryptase (10 nm) was measured as a function of peptide substrate concentration (0–2 mm).
Hydrolysis of Fibronectin and Fibrinogen by Oxidized and Reduced βII-Tryptase
βII-Tryptase was prepared as described for the chromogenic substrate assay. Any unpaired cysteines in the thioredoxin were alkylated with 25 mm iodoacetamide for 1 h to prevent thioredoxin from reducing disulfide bonds in fibronectin or fibrinogen. Alkylation of reduced βII-tryptase with iodoacetamide had no effect on activity in the chromogenic substrate assay (data not shown). Oxidized or reduced βII-tryptase (20 nm) was incubated with fibronectin (Sigma-Aldrich; 10 mg/ml) or fibrinogen (Sigma-Aldrich; 5 mg/ml) at room temperature for 21 h in either 50 mm HEPES, pH 7.4 buffer containing 30 mm NaCl, 10% glycerol, and 1 mg/ml PEG 6000 or 50 mm MES, pH 6.2 buffer containing 30 mm NaCl, 10% glycerol, and 1 mg/ml PEG 6000. Aliquots were removed at various time points, and cleavage of fibronectin was measured by resolving the reactions on SDS-PAGE and staining with silver (Thermo Scientific). βII-Tryptase is not visible in the stained gels because of the low concentration, and thioredoxin (17.2 kDa) runs significantly below the fibronectin (30–50 kDa) and fibrinogen (200–400 kDa) peptides and does not interfere with interpretation of protein cleavage.
RESULTS
Native Human β-Tryptase and the Equivalent Murine Enzyme Contain a Reduced Disulfide Bond
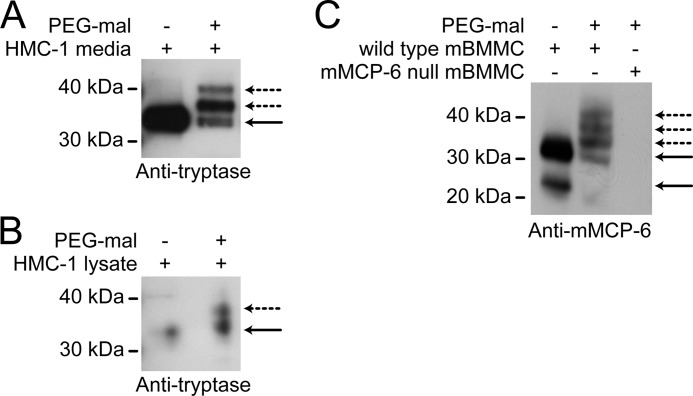
The human mast cell line, HMC-1, produces, stores, and secretes β-tryptase (18). We confirmed secretion of the active enzyme by chromogenic substrate assays of conditioned medium from serum-deprived cells (not shown). Conditioned medium and lysate of HMC-1 cells was incubated with a branched polyethylene glycol-maleimide (three 12-PEG units). Maleimides specifically react with thiolates at neutral pH (19). This alkylator adds a mass of ∼2.5 kDa per labeled thiol, and incorporation is measured by increase in molecular mass by SDS-PAGE and immunoblotting of the protein of interest. Both the secreted (Fig. 1A) and lysate (Fig. 1B) β-tryptase incorporated up to two molecules of PEG-maleimide, indicating the presence of two unpaired cysteine thiols in the enzyme. The crystal structure of βII-tryptase shows that all eight cysteines in each monomer participate in a disulfide bond (20). This result indicates that β-tryptase stored and secreted by HMC-1 cells contains a reduced disulfide bond. The majority of the β-tryptase was labeled with PEG-maleimide, indicating that most of the secreted enzyme was in the reduced state. This finding suggested that β-tryptase is post-translationally regulated by a redox active disulfide bond.
FIGURE 1.
Native human β-tryptase and the equivalent murine enzyme contain a reduced disulfide bond. A and B, conditioned medium (A) and lysate (B) of HMC-1 cells were labeled with the PEG-tagged thiol specific alkylator, PEG-maleimide (PEG-mal). The labeled samples were resolved on SDS-PAGE and immunoblotted for β-tryptase. The PEG-maleimide adds a mass of ∼2.5 kDa for each labeled thiol in β-tryptase. The enzyme in both milieu incorporated PEG-maleimide, indicating the presence of unpaired cysteine thiols. The solid lines are unlabeled β-tryptase, and the dotted lines are the PEGylated protein. C, wild type mMCP-6 or mMCP-6 null bone marrow mast cell (mBMMC) lysates were labeled with PEG-maleimide and a sample resolved on SDS-PAGE and immunoblotted for mMCP-6. The mMCP-6 labels with PEG-maleimide. As a control, no mMCP-6 was detected in the mouse bone marrow mast cell lysate of mMCP-6 null mice. The solid lines are unlabeled β-tryptase, and the dotted lines are the PEGylated protein.
The murine equivalent of β-tryptase is mMCP-6, which shares 77% sequence identity to human βII-tryptase and has similar substrate specificity (3, 21). mMCP-6 contains the same conserved disulfide bonds as human βII-tryptase. Bone marrow mast cells were harvested from wild type and mMCP-6 null C57BL/6 mice, and the redox state of mMCP-6 in bone marrow mast cell lysate was measured by labeling with PEG-maleimide. The majority of the mMCP-6 is reduced (Fig. 1C), which is consistent with the nature of the enzyme stored and secreted by human mast cells (Fig. 1, A and B). No bands were observed in the mMCP-6 null mice, indicating that the antibody is specific for mMCP-6.
The Cys220–Cys248 Disulfide Bond Is Reduced in βII-Tryptase
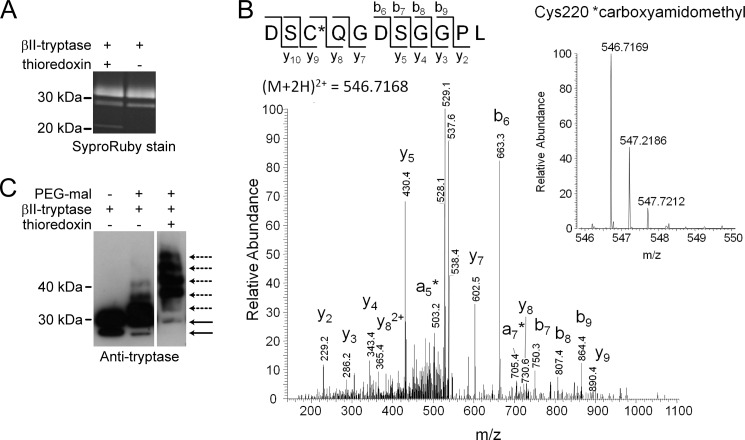
The identity of the redox active disulfide bond in βII-tryptase was determined using recombinant enzyme produced in yeast cells and mass spectrometry. Differential cysteine labeling allows identification of the individual cysteines that are unpaired and those that participate in a disulfide bond. The unpaired cysteines were alkylated with iodoacetamide and the disulfide-bonded cysteines with methyl methanethiolsulfonate following reduction with DTT. The ratio of carboxyamidomethyl (the iodoacetamide adduct) to methyldisulfide (the methyl methanethiolsulfonate adduct) labeling represents the fraction of the cysteine in the population that is in the reduced state. We were able to detect seven of the eight cysteines in βII-tryptase and so could measure the redox state of all four disulfide bonds in the enzyme.
Recombinant βII-tryptase appears as two bands of ∼34 and 30 kDa on SDS-PAGE because of different glycosylation patterns (3) (Fig. 2A). The Cys220–Cys248 disulfide bond was reduced in ∼30% of βII-tryptase molecules in the preparation, which increased to ∼80% following incubation of 1 μm enzyme with 0.5 μm thioredoxin (Fig. 2B and Table 1). Thioredoxin is a relevant protein reductant in the cell and in inflammatory environments where β-tryptase is found. Thioredoxin gene expression is up-regulated, and the protein is secreted by immune cells during inflammation, leading to a high local concentration and also elevated blood levels (22). High serum levels of thioredoxin have been observed in patients with asthma, rheumatoid arthritis, and heart failure (23, 24), and serum levels correlate with disease activity in rheumatoid arthritis (25, 26). The ∼50% increase in reduction of the Cys220–Cys248 disulfide bond in an incubation of 0.5 mol of thioredoxin/mol of βII-tryptase indicates that the reaction is stoichiometric. Thioredoxin did not influence the redox state of the other three disulfide bonds in βII-tryptase.
FIGURE 2.
The Cys220–Cys248 disulfide bond is reduced in βII-tryptase. A, the SYPRO Ruby-stained βII-tryptase used for mass spectrometry analysis. B, tandem mass spectrum of the DSCQGDSGGPL peptide from βII-tryptase showing the reduced form of Cys220 (underlined) labeled with the IAM adduct, carboxyamidomethyl. The accurate mass spectrum of the peptide is shown in the inset (observed [M + 2H]2+ = 546.7169 m/z; expected [M+2H]2+ = 546.7168 m/z). C, βII-tryptase (1 μm) was incubated without or with thioredoxin (1 μm) and then labeled with PEG-maleimide. The labeled sample was resolved on SDS-PAGE and immunoblotted for βII-tryptase. The solid lines are unlabeled β-tryptase, and the dotted lines are the PEGylated protein.
TABLE 1.
The Cys220–Cys248 disulfide bond in βII-tryptase is reduced in a fraction of the molecules, and the oxidized fraction can be reduced by thioredoxin
The fraction of peptides containing Cys220 or Cys248 that labeled with IAM increased upon addition of thioredoxin (bold face type), indicating reduction of the Cys220–Cys248 disulfide bond. Thioredoxin selectively reduced this bond because there was no appreciable change in the IAM labeling of the other six cysteines in the protease (Cys59–Cys75, Cys155–Cys230, and Cys188–Cys211). MMTS, methyl methanethiolsulfonate.
| Digestion | βII-Tryptase peptide | Cys | No thioredoxin |
With thioredoxin |
||||
|---|---|---|---|---|---|---|---|---|
| Abundance |
Percentage reduced | Abundance |
Percentage reduced | |||||
| IAM adduct | MMTS adduct | IAM adduct | MMTS adduct | |||||
| Chymotrypsin | CGGSLIHPQW | 59 | 60,999 | 287,060 | 18 | 402,680 | 127,741 | 25 |
| Trypsin | TAAHCVGPDVK | 75 | 28,314 | 600,511 | 5 | 18,112 | 384,333 | 5 |
| Trypsin | VPIMENHICDAK | 188 | 5,788,350 | 23,433,269 | 20 | 4,242,676 | 14,492,330 | 23 |
| Trypsin | IVRDDMLCAGNTR | 211 | 1,580,273 | 9,798,215 | 14 | 1,067,604 | 5,531,002 | 16 |
| Trypsin | DSCQGDSGGPL | 220 | 146,813 | 474,108 | 24 | 176,254 | 43,489 | 80 |
| Trypsin | DSCQGDSGGPLVCK | 230 | 775,247 | 11,931,392 | 6 | 433,216 | 6,693,478 | 6 |
| Chymotrypsin | GEGCAQPNRPGIY | 248 | 643,460 | 1,302,075 | 33 | 1,885,816 | 354,137 | 84 |
To test the veracity of the mass spectrometry analysis, the reduced βII-tryptase was labeled with PEG-maleimide. A fraction of the βII-tryptase incorporated two molecules of PEG-maleimide, in accordance with partial reduction of the Cys220–Cys248 disulfide bond, and incubation with a molar equivalent of thioredoxin fully reduced the enzyme, which is consistent with the mass spectrometry analysis (Fig. 2C).
Chemical Properties of the βII-Tryptase Cys220–Cys248 Disulfide Bond
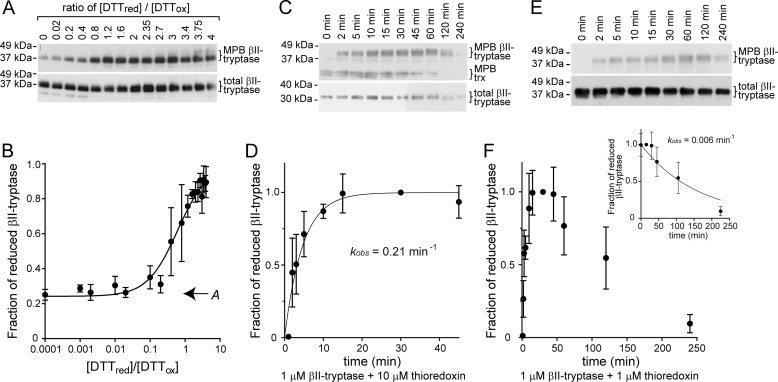
The redox potential and rate of reduction of the βII-tryptase Cys220–Cys248 disulfide bond by thioredoxin were measured. The standard redox potential of the Cys220–Cys248 disulfide bond was determined using oxidized and reduced DTT. The MPB labeling of fully reduced βII-tryptase was used to calculate the ratio of reduced to oxidized βII-tryptase as a function of the ratio of reduced to oxidized DTT (Fig. 3A). A standard redox potential of −301 mV was calculated (Fig. 3B). Analysis of the DTT titration results indicated that 24% of the βII-tryptase molecules were already reduced (variable A in Equation 1; Fig. 3B). This result is consistent with the fraction of reduced βII-tryptase molecules in the preparation measured by mass spectrometry (Table 1) and PEG-maleimide labeling (Fig. 2C).
FIGURE 3.
Chemical properties of the βII-tryptase Cys220–Cys248 disulfide bond. A, βII-tryptase was incubated with 0.5 mm oxidized DTT (DTTox) and varying concentrations of reduced DTT (DTTred). Reduced protein was labeled with maleimide-biotin (MPB) and resolved on SDS-PAGE, and the MPB label was visualized by blotting with streptavidin-peroxidase. The positions of molecular mass standards in kDa are shown on the left. B, plot of the ratio of reduced to oxidized βII-tryptase as a function of the ratio of reduced to oxidized DTT from experiments of the type shown in A. The solid line represents the best nonlinear least squares fit of the data to Equation 1. The calculated equilibrium constant is 0.62 ± 0.13, and offset A is 0.24 ± 0.04 (indicated on graph). From the Nernst equation, the standard redox potential of the Cys220–Cys248 disulfide is −301 mV. The data points and errors are the means ± S.E. of three independent experiments. C, rate of reduction of the Cys220–Cys248 βII-tryptase disulfide bond by thioredoxin (trx). βII-Tryptase (1 μm) was incubated with thioredoxin (10 μm), and the reduced enzyme was detected by labeling with MPB at discrete times as described above. The positions of molecular mass standards in kDa are shown at the left. D, plot of the fraction of reduced βII-tryptase as a function of time from experiments of the type shown in C. The solid line represents the best nonlinear least squares fit of the data to a single exponential. The calculated kobs for the pseudo-first order reaction is 0.21 ± 0.05 min−1. The data points and errors are the means ± S.E. of three independent experiments. E, rate of oxidation of the Cys220 and Cys248 thiols. βII-Tryptase (1 μm) was incubated with thioredoxin (1 μm), and the reduced enzyme was detected by labeling with MPB at discrete times as described above. The positions of molecular mass standards in kDa are shown at the left. F, plot of the fraction of reduced βII-tryptase as a function of time from experiments of the type shown in E. The inset is the best nonlinear least squares fit of the oxidation data to a single exponential decay. The calculated kobs for the reaction is 0.006 ± 0.002 min−1. The data points and errors are the means ± S.E. of three independent experiments.
The rate of reduction of the βII-tryptase Cys220–Cys248 disulfide bond by thioredoxin was measured under pseudo-first order conditions (Fig. 3C). A kobs of 0.21 ± 0.05 min−1 was calculated, which equates to a second order rate constant of 350 m−1 s−1 (Fig. 3D). The rate of oxidation of the Cys220 and Cys248 dithiol was calculated from reactions of equimolar βII-tryptase and thioredoxin (Fig. 3E). The bond was reduced after ∼30 min, and the reduced thioredoxin was consumed in the reaction. Oxidation of the Cys220 and Cys248 thiols in the enzyme exposed to ambient air was monitored with time. The kobs for the oxidation was 0.006 ± 0.002 min−1 (Fig. 3F, inset), which is 35-fold slower than the rate of reduction of the bond by a 10-fold molar excess of thioredoxin.
Reduction of the Cys220–Cys248 βII-Tryptase Disulfide Bond Does Not Influence Tetramer Formation
β-Tryptase forms a ∼134-kDa homotetramer that is stored in mast cells as the fully active form rather than as a pro-enzyme, both of which are unique traits among the S1A proteases. The crystal structure of β-tryptase is a tetramer composed of four quasiequivalent monomers arranged in a square flat ring (20). In the tetramer, all of the active sites face into the central pore (20), limiting both substrate and inhibitor access (27). Whether or not the β-tryptase monomer possesses enzyme activity is debated.
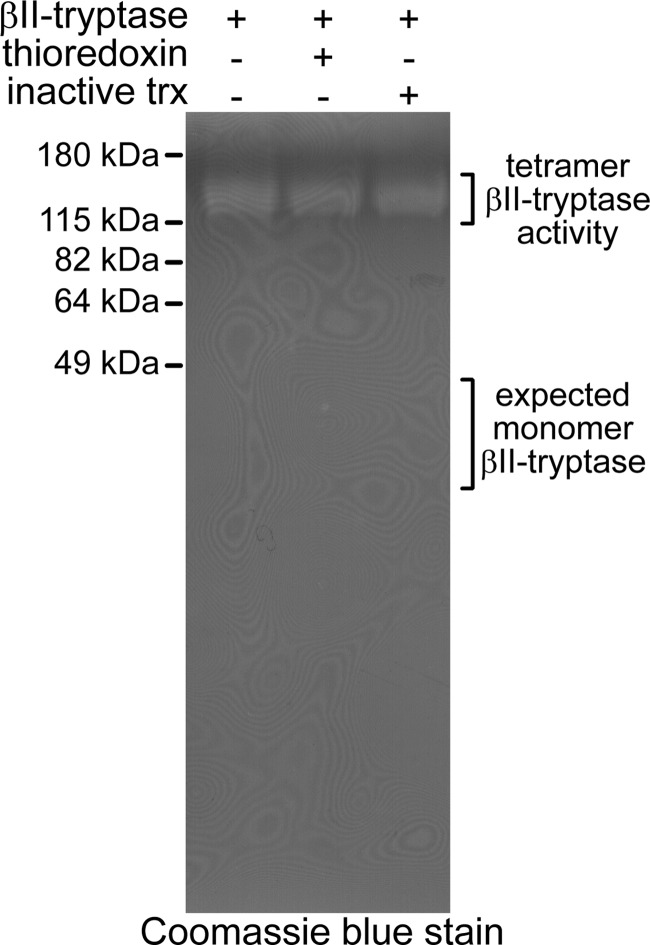
The effect of thioredoxin reduction of the Cys220–Cys248 disulfide bond on tetramer formation was assessed using gelatin zymography. The zymogram separates the monomers from tetramers in mild denaturing conditions, and enzyme activity is detected from degradation of the gelatin embedded in the gel. Only the tetrameric βII-tryptase is active in this assay, and thioredoxin treatment had no obvious effect on tetramer formation or gelatinase activity (Fig. 4).
FIGURE 4.
Reduction of the Cys220–Cys248 βII-tryptase disulfide bond does not influence tetramer formation. βII-Tryptase (1 μm) was incubated without or with thioredoxin (1 μm), and the enzyme was resolved on a zymogram gelatin gel. βII-Tryptase activity was observed in the region between 115 and 180 kDa, indicating that the βII-tryptase tetramer (134 kDa) was the source of proteolytic activity. No proteolytic activity was seen in the region of the βII-tryptase monomer (34 kDa).
Oxidized and Reduced βII-Tryptase Cleave a Small Peptide Substrate with Different Efficiency
The Cys220–Cys248 disulfide bond links two β-loop structures that define the rim of the active site pocket of βII-tryptase (20). Structural changes to this region have previously been shown to alter enzymatic activity of the tryptases (28–30). Thus, it was anticipated that cleavage of this disulfide bond would alter the S1 pocket region and influence the catalytic properties of the enzyme. This was tested using the chromogenic substrate, pyro-Glu-Pro-Arg-p-nitroanilide.
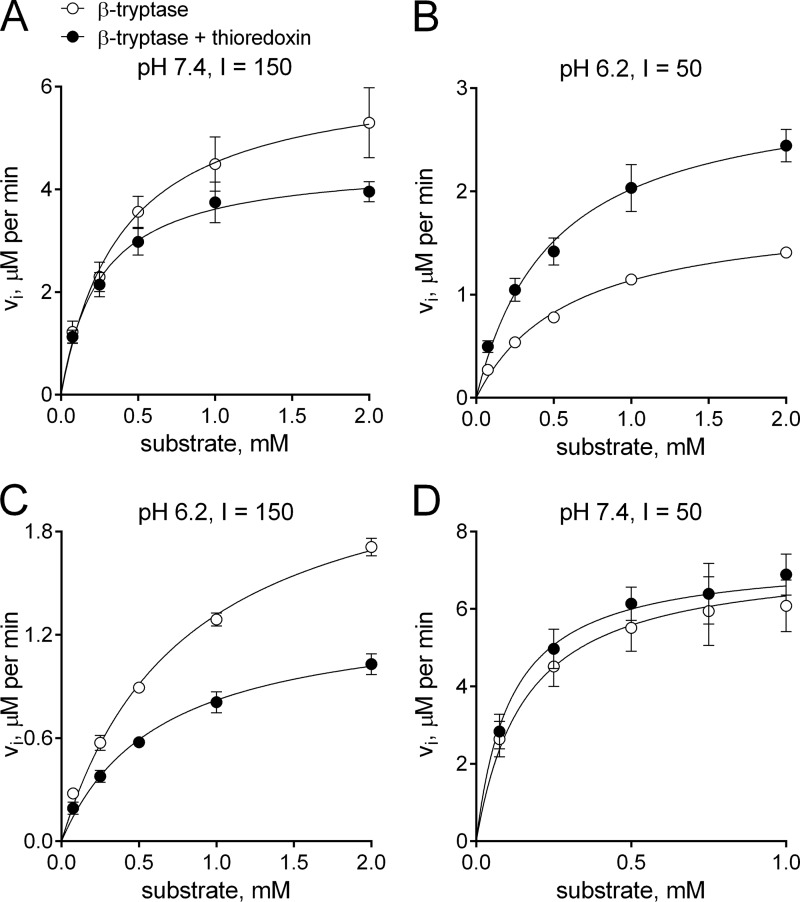
We tested the activity of βII-tryptase at neutral pH (7.4) and at pH 6.2 because the intracellular pH of mast cells and granules, where βII-tryptase is stored, has been found to range from 5.2 to 6.5 (31–33). The enzyme also functions in acidic inflammatory environments such as the synovial fluid of rheumatoid arthritic joints (34, 35) and the airways of asthmatics (36, 37). Synovial fluid acidosis positively correlates with radiological knee joint destruction in patients with rheumatoid arthritis (35), and the average pH of airway vapor in patients with acute asthma is 5.23 (36). Acidic pH has also been shown to alter β-tryptase activity toward various substrates (38–41), making it an important variable to test. Because ionic strength has been found to influence β-tryptase activity and the equilibrium between monomer and tetramer (38–41), we also selected two different ionic strengths for our assays. The catalytic efficiency of oxidized versus reduced βII-tryptase was therefore tested at both neutral (7.4) and acidic (pH 6.2) pH and also at physiological (I 150) and low (I 50) ionic strength.
There was a small difference in the maximal velocity (Vmax) of the oxidized versus reduced enzyme at neutral pH and physiological ionic strength (Fig. 5A) but no difference at low ionic strength (Fig. 5B). The difference in Vmax was more pronounced at pH 6.2 at either ionic strength (Fig. 5, C and D). There was no significant change in Km at either pH or ionic strength (Table 2).
FIGURE 5.
Oxidized and reduced βII-tryptase cleave a small peptide substrate with different efficiency. The initial rate of hydrolysis of pyro-Glu-Pro-Arg-p-nitroanilide by oxidized or reduced βII-tryptase was measured at pH 7.4 (A and D) or 6.2 (B and C) in low (I = 50) or physiological ionic strength buffer (I = 150). There is a small difference in the maximal velocity (Vmax) of the oxidized versus reduced enzyme at neutral pH and physiological ionic strength (A), but no difference at low ionic strength (D). The difference in Vmax was more pronounced at pH 6.2 at either ionic strength (B and C). There was no significant change in the Michaelis constant (Km) at either pH or ionic strength (Table 2). The data points and errors are the means ± S.E. of three determinations, and the solid lines represent the nonlinear least squares fit of the data to the Michaelis-Menten competitive substrate inhibition model.
TABLE 2.
Kinetic parameters for hydrolysis of pyro-Glu-Pro-Arg-p-nitroanilide by oxidized and reduced βII-tryptase
The values were determined in experiments of the type shown in Fig. 4. Errors are ± S.E.
| Conditions | Enzyme form | Vmax | Km |
|---|---|---|---|
| m/s | μm | ||
| pH 6.2, low ionic strength (50) | βII-Tryptase | 1.8 ± 0.1 | 605 ± 69 |
| βII-Tryptase + thioredoxin | 3.0 ± 0.3a | 495 ± 118 | |
| pH 7.4, low ionic strength (50) | βII-Tryptase | 6.9 ± 0.7 | 125 ± 52 |
| βII-Tryptase + thioredoxin | 7.7 ± 0.6 | 132 ± 42 | |
| pH 6.2, high ionic strength (150) | βII-Tryptase | 2.4 ± 0.1 | 796 ± 87 |
| βII-Tryptase + thioredoxin | 1.3 ± 0.1b | 634 ± 119 | |
| pH 7.4, high ionic strength (150) | βII-Tryptase | 6.3 ± 0.7 | 394 ± 120 |
| βII-Tryptase + thioredoxin | 4.5 ± 0.3c | 254 ± 59 |
a p ≤ 0.01.
b p ≤ 0.001.
c p ≤ 0.05.
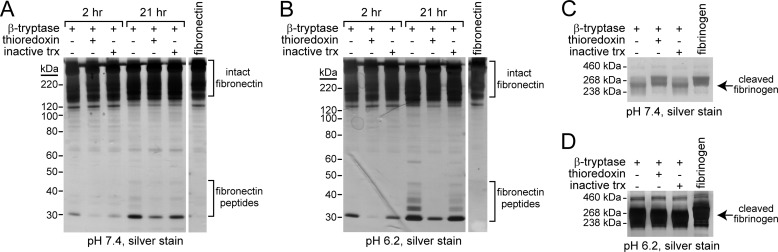
Oxidized and Reduced βII-Tryptase Cleave Macromolecular Substrates with Different Efficiency and Specificity
The redox state of the β-tryptase Cys220–Cys248 disulfide bond also influences cleavage of the macromolecular substrates fibronectin and fibrinogen. βII-Tryptase cleaves the large substrate fibronectin into multiple, smaller peptides ranging from ∼30–50 kDa in size, which can be visualized using SDS-PAGE and silver staining. βII-Tryptase has also been found to cleave fibrinogen, which can be visualized as a shift from a molecular mass of ∼340 kDa to a slightly smaller protein ∼238–268 kDa on SDS-PAGE (42).
Both the rate of cleavage of fibronectin and the sites of cleavage differ for the oxidized and reduced enzyme, which is more pronounced at acidic pH. βII-Tryptase cleaves fibronectin more rapidly and at more sites at pH 6.2 (Fig. 6B) than pH 7.4 (Fig. 6A), and there is significantly less fibronectin cleavage by reduced βII-tryptase at both pH. Incubation of βII-tryptase with redox-inactive thioredoxin mimicked the results in the absence of thioredoxin. This experiment controlled for the possibility that thioredoxin was acting as a competitive substrate for βII-tryptase in the assay and so biasing the results. Reduced βII-tryptase was also less efficient at cleaving fibrinogen at neutral pH (Fig. 6C), but both forms of the enzymes were equally efficient at pH 6.2 (Fig. 6D). βII-Tryptase has been observed to cleave fibrinogen at a faster rate in acidic pH (43).
FIGURE 6.
Oxidized and reduced βII-tryptase cleave macromolecular substrates with different efficiency and specificity. A and B, oxidized or reduced β-tryptase was incubated with fibronectin, and the cleavage products were resolved on SDS-PAGE and silver-stained. As a control for the possible effects of thioredoxin on the substrate, βII-tryptase was also incubated with a redox-inactive form of thioredoxin (inactive trx). Oxidized and reduced βII-tryptase produce different fibronectin peptides at both pH 7.4 (A) and 6.2 (B). Intact fibronectin can be seen near the top of the gel, whereas cleaved portions of fibronectin are ∼30–40 kDa in size. C and D, oxidized or reduced β-tryptase was incubated with fibrinogen, and the cleavage products were resolved on SDS-PAGE and silver-stained. Oxidized but not reduced βII-tryptase cleaves fibrinogen at pH 7.4. There is little to no difference in the fibrinogen cleavage products produced by the oxidized and reduced forms of βII-tryptase at pH 6.2.
DISCUSSION
The tryptases belong to the serine protease subfamily S1A, a group of trypsin-like, neutral serine proteases categorized by their function: coagulation and immunity, digestion, tryptase, kallikrein, granzymes, and matriptase (44). A global alignment of 79 human S1A proteases found that the Cys191–Cys220 disulfide bond (chymotrypsin numbering) is conserved in all forms of human tryptase and in all S1A serine proteases, except for the cathepsins and granzymes, which lack the disulfide bond (44).
The structural/functional role of the Cys191–Cys220 disulfide bond has been investigated in some S1A proteases. In coagulation factor XII, the disulfide bond (Cys540–Cys571 in factor XII) is essential for activity. In the rare blood coagulation disorder known as Hageman trait, Cys571 is mutated to a serine in factor XII, which ablates the disulfide bond and inactivates the enzyme (45). Elimination of the disulfide bond in the blood coagulation protease thrombin (Cys540–Cys571 in thrombin) results in a ∼100-fold loss of activity toward peptide substrates (46). The disulfide bond in trypsin (Cys173–Cys197 in trypsin) is also important for activity, although there are conflicting reports on the nature of the change in activity upon elimination of the bond. One study found that ablation of the disulfide bond has no effect on the function, structure, Km, or stability of the enzyme but does affect the kcat for hydrolysis of peptide substrates (47). Another study reported that eliminating the disulfide bond increases the Km for peptide substrates and shifts substrate specificity from trypsin-like to chymotrypsin-like (48).
Some disulfide bonds, like some peptide bonds, are cleaved in the mature protein. These disulfide bonds are known as allosteric disulfides (49, 50). The bonds are reductively cleaved by oxidoreductases or by thiol-disulfide exchange to control protein function (51, 52). Reduction of an allosteric disulfide bond results in a functional change at another site in the protein, and changes in ligand binding, substrate hydrolysis, proteolysis, or oligomer formation have been documented (reviewed in Refs. 49 and 50). Approximately 30 mammalian, plant, bacterial, or viral proteins have been found to contain allosteric disulfides so far, and a few have been linked to unwanted thrombosis (β2-glycoprotein I and factor XI) and impaired blood pressure regulation (angiotensinogen) in humans. The findings reported herein indicate that the β-tryptase Cys220–Cys248 disulfide is an allosteric bond.
A pattern is appearing in the configurations of allosteric disulfides and the secondary structures that they link (49, 50). The disulfide bond, or cystine residue, is defined by five angles and different combinations of these angles define 20 possible cystine configurations (51, 52). Three of the 20 disulfide bond configurations are emerging as allosteric configurations: the −RHStaple, −LHHook and −/+RHHook bonds (49). The most common secondary structures linked by allosteric disulfides are β-strands and/or β-loops (49). The β-tryptase Cys220–Cys248 disulfide exists in two different configurations in the crystal structure of the tetramer (20), −RHHook and −RHSpiral, and the bond links two extended β-loops (53). The disulfide is also exposed to solvent (Fig. 7), in particular the Cys220 residue, which is likely an important factor in the accessibility of this bond to thioredoxin.
FIGURE 7.
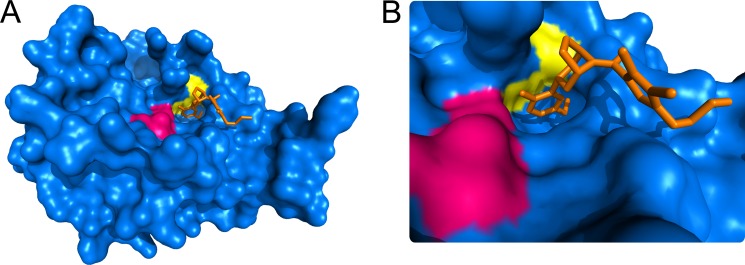
A, surface rendering of the βII-tryptase monomer showing the active site residues in magenta, the Cys220–Cys248 disulfide bond in yellow, and an inhibitor bound in the active site in orange (Protein Data Bank identifier 3V7T) (62). B, close-up of the active site. The structure was generated using PyMOL v0.99 (DeLano Scientific LLC).
Reduction of the Cys220–Cys248 disulfide bond is predicted to alter the shape of the active site pocket, which would explain the altered substrate specificity and catalytic efficiency of the reduced enzyme. The two β-loops linked by the Cys220–Cys248 disulfide bond shape the S1 substrate pocket that binds the residue immediately N-terminal of the target peptide bond (the P1 position). The importance of the S1 substrate pocket is demonstrated in the tryptase isoform, α-tryptase. Despite α-tryptase being 92% identical to βII-tryptase, it is completely inactive because of a single amino acid change that distorts the S1 pocket and blocks substrate binding (28–30). Distortion of the βII-tryptase S1 pocket is likely to be occurring when the Cys220–Cys248 disulfide bond is reduced, leading to the changes we observed.
Thioredoxin is a potentially relevant βII-tryptase reductant in vivo but may not be the only one. Both βII-tryptase and thioredoxin are released into the extracellular space during physiological inflammation and are found in synovial fluid in rheumatoid arthritis and bronchoalveolar fluid in asthma (10, 11, 23–26, 54). Thioredoxin concentrations up to 100 nm have been reported in synovial fluid of rheumatoid arthritic joints (25, 54). Other oxidoreductases are secreted during inflammation and other conditions, including protein-disulfide isomerase (55) and γ interferon-inducible lysosomal thiolreductase (GILT) (56, 57). It is possible that protein-disulfide isomerase and GILT play a role in regulating βII-tryptase through redox control of the Cys220–Cys248 allosteric disulfide bond. Interestingly, GILT is optimally active in acidic pH (58), which is an environment in which β-tryptase functions (34–37).
The finding that βII-tryptase is controlled by an allosteric disulfide bond is informative, since very little is known about the regulation of βII-tryptase activity because no endogenous inhibitors have been identified. β-Tryptase forms a homotetramer held together by polar and hydrophobic interactions that is further stabilized by heparin proteoglycans (20, 38, 39, 41, 59, 60). Our results indicate that the disulfide bond is reduced in the tetramer in the absence of heparin. How heparin influences the equilibrium between oxidized and reduced βII-tryptase and the activities of these two forms will be the subject of future studies.
There is precedence for allosteric regulation of a S1A protease. The blood coagulation protease, thrombin, exists in three forms: a low activity sodium-free form, a high activity sodium-bound form, and a form that is inactive and unable to bind sodium (2, 61). Conversion of the inactive to highly active form occurs very rapidly, and modulation of this equilibrium is thought to regulate thrombin activity in vivo. Redox control of the conserved Cys191–Cys220 disulfide bond in S1A proteases is possibly a general mechanism of allosteric regulation of this family of enzymes, which is predicted to influence their function in a number of important biological processes.
Acknowledgments
We thank Dr. Jason Wong for assistance with the design and analysis of the mass spectrometry experiments and Dr. Richard L Stevens for providing the mMCP-6 null mice.
This work was supported by grants from the National Health and Medical Research Council. Mass spectrometric analysis for this work was carried out at the Bioanalytical Mass Spectrometry Facility, University of New South Wales and was supported in part by infrastructure funding from the New South Wales Government as part of its co-investment in the National Collaborative Research Infrastructure Strategy.
- mMCP-6
- mouse mast cell protease 6
- IAM
- iodoacetamide
- IPTG
- isopropyl β-d-1-thiogalactopyranoside
- MPB
- 3-(N-maleimido-propionyl)biocytin
- GILT
- γ interferon-inducible lysosomal thiolreductase.
REFERENCES
- 1. Rawlings N. D., Morton F. R., Kok C. Y., Kong J., Barrett A. J. (2008) MEROPS. The peptidase database. Nucleic Acids Res. 36, D320–D325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Di Cera E. (2009) Serine proteases. IUBMB Life 61, 510–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fiorucci L., Ascoli F. (2004) Mast cell tryptase, a still enigmatic enzyme. Cell. Mol. Life Sci. 61, 1278–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Trivedi N. N., Tamraz B., Chu C., Kwok P. Y., Caughey G. H. (2009) Human subjects are protected from mast cell tryptase deficiency despite frequent inheritance of loss-of-function mutations. J. Allergy Clin. Immunol. 124, 1099–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Miller J. S., Westin E. H., Schwartz L. B. (1989) Cloning and characterization of complementary DNA for human tryptase. J. Clin. Invest. 84, 1188–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vanderslice P., Ballinger S. M., Tam E. K., Goldstein S. M., Craik C. S., Caughey G. H. (1990) Human mast cell tryptase. Multiple cDNAs and genes reveal a multigene serine protease family. Proc. Natl. Acad. Sci. U.S.A. 87, 3811–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cairns J. A. (2005) Inhibitors of mast cell tryptase β as therapeutics for the treatment of asthma and inflammatory disorders. Pulm. Pharmacol. Ther. 18, 55–66 [DOI] [PubMed] [Google Scholar]
- 8. Castells M., Schwartz L. B. (1988) Tryptase levels in nasal-lavage fluid as an indicator of the immediate allergic response. J. Allergy Clin. Immunol. 82, 348–355 [DOI] [PubMed] [Google Scholar]
- 9. Jarjour N. N., Calhoun W. J., Schwartz L. B., Busse W. W. (1991) Elevated bronchoalveolar lavage fluid histamine levels in allergic asthmatics are associated with increased airway obstruction. Am. Rev. Respir. Dis. 144, 83–87 [DOI] [PubMed] [Google Scholar]
- 10. Buckley M. G., Walters C., Wong W. M., Cawley M. I., Ren S., Schwartz L. B., Walls A. F. (1997) Mast cell activation in arthritis. Detection of α- and β-tryptase, histamine and eosinophil cationic protein in synovial fluid. Clin. Sci. 93, 363–370 [DOI] [PubMed] [Google Scholar]
- 11. McNeil H. P., Shin K., Campbell I. K., Wicks I. P., Adachi R., Lee D. M., Stevens R. L. (2008) The mouse mast cell-restricted tetramer-forming tryptases mouse mast cell protease 6 and mouse mast cell protease 7 are critical mediators in inflammatory arthritis. Arthritis Rheum. 58, 2338–2346 [DOI] [PubMed] [Google Scholar]
- 12. Thakurdas S. M., Melicoff E., Sansores-Garcia L., Moreira D. C., Petrova Y., Stevens R. L., Adachi R. (2007) The mast cell-restricted tryptase mMCP-6 has a critical immunoprotective role in bacterial infections. J. Biol. Chem. 282, 20809–20815 [DOI] [PubMed] [Google Scholar]
- 13. Schwertassek U., Balmer Y., Gutscher M., Weingarten L., Preuss M., Engelhard J., Winkler M., Dick T. P. (2007) Selective redox regulation of cytokine receptor signaling by extracellular thioredoxin-1. EMBO J. 26, 3086–3097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schwertassek U., Weingarten L., Dick T. P. (2007) Identification of redox-active cell-surface proteins by mechanism-based kinetic trapping. Sci STKE 2007, pl8. [DOI] [PubMed] [Google Scholar]
- 15. Ganderton T., Wong J. W., Schroeder C., Hogg P. J. (2011) Lateral self-association of VWF involves the Cys2431-Cys2453 disulfide/dithiol in the C2 domain. Blood 118, 5312–5318 [DOI] [PubMed] [Google Scholar]
- 16. Gatlin C. L., Kleemann G. R., Hays L. G., Link A. J., Yates J. R., 3rd. (1998) Protein identification at the low femtomole level from silver-stained gels using a new fritless electrospray interface for liquid chromatography-microspray and nanospray mass spectrometry. Anal. Biochem. 263, 93–101 [DOI] [PubMed] [Google Scholar]
- 17. Rothwarf D. M., Scheraga H. A. (1992) Equilibrium and kinetic constants for the thiol-disulfide interchange reaction between glutathione and dithiothreitol. Proc. Natl. Acad. Sci. U.S.A. 89, 7944–7948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nilsson G., Blom T., Kusche-Gullberg M., Kjellén L., Butterfield J. H., Sundström C., Nilsson K., Hellman L. (1994) Phenotypic characterization of the human mast-cell line HMC-1. Scand. J. Immunol. 39, 489–498 [DOI] [PubMed] [Google Scholar]
- 19. Torchinski M. (1974) Thiol and Disulfide Groups of Proteins, Consultants Bureau, New York [Google Scholar]
- 20. Pereira P. J., Bergner A., Macedo-Ribeiro S., Huber R., Matschiner G., Fritz H., Sommerhoff C. P., Bode W. (1998) Human β-tryptase is a ring-like tetramer with active sites facing a central pore. Nature 392, 306–311 [DOI] [PubMed] [Google Scholar]
- 21. Huang C., Friend D. S., Qiu W. T., Wong G. W., Morales G., Hunt J., Stevens R. L. (1998) Induction of a selective and persistent extravasation of neutrophils into the peritoneal cavity by tryptase mouse mast cell protease 6. J. Immunol. 160, 1910–1919 [PubMed] [Google Scholar]
- 22. Lillig C. H., Holmgren A. (2007) Thioredoxin and related molecules. From biology to health and disease. Antioxid. Redox Signal. 9, 25–47 [DOI] [PubMed] [Google Scholar]
- 23. Yamada Y., Nakamura H., Adachi T., Sannohe S., Oyamada H., Kayaba H., Yodoi J., Chihara J. (2003) Elevated serum levels of thioredoxin in patients with acute exacerbation of asthma. Immunol. Lett. 86, 199–205 [DOI] [PubMed] [Google Scholar]
- 24. Burke-Gaffney A., Callister M. E., Nakamura H. (2005) Thioredoxin. Friend or foe in human disease? Trends Pharmacol. Sci. 26, 398–404 [DOI] [PubMed] [Google Scholar]
- 25. Jikimoto T., Nishikubo Y., Koshiba M., Kanagawa S., Morinobu S., Morinobu A., Saura R., Mizuno K., Kondo S., Toyokuni S., Nakamura H., Yodoi J., Kumagai S. (2002) Thioredoxin as a biomarker for oxidative stress in patients with rheumatoid arthritis. Mol. Immunol. 38, 765–772 [DOI] [PubMed] [Google Scholar]
- 26. Lemarechal H., Allanore Y., Chenevier-Gobeaux C., Ekindjian O. G., Kahan A., Borderie D. (2006) High redox thioredoxin but low thioredoxin reductase activities in the serum of patients with rheumatoid arthritis. Clin. Chim. Acta 367, 156–161 [DOI] [PubMed] [Google Scholar]
- 27. Alter S. C., Kramps J. A., Janoff A., Schwartz L. B. (1990) Interactions of human mast cell tryptase with biological protease inhibitors. Arch. Biochem. Biophys. 276, 26–31 [DOI] [PubMed] [Google Scholar]
- 28. Huang C. (1999) Human tryptases α and β/II are functionally distinct due, in part, to a single amino acid difference in one of the surface loops that forms the substrate-binding cleft. J. Biol. Chem. 274, 19670–19676 [DOI] [PubMed] [Google Scholar]
- 29. Marquardt U., Zettl F., Huber R., Bode W., Sommerhoff C. (2002) The crystal structure of human α1-tryptase reveals a blocked substrate-binding region. J. Mol. Biol. 321, 491–502 [DOI] [PubMed] [Google Scholar]
- 30. Selwood T., Wang Z. M., McCaslin D. R., Schechter N. M. (2002) Diverse stability and catalytic properties of human tryptase α and β isoforms are mediated by residue differences at the S1 pocket. Biochemistry 41, 3329–3340 [DOI] [PubMed] [Google Scholar]
- 31. De Young M. B., Nemeth E. F., Scarpa A. (1987) Measurement of the internal pH of mast cell granules using microvolumetric fluorescence and isotopic techniques. Arch. Biochem. Biophys. 254, 222–233 [DOI] [PubMed] [Google Scholar]
- 32. Johnson R. G., Carty S. E., Fingerhood B. J., Scarpa A. (1980) The internal pH of mast cell granules. FEBS Lett. 120, 75–79 [DOI] [PubMed] [Google Scholar]
- 33. Lagunoff D., Rickard A. (1983) Evidence for control of mast cell granule protease in situ by low pH. Exp. Cell Res. 144, 353–360 [DOI] [PubMed] [Google Scholar]
- 34. Farr M., Garvey K., Bold A. M., Kendall M. J., Bacon P. A. (1985) Significance of the hydrogen ion concentration in synovial fluid in rheumatoid arthritis. Clin. Exp. Rheumatol. 3, 99–104 [PubMed] [Google Scholar]
- 35. Geborek P., Saxne T., Pettersson H., Wollheim F. A. (1989) Synovial fluid acidosis correlates with radiological joint destruction in rheumatoid arthritis knee joints. J. Rheumatol. 16, 468–472 [PubMed] [Google Scholar]
- 36. Hunt J. F., Fang K., Malik R., Snyder A., Malhotra N., Platts-Mills T. A., Gaston B. (2000) Endogenous airway acidification. Implications for asthma pathophysiology. Am. J. Respir. Crit. Care Med. 161, 694–699 [DOI] [PubMed] [Google Scholar]
- 37. Kodric M., Shah A. N., Fabbri L. M., Confalonieri M. (2007) An investigation of airway acidification in asthma using induced sputum. A study of feasibility and correlation. Am. J. Respir. Crit. Care Med. 175, 905–910 [DOI] [PubMed] [Google Scholar]
- 38. Fajardo I., Pejler G. (2003) Formation of active monomers from tetrameric human β-tryptase. Biochem. J. 369, 603–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fukuoka Y., Schwartz L. B. (2004) Human β-tryptase. Detection and characterization of the active monomer and prevention of tetramer reconstitution by protease inhibitors. Biochemistry 43, 10757–10764 [DOI] [PubMed] [Google Scholar]
- 40. Fukuoka Y., Schwartz L. B. (2006) The B12 anti-tryptase monoclonal antibody disrupts the tetrameric structure of heparin-stabilized β-tryptase to form monomers that are inactive at neutral pH and active at acidic pH. J. Immunol. 176, 3165–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hallgren J., Lindahl S., Pejler G. (2005) Structural requirements and mechanism for heparin-dependent activation and tetramerization of human βI- and βII-tryptase. J. Mol. Biol. 345, 129–139 [DOI] [PubMed] [Google Scholar]
- 42. Schwartz L. B., Bradford T. R., Littman B. H., Wintroub B. U. (1985) The fibrinogenolytic activity of purified tryptase from human lung mast cells. J. Immunol. 135, 2762–2767 [PubMed] [Google Scholar]
- 43. Ren S., Lawson A. E., Carr M., Baumgarten C. M., Schwartz L. B. (1997) Human tryptase fibrinogenolysis is optimal at acidic pH and generates anticoagulant fragments in the presence of the anti-tryptase monoclonal antibody B12. J. Immunol. 159, 3540–3548 [PubMed] [Google Scholar]
- 44. Yousef G. M., Elliott M. B., Kopolovic A. D., Serry E., Diamandis E. P. (2004) Sequence and evolutionary analysis of the human trypsin subfamily of serine peptidases. Biochim. Biophys. Acta 1698, 77–86 [DOI] [PubMed] [Google Scholar]
- 45. Miyata T., Kawabata S., Iwanaga S., Takahashi I., Alving B., Saito H. (1989) Coagulation factor XII (Hageman factor) Washington D.C. Inactive factor XIIa results from Cys-571–Ser substitution. Proc. Natl. Acad. Sci. U.S.A. 86, 8319–8322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bush-Pelc L. A., Marino F., Chen Z., Pineda A. O., Mathews F. S., Di Cera E. (2007) Important role of the Cys-191 Cys-220 disulfide bond in thrombin function and allostery. J. Biol. Chem. 282, 27165–27170 [DOI] [PubMed] [Google Scholar]
- 47. Wang E. C., Hung S. H., Cahoon M., Hedstrom L. (1997) The role of the Cys191-Cys220 disulfide bond in trypsin. New targets for engineering substrate specificity. Protein Eng. 10, 405–411 [DOI] [PubMed] [Google Scholar]
- 48. Várallyay E., Lengyel Z., Gráf L., Szilágyi L. (1997) The role of disulfide bond C191-C220 in trypsin and chymotrypsin. Biochem. Biophys. Res. Commun. 230, 592–596 [DOI] [PubMed] [Google Scholar]
- 49. Cook K. M., Hogg P. J. (2013) Post-translational control of protein function by disulfide bond cleavage. Antioxid. Redox Signal. 18, 1987–2015 [DOI] [PubMed] [Google Scholar]
- 50. Hogg P. J. (2013) Targeting allosteric disulphide bonds in cancer. Nat. Rev. Cancer 13, 425–431 [DOI] [PubMed] [Google Scholar]
- 51. Schmidt B., Ho L., Hogg P. J. (2006) Allosteric disulfide bonds. Biochemistry 45, 7429–7433 [DOI] [PubMed] [Google Scholar]
- 52. Schmidt B., Hogg P. J. (2007) Search for allosteric disulfide bonds in NMR structures. BMC Struct. Biol. 7, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wong J. W., Hogg P. J. (2010) Analysis of disulfide bonds in protein structures. J. Thromb. Haemost. 8, 2345. [DOI] [PubMed] [Google Scholar]
- 54. Xu S. Z., Sukumar P., Zeng F., Li J., Jairaman A., English A., Naylor J., Ciurtin C., Majeed Y., Milligan C. J., Bahnasi Y. M., Al-Shawaf E., Porter K. E., Jiang L. H., Emery P., Sivaprasadarao A., Beech D. J. (2008) TRPC channel activation by extracellular thioredoxin. Nature 451, 69–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jiang X. M., Fitzgerald M., Grant C. M., Hogg P. J. (1999) Redox control of exofacial protein thiols/disulfides by protein disulfide isomerase. J. Biol. Chem. 274, 2416–2423 [DOI] [PubMed] [Google Scholar]
- 56. Lackman R. L., Jamieson A. M., Griffith J. M., Geuze H., Cresswell P. (2007) Innate immune recognition triggers secretion of lysosomal enzymes by macrophages. Traffic 8, 1179–1189 [DOI] [PubMed] [Google Scholar]
- 57. Metcalfe C., Cresswell P., Ciaccia L., Thomas B., Barclay A. N. (2011) Labile disulfide bonds are common at the leucocyte cell surface. Open Biol. 1, 110010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Phan U. T., Arunachalam B., Cresswell P. (2000) γ-Interferon-inducible lysosomal thiol reductase (GILT). Maturation, activity, and mechanism of action. J. Biol. Chem. 275, 25907–25914 [DOI] [PubMed] [Google Scholar]
- 59. Alter S. C., Metcalfe D. D., Bradford T. R., Schwartz L. B. (1987) Regulation of human mast cell tryptase. Effects of enzyme concentration, ionic strength and the structure and negative charge density of polysaccharides. Biochem. J. 248, 821–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schwartz L. B., Bradford T. R. (1986) Regulation of tryptase from human lung mast cells by heparin. Stabilization of the active tetramer. J. Biol. Chem. 261, 7372–7379 [PubMed] [Google Scholar]
- 61. Di Cera E. (2008) Thrombin. Mol. Aspects Med. 29, 203–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Liang G., Choi-Sledeski Y. M., Shum P., Chen X., Poli G. B., Kumar V., Minnich A., Wang Q., Tsay J., Sides K., Kang J., Zhang Y. (2012) A β-tryptase inhibitor with a tropanylamide scaffold to improve in vitro stability and to lower hERG channel binding affinity. Bioorg. Med. Chem. Lett. 22, 1606–1610 [DOI] [PubMed] [Google Scholar]