Abstract
Interferon regulatory factor-1 (IRF-1), a transcription factor and tumor suppressor involved in cell growth regulation and immune responses, has been shown to be induced by all-trans retinoic acid (ATRA). However, the factors controlling the cellular location and activity of IRF-1 are not well understood. In this study, we examined the expression of IRF-1 and its nuclear localization, DNA-binding activity, and target gene expression in human mammary epithelial MCF10A cells, a model of breast epithelial cell differentiation and carcinogenesis. Following initial treatment with ATRA, IRF-1 mRNA and protein were induced within 2 hrs, reached a peak (>30-fold induction) at 8 hrs, and declined afterwards. IRF-1 protein was predominantly cytoplasmic during this treatment. Although a second dose of ATRA or Am580 (a related retinoid selective for retinoic acid receptor-α [RARα]), given 16 hrs after the first dose, restimulated IRF-1 mRNA and protein levels to a similar level to that obtained by the first dose, IRF-1 was predominantly concentrated in the nucleus after restimulation. ATRA and Am580 also increased nuclear RARα, whereas retinoid × receptor-α (RXRα)—a dimerization partner for RARα, was localized to the nucleus upon second exposure to ATRA. However, ATRA and Am580 did not regulate the expression or activation of signal transducer and activator of transcription-1 (STAT-1), a transcription factor capable of inducing the expression of IRF-1, indicating an STAT-1–independent mechanism of regulation by ATRA and Am580. The increase in nuclear IRF-1 after retinoid restimulation was accompanied by enhanced binding to an IRF-E DNA response element, and elevated expression of an IRF-1 target gene, 2′ ,5′-oligoadenylate synthetase-2. The dual effect of retinoids in increasing IRF-1 mRNA and protein and in augmenting the nuclear localization of IRF-1 protein may be essential for maximizing the tumor suppressor activity and the immunosurveillance functions of IRF-1 in breast epithelial cells.
Keywords: retinoic acid, IRF-1, nuclear localization, restimulation, breast cancer
Introduction
Vitamin A, a fat-soluble vitamin, and its hormonal metabolite, retinoic acid, are well known as important regulators of cell growth and differentiation (1) and immune responses (2). Retinoids induce gene transactivation by binding to nuclear receptor proteins, retinoic acid receptor (RAR) and retinoid × receptor (RXR), which form heterodimers that bind to retinoic acid response elements (RAREs), typically located in the promoter region of target genes (3). In the presence of retinoids (ligands), the receptor proteins recruit coactivators and histone acetyltransferase required for initiation of transcription (4, 5). All-trans retinoic acid (ATRA), a potent natural metabolite of vitamin A, can interact with RARα, RARβ, or RARγ, and thereby is implicated in the transcriptional regulation of many genes. Several retinoid analogs also possess receptor selectivity and display some of the actions of ATRA (6).
One of the known targets of ATRA is interferon regulatory factor-1 (IRF-1) (7), an important factor for immune responses and a putative tumor suppressor gene. IRF-1 was discovered in studies of virus-induced interferon (IFN)α/β gene regulation (8). The antiviral functions of IRF-1 were thereafter recognized (9). Additionally, IRF-1 is capable of regulating cell growth and apoptosis (10, 11), and its tumor suppressor activity has been demonstrated in oncogenic transformation of primary IRF-1−/− mouse embryonic fibroblasts (12, 13). Interestingly, IRF-1 expression was recently found to be negatively correlated with breast tumor progression (14), indicating the importance of maximizing the tumor suppressor activity of IRF-1 in the maintenance of breast cancer.
In our previous studies, ATRA was shown to synergize with IFNγ, a known inducer of IRF-1, to increase the level of IRF-1 and its functionality in lung epithelial carcinoma A549 cells (15). However, in some cells ATRA by itself can also induce IRF-1, as shown in promyelocytic leukemia NB4 cells bearing a natural mutation of RARα, in which ATRA rapidly upregulated IRF-1 mRNA nearly 8-fold (7). A 3-fold increase of IRF-1 protein levels by ATRA was also found in cervical squamous carcinoma SiHa cells (16). A number of studies have investigated the mechanisms of ATRA-mediated activation of the IRF-1 promoter; although a functional RARE has not yet been identified, various investigators have suggested the involvement of other transcription factors in the regulation of IRF-1, such as signal transducer and activator of transcription (STAT)-1 and nuclear factor-κB (16-18). However, it has not been examined whether ATRA and related retinoids can regulate the subcellular localization of IRF-1 in addition to increasing IRF-1 expression. The nuclear localization of IRF-1 is presumably essential for its transactivation activity, and therefore for the regulation by IRF-1 of immune function– and/or apoptosis-related target genes.
In the present study, we hypothesized that ATRA modulates both gene expression and nuclear localization of IRF-1. Using a human mammary epithelial cell line, MCF10A, we examined the effects of sequential treatments of ATRA on IRF-1 expression, localization, and DNA-binding activity. MCF10A is a nontumorigenic mammary epithelial cell line that, although having characteristics of normal breast epithelium (19), is also readily transformed (20). The ability to maintain effective immunity, in which IRF-1 is involved, is likely to be critical for cancer prevention (21, 22). Moreover, breast cancer progression in MCF10A cells is associated with alterations in retinoid receptors, which can be reversed by ATRA (23). Thus, in the current study we chose MCF10A cells as a cancer-susceptible model in which to examine the impact of retinoids on IRF-1 expression, localization, and signaling. The results suggest that ATRA and related retinoids induce the level of IRF-1 mRNA without regulating STAT-1 activation, and also augment the nuclear localization of IRF-1 protein. These actions of ATRA could be essential for maximizing the tumor suppressor activity and the immunosurveillance functions of IRF-1 in breast epithelial cells.
Materials and Methods
Reagents, Antibodies, and Cell Culture
ATRA (prepared in ethanol and stored at −20°C), 9-cis-retinoic acid (9cRA), retinyl trimethoxybenzyl ether (RTMBE), and actinomycin D (AD) were obtained from Sigma-Aldrich (St. Louis, MO). Receptor-selective retinoids were provided by Michael Klaus, Hoffmann-La Roche (Nutley, NJ). They include Am580 (RARα agonist), Ro19–0645 (RARβ agonist), CD437 (RARγ agonist), Ro25–7386 (RXR pan-agonist), and Ro41–5253 (RARα antagonist). IRF-1 polyclonal antibody and IRF-1 and STAT-1 consensus gel shift oligonucleotides were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). IRF-1 and STAT-1 monoclonal antibodies were obtained from Transduction Laboratory (Lexington, KY). Polyclonal antibodies against phospho-STAT-1 at Tyr-701 and Ser-727 were obtained from Cell Signaling Technology (Beverly, MA). Recombinant human IFNγ was obtained from PreproTech Inc. (Rocky Hill, NJ). Alexa Fluor 568-conjugated anti-rabbit IgG and To-Pro.3 iodide were obtained from Molecular Probes (Eugene, OR). MCF10A cells were maintained in DMEM/F-12 medium (GIBCO/Invitrogen, Carlsbad, CA) supplemented with 5% heat-inactivated horse serum (GIBCO), 10 μg/ml human insulin (Sigma), 10 ng/ml epidermal growth factor (Invitrogen), 100 ng/ml cholera toxin (Sigma), and 0.5 μg/ml hydrocortisone (Sigma) at 37°C in a 5% CO2-air incubator. In most experiments, the cells were plated at approximately 70% confluency, allowed to attach in complete medium, and adjusted to low-serum medium (0.5% horse serum) for 16–24 hrs before the addition of stimuli. For immunofluorescence experiments, the cells were plated at 20% confluency and grown overnight before use.
Preparation of Whole-Cell and Nuclear Extracts
For extraction of whole-cell lysates, MCF10A cells were lysed in RIPA buffer (1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS] in phosphate-buffered saline [PBS]) containing 10% (v/v) protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN) and 1 mM sodium orthovanadate as phosphatase inhibitor (24). Whole-cell lysates were obtained by centrifugation at 13,000 g for 15 mins at 4°C. To obtain nuclear extract, cells were homogenized in a hypotonic buffer (10 mM HEPES pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.2 mM phenylmethylsulfonyl fluoride [PMSF], 0.5 mM DTT, 1 mM sodium orthovanadate, and 0.5% Nonidet P-40). After centrifugation at 2500 g at 4°C for 5 mins, the supernatant (cytoplasmic fraction) was removed. Pellets were washed once with hypotonic buffer containing no detergent, and hypertonic buffer (final concentrations: 20 mM HEPES pH 7.9, 10% glycerol, 1.5 mM MgCl2, 400 mM KCl, 0.2 mM EDTA, 0.2 mM PMSF, 0.5 mM DTT, and 1 mM sodium orthovanadate) was added to extract nuclear proteins. After a 30-min incubation on ice, the mixture was centrifuged at 13,000 g for 30 mins. The supernatant was then collected as the nuclear extract (24). Protein concentrations of whole-cell and nuclear extracts were determined using Bio-Rad protein assay (Hercules, CA).
Immunoblotting
Whole-cell lysates (25 μg) or nuclear extracts (5 μg) were denatured and separated by polyacrylamide gel electrophoresis. After separation, proteins were electrophonically transferred to nitrocellulose membranes, which were then sequentially incubated in primary antibody and horseradish peroxidase (HRP)–conjugated secondary antibody (24). Detection of the HRP conjugate was done using the ECL system (Pierce Biotechnology, Rockford, IL). For equal loading controls, the membranes were blotted with β-actin antibody (Santa Cruz Biotechnology) for comparing whole-cell proteins or histones (H1 and core proteins) antibody (CHEMICON International, Inc., Temecula, CA) for nuclear proteins.
Reversed Transcription (RT)-Polymerase Chain Reaction (PCR)
Total cellular RNA was isolated using Qiagen RNeasy Kit (Qiagen Inc., Valencia, CA) according to the manufacturer’s instructions. Total RNA (0.5 μg) was subjected to RT, and one-tenth of the reaction mixture was used for PCR analysis. A pair of primers was designed to detect differential expression of IRF-1 mRNAs: 5′-GGC TGG GAC ATC AAC AAG GAT G-3′ (forward) and 5′-GAG CTG CTG AGT CCA TCA GAG AA-3′ (reverse), amplicon size 330 base pairs (bp). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was employed as an internal control: 5′-TGA AGG TCG GAG TCA ACG GAT TTG GT-3′ (forward) and 5′-CAT GTG GGC CAT GAG GTC CAC CAC-3′ (reverse), amplicon size 980 bp. Primer sequences for IRF-1 target gene 2′-5′ oligoadenylate synthetase-2 (OAS-2) were: 5′-CCA GGA GAA GCT GTG TAT CT-3′ (forward) and 5′-GTC TTC AGA GCT GTG CCT TT-3′ (reverse), amplicon size 440 bp. During PCR amplification, 0.5 μCi of [α-33P]dATP was added to each reaction as described previously (24). PCR products were separated on a 5% native polyacrylamide gel. The gel was then dried and exposed to Kodak Biomax MS film (Eastman Kodak Company, Rochester, NY). Individual bands were cut from the dried gel and counted in 3 ml of ScintiVerse scintillation fluid (Fisher Scientific, Fair Lawn, NJ) by using a liquid scintillation spectrometer (Beckman Instruments, Irvine, CA) to quantify relative gene expression levels.
Confocal Microscopy
MCF10A cells were plated in Lab-Tek two-well chambered coverglasses (Nalge Nunc International, Rochester, NY) at 20% confluency. After the treatments, cells were washed twice in PBS and immediately fixed with 3.7% formaldehyde (w/v) in PBS for 20 mins at room temperature. They were then permeabilized with 0.2% Triton X-100 in PBS for 5 mins and the reactions were quenched with freshly prepared 0.1% sodium borohydride in PBS for 5 mins. Afterwards, the fixed and permeabilized cell monolayer was sequentially incubated in blocking buffer containing 10% FBS, 1% BSA, 0.02% NaN3 in PBS, anti-IRF-1 polyclonal antibody in 1% BSA in PBS, and Alexa Fluor 568-labeled anti-rabbit secondary antibody and To-Pro.3 iodide in 1% BSA in PBS in the dark. After washing the monolayer twice in PBS, a drop of SlowFade Gold antifade reagent (Invitrogen, Eugene, OR) and a coverslip were mounted onto the cells. Samples were visualized under the Olympus Fluoview 300 Confocal Laser Scanning Microscope (Olympus America Inc., Melville, NY). Images were analyzed by the Fluoview software. To quantify nuclear fluorescence, a small circle (5 μM in diameter) was applied to a total of 100 nuclei (excluding the nucleoli) in each treatment; the fluorescence intensity in the circle was then read by the software and analyzed as an index of nuclear protein expression.
DNA-Binding or Electrophoretic Mobility Shift Assay (EMSA)
Nuclear extract was prepared as described earlier, aliquoted, and stored at −80°. For each EMSA reaction, 5 μg of nuclear protein was incubated with 15,000 cpm of [γ-32P]ATP-labeled consensus IRF-1 gel shift oligonucleotide (Santa Cruz Biotechnology) for 30 mins on ice. For competition or supershift assay, unlabeled IRF-1 consensus or mutant oligonucleotides (50×), or anti-IRF-1 monoclonal antibody (1 μl) were incubated with nuclear extracts for 10 mins on ice prior to the addition of radiolabeled oligonucleotides. Reaction mixtures were then separated on a 5% native polyacrylamide gel. After electrophoresis, the gel was dried and subjected to autoradiography (24).
Statistical Analysis
Statistical analysis was performed by using SuperANOVA software (Abacus Concepts, Berkeley, CA) for one-way analysis of variance (ANOVA), two-way ANOVA, and simple regression. All data, unless specified, are shown as the mean + SEM, and difference was considered statistically significant P value was less than 0.05.
Results
ATRA Induces IRF-1 in MCF10A Cells
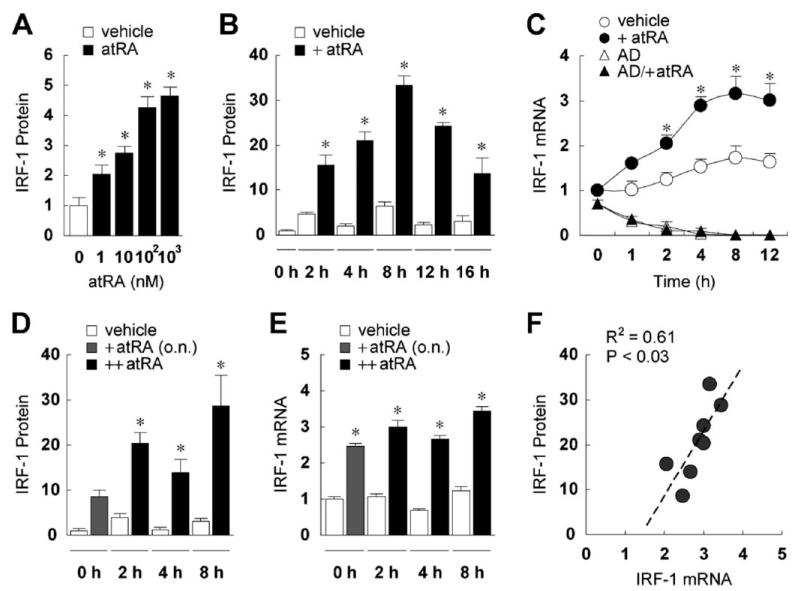
A single dose of ATRA rapidly induced IRF-1 in MCF10A cells. A titration analysis identified an optimal concentration of 0.1 μM of ATRA (Fig. 1A), which was used in subsequent experiments. IRF-1 protein was increased within 2 hrs of ATRA stimulation, reached a maximum at 8 hrs, and declined afterwards (Fig. 1B). The maximal induction of IRF-1 protein was over 30-fold, and the level remained elevated over the initial level 16 hrs after treatment. The increase in IRF-1 protein was concordant with an increase in IRF-1 mRNA (Fig. 1C), which started to rise within 1 hr after ATRA treatment, increased about 3-fold from 4 to 12 hrs, and was blocked by the transcription inhibitor actinomycin D, indicating that ATRA may activate IRF-1 gene transcription. As anticipated, the IRF-1 message level fluctuated in cells treated with vehicle, consistent with reports that IRF-1 is regulated by progression of the cell cycle (22). Despite this fluctuation, ATRA significantly induced IRF-1 at each time examined between 2 and 12 hrs. These results indicate that ATRA alone can rapidly induce both IRF-1 mRNA and protein expression in MCF10A cells.
Figure 1.
ATRA induces IRF-1 in MCF10A cells. (A) MCF10A cells were treated with vehicle or ATRA (1, 10, 100, or 1000 nM) overnight, and then lysed and assayed by immunoblotting for IRF-1 expression. The membranes were also blotted for β-actin to assure equal loading of proteins (data not shown). Fold increases of IRF-1 protein over the vehicle-treated level are shown (n = 3). ATRA-treated cells had significantly higher levels of IRF-1 protein as determined by one-way ANOVA (*). (B) Cells were treated with vehicle or ATRA (0.1 μM), cultured for 2–16 hrs, and then lysed and assayed by immunoblotting. Fold increases of IRF-1 protein compared to the untreated level at time 0 hrs are shown (n = 3). Two-way ANOVA showed ATRA and time as significant factors. For all time points, +ATRA was significantly different from vehicle (*). (C)Cells were treated with vehicle or ATRA (0.1 μM) for 1–12 hrs and RT-PCR was performed to assess the mRNA levels of IRF-1 and GAPDH. In some experiments, cells were pretreated with AD (5 μg/ml) 1 hr prior to the addition of ATRA. After normalization by GAPDH mRNA, fold increases of IRF-1 mRNA compared to that of vehicle-treated cells at 0 hrs are shown (n = 3). Two-way ANOVA showed treatment and time as significant factors. The asterisks indicate significant differences between +ATRA and vehicle groups. (D) Cells were treated with ATRA (0.1 μM) overnight (o.n.), followed by a second dose of the same concentration at time 0 hrs, shown in the figure as ++ATRA. Cells were then cultured for 2, 4, or 8 hrs, and then lysed and assayed by immunoblotting for IRF-1 expression. Fold increases of IRF-1 protein compared to vehicle at time 0 hrs are shown (n = 4). Two-way ANOVA showed ATRA and time as significant factors. For 2, 4, and 8 hrs, ++ATRA was significantly different from vehicle (*). (E) Cells were treated the same as in (D). RT-PCR was performed to assess the mRNA levels of IRF-1 and GAPDH. After normalization by GAPDH mRNA, fold increases of IRF-1 mRNA compared to vehicle-treated cells at 0 hrs are shown (n = 3). Two-way ANOVA showed treatment and time as significant factors. The asterisks indicate significant differences between ++ATRA and vehicle groups. (F) Regression analysis showed significant correlation between fold increases of IRF-1 mRNA and protein, regardless of +ATRA or ++ATRA.
IRF-1 is a transcription factor with a half-life of less than 1 hr (25). Because the increase of IRF-1 protein in MCF10A cells was transient, with a marked reduction after it peaked at 8 hrs, and considering that ATRA is readily metabolized, we next tested the effect of re-exposing the cells to a second dose of ATRA, of the same concentration, 16 hrs after the first dose. The re-exposure induced a second wave of IRF-1 protein (Fig. 1D). At 2 and 8 hrs after the second dose, the ATRA-treated cells had significantly higher levels of IRF-1 protein as compared to the residual level of the first dose (16 hrs after initial application). Similarly, IRF-1 mRNA was also increased after reexposure (Fig. 1E). Correlation analysis between IRF-1 mRNA and protein (R2 = 0.61; Fig. 1F) suggests that production of nascent transcripts is required for IRF-1 protein induction. Notably, however, the second dose of ATRA did not further increase the maximal induction of either IRF-1 mRNA or protein as compared to the first dose, indicating that previous exposure to ATRA did not “prime” or potentiate the response of IRF-1 to this retinoid.
IRF-1 Is Induced by an RARα-Selective Retinoid, Am580
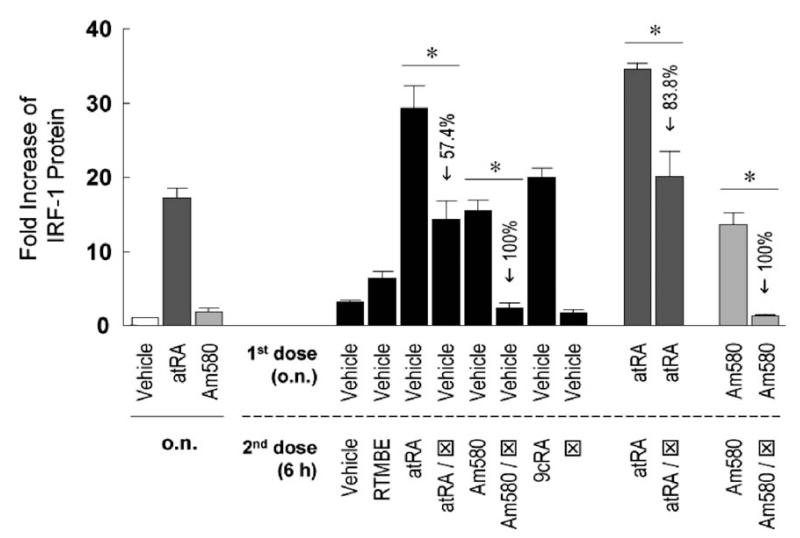
ATRA is a pan-agonist of all RARs; thus, we asked which receptor family members may mediate the effect of ATRA on IRF-1 induction in MCF10A cells. Am580, a ligand specific for RARα, increased IRF-1 protein expression (Fig. 2), indicating that RARα is capable of mediating the effect of ATRA on IRF-1 induction. This was confirmed by experiments using cotreatments between the agonists (ATRA or Am580) and an RARα antagonist (Fig. 2), wherein the increase of IRF-1 level was either partially (in the case of ATRA) or completely (in the case of Am580) blocked by the antagonist. However, the Am580-mediated induction was relatively smaller than that of ATRA. Because the same dosage (0.1 μM) was given, which is higher than the Kd values of ATRA and Am580 for RARα (0.2 nM in Ref. 26 and 36 nM in Ref. 27, respectively), it is unlikely that the differences in IRF-1 induction are because of differences in receptor affinities. On the other hand, ligands for RARβ and RARγ also induced IRF-1. The fold increases of IRF-1 protein were 12.4 ± 1.0 for RARβ and 11.8 ± 1.4 for RARγ agonists, comparable to 15.5 ± 1.5 for Am580 (Fig. 2). In addition, 9cRA also induced IRF-1; because additional ligation with RXR did not further increase the effects of RAR agonists (data not shown), 9cRA may act through RARs, but not through RXRs. The negative control, RTMBE, a retinoid analog without known binding activity to retinoid receptors (28), did not significantly increase IRF-1 protein level. In summary, it is likely that ATRA increases IRF-1 protein though each of the RARs, RARα, RARβ, and RARγ, although RARα alone can effectively increase IRF-1 expression in MCF10A cells.
Figure 2.
The RARα-selective ligand Am580 induces IRF-1. MCF10A cells were treated with vehicle, ATRA (0.1 μM), or Am580 (0.1 μM) o.n., followed by a second dose of different retinoids of the same concentration (except for RARα antagonist ⊠, which was 5 μM) and further incubation of 6 hrs. Cells were lysed and assayed by immunoblotting for IRF-1 protein expression. Fold increases of IRF-1 protein compared to the vehicle at time 0 hrs are shown (n = 3 or 6). One-way ANOVA was performed and outcome was as follows. Compared to vehicle, ATRA induced a significantly higher level of IRF-1 after o.n. treatment, whereas Am580 did not. When vehicle was the first dose (black bars), ATRA, ATRA/, Am580, and 9cRA as the second dose induced significantly higher levels of IRF-1 protein. When ATRA or Am580 was the first dose (darker or lighter gray bars, respectively), restimulation with the same retinoid further increased IRF-1 level significantly. The inhibitory effects of ⊠ were significant in all cases (*), with the percentages of reduction indicated in the figure.
ATRA-Mediated Induction of IRF-1 is STAT-1-Independent
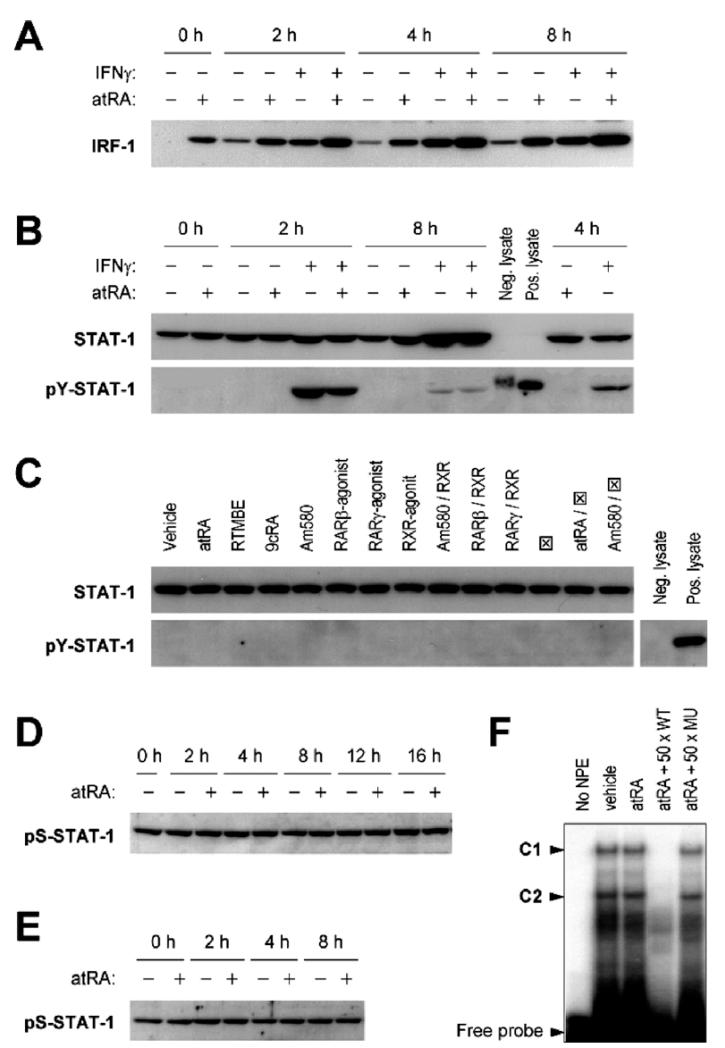
Because ATRA increases IFNγ-induced STAT-1 activation in human lung epithelial A549 cells (15), we examined whether ATRA could induce IRF-1 through the functions of STAT-1 in MCF10A cells. Unlike in A549 cells, where ATRA and IFNγ synergistically induced IRF-1 protein, these two stimuli acted additively to increase IRF-1 in MCF10A cells (Fig. 3A), suggesting that ATRA and IFNγ may affect IRF-1 transcription through different mechanisms. Further, we found that ATRA pretreatment did not potentiate IFNγ-induced tyrosine phosphorylation of STAT-1; rather, it decreased STAT-1 tyrosine phosphorylation at 2 hrs, indicating that the increase in IRF-1 protein by the combination treatment of ATRA and IFNγ was not caused by an increase in STAT-1 activation (Fig. 3B). In addition, none of the receptor-selective retinoids affected STAT-1 tyrosine phosphorylation (Fig. 3C). Because serine phosphorylation is required for transcriptional functions of STAT-1, we also tested serine-phosphorylated STAT-1. Serine-727 of STAT-1 is constitutively phosphorylated in MCF10A cells, and neither one dose (Fig. 3D) nor sequential doses (Fig. 3E) of ATRA altered the level of serine phosphorylation. Finally, the DNA-binding activity of STAT-1 to a consensus GAS element was also unaffected after ATRA treatment (Fig. 3F). These results suggest that ATRA-mediated induction of IRF-1 in MCF10A cells is STAT-1–independent.
Figure 3.
ATRA does not affect the expression, phosphorylation, or DNA-binding activity of STAT-1 to induce IRF-1. (A) MCF10A cells were pretreated with ATRA (0.1 μM) o.n., and then treated with ATRA and IFNγ (0.5 U/ml) for 2, 4, and 8 hrs. Cell lysates were prepared and assayed for IRF-1 protein by Western blot. The membranes were also stained by Ponceau S to assure equal loading of proteins. (B) Cells were pretreated with ATRA (0.1 μM) o.n., and then treated with ATRA and IFNγ (0.5 U/ml) for indicated times. Western blots of STAT-1 and pY-STAT-1 are shown. Positive (IFNα-treated HeLa) and negative (untreated HeLa) control lysates were included to test the specificity of pY-STAT-1 antibody. (C) Cells were treated with different retinoids (0.1 μM, except for RARα antagonist, which was 50× in excess) for 6 hrs. Cell lysates were prepared and assayed for STAT-1 and pY-STAT-1 by Western blot. RTMBE was used as a negative control. RARα antagonist is shown as ⊠. (D) Cells were treated with vehicle or ATRA (0.1 μM), cultured for 2–16 hrs, and then lysed and assayed by Western blot for pS-STAT-1. (E) Cells were pretreated with ATRA (0.1 μM) o.n., and then treated with a second dose for 2, 4, and 8 hrs. Cell lysates were prepared and assayed for pS-STAT-1 by Western blot. (F) Cells were treated with vehicle or ATRA (0.5 μM) for 30 mins and NPEs were prepared for EMSA experiments. NPEs were incubated with [γ-32P]ATP-labeled GAS/ISRE consensus oligonucleotide with or without unlabeled competitor DNA (wild-type [WT] or mutant [MU]). Protein-DNA complexes were separated on a 5% PAGE gel and two STAT-1–related complexes (C1 and C2) were identified.
ATRA Increases Nuclear Localization of RARα and RXRα
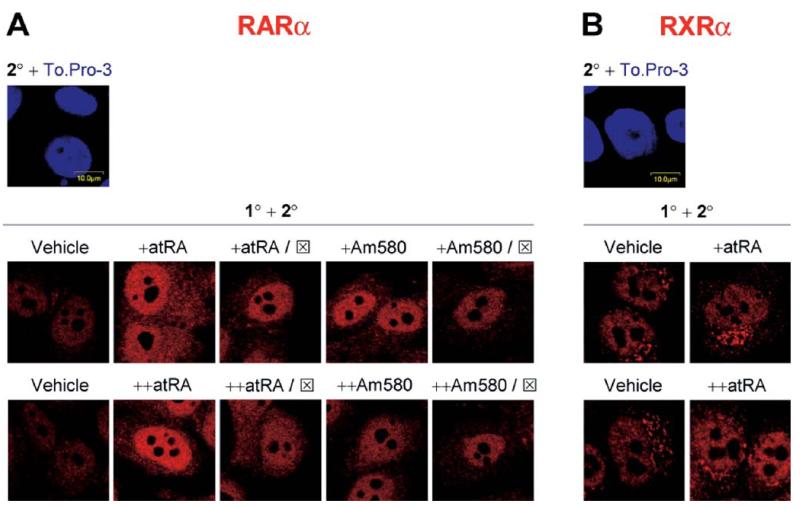
Because each of the ligands for RARα (ATRA, Am580, and 9cRA) induced IRF-1, we next tested the expression and/or localization of RARα and its dimerization partner, RXRα. The basal expression of RARα in MCF10A cells is low and mostly within the nucleus. The initial dose of ATRA increased the nuclear intensity of RARα, and the intensity was similar after the cells were exposed to a second dose of ATRA (Fig. 4A). Co-incubation of the initial dose of ATRA with RARα antagonist reduced the overall fluorescence; however, RARα still resided in the nucleus. In contrast, the RARα antagonist blocked the effect of the second dose of ATRA, with RARα showing a diffuse localization over the nucleus and the cytoplasm, indicating that antagonism may affect the localization of this receptor. Although Am580 slightly increased both nuclear and cytoplasmic RARα, whereas the antagonist partially decreased the effect of Am580 on the overall expression of RARα, Am580 did not alter the relative distribution of RARα between the nucleus and the cytoplasm. Together, these results indicate that whereas both ATRA and Am580 induce RARα expression, ATRA may have an additional effect on the localization of RARα.
Figure 4.
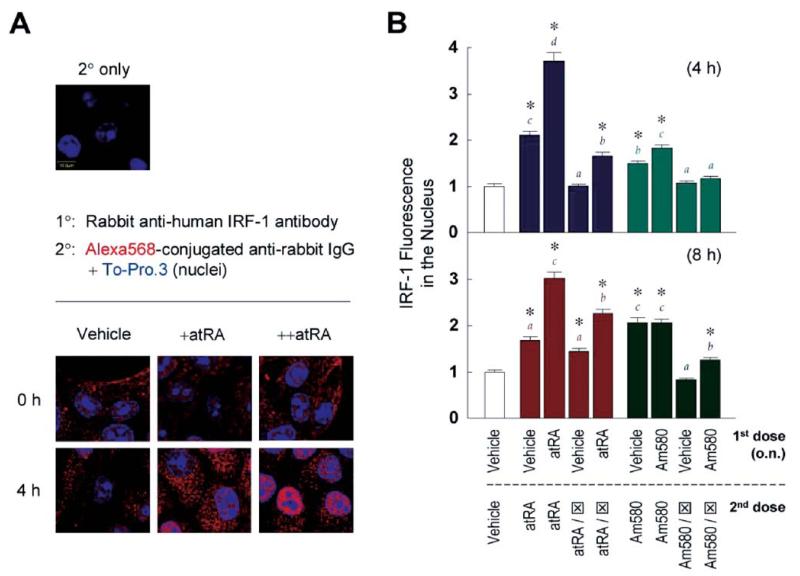
ATRA increases nuclear localization of RARα and RXRα. MCF10A cells were treated with one dose (o.n.) or two doses (o.n., and then for 3 hrs) of vehicle, ATRA (0.1 μM), or Am580 (0.1 μM). In some experiments, RARα antagonist (⊠; 5 μM) was given with the retinoids (in the case of a single dose, or +), or with the second dose (in the case of two doses, or ++). Cells were fixed, permeabilized, and stained with primary (1°) and secondary (2°) antibodies as described in Materials and Methods. Representative confocal images from at least 10 microscopic fields are shown, with red fluorescence indicating RARα (A) or RXRα (B).
RXRα showed a different distribution pattern in untreated cells. RXRα was localized mostly in the nucleus; however, a cluster of fluorescence was visible adjacent to the nucleus (Fig. 4B). Stimulation of the cells with the first dose of ATRA increased the fluorescence intensity in the cytoplasm, including the perinuclear cluster. Interestingly, nuclear and perinuclear staining of RXRα were increased after the cells were treated with two sequential doses of ATRA. The second dose of ATRA may have assisted in the transport of RXRα from the cytoplasmic cluster to the nucleus. The identity of the cluster and the role of ATRA in RXRα translocation require further investigation. Nevertheless, ATRA regulated the expression and/or localization of RARα and RXRα.
Restimulation with ATRA Increases Nuclear Localization of IRF-1
Whereas the experiments shown in Figure 1 showed that restimulation with ATRA induces a second wave of IRF-1 mRNA and protein without elevating the maximal levels, it was unknown whether ATRA also alters the localization of IRF-1. As shown in Figure 5A, the initial dose increased mostly the cytoplasmic level of IRF-1. In contrast, IRF-1 fluorescence was concentrated in the nucleus by sequential doses of ATRA. Quantification of nuclear IRF-1 fluorescence revealed modest increases with the first dose of ATRA at both 4 and 8 hrs; however, the second dose markedly elevated these levels (Fig. 5B). Coincubation with the RARα antagonist partially decreased the effects of ATRA, indicating a role of RARα in regulating IRF-1 localization. Similar results were obtained using Am580 as the stimulus, although a significant difference between the first and second exposures was observed only at 4 hrs. Collectively, these results suggest a novel role of ATRA in regulating IRF-1 localization.
Figure 5.
Restimulation with ATRA increases nuclear localization of IRF-1. (A) MCF10A cells were treated with vehicle or ATRA (0.1 μM) o.n., followed by a dose of ATRA and incubation for 4 hrs, shown in the figure as +ATRA and ++ATRA, respectively. Primary(1° and secondary (2°) antibodies were used as described in Materials and Methods, and confocal microscopy was performed. Overlays of Alexa Fluor 568 (IRF-1; red) and To-Pro.3 (nuclei; blue) are shown. (B) Cells were treated with vehicle, ATRA (0.1 μM) or Am580 (0.1 μM) o.n., followed by a second dose of different retinoids of the same concentration (except for RARα antagonist ⊠, which was 5 μM) and further incubation of 4 hrs (upper panel) or 8 hrs (lower panel). Confocal microscopic images were quantified by the Fluoview software, and averages of IRF-1 fluorescence from 100 nuclei per treatment were plotted. One-way ANOVA was performed. For each time point, asterisks indicate significant differences compared to the level caused by two doses of vehicle. In addition, four groups of results were color-coded, and different letters of matching colors above the bars indicate significant differences between treatments.
Restimulation with ATRA Increases Nuclear IRF-1 and DNA-Binding Activity of IRF-1
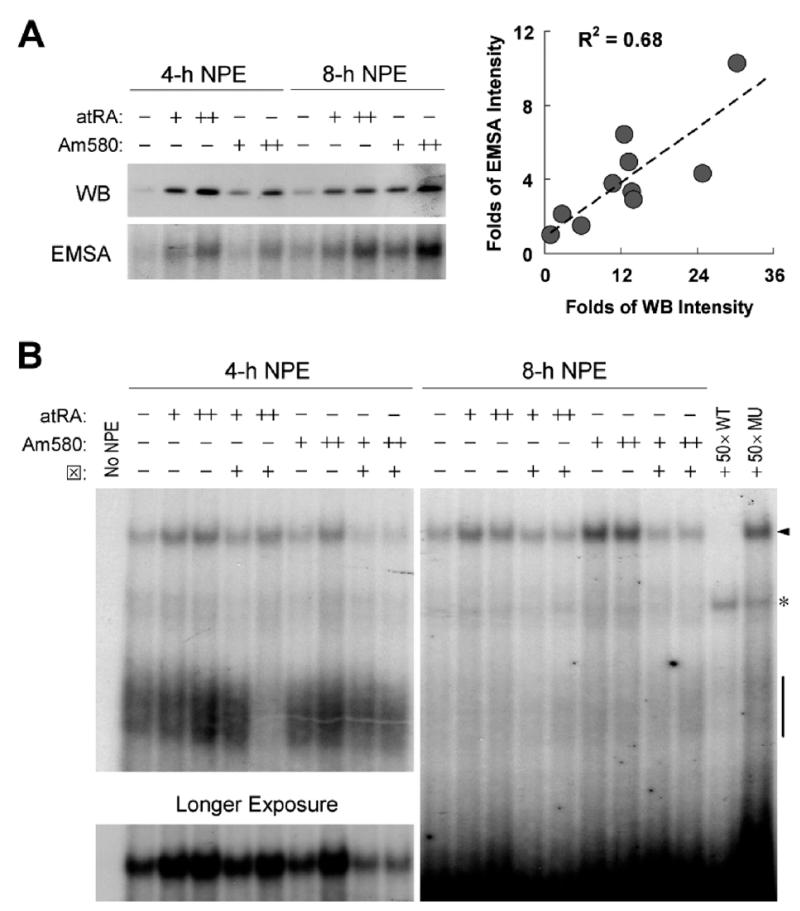
ATRA-mediated nuclear localization of IRF-1 was confirmed by immunoblot analysis of nuclear protein extracts (NPEs) and by DNA-binding experiments. The initial exposure of MCF10A cells to ATRA increased both IRF-1 nuclear expression and DNA-binding activity to an IRF-E consensus element compared to untreated cells; however, a second dose of ATRA further increased both levels (Fig. 6A). In addition, the increase of nuclear IRF-1 was highly correlated with that of IRF-1 DNA-binding activity. Sequential doses of Am580 functioned similarly to ATRA, although the peak of Am580-induced IRF-1 nuclear expression and DNA-binding activity was later than that seen after ATRA, with higher responses at 8 hrs (Fig. 6A). Unlike ATRA, which is unstable and readily degraded in aqueous solution, Am580 is very stable (29) and the lack of its catabolism may contribute to sustained effects of this compound. As anticipated, the RARα antagonist blocked the effects of ATRA and Am580 on DNA-binding activity of IRF-1 (Fig. 6B). These results agreed very well with those obtained by immunofluorescence staining, indicating that sequential doses of ATRA or Am580 increased IRF-1 protein in the nucleus and its DNA-binding activity.
Figure 6.
Restimulation with ATRA increases nuclear IRF-1 and DNA-binding activity of IRF-1. MCF10A cells were treated with vehicle, ATRA (0.1 μM), or Am580 (0.1 μM) o.n., followed by a second dose of different retinoids of the same concentration (except for RARα antagonist ⊠, which was 5 μM) and further incubation of 4 or 8 hrs. (A) Immunoblotting of NPEs and EMSA was performed as described in Materials and Methods. For EMSA, NPEs were incubated with an anti–IRF-1 monoclonal antibody prior to the addition of [γ-32P]ATP-labeled IRF-1 consensus oligonucleotide. Protein–DNA complexes were resolved using a 5% PAGE gel. Only the supershifted bands are shown. (B) NPEs were incubated with an anti-IRF-1 monoclonal antibody prior to the addition of [γ-32P]ATP-labeled IRF-1/IRF-E consensus oligonucleotide with or without unlabeled competitor DNA (WT or MU; 50× molar excess). Protein-DNA complexes were resolved using a 5% PAGE gel. The arrow indicates complexes supershifted by IRF-1 antibody, with the 4-hr supershifted bands exposed longer and shown as well. Original IRF-1/IRF-E-containing complexes are pointed out by a short segment. A nonspecific band is also shown (*).
Restimulation with ATRA Increases Transcription of IRF-1 Target Gene OAS-2
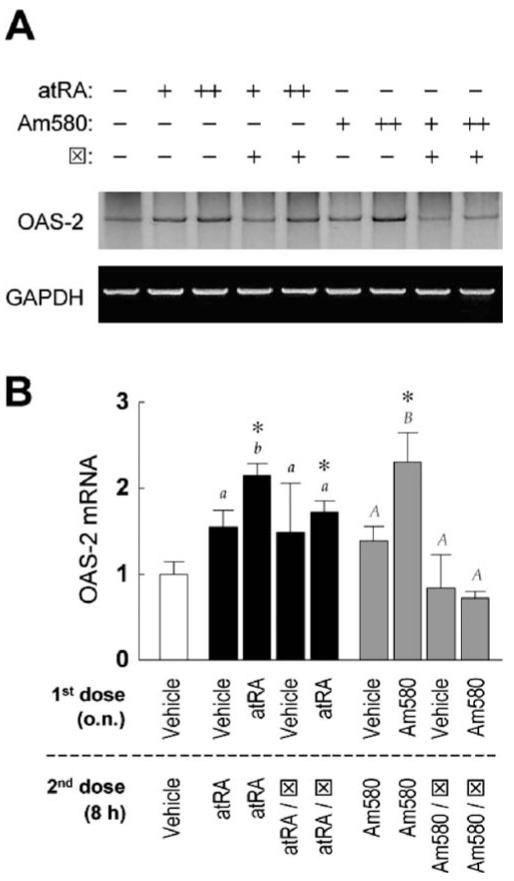
As additional evidence that retinoid-induced nuclear localization of IRF-1 could be related to an augmented functionality of this transcription factor, we next tested the expression of an IRF-1 target gene, OAS-2. Sequential doses of ATRA or Am580 significantly enhanced OAS-2 mRNA (Fig. 7). The effect of Am580 on OAS-2 was completely inhibited by the RARαantagonist, indicating a role of RARα in regulating the transcriptional function of IRF-1. Because the RARα antagonist partially downregulated the effect of ATRA, other factors (such as RARβ or RARγ) may also confer the ability of ATRA to increase IRF-1–mediated gene transactivation.
Figure 7.
Restimulation with ATRA increases transcription of OAS-2, an IRF-1 target gene. MCF10A cells were treated with vehicle, ATRA (0.1 μM), or Am580 (0.1 μM) o.n., followed by a second dose of different retinoids of the same concentration (except for RARα antagonist ⊠, which was 5 μM) and further incubation of 8 hrs. (A) Total RNA was isolated and transcript levels of OAS-2 and GAPDH was measured by RT-PCR. (B) Fold increases of OAS-2 mRNA after normalization are shown (n = 3 or 6). One way ANOVA was performed. The asterisks indicate significant differences compared to the level caused by two doses of vehicle. In addition, two groups of results were coded as black or gray, with regular or capitalized letters indicating significant differences, respectively.
Discussion
RF-1 is well known to be regulated by interferons (30), but its expression is also regulated by a number of other factors (22), which include protein hormones such as prolactin (31) and growth hormone (32), and lipid hormones such as retinoids (7). The expression of IRF-1 is essential for the maintenance of normal immune competence, and, because of its tumor suppressor function, it is likely to play a key role in preventing the transition from the normal state to the carcinogenic state in readily transformed cells. The MCF10A cell line is of fibrocystic breast origin, a condition with increased risk for progression to breast cancer (33). In our previous study of A549 lung epithelial cells, ATRA increased IFNγ-induced expression of IRF-1 by affecting multiple components of the IFNγ signaling pathway, including activation of STAT-1 (15). However, having observed that MCF10A cells respond robustly to ATRA in the absence of IFNγ, we focused in the present study on ATRA itself and investigated whether it independently regulates the expression, subcellular localization, and functional activity of IRF-1. In MCF10A cells, ATRA alone increased IRF-1 expression (Fig. 1), consistent with the observations in other cell lines (7, 17, 18). Unlike prolactin and growth hormone that induce IRF-1 via STAT-dependent pathways (32, 34, 35), ATRA-mediated induction of IRF-1 is STAT-1–independent, because neither expression, tyrosine/serine phosphorylation, nor DNA-binding activity of STAT-1 was affected by ATRA (Fig. 3), indicating that this hormone may affect IRF-1 transcription through mechanisms distinct from those engaged by protein hormones.
A main objective of the present study was to determine whether ATRA, in addition to its role in inducing IRF-1, also modulates IRF-1 localization as one way of regulating the transcriptional activity of IRF-1. IRF-1 has been shown to reside primarily in the cytoplasm in a number of breast cancer cell lines, whereas IFNγ simulation was shown to induce its nuclear localization (36). However, the mechanism by which IFNγ exerts such an effect and whether ATRA could function similarly are unknown. In the present study, we found that although an initial exposure of cells to ATRA increased mostly cytoplasmic IRF-1 (Fig. 5A), restimulation of MCF10A cells with a second dose of ATRA increased the nuclear localization of IRF-1 (Figs. 5 and 6). The increase in nuclear level of IRF-1 was apparently because of accumulation in the nucleus rather than an increase of whole-cell expression, because the second exposure to ATRA induced a similar level of IRF-1 to that induced by the first dose (Fig. 1). These results of these experiments suggest a novel role of ATRA in nucleocytoplasmic transport of IRF-1.
Nuclear import of proteins is thought to be mediated by the large nuclear pore complex (NPC; Ref. 37). The NPC provides passive diffusion channels for proteins smaller than 50–60 kDa, but in most cases, even small proteins are imported by an energy-dependent and receptor-mediated process. The nuclear import receptors recognize the nuclear localization signal (NLS) present in most nuclear proteins. The export of proteins out of the nucleus, on the other hand, is mediated by recognition of the nuclear export signal (NES) by export receptors, such as CRM1 (38).
IRF-1 possesses a putative NLS (39), whereas no NES has been identified. IRF-1 can be serine-phosphorylated (40), although a direct correlation between IRF-1 phosphorylation and transcriptional activity is yet to be established. In the present study, we have observed that ATRA increases the content of IRF-1 in the nucleus. The mechanism of ATRA-mediated regulation of IRF-1 nuclear transport is still unclear; however, we hypothesize that ATRA may function through the shuttling of retinoid receptors (RAR and RXR). One of these, RXRα, has been shown to shuttle between the nucleus and the cytoplasm ligand-dependently (41). RXRα has been shown to act as a nuclear export carrier for TR3/Nur77, directing this orphan receptor to the mitochondria, a process crucial for the effect of TR3 in inducing apoptosis (41). In our studies that tracked the localization of RXRα in MCF10A cells, the sequential exposure of cells to ATRA induced the translocation of RXRα from a cytoplasmic cluster to the nucleus (Fig. 4B). Thus, we hypothesize that ATRA-mediated relocation of RXRα in MCF10A cells may contribute to the movement of IRF-1, either through a direct interaction between the two proteins or by other, more complex, processes. Because ATRA is solely a ligand for RARs, and not for RXRα, we speculate that increased nuclear localization of RARα, observed after ATRA treatment (Fig. 4A), may assist in nuclear sequestration of RXRα. Furthermore, nuclear localization of both IRF-1 and RXRα was increased by sequential doses of ATRA, but not by one dose, indicating that previous exposure to ATRA may be required. The mechanism and significance of this effect need further investigation; however, it can be hypothesized that the initial dose of ATRA may affect the production and/or activation of a yet-to-be-identified transporter, which mediates the nuclear import, export, and/or nuclear retention of IRF-1 when re-exposed to ATRA. Interestingly, a role of nuclear import and export factors in the differential nuclear localization of the vitamin D receptor (VDR), RXR, and the VDR-RXR complex has recently been reported (42). In our studies, nuclear localization of IRF-1, as visualized by confocal microscopy (Fig. 5), was confirmed by observations that both nuclear expression and DNA-binding activity of IRF-1 were increased by sequential treatments with ATRA (Fig. 6).
Recently, the subcellular localization of IRF-5, another member of the IRF protein family, was reported to be regulated by a CRM1-dependent nuclear export pathway (43). Mutation of the NES of IRF-5 results in nuclear accumulation of this protein, potentially increasing transactivation functions of IRF-5 (44). The importance of nuclear import and sequestration in transcriptional regulation has also been reported. For instance, nuclear localization of IRF-3 appears to be initiated by its NLS (45), which is constitutively active and shuttles IRF-3 between the nucleus and the cytoplasm; upon virus infection, IRF-3 is serine-phosphorylated and bound to CREB-binding protein or p300 that stabilizes nuclear retention of IRF-3. How IRF-1 subcellular localization is regulated requires further investigation.
Increased nuclear localization could potentially enhance the transcriptional functions of IRF-1, inducing genes important for strengthening immune responses and/or promoting cancer prevention. ATRA-induced nuclear IRF-1 was apparently transcriptionally active, because expression of IRF-1 target gene OAS-2 was enhanced (Fig. 7). OAS-2 is a known IFN-stimulated gene that promotes an RNase L-mediated antiviral response (46) and tumor suppression (47). The tumor suppressor functions of RNase L can also amplify the apoptotic signals generated by another IRF-1 target gene, tumor necrosis factor-related apoptosis inducing ligand (TRAIL) (48). Thus, ATRA-induced IRF-1 nuclear localization and transcriptional activity may be important for the generation of a multicomponent network containing OAS-2, RNase L, TRAIL, and other IRF-1–responsive genes that may function in concert to induce apoptosis of cancer cells. Similar effects were found in the case of Am580, an ATRA analog specific for RARα, suggesting possible implication of this synthetic, yet more stable, retinoid in therapeutic practices.
In summary, the present study has provided evidence for dual regulatory roles of ATRA on IRF-1 expression and nuclear localization. Using RARα and RXRα as mediators, ATRA can induce both IRF-1 mRNA and protein. In addition, sequential treatments of ATRA can also increase IRF-1 nuclear localization, DNA-binding activity, and transcription of the IRF-1 target gene OAS-2 (Fig. 8). Such effects may not be seen in a chronic exposure model, because IRF-1 expression and nuclear localization may already both be increased by repeated dosing with ATRA. However, our study using sequential treatment has revealed the possibility of separate mechanisms of ATRA actions on IRF-1. In the regulation of IRF-1 gene expression, ATRA appears to facilitate the shuttling of the RARα/RXRα heterodimer from the cytoplasm to the nucleus, leading to yet-to-be-characterized changes that can activate the IRF-1 promoter. Sequential treatments of ATRA, on the other hand, increase IRF-1 nuclear localization. Alterations in the functions of transport proteins responsible for nuclear translocation and/or sequestration of IRF-1 are possible mechanisms that require further exploration. Together, these results suggest that ATRA and related retinoids play dual roles in inducing the level of IRF-1 mRNA and protein, and in augmenting the nuclear localization of IRF-1 protein, both of which could be essential for full expression of the tumor suppressor activity and the immunosurveillance functions of IRF-1 in breast epithelial cells. The results therefore suggest that retinoids may reduce or delay the progression from the premalignant state to the transformed tumorigenic state, in part through the regulation of IRF-1.
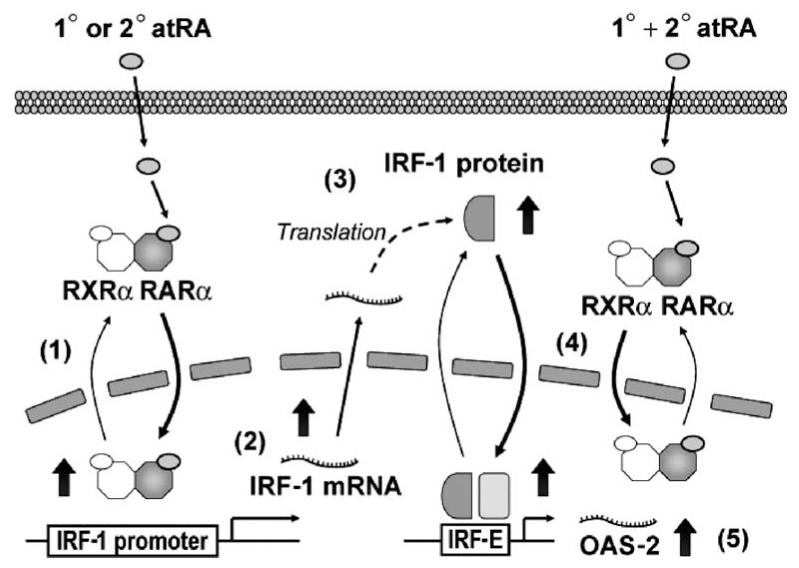
Figure 8.
Working model of ATRA-mediated regulation of IRF-1 expression and localization. In MCF10A cells, the 1° dose of ATRA initiates the shuttling of RARα and its dimerization partner, RXRα, from the cytoplasm to the nucleus in a ligand-dependent manner (Ref. 1; Fig. 4), thereby enhancing the transactivation activity of the heterodimer, which is proposed to lead to yet-to-be-characterized changes that activate the IRF-1 promoter. IRF-1 is induced (Ref. 2; Fig. 1C and E) and translated (Ref. 3; Fig. 1B and D). Sequential doses (1° + 2°) of ATRA increase the nuclear localization of IRF-1 (Ref. 4; Figs. 5 and 6), possibly assisted by the shuttling of RARα/RXRα. Increased nuclear localization and DNA-binding activity of IRF-1 is followed by augmented transcription of IRF-1 target genes, such as OAS-2 (Ref. 5; Fig. 7).
Acknowledgments
This work was supported by grants DK 41479 and CA 90214 from the National Institutes of Health, and by Tobacco Settlement Funds awarded to The Pennsylvania State University.
We are grateful to Elaine Kunze, Center for Quantitative Cell Analysis, The Pennsylvania State University, for technical assistance in confocal microscopy; and to Dr. Judith Weisz, Hershey Medical Center, for facilitating our use of the MCF10A cell line.
References
- 1.Altucci L, Gronemeyer H. The promise of retinoids to fight against cancer. Nat Rev Cancer. 2001;1:181–193. doi: 10.1038/35106036. [DOI] [PubMed] [Google Scholar]
- 2.Ross AC, Stephensen CB. Vitamin A and retinoids in antiviral responses. FASEB J. 1996;10:979–985. [PubMed] [Google Scholar]
- 3.Gronemeyer H, Gustafsson JA, Laudet V. Principles for modulation of the nuclear receptor superfamily. Nat Rev Drug Discov. 2004;3:950–964. doi: 10.1038/nrd1551. [DOI] [PubMed] [Google Scholar]
- 4.Wei LN. Retinoids and receptor interacting protein 140 (RIP140) in gene regulation. Curr Med Chem. 2004;11:1527–1532. doi: 10.2174/0929867043365017. [DOI] [PubMed] [Google Scholar]
- 5.Privalsky ML. The role of corepressors in transcriptional regulation by nuclear hormone receptors. Annu Rev Physiol. 2004;66:315–360. doi: 10.1146/annurev.physiol.66.032802.155556. [DOI] [PubMed] [Google Scholar]
- 6.Kagechika H, Shudo K. Synthetic retinoids: recent developments concerning structure and clinical utility. J Med Chem. 2005;48:5875–5883. doi: 10.1021/jm0581821. [DOI] [PubMed] [Google Scholar]
- 7.Matikainen S, Ronni T, Hurme M, Pine R, Julkunen I. Retinoic acid activates interferon regulatory factor-1 gene expression in myeloid cells. Blood. 1996;88:114–123. [PubMed] [Google Scholar]
- 8.Miyamoto M, Fujita T, Kimura Y, Maruyama M, Harada H, Sudo Y, Miyata T, Taniguchi T. Regulated expression of a gene encoding a nuclear factor, IRF-1, that specifically binds to IFN-beta gene regulatory elements. Cell. 1988;54:903–913. doi: 10.1016/s0092-8674(88)91307-4. [DOI] [PubMed] [Google Scholar]
- 9.Kimura T, Nakayama K, Penninger J, Kitagawa M, Harada H, Matsuyama T, Tanaka N, Kamijo R, Vilcek J, Mak TW, Taniguchi T. Involvement of the IRF-1 transcription factor in antiviral responses to interferons. Science. 1994;264:1921–1924. doi: 10.1126/science.8009222. [DOI] [PubMed] [Google Scholar]
- 10.Harada H, Taniguchi T, Tanaka N. The role of interferon regulatory factors in the interferon system and cell growth control. Biochimie. 1998;80:641–650. doi: 10.1016/s0300-9084(99)80017-0. [DOI] [PubMed] [Google Scholar]
- 11.Romeo G, Fiorucci G, Chiantore MV, Percario ZA, Vannucchi S, Affabris E. IRF-1 as a negative regulator of cell proliferation. J Interferon Cytokine Res. 2002;22:39–47. doi: 10.1089/107999002753452647. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka N, Ishihara M, Kitagawa M, Harada H, Kimura T, Matsuyama T, Lamphier MS, Aizawa S, Mak TW, Taniguchi T. Cellular commitment to oncogene-induced transformation or apoptosis is dependent on the transcription factor IRF-1. Cell. 1994;77:829–839. doi: 10.1016/0092-8674(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 13.Bouker KB, Skaar TC, Riggins RB, Harburger DS, Fernandez DR, Zwart A, Wang A, Clarke R. Interferon regulatory factor-1 (IRF-1) exhibits tumor suppressor activities in breast cancer associated with caspase activation and induction of apoptosis. Carcinogenesis. 2005;26:1527–1535. doi: 10.1093/carcin/bgi113. [DOI] [PubMed] [Google Scholar]
- 14.Connett JM, Badri L, Giordano TJ, Connett WC, Doherty GM. Interferon regulatory factor 1 (IRF-1) and IRF-2 expression in breast cancer tissue microarrays. J Interferon Cytokine Res. 2005;25:587–594. doi: 10.1089/jir.2005.25.587. [DOI] [PubMed] [Google Scholar]
- 15.Luo XM, Ross AC. Physiological and receptor-selective retinoids modulate interferon-gamma signaling by increasing the expression, nuclear localization, and functional activity of interferon regulatory factor-1. J Biol Chem. 2005;280:36228–36236. doi: 10.1074/jbc.M505749200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arany I, Whitehead WE, Grattendick KJ, Ember IA, Tyring SK. Suppression of growth by all-trans retinoic acid requires prolonged induction of interferon regulatory factor 1 in cervical squamous carcinoma (SiHa) cells. Clin Diagn Lab Immunol. 2002;9:1102–1106. doi: 10.1128/CDLI.9.5.1102-1106.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pelicano L, Li F, Schindler C, Chelbi-Alix MK. Retinoic acid enhances the expression of interferon-induced proteins: evidence for multiple mechanisms of action. Oncogene. 1997;15:2349–2359. doi: 10.1038/sj.onc.1201410. [DOI] [PubMed] [Google Scholar]
- 18.Percario ZA, Giandomenico V, Fiorucci G, Chiantore MV, Vannucchi S, Hiscott J, Affabris E, Romeo G. Retinoic acid is able to induce interferon regulatory factor 1 in squamous carcinoma cells via a STAT-1 independent signalling pathway. Cell Growth Differ. 1999;10:263–270. [PubMed] [Google Scholar]
- 19.Soule HD, Maloney TM, Wolman SR, Peterson WD, Jr, Brenz R, McGrath CM, Russo J, Pauley RJ, Jones RF, Brooks SC. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990;50:6075–6086. [PubMed] [Google Scholar]
- 20.Fernandez SV, Russo IH, Russo J. Estradiol and its metabolites 4-hydroxyestradiol and 2-hydroxyestradiol induce mutations in human breast epithelial cells. Int J Cancer. 2005 doi: 10.1002/ijc.21590. (article online in advance of print) [DOI] [PubMed] [Google Scholar]
- 21.Tanaka N, Taniguchi T. The interferon regulatory factors and oncogenesis. Semin Cancer Biol. 2000;10:73–81. doi: 10.1006/scbi.2000.0310. [DOI] [PubMed] [Google Scholar]
- 22.Kroger A, Koster M, Schroeder K, Hauser H, Mueller PP. Activities of IRF-1. J Interferon Cytokine Res. 2002;22:5–14. doi: 10.1089/107999002753452610. [DOI] [PubMed] [Google Scholar]
- 23.Peng X, Yun D, Christov K. Breast cancer progression in MCF10A series of cell lines is associated with alterations in retinoic acid and retinoid × receptors and with differential response to retinoids. Int J Oncol. 2004;25:961–971. [PubMed] [Google Scholar]
- 24.Chen Q, Ma Y, Ross AC. Opposing cytokine-specific effects of all trans-retinoic acid on the activation and expression of signal transducer and activator of transcription (STAT)-1 in THP-1 cells. Immunology. 2002;107:199–208. doi: 10.1046/j.1365-2567.2002.01485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watanabe N, Sakakibara J, Hovanessian AG, Taniguchi T, Fujita T. Activation of IFN-beta element by IRF-1 requires a posttranslational event in addition to IRF-1 synthesis. Nucleic Acids Res. 1991;19:4421–4428. doi: 10.1093/nar/19.16.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allenby G, Bocquel MT, Saunders M, Kazmer S, Speck J, Rosenberger M, Lovey A, Kastner P, Grippo JF, Chambon P, et al. Retinoic acid receptors and retinoid × receptors: interactions with endogenous retinoic acids. Proc Natl Acad Sci U S A. 1993;90:30–34. doi: 10.1073/pnas.90.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin B, Bernardon JM, Cavey MT, Bernard B, Carlavan I, Charpentier B, Pilgrim WR, Shroot B, Reichert U. Selective synthetic ligands for human nuclear retinoic acid receptors. Skin Pharmacol. 1992;5:57–65. doi: 10.1159/000211018. [DOI] [PubMed] [Google Scholar]
- 28.Sani BP, Zhang X, Hill DL, Shealy YF. Retinyl methyl ether: binding to transport proteins and effect on transcriptional regulation. Biochem Biophys Res Commun. 1996;223:293–298. doi: 10.1006/bbrc.1996.0887. [DOI] [PubMed] [Google Scholar]
- 29.Gianni M, Li Calzi M, Terao M, Guiso G, Caccia S, Barbui T, Rambaldi A, Garattini E. AM580, a stable benzoic derivative of retinoic acid, has powerful and selective cyto-differentiating effects on acute promyelocytic leukemia cells. Blood. 1996;87:1520–1531. [PubMed] [Google Scholar]
- 30.Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- 31.Yu-Lee LY. Prolactin modulation of immune and inflammatory responses. Recent Prog Horm Res. 2002;57:435–455. doi: 10.1210/rp.57.1.435. [DOI] [PubMed] [Google Scholar]
- 32.Le Stunff C, Rotwein P. Growth hormone stimulates interferon regulatory factor-1 gene expression in the liver. Endocrinology. 1998;139:859–866. doi: 10.1210/endo.139.3.5819. [DOI] [PubMed] [Google Scholar]
- 33.Tait L, Soule HD, Russo J. Ultrastructural and immunocytochemical characterization of an immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990;50:6087–6094. [PubMed] [Google Scholar]
- 34.Yu-Lee LY, Hrachovy JA, Stevens AM, Schwarz LA. Interferon-regulatory factor 1 is an immediate-early gene under transcriptional regulation by prolactin in Nb2 T cells. Mol Cell Biol. 1990;10:3087–3094. doi: 10.1128/mcb.10.6.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilmour KC, Reich NC. Receptor to nucleus signaling by prolactin and interleukin 2 via activation of latent DNA-binding factors. Proc Natl Acad Sci U S A. 1994;91:6850–6854. doi: 10.1073/pnas.91.15.6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Connett JM, Hunt SR, Hickerson SM, Wu SJ, Doherty GM. Localization of IFN-gamma-activated Stat1 and IFN regulatory factors 1 and 2 in breast cancer cells. J Interferon Cytokine Res. 2003;23:621–630. doi: 10.1089/107999003322558755. [DOI] [PubMed] [Google Scholar]
- 37.Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 38.Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 39.Schaper F, Kirchhoff S, Posern G, Koster M, Oumard A, Sharf R, Levi BZ, Hauser H. Functional domains of interferon regulatory factor I (IRF-1) Biochem J. 1998;335:147–157. doi: 10.1042/bj3350147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin R, Hiscott J. A role for casein kinase II phosphorylation in the regulation of IRF-1 transcriptional activity. Mol Cell Biochem. 1999;191:169–180. [PubMed] [Google Scholar]
- 41.Lin XF, Zhao BX, Chen HZ, Ye XF, Yang CY, Zhou HY, Zhang MQ, Lin SC, Wu Q. RXRαlpha acts as a carrier for TR3 nuclear export in a 9-cis retinoic acid-dependent manner in gastric cancer cells. J Cell Sci. 2004;117:5609–5621. doi: 10.1242/jcs.01474. [DOI] [PubMed] [Google Scholar]
- 42.Yasmin R, Williams RM, Xu M, Noy N. Nuclear import of the retinoid × receptor, the vitamin D receptor, and their mutual heterodimer. J Biol Chem. 2005;280:40152–40160. doi: 10.1074/jbc.M507708200. [DOI] [PubMed] [Google Scholar]
- 43.Lin R, Yang L, Arguello M, Penafuerte C, Hiscott J. A CRM1-dependent nuclear export pathway is involved in the regulation of IRF-5 subcellular localization. J Biol Chem. 2005;280:3088–3095. doi: 10.1074/jbc.M408452200. [DOI] [PubMed] [Google Scholar]
- 44.Barnes BJ, Kellum MJ, Pinder KE, Frisancho JA, Pitha PM. Interferon regulatory factor 5, a novel mediator of cell cycle arrest and cell death. Cancer Res. 2003;63:6424–6431. [PubMed] [Google Scholar]
- 45.Kumar KP, McBride KM, Weaver BK, Dingwall C, Reich NC. Regulated nuclear-cytoplasmic localization of interferon regulatory factor 3, a subunit of double-stranded RNA-activated factor 1. Mol Cell Biol. 2000;20:4159–4168. doi: 10.1128/mcb.20.11.4159-4168.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Behera AK, Kumar M, Lockey RF, Mohapatra SS. 2′-5′ oligoadenylate synthetase plays a critical role in interferon-gamma inhibition of respiratory syncytial virus infection of human epithelial cells. J Biol Chem. 2002;277:25601–25608. doi: 10.1074/jbc.M200211200. [DOI] [PubMed] [Google Scholar]
- 47.Mullan PB, Hosey AM, Buckley NE, Quinn JE, Kennedy RD, Johnston PG, Harkin DP. The 2,5 oligoadenylate synthetase/RNaseL pathway is a novel effector of BRCA1- and interferon-gamma–mediated apoptosis. Oncogene. 2005;24:5492–5501. doi: 10.1038/sj.onc.1208698. [DOI] [PubMed] [Google Scholar]
- 48.Malathi K, Paranjape JM, Ganapathi R, Silverman RH. HPC1/RNA-SEL mediates apoptosis of prostate cancer cells treated with 2′,5′-oligoadenylates, topoisomerase I inhibitors, and tumor necrosis factor-related apoptosis-inducing ligand. Cancer Res. 2004;64:9144–9151. doi: 10.1158/0008-5472.CAN-04-2226. [DOI] [PubMed] [Google Scholar]