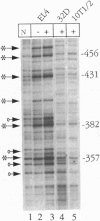
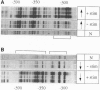
Abstract
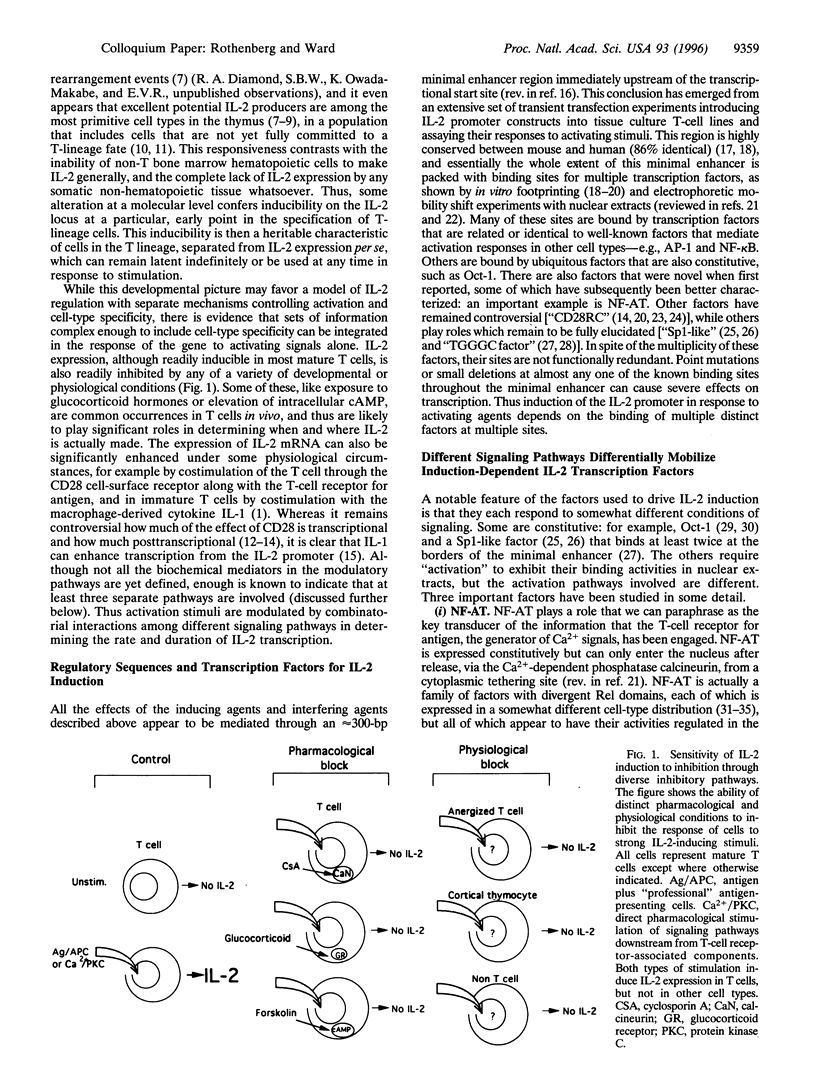
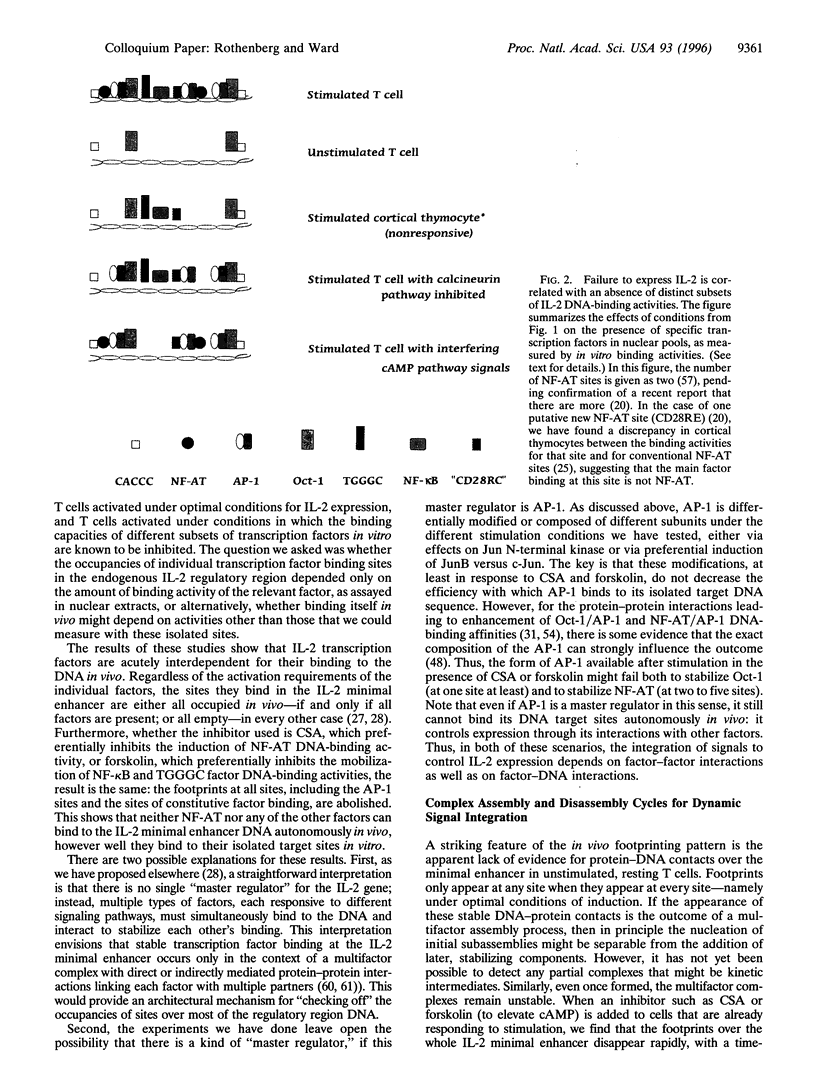
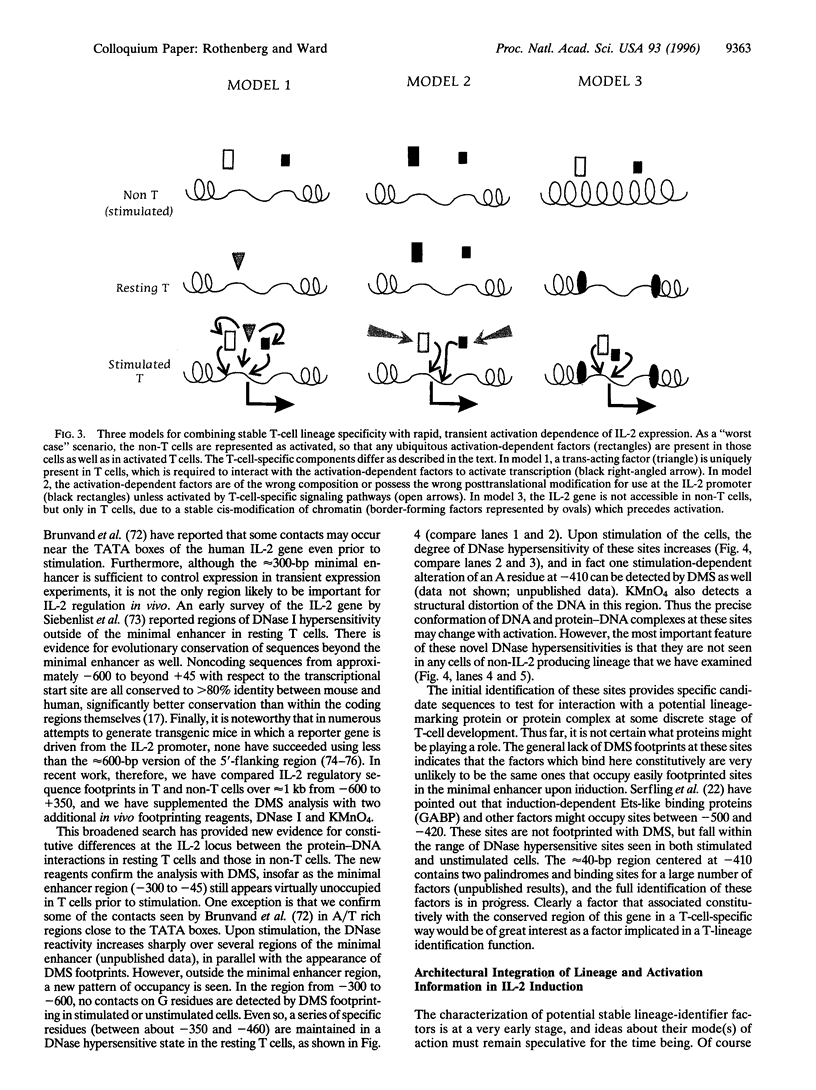
The interleukin 2 (IL-2) gene is subject to two types of regulation: its expression is T-lymphocyte-specific and it is acutely dependent on specific activation signals. The IL-2 transcriptional apparatus integrates multiple types of biochemical information in determining whether or not the gene will be expressed, using multiple diverse transcription factors that are each optimally activated or inhibited by different signaling pathways. When activation of one or two of these factors is blocked IL-2 expression is completely inhibited. The inability of the other, unaffected factors to work is explained by the striking finding that none of the factors interacts stably with its target site in the IL-2 enhancer unless all the factors are present. Coordinate occupancy of all the sites in the minimal enhancer is apparently maintained by continuous assembly and disassembly cycles that respond to the instantaneous levels of each factor in the nuclear compartment. In addition, the minimal enhancer undergoes specific increases in DNase I accessibility, consistent with dramatic changes in chromatin structure upon activation. Still to be resolved is what interaction(s) conveys T-lineage specificity. In the absence of activating signals, the minimal IL-2 enhancer region in mature T cells is apparently unoccupied, exactly as in non-T lineage cells. However, in a conserved but poorly studied upstream region, we have now mapped several novel sites of DNase I hypersensitivity in vivo that constitutively distinguish IL-2 producer type T cells from cell types that cannot express IL-2. Thus a distinct domain of the IL-2 regulatory sequence may contain sites for competence- or lineage-marking protein contacts.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams C. C., Workman J. L. Binding of disparate transcriptional activators to nucleosomal DNA is inherently cooperative. Mol Cell Biol. 1995 Mar;15(3):1405–1421. doi: 10.1128/mcb.15.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Algarté M., Lécine P., Costello R., Plet A., Olive D., Imbert J. In vivo regulation of interleukin-2 receptor alpha gene transcription by the coordinated binding of constitutive and inducible factors in human primary T cells. EMBO J. 1995 Oct 16;14(20):5060–5072. doi: 10.1002/j.1460-2075.1995.tb00188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardavin C., Wu L., Li C. L., Shortman K. Thymic dendritic cells and T cells develop simultaneously in the thymus from a common precursor population. Nature. 1993 Apr 22;362(6422):761–763. doi: 10.1038/362761a0. [DOI] [PubMed] [Google Scholar]
- Brabletz T., Pietrowski I., Serfling E. The immunosuppressives FK 506 and cyclosporin A inhibit the generation of protein factors binding to the two purine boxes of the interleukin 2 enhancer. Nucleic Acids Res. 1991 Jan 11;19(1):61–67. doi: 10.1093/nar/19.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresnick E. H., Felsenfeld G. Dual promoter activation by the human beta-globin locus control region. Proc Natl Acad Sci U S A. 1994 Feb 15;91(4):1314–1317. doi: 10.1073/pnas.91.4.1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brombacher F., Schäfer T., Weissenstein U., Tschopp C., Andersen E., Bürki K., Baumann G. IL-2 promoter-driven lacZ expression as a monitoring tool for IL-2 expression in primary T cells of transgenic mice. Int Immunol. 1994 Feb;6(2):189–197. doi: 10.1093/intimm/6.2.189. [DOI] [PubMed] [Google Scholar]
- Brooks J. W., Yoza B. K., Mizel S. B. Interleukin 1 activation of the AP-1 transcription complex in murine T cells is regulated at the level of Jun B protein accumulation. Mol Immunol. 1995 Aug;32(11):779–788. doi: 10.1016/0161-5890(95)00055-j. [DOI] [PubMed] [Google Scholar]
- Brunvand M. W., Krumm A., Groudine M. In vivo footprinting of the human IL-2 gene reveals a nuclear factor bound to the transcription start site in T cells. Nucleic Acids Res. 1993 Oct 11;21(20):4824–4829. doi: 10.1093/nar/21.20.4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunvand M. W., Schmidt A., Siebenlist U. Nuclear factors interacting with the mitogen-responsive regulatory region of the interleukin-2 gene. J Biol Chem. 1988 Dec 15;263(35):18904–18910. [PubMed] [Google Scholar]
- Cardell S., Sander B. Interleukin 2, 4 and 5 are sequentially produced in mitogen-stimulated murine spleen cell cultures. Eur J Immunol. 1990 Feb;20(2):389–395. doi: 10.1002/eji.1830200223. [DOI] [PubMed] [Google Scholar]
- Chen D., Rothenberg E. V. Interleukin 2 transcription factors as molecular targets of cAMP inhibition: delayed inhibition kinetics and combinatorial transcription roles. J Exp Med. 1994 Mar 1;179(3):931–942. doi: 10.1084/jem.179.3.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Rothenberg E. V. Molecular basis for developmental changes in interleukin-2 gene inducibility. Mol Cell Biol. 1993 Jan;13(1):228–237. doi: 10.1128/mcb.13.1.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civil A., Verweij C. L. Regulation of IL2 gene transcription via the T-cell accessory molecule CD28. Res Immunol. 1995 Mar-Apr;146(3):158–164. doi: 10.1016/0923-2494(96)80250-1. [DOI] [PubMed] [Google Scholar]
- Ellis J., Tan-Un K. C., Harper A., Michalovich D., Yannoutsos N., Philipsen S., Grosveld F. A dominant chromatin-opening activity in 5' hypersensitive site 3 of the human beta-globin locus control region. EMBO J. 1996 Feb 1;15(3):562–568. [PMC free article] [PubMed] [Google Scholar]
- Emmel E. A., Verweij C. L., Durand D. B., Higgins K. M., Lacy E., Crabtree G. R. Cyclosporin A specifically inhibits function of nuclear proteins involved in T cell activation. Science. 1989 Dec 22;246(4937):1617–1620. doi: 10.1126/science.2595372. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin as an essential part of the transcriptional mechanism. Nature. 1992 Jan 16;355(6357):219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- Frantz B., Nordby E. C., Bren G., Steffan N., Paya C. V., Kincaid R. L., Tocci M. J., O'Keefe S. J., O'Neill E. A. Calcineurin acts in synergy with PMA to inactivate I kappa B/MAD3, an inhibitor of NF-kappa B. EMBO J. 1994 Feb 15;13(4):861–870. doi: 10.1002/j.1460-2075.1994.tb06329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser J. D., Irving B. A., Crabtree G. R., Weiss A. Regulation of interleukin-2 gene enhancer activity by the T cell accessory molecule CD28. Science. 1991 Jan 18;251(4991):313–316. doi: 10.1126/science.1846244. [DOI] [PubMed] [Google Scholar]
- Garrity P. A., Chen D., Rothenberg E. V., Wold B. J. Interleukin-2 transcription is regulated in vivo at the level of coordinated binding of both constitutive and regulated factors. Mol Cell Biol. 1994 Mar;14(3):2159–2169. doi: 10.1128/mcb.14.3.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity P. A., Wold B. J. Effects of different DNA polymerases in ligation-mediated PCR: enhanced genomic sequencing and in vivo footprinting. Proc Natl Acad Sci U S A. 1992 Feb 1;89(3):1021–1025. doi: 10.1073/pnas.89.3.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giese K., Kingsley C., Kirshner J. R., Grosschedl R. Assembly and function of a TCR alpha enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 1995 Apr 15;9(8):995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- Godfrey D. I., Kennedy J., Suda T., Zlotnik A. A developmental pathway involving four phenotypically and functionally distinct subsets of CD3-CD4-CD8- triple-negative adult mouse thymocytes defined by CD44 and CD25 expression. J Immunol. 1993 May 15;150(10):4244–4252. [PubMed] [Google Scholar]
- Godfrey D. I., Zlotnik A. Control points in early T-cell development. Immunol Today. 1993 Nov;14(11):547–553. doi: 10.1016/0167-5699(93)90186-O. [DOI] [PubMed] [Google Scholar]
- Granelli-Piperno A. In situ hybridization for interleukin 2 and interleukin 2 receptor mRNA in T cells activated in the presence or absence of cyclosporin A. J Exp Med. 1988 Nov 1;168(5):1649–1658. doi: 10.1084/jem.168.5.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grilli M., Chiu J. J., Lenardo M. J. NF-kappa B and Rel: participants in a multiform transcriptional regulatory system. Int Rev Cytol. 1993;143:1–62. doi: 10.1016/s0074-7696(08)61873-2. [DOI] [PubMed] [Google Scholar]
- Hentsch B., Mouzaki A., Pfeuffer I., Rungger D., Serfling E. The weak, fine-tuned binding of ubiquitous transcription factors to the Il-2 enhancer contributes to its T cell-restricted activity. Nucleic Acids Res. 1992 Jun 11;20(11):2657–2665. doi: 10.1093/nar/20.11.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho A. M., Jain J., Rao A., Hogan P. G. Expression of the transcription factor NFATp in a neuronal cell line and in the murine nervous system. J Biol Chem. 1994 Nov 11;269(45):28181–28186. [PubMed] [Google Scholar]
- Ho S. N., Thomas D. J., Timmerman L. A., Li X., Francke U., Crabtree G. R. NFATc3, a lymphoid-specific NFATc family member that is calcium-regulated and exhibits distinct DNA binding specificity. J Biol Chem. 1995 Aug 25;270(34):19898–19907. doi: 10.1074/jbc.270.34.19898. [DOI] [PubMed] [Google Scholar]
- Hoey T., Sun Y. L., Williamson K., Xu X. Isolation of two new members of the NF-AT gene family and functional characterization of the NF-AT proteins. Immunity. 1995 May;2(5):461–472. doi: 10.1016/1074-7613(95)90027-6. [DOI] [PubMed] [Google Scholar]
- Hsueh Y. P., Lai M. Z. c-Jun N-terminal kinase but not mitogen-activated protein kinase is sensitive to cAMP inhibition in T lymphocytes. J Biol Chem. 1995 Jul 28;270(30):18094–18098. doi: 10.1074/jbc.270.30.18094. [DOI] [PubMed] [Google Scholar]
- Jain J., Loh C., Rao A. Transcriptional regulation of the IL-2 gene. Curr Opin Immunol. 1995 Jun;7(3):333–342. doi: 10.1016/0952-7915(95)80107-3. [DOI] [PubMed] [Google Scholar]
- Kamps M. P., Corcoran L., LeBowitz J. H., Baltimore D. The promoter of the human interleukin-2 gene contains two octamer-binding sites and is partially activated by the expression of Oct-2. Mol Cell Biol. 1990 Oct;10(10):5464–5472. doi: 10.1128/mcb.10.10.5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S. M., Beverly B., Tran A. C., Brorson K., Schwartz R. H., Lenardo M. J. Transactivation by AP-1 is a molecular target of T cell clonal anergy. Science. 1992 Aug 21;257(5073):1134–1138. doi: 10.1126/science.257.5073.1134. [DOI] [PubMed] [Google Scholar]
- Kang S. M., Tran A. C., Grilli M., Lenardo M. J. NF-kappa B subunit regulation in nontransformed CD4+ T lymphocytes. Science. 1992 Jun 5;256(5062):1452–1456. doi: 10.1126/science.1604322. [DOI] [PubMed] [Google Scholar]
- Kolesnick R., Golde D. W. The sphingomyelin pathway in tumor necrosis factor and interleukin-1 signaling. Cell. 1994 May 6;77(3):325–328. doi: 10.1016/0092-8674(94)90147-3. [DOI] [PubMed] [Google Scholar]
- Lai J. H., Horvath G., Subleski J., Bruder J., Ghosh P., Tan T. H. RelA is a potent transcriptional activator of the CD28 response element within the interleukin 2 promoter. Mol Cell Biol. 1995 Aug;15(8):4260–4271. doi: 10.1128/mcb.15.8.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehming N., Thanos D., Brickman J. M., Ma J., Maniatis T., Ptashne M. An HMG-like protein that can switch a transcriptional activator to a repressor. Nature. 1994 Sep 8;371(6493):175–179. doi: 10.1038/371175a0. [DOI] [PubMed] [Google Scholar]
- Lindstein T., June C. H., Ledbetter J. A., Stella G., Thompson C. B. Regulation of lymphokine messenger RNA stability by a surface-mediated T cell activation pathway. Science. 1989 Apr 21;244(4902):339–343. doi: 10.1126/science.2540528. [DOI] [PubMed] [Google Scholar]
- Masuda E. S., Naito Y., Tokumitsu H., Campbell D., Saito F., Hannum C., Arai K., Arai N. NFATx, a novel member of the nuclear factor of activated T cells family that is expressed predominantly in the thymus. Mol Cell Biol. 1995 May;15(5):2697–2706. doi: 10.1128/mcb.15.5.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila P. S., Ullman K. S., Fiering S., Emmel E. A., McCutcheon M., Crabtree G. R., Herzenberg L. A. The actions of cyclosporin A and FK506 suggest a novel step in the activation of T lymphocytes. EMBO J. 1990 Dec;9(13):4425–4433. doi: 10.1002/j.1460-2075.1990.tb07893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey P. G., Luo C., Kerppola T. K., Jain J., Badalian T. M., Ho A. M., Burgeon E., Lane W. S., Lambert J. N., Curran T. Isolation of the cyclosporin-sensitive T cell transcription factor NFATp. Science. 1993 Oct 29;262(5134):750–754. doi: 10.1126/science.8235597. [DOI] [PubMed] [Google Scholar]
- Minasi L. E., Kamogawa Y., Carding S., Bottomly K., Flavell R. A. The selective ablation of interleukin 2-producing cells isolated from transgenic mice. J Exp Med. 1993 May 1;177(5):1451–1459. doi: 10.1084/jem.177.5.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore T. A., Zlotnik A. T-cell lineage commitment and cytokine responses of thymic progenitors. Blood. 1995 Sep 1;86(5):1850–1860. [PubMed] [Google Scholar]
- Mouzaki A., Rungger D. Properties of transcription factors regulating interleukin-2 gene transcription through the NFAT binding site in untreated or drug-treated naive and memory T-helper cells. Blood. 1994 Oct 15;84(8):2612–2621. [PubMed] [Google Scholar]
- Mueller P. R., Salser S. J., Wold B. Constitutive and metal-inducible protein:DNA interactions at the mouse metallothionein I promoter examined by in vivo and in vitro footprinting. Genes Dev. 1988 Apr;2(4):412–427. doi: 10.1101/gad.2.4.412. [DOI] [PubMed] [Google Scholar]
- Neumann M., Grieshammer T., Chuvpilo S., Kneitz B., Lohoff M., Schimpl A., Franza B. R., Jr, Serfling E. RelA/p65 is a molecular target for the immunosuppressive action of protein kinase A. EMBO J. 1995 May 1;14(9):1991–2004. doi: 10.1002/j.1460-2075.1995.tb07191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northrop J. P., Crabtree G. R., Mattila P. S. Negative regulation of interleukin 2 transcription by the glucocorticoid receptor. J Exp Med. 1992 May 1;175(5):1235–1245. doi: 10.1084/jem.175.5.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northrop J. P., Ho S. N., Chen L., Thomas D. J., Timmerman L. A., Nolan G. P., Admon A., Crabtree G. R. NF-AT components define a family of transcription factors targeted in T-cell activation. Nature. 1994 Jun 9;369(6480):497–502. doi: 10.1038/369497a0. [DOI] [PubMed] [Google Scholar]
- Novak T. J., Chen D., Rothenberg E. V. Interleukin-1 synergy with phosphoinositide pathway agonists for induction of interleukin-2 gene expression: molecular basis of costimulation. Mol Cell Biol. 1990 Dec;10(12):6325–6334. doi: 10.1128/mcb.10.12.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak T. J., White P. M., Rothenberg E. V. Regulatory anatomy of the murine interleukin-2 gene. Nucleic Acids Res. 1990 Aug 11;18(15):4523–4533. doi: 10.1093/nar/18.15.4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parekh B. S., Hatfield G. W. Transcriptional activation by protein-induced DNA bending: evidence for a DNA structural transmission model. Proc Natl Acad Sci U S A. 1996 Feb 6;93(3):1173–1177. doi: 10.1073/pnas.93.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeuffer I., Klein-Hessling S., Heinfling A., Chuvpilo S., Escher C., Brabletz T., Hentsch B., Schwarzenbach H., Matthias P., Serfling E. Octamer factors exert a dual effect on the IL-2 and IL-4 promoters. J Immunol. 1994 Dec 15;153(12):5572–5585. [PubMed] [Google Scholar]
- Rao A. NF-ATp: a transcription factor required for the co-ordinate induction of several cytokine genes. Immunol Today. 1994 Jun;15(6):274–281. doi: 10.1016/0167-5699(94)90007-8. [DOI] [PubMed] [Google Scholar]
- Reitman M., Lee E., Westphal H., Felsenfeld G. An enhancer/locus control region is not sufficient to open chromatin. Mol Cell Biol. 1993 Jul;13(7):3990–3998. doi: 10.1128/mcb.13.7.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T., Yang-Snyder J. A., Rothenberg E. V., Carding S. R. Regulated expression and function of CD122 (interleukin-2/interleukin-15R-beta) during lymphoid development. Blood. 1996 Jan 1;87(1):190–201. [PubMed] [Google Scholar]
- Riegel J. S., Corthesy B., Flanagan W. M., Crabtree G. R. Regulation of the interleukin-2 gene. Chem Immunol. 1992;51:266–298. [PubMed] [Google Scholar]
- Rincon M., Flavell R. A. Regulation of AP-1 and NFAT transcription factors during thymic selection of T cells. Mol Cell Biol. 1996 Mar;16(3):1074–1084. doi: 10.1128/mcb.16.3.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincón M., Flavell R. A. AP-1 transcriptional activity requires both T-cell receptor-mediated and co-stimulatory signals in primary T lymphocytes. EMBO J. 1994 Sep 15;13(18):4370–4381. doi: 10.1002/j.1460-2075.1994.tb06757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano-Spica V., Georgiou P., Suzuki H., Papas T. S., Bhat N. K. Role of ETS1 in IL-2 gene expression. J Immunol. 1995 Mar 15;154(6):2724–2732. [PubMed] [Google Scholar]
- Rooney J. W., Sun Y. L., Glimcher L. H., Hoey T. Novel NFAT sites that mediate activation of the interleukin-2 promoter in response to T-cell receptor stimulation. Mol Cell Biol. 1995 Nov;15(11):6299–6310. doi: 10.1128/mcb.15.11.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg E. V., Chen D., Diamond R. A. Functional and phenotypic analysis of thymocytes in SCID mice. Evidence for functional response transitions before and after the SCID arrest point. J Immunol. 1993 Oct 1;151(7):3530–3546. [PubMed] [Google Scholar]
- Rothenberg E. V., Diamond R. A., Chen D. Programming for recognition and programming for response. Separate developmental subroutines in the murine thymus. Thymus. 1994;22(4):215–244. [PubMed] [Google Scholar]
- Rothenberg E. V., Diamond R. A., Pepper K. A., Yang J. A. IL-2 gene inducibility in T cells before T cell receptor expression. Changes in signaling pathways and gene expression requirements during intrathymic maturation. J Immunol. 1990 Mar 1;144(5):1614–1624. [PubMed] [Google Scholar]
- Rothenberg E. V. The development of functionally responsive T cells. Adv Immunol. 1992;51:85–214. doi: 10.1016/s0065-2776(08)60487-3. [DOI] [PubMed] [Google Scholar]
- Sauer F., Hansen S. K., Tjian R. Multiple TAFIIs directing synergistic activation of transcription. Science. 1995 Dec 15;270(5243):1783–1788. doi: 10.1126/science.270.5243.1783. [DOI] [PubMed] [Google Scholar]
- Sen J., Venkataraman L., Shinkai Y., Pierce J. W., Alt F. W., Burakoff S. J., Sen R. Expression and induction of nuclear factor-kappa B-related proteins in thymocytes. J Immunol. 1995 Apr 1;154(7):3213–3221. [PubMed] [Google Scholar]
- Serfling E., Avots A., Neumann M. The architecture of the interleukin-2 promoter: a reflection of T lymphocyte activation. Biochim Biophys Acta. 1995 Sep 19;1263(3):181–200. doi: 10.1016/0167-4781(95)00112-t. [DOI] [PubMed] [Google Scholar]
- Serfling E., Barthelmäs R., Pfeuffer I., Schenk B., Zarius S., Swoboda R., Mercurio F., Karin M. Ubiquitous and lymphocyte-specific factors are involved in the induction of the mouse interleukin 2 gene in T lymphocytes. EMBO J. 1989 Feb;8(2):465–473. doi: 10.1002/j.1460-2075.1989.tb03399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J. P., Utz P. J., Durand D. B., Toole J. J., Emmel E. A., Crabtree G. R. Identification of a putative regulator of early T cell activation genes. Science. 1988 Jul 8;241(4862):202–205. doi: 10.1126/science.3260404. [DOI] [PubMed] [Google Scholar]
- Siebenlist U., Durand D. B., Bressler P., Holbrook N. J., Norris C. A., Kamoun M., Kant J. A., Crabtree G. R. Promoter region of interleukin-2 gene undergoes chromatin structure changes and confers inducibility on chloramphenicol acetyltransferase gene during activation of T cells. Mol Cell Biol. 1986 Sep;6(9):3042–3049. doi: 10.1128/mcb.6.9.3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerka C., Decker E. L., Zipfel P. F. A regulatory element in the human interleukin 2 gene promoter is a binding site for the zinc finger proteins Sp1 and EGR-1. J Biol Chem. 1995 Sep 22;270(38):22500–22506. doi: 10.1074/jbc.270.38.22500. [DOI] [PubMed] [Google Scholar]
- Strubin M., Newell J. W., Matthias P. OBF-1, a novel B cell-specific coactivator that stimulates immunoglobulin promoter activity through association with octamer-binding proteins. Cell. 1995 Feb 10;80(3):497–506. doi: 10.1016/0092-8674(95)90500-6. [DOI] [PubMed] [Google Scholar]
- Su B., Jacinto E., Hibi M., Kallunki T., Karin M., Ben-Neriah Y. JNK is involved in signal integration during costimulation of T lymphocytes. Cell. 1994 Jun 3;77(5):727–736. doi: 10.1016/0092-8674(94)90056-6. [DOI] [PubMed] [Google Scholar]
- Sundstedt A., Sigvardsson M., Leanderson T., Hedlund G., Kalland T., Dohlsten M. In vivo anergized CD4+ T cells express perturbed AP-1 and NF-kappa B transcription factors. Proc Natl Acad Sci U S A. 1996 Feb 6;93(3):979–984. doi: 10.1073/pnas.93.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski P., Levine M. Multiple modes of dorsal-bHLH transcriptional synergy in the Drosophila embryo. EMBO J. 1995 May 15;14(10):2229–2238. doi: 10.1002/j.1460-2075.1995.tb07217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanos D., Maniatis T. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell. 1995 Dec 29;83(7):1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- Thompson C. B., Wang C. Y., Ho I. C., Bohjanen P. R., Petryniak B., June C. H., Miesfeldt S., Zhang L., Nabel G. J., Karpinski B. cis-acting sequences required for inducible interleukin-2 enhancer function bind a novel Ets-related protein, Elf-1. Mol Cell Biol. 1992 Mar;12(3):1043–1053. doi: 10.1128/mcb.12.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuruta L., Lee H. J., Masuda E. S., Koyano-Nakagawa N., Arai N., Arai K., Yokota T. Cyclic AMP inhibits expression of the IL-2 gene through the nuclear factor of activated T cells (NF-AT) site, and transfection of NF-AT cDNAs abrogates the sensitivity of EL-4 cells to cyclic AMP. J Immunol. 1995 May 15;154(10):5255–5264. [PubMed] [Google Scholar]
- Ullman K. S., Flanagan W. M., Edwards C. A., Crabtree G. R. Activation of early gene expression in T lymphocytes by Oct-1 and an inducible protein, OAP40. Science. 1991 Oct 25;254(5031):558–562. doi: 10.1126/science.1683003. [DOI] [PubMed] [Google Scholar]
- Ullman K. S., Northrop J. P., Admon A., Crabtree G. R. Jun family members are controlled by a calcium-regulated, cyclosporin A-sensitive signaling pathway in activated T lymphocytes. Genes Dev. 1993 Feb;7(2):188–196. doi: 10.1101/gad.7.2.188. [DOI] [PubMed] [Google Scholar]
- Umlauf S. W., Beverly B., Lantz O., Schwartz R. H. Regulation of interleukin 2 gene expression by CD28 costimulation in mouse T-cell clones: both nuclear and cytoplasmic RNAs are regulated with complex kinetics. Mol Cell Biol. 1995 Jun;15(6):3197–3205. doi: 10.1128/mcb.15.6.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacca A., Felli M. P., Farina A. R., Martinotti S., Maroder M., Screpanti I., Meco D., Petrangeli E., Frati L., Gulino A. Glucocorticoid receptor-mediated suppression of the interleukin 2 gene expression through impairment of the cooperativity between nuclear factor of activated T cells and AP-1 enhancer elements. J Exp Med. 1992 Mar 1;175(3):637–646. doi: 10.1084/jem.175.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga-Weisz P. D., Becker P. B. Transcription factor-mediated chromatin remodelling: mechanisms and models. FEBS Lett. 1995 Aug 1;369(1):118–121. doi: 10.1016/0014-5793(95)00549-o. [DOI] [PubMed] [Google Scholar]
- Wallrath L. L., Lu Q., Granok H., Elgin S. C. Architectural variations of inducible eukaryotic promoters: preset and remodeling chromatin structures. Bioessays. 1994 Mar;16(3):165–170. doi: 10.1002/bies.950160306. [DOI] [PubMed] [Google Scholar]
- Williams T. M., Moolten D., Burlein J., Romano J., Bhaerman R., Godillot A., Mellon M., Rauscher F. J., 3rd, Kant J. A. Identification of a zinc finger protein that inhibits IL-2 gene expression. Science. 1991 Dec 20;254(5039):1791–1794. doi: 10.1126/science.1840704. [DOI] [PubMed] [Google Scholar]
- Wolffe A. P. Nucleosome positioning and modification: chromatin structures that potentiate transcription. Trends Biochem Sci. 1994 Jun;19(6):240–244. doi: 10.1016/0968-0004(94)90148-1. [DOI] [PubMed] [Google Scholar]
- Yang-Snyder J. A., Rothenberg E. V. Developmental and anatomical patterns of IL-2 gene expression in vivo in the murine thymus. Dev Immunol. 1993;3(2):85–102. doi: 10.1155/1993/86096. [DOI] [PMC free article] [PubMed] [Google Scholar]