Abstract
Age-related changes in splice-forms of LMNA, which encodes the nuclear lamina proteins lamin A/C, have not been investigated in skeletal muscle. In the rare premature ageing disease, Hutchinson-Gilford progeria syndrome (HGPS), de novo point mutations in LMNA activate a cryptic splice site in exon 11, resulting in a 150 base deletion in LMNA mRNA and accumulation of a truncated protein isoform, progerin. The LMNA Δ150 progerin transcript has also been found in trace quantities in tissues of healthy people and its implication in ‘natural’ ageing has been proposed. We therefore investigated the expression of progerin and lamin A/C in normal human and mouse skeletal muscles of different ages. LMNA Δ150 was detected in most muscle samples from healthy individuals aged 16-71 years, but was not present in any mouse muscle samples up to the age of 18 months. Real time qPCR of human muscle samples showed that there was an age-related increase in both the full length lamin A and LMNA Δ150 transcripts, whereas their protein levels did not change significantly with age. These findings indicate that there is a basal level of mis-splicing during LMNA expression that does not change with ageing in human muscle, but at levels that do not result in increased aberrant protein. The significance of these findings in the pathophysiology of muscle ageing is uncertain and warrants further investigation.
Keywords: Human, mouse, skeletal muscle, lamin A/C, progerin, ageing
Introduction
The LMNA gene encodes the nuclear lamina proteins, lamin A and C, through alternative splicing involving exon 10 and terminal exon usage. Apart from providing mechanical support to maintain nuclear shape, the lamina network has a number of other roles, such as interacting with heterochromatin to localize it to the periphery of the nucleus [1], controlling mitosis through interaction with cell division regulators [2], and being involved in initiation of DNA replication and transcription [3,4]. Mutations in LMNA are associated with a heterogeneous group of disorders, collectively known as the ‘laminopathies’, which include the premature ageing disease Hutchinson-Gilford Progeria Syndrome (HGPS), a number of different forms of muscular dystrophy, Charcot-Marie-Tooth disease type 2B1, Dunnigan-type familial partial lipodystrophy, and mandibuloacral dysplasia [5,6]. Most patients with HGPS carry a heterozygous de novo synonymous substitution in LMNA (1824C>T, G608G) that activates a nearby cryptic donor splice site within exon 11, resulting in the production of an internally truncated LMNA mRNA missing 150 bases (LMNA Δ150) [7,8]. It is uncertain how the encoded protein ‘progerin’ causes accelerated tissue ageing, but the de novo dominant mode of inheritance and the failure to rescue the phenotype by increasing wild-type lamin A expression suggests a dominant-negative effect [9].
Progerin is also present in trace amounts in some normal tissues such as skin, liver, heart and blood vessels, and it has been suggested that its accumulation may play a role in the normal ageing process in these tissues [10,11]. However, previous studies have not investigated the expression of progerin in skeletal muscle, and it remains unproven whether progerin expression in normal tissues is age-related or contributes to cellular ageing.
In this study we investigated the changes in LMNA splicing and expression of the different isoforms in human and mouse skeletal muscle using RT-PCR, real-time qPCR, immunoblotting and confocal microscopy, and we compared the levels of progerin expression with those in HGPS cell lines.
Materials and methods
Tissue samples
Tissue samples from the vastus lateralis muscle were obtained from otherwise healthy individuals, aged 16 to 71 years (n=18, 10 male), undergoing evaluation for malignant hyperthermia (MH) susceptibility. All these individuals were subsequently classified as MH-negative after in vitro contracture testing. Surplus material from muscle biopsies was stored in the Department of Anatomical Pathology at Royal Perth Hospital and was provided after informed consent. All biopsies showed normal muscle histology.
Muscle samples were also obtained from the triceps brachii, quadriceps femoris and tibialis anterior muscles of wild-type C57BL6/SJL mice aged 6, 9, 12 and 18 months (n=13, 9 males). Additional heart and liver tissues were obtained from the same mouse colony of the same age range (n=8, 6 males). Tissues were snap-frozen in isopentane chilled with liquid nitrogen. All muscle samples were stored at -80°C before use. Sections 8 μm thick for immunohistological studies and immunoblotting were cut using a Leica CM1900 cryostat (Leica Microsystems, North Ryde, Australia).
Ethical approval for the studies was obtained from the Royal Perth Hospital Human Research Ethics Committee and the University of Western Australia Animal Experimentation Committee.
Cell culture
Primary HGPS fibroblasts were obtained from Coriell cell repositories (Coriell Institute for Medical Research, Camden, NJ, Cat # AG03513). Cells were proliferated in Dulbecco’s Modified Eagle Medium (Gibco, Mulgrave, Australia) supplemented with 15% fetal calf serum, 10 U/ml penicillin, 10 mg/ml streptomycin, and 250 ng/ml Amphotericin B (Sigma Aldrich, Sydney, Australia) in a 37°C incubator with 5% CO2.
RNA extraction and reverse-transcription polymerase chain analysis (RT-PCR)
RNA was extracted from HGPS fibroblast cultures, human muscle specimens, and muscle, heart and liver tissues from wild-type mice, using Trizol (Invitrogen, Mulgrave, Australia) according to the manufacturer’s instructions. RNA pellets were resuspended in RNase-free water and purity and concentration estimated from absorbance reading using a Nano-drop spectrophotometer (Thermo Scientific, Scoresby, Australia).
100 ng of total RNA was used as template in a one-step RT-PCR with Superscript III (Invitrogen), using human specific primers located in exons 7 and 12, or murine specific primers annealing to the exon 9/10 junction and exon 12 for detection of both LMNA and LMNA Δ150 transcripts (Table 1). Reverse transcriptase-amplification reactions were incubated in a G-Storm GS1 thermocycler (GeneWorks, Hindmarsh, Australia) using the following conditions: 55°C for 30 min, 95°C for 10 min, 35 cycles of 94°C for 30 sec, 60°C for 1 min, 68°C for 2 min. Amplicons were separated on 2% agarose gels and, after staining with ethidium bromide, images were captured using the Chemi-Smart 3000 gel documentation system (Vilber Lourmat, Marne-la-Vallée, France).
Table 1.
Primers for RT-PCR
| Species | Primer sequences (5’→3’) | |
|---|---|---|
| Human | AACTGGAGTCCACTGAGA | Ex7 forward |
| AGATTACATGATGCTGCAGTT | Ex12 reverse | |
| Mouse | ATCAACTCCACTGGAGAAGAAGT | Ex9/10 forward |
| CAGACAGGAGGTGGCATGT | Ex12 reverse | |
Real time polymerase chain analysis (qRT-PCR)
TaqMan qRT-PCR assays (TaqMan, Mulgrave, Australia) were used to detect LMNA, LMNA Δ150 and LMNC transcripts [11]. A commercially available assay (Applied Biosystems, Mulgrave, Australia) targeting human RPLP0, was included as an endogenous control. RNA was reverse-transcribed using the Superscript II cDNA synthesis kit (Invitrogen) with 50 ng random hexamers. Ten μl real-time reactions with 5 μl of TaqMan Universal Master Mix were performed in MicroAmp 384-well plate in the 7900HT Real-Time PCR system (Applied Biosystems). To increase the detection capability, 4.8 μl cDNA was used for LMNA Δ150 reactions and 2.5 μl for LMNA and RPLP0 reactions. For LMNA Δ150 assays: 200 nM MGB probe, 50 nM forward primer and 100 nM reverse primer were used, whereas for LMNA and LMNC assays: 200 nM MGB probe, 500 nM forward primer and 500 nM reverse primer were used [11]. Reactions were carried out in triplicate and end-point products were fractionated on 2% agarose gels and sequenced to confirm amplicon identities.
Standard curves for all three transcripts were obtained by diluting cDNA, prepared from RNA extracted from cultured HGPS fibroblasts. The detectable range was over a 5-log dilution, and the efficiencies were calculated to be 102.9%, 97.8%, 102.0% and 91.4% for LMNA Δ150, LMNA, LMNC and RPLP0, respectively.
Western blots
Western blotting was performed based on the protocols of Cooper et al [12]. Muscle tissues (10 mg) from 12 normal individuals were cryosectioned and suspended in 200 μl treatment buffer containing 125 mM Tris-HCl (pH 6.8), 15% SDS (w/v), 10% glycerol (v/v), 0.5 mM phenylmethylsulfonyl fluoride and 9 μl protease inhibitor cocktail (Sigma Aldrich). After sonication, 6 pulses of 1 sec duration at a setting of 30/100 on the Sonics sonicator (Sonics, Newtown, CT), the homogenates were stored at -80°C. The samples were reduced by adding dithiothreitol to a final concentration of 50 mM and mixed with 0.4 μg/ml bromophenol blue before use. The protein extracts were incubated at 95°C for 5 min and centrifuged at 14,000 rpm for 5 min. Protein extracts (15 μl) were separated on NuPAGE 4-12% Bis-Tris gels (Life Technologies, Mulgrave, Australia) and electro-transferred to polyvinylidene fluoride membranes (Pall, Melbourne, Australia). After immersion in 5% skim milk blocking solution for 30 min, membranes were incubated with primary antibodies (anti-lamin A/C polyclonal, Santa Cruz, Dallas, TX, 1:100; anti-lamin B2, Santa Cruz, 1:100) overnight at 4°C. After subsequent washing, membranes were incubated with with HRP labeled goat anti rabbit immunoglobulins (Dako, 1:2,000) for 1 hr and incubated with chemiluminescent substrate for 5 min. Images were captured by a Chemi-Smart 3000 gel documentation system (Vilber Lourmat) using Chemi-capt software and image analysis was performed using Bio-1D software.
Confocal microscopy
Immunohistochemistry for progerin was performed on human muscle sections (n=6, 16 to 70 yr) and HGPS fibroblast cultures. Briefly, endogenous peroxidase was blocked in 1% (v/v) H2O2 in methanol, followed by incubation with a specific anti-progerin antibody (Abcam, Sapphire Bioscience, Waterloo, Australia, 1:50) overnight at 4°C. After washing by PBS, sections and cultures were incubated with Alexor Fluor 488 goat anti-mouse immunoglobulin (Invitrogen, 1:400) for 1 hr at room temperature, and then counterstained with Hoechst 33342 (Sigma Aldrich, 1:4,000) for 5 min. After rinsing with PBS, slides were viewed under a Nikon A1Si laser scanning confocal microscope (Coherent Scientific, Hilton, Australia).
Data analysis
Data were analyzed by the Sequence Detector System (SDS) software 1.3.1 (Applied Biosystems). A standard curve analysis method was used to calculate the fold difference between samples and the calibrator sample (normal 25 years old). Linear regression was used to test the correlation between age and the levels of LMNA, LMNC and LMNA Δ150 transcripts, and the LMNA/LMNA Δ150 and LMNA/LMNC ratios. Independent t test was used to test the difference in LMNA Δ150 and LMNA levels between males and females. P values <0.05 were considered statistically significant.
Results
RT-PCR of human muscle
An amplicon representing the LMNA transcript was ubiquitously detected in human samples by RT-PCR. Upon increasing amplification sensitivity to detect less abundant transcripts, an amplicon 150 bp smaller than the expected full length LMNA product was also detected in most of the human muscle samples (Figure 1). DNA sequencing confirmed this shorter amplicon to represent the LMNA Δ150 transcript.
Figure 1.
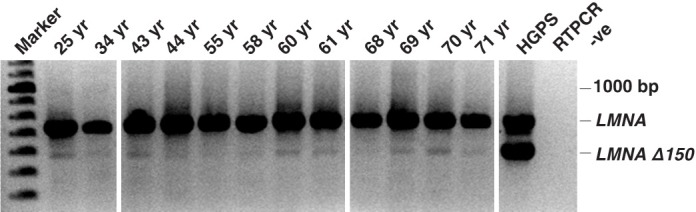
High sensitivity RT-PCR clearly indicated the presence of the LMNA Δ150 transcript in most normal human skeletal muscle.
qRT-PCR analysis of human muscle
Quantitative real time-PCR analyses employing validated assays specific for LMNA, LMNA Δ150, LMNC, and RPLP0 transcripts were undertaken [11]. There was a moderate correlation between age and levels of both LMNA (r2=0.624, p<0.0001) and LMNA Δ150 (r2=0.593, p=0.001) in the human muscle samples (Figure 2A, 2B). There was a subtle decrease in the ratio of LMNA/LMNA Δ150 transcripts, although the statistical significance of this trend is limited by the number of samples at each time point available for evaluation (r2=0.565, p=0.035), In contrast, no correlation was found between age and LMNC levels (Figure 2C) or LMNA/LMNC ratios (p=0.244, 0.088, r2=0.02, 0.241). The LMNA level was 900 to 2,400 times that of LMNA Δ150 (1,200 times on average). There was no significant difference in LMNA Δ150 or LMNA levels between male and female muscle samples (p=0.866, 0.221). Based upon the qPCR data, the level of progerin transcript in the human muscle samples was estimated to be 150-1400 times lower than that detected in HGPS fibroblasts.
Figure 2.
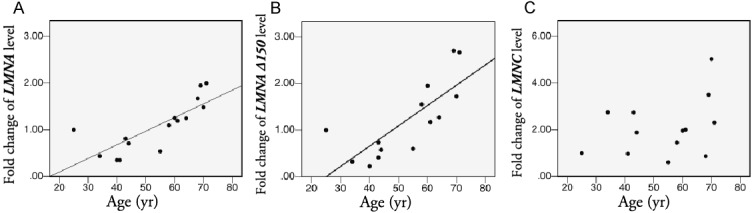
Relative quantification analysis by qRT-PCR. Samples with threshold cycle (Ct) numbers that fell outside the dynamic range of the assays were excluded from the analysis. As a result, 14, 16 and 13 normal samples were used for analysis of LMNA, LMNA Δ150, LMNC levels respectively. There is an age-dependent increase of wild-type LMNA (A) and LMNA Δ150 (B), but not LMNC (C), transcript levels in human skeletal muscle biopsies. The youngest sample in the qRT-PCR study (25 yr old) was used as the calibrator sample.
Detection of progerin protein
Western blotting of protein extracts from the HGPS cells readily demonstrated the presence of all three target proteins, lamin A, progerin and lamin C, when probed with the lamin A/C antibody (Figure 3). A faint progerin band was also detected in all of the human muscle samples. Although low levels of progerin could be detected in most samples, there was considerable variation between individuals. Densitometry analysis did not show any significant correlation between age and progerin or lamin A levels, or lamin A/progerin ratios as in the transcript studies (results not shown).
Figure 3.
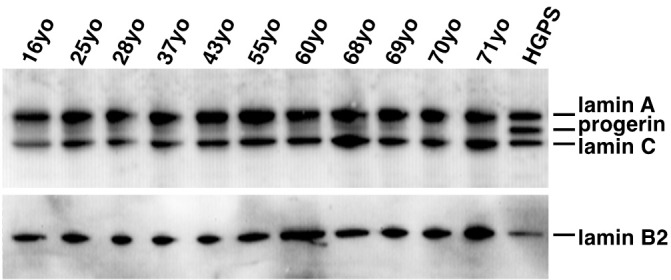
Lamin A and C proteins are readily demonstrated in extracts from all muscle samples. Trace amounts of progerin in normal human muscle are detected in all samples as well. HGPS fibroblast cultures were included as a positive control for progerin. Probed with Santa Cruz polyclonal anti-lamin A/C antibody.
Immunohistochemistry
Immunohistochemistry employing the progerin specific antibody [13] did not show any progerin positive staining myonuclei in human muscle sections (0/6,054 nuclei counted), whereas 25.4% of HGPS fibroblast nuclei (26/95) were positive for progerin on confocal microscopy (Figure 4). This indicates that progerin detected in normal skeletal muscle is diffusely expressed at very low levels, rather than being concentrated in a few myonuclei.
Figure 4.
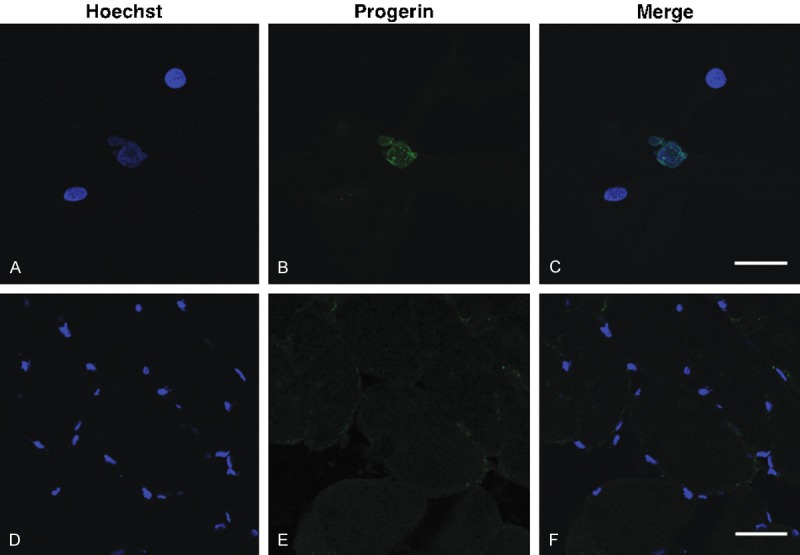
Immunohistochemistry probed with progerin specific antibody. A proportion of HGPS fibroblasts demonstrated immunoreactivity for progerin (A-C). Muscle sections from a 68 yr normal individual did not show any progerin-positive nuclei (D-F).
Mouse muscle studies
Although Lmna was consistently found in all mouse muscle samples, no Lmna Δ150 bands were detected in any of the murine samples, even after increasing the rounds of PCR to enhance sensitivity (Figure 5). Aged mouse heart and liver tissues were also assessed for progerin expression to ascertain if inappropriate Lmna splicing occurs in these murine tissues. As in murine muscle, no Lmna Δ150 amplicon was generated from RNA extracted from the heart or liver (data not shown). Since the Lmna Δ150 transcript could not be detected, immunoblotting for progerin was not performed.
Figure 5.
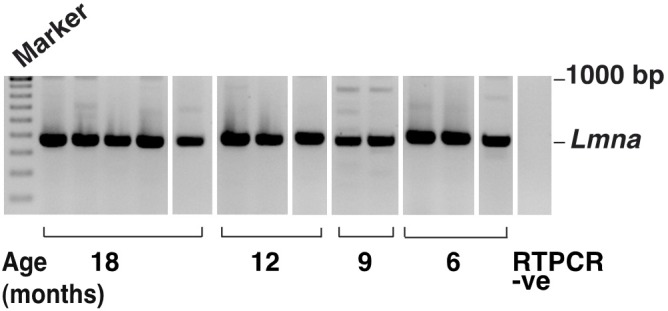
Lmna transcript (481 bases) was present in mouse muscle, but the Lmna Δ150 product was not detected using highly sensitive RT-PCR conditions. Several smaller bands were sporadically amplified and DNA sequencing indicated these to be RT-PCR artifacts.
Discussion
This study investigated the changes in expression of LMNA in skeletal muscle obtained from individuals aged between 16 to 71 years. There appears to be an age-related increase in expression of the full length lamin A transcript. The increase in the progerin transcript is in line with the elevated LMNA expression and corresponds with elevated transcription rather than an increase in splicing errors in human skeletal muscle. The age-dependent increase in progerin transcript in muscle samples does not translate into increased accumulation the progerin protein in older individuals. Although extremely low levels of progerin could be detected in the majority of samples, there was considerable variation in progerin levels between individuals when compared to lamin A/C. This work is consistent with previous studies in more rapidly dividing tissues such as skin and liver, in which no obvious correlation was found between age and the levels of LMNA Δ150 or progerin protein [10,14].
Progerin is generated through inappropriate alternative splicing of LMNA exon 11, and is 50 amino acids shorter than the normal lamin A. The deleted domain includes an endoproteolytic site for post-translational modification of prelamin A, the precursor of lamin A [15]. As a result, instead of the last 18 amino acids being cleaved as normally occurs, progerin retains a farnesyl and a methyl group in the carboxyl terminal and the truncated protein remains permanently inserted into the nuclear lamina [16]. During mitosis, phosphorylation of lamin A causes it to separate from the nuclear membrane and become soluble in the cytosol [17]. In contrast, progerin fails to dissociate from the nuclear membrane, interfering with normal nuclear splitting and resulting in dysmorphic nuclear shapes. In dividing cells, nuclei containing excessive progerin show dramatically reduced lamin A foci in the nucleoplasm and segregation of chromosomes during mitosis [18]. The Notch cell fate determination pathway and the p53 tumour suppression pathway are both activated in progerin-overexpressing cells as well as in a HGPS mouse model [10,19,20], suggesting that progerin can also interfere with cell differentiation. Although muscle weakness is not a prominent symptom of HGPS, impaired muscle function has been noted in affected individuals and in a mouse model of HGPS [21,22]. In accordance with these findings, transcriptome studies have shown altered gene expression, particularly in tissues of mesodermal origin [23], including genes involved in muscle development, differentiation and muscle contraction (MEOX2, EYA2, ACTA2).
Our finding that lamin A transcript levels increase with age in human skeletal muscles is in accord with the finding of Wegner et al., who reported that LMNA transcript levels were higher in older human muscles [24], and proposed that this may represent a compensatory mechanism for deteriorating nuclear function in muscle with ageing [25]. Whether the associated up-regulation of the alternatively spliced progerin isoform further exacerbates the effects of ageing is uncertain. Since the increment trends are very similar between LMNA and LMNA Δ150 transcripts, it does not appear likely that splicing errors increase with age in human skeletal muscle. The variable progerin levels in different individuals may reflect differences in their genetic background and splicing machinery. In addition to the 150-1400x difference in LMNA Δ150 levels between normal muscle and HGPS fibroblasts, a similar fold difference (110-1200) was also demonstrated when digital droplet PCR [26] was used to analyze progerin transcript levels (data not shown).
In contrast to human muscle, we did not detect progerin transcripts in mouse skeletal muscles up to 18 months of age, or in other murine tissues such as liver and heart. In a separate study, we were unable to induce expression of Lmna Δ150 in mouse myoblast cultures using splice switching antisense oligomers (unpublished data), whereas this splice switching was readily achieved in human muscle cultures (manuscript in preparation). We hypothesize that these results suggest that the processing of murine Lmna pre-mRNA is more robust and less prone to splicing errors than the human equivalent. An in silico evaluation of the splice strength for the wild-type donor splice site (lamin A) and the alternative splice site (progerin) showed that they are identical in the human and the mouse (http://www.umd.be/HSF/). Nevertheless, splicing requires more than the donor or acceptor splice sites and prediction of exonic splicing enhancers (ESEs) in human and mouse exon 11 shows several different potential binding sites for splicing factors, such as the lack of putative binding motifs for serine/arginine splicing factor 6 around the alternative splice site in the mouse Lmna exon 11 (data not shown, http://rulai.cshl.edu/cgibin/tools/ESE3/esefinder.cgi). Hence the differences in lamin A/C splicing found in human and mouse muscle may be due to the presence of different ESEs that strengthen the recognition of the normal mouse Lmna exon 11 donor site, and/or silencers that weaken selection of the alternative cryptic donor splice site.
Conclusions
In conclusion, our findings indicate that there is increased expression of LMNA in human skeletal muscle, presumably through increased transcription, and this leads to a corresponding increase in the LMNA Δ150 progerin transcript. Nevertheless, levels of the progerin transcript are extremely low and are not translated into a corresponding increase in the protein. Usage of the HGPS cryptic splice site in LMNA exon 11 with ageing reflects a background splicing error, which does not increase with age and only trace quantities of progerin protein accumulate. Further studies are required to determine the significance of these findings in the ageing process in skeletal muscle.
Acknowledgements
The authors thank Dr Gisèle Bonne for her helpful discussions and advice, and Ms Abbie Adams for technical support with the animal work. This work was supported by the Neuromuscular Foundation of Western Australia. Dr Yue-Bei Luo was supported by a China Scholarship Council-University of Western Australia joint PhD scholarship. Dr Chalermchai Mitrpant was partly supported by a Chalermphrakiat grant, Faculty of Medicine, Siriraj Hospital, Mahidol University.
Disclosure of conflict of interest
None.
References
- 1.Mounkes LC, Burke B, Stewart CL. The A-type lamins: nuclear structural proteins as a focus for muscular dystrophy and cardiovascular diseases. Trends Cardiovasc Med. 2001;11:280–285. doi: 10.1016/s1050-1738(01)00126-8. [DOI] [PubMed] [Google Scholar]
- 2.Boban M, Braun J, Foisner R. Lamins: ‘structure goes cycling’. Biochem Soc Trans. 2010;38:301–6. doi: 10.1042/BST0380301. [DOI] [PubMed] [Google Scholar]
- 3.Kennedy BK, Barbie DA, Classon M, Dyson N, Harlow E. Nuclear organization of DNA replication in primary mammalian cells. Genes Dev. 2000;14:2855–2868. doi: 10.1101/gad.842600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumaran RI, Muralikrishna B, Parnaik VK. Lamin A/C speckles mediate spatial organization of splicing factor compartments and RNA polymerase II transcription. J Cell Biol. 2002;159:783–793. doi: 10.1083/jcb.200204149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broers JL, Ramaekers FC, Bonne G, Yaou RB, Hutchison CJ. Nuclear lamins: laminopathies and their role in premature ageing. Physiol Rev. 2006;86:967–1008. doi: 10.1152/physrev.00047.2005. [DOI] [PubMed] [Google Scholar]
- 6.Quijano-Roy S, Mbieleu B, Bonnemann CG, Jeannet PY, Colomer J, Clarke NF, Cuisset JM, Roper H, De Meirleir L, D’Amico A, Ben Yaou R, Nascimento A, Barois A, Demay L, Bertini E, Ferreiro A, Sewry CA, Romero NB, Ryan M, Muntoni F, Guicheney P, Richard P, Bonne G, Estournet B. De novo LMNA mutations cause a new form of congenital muscular dystrophy. Ann Neurol. 2008;64:177–186. doi: 10.1002/ana.21417. [DOI] [PubMed] [Google Scholar]
- 7.Reddel CJ. Lamin A expression levels are unperturbed at the normal and mutant alleles but display partial splice site selection in Hutchinson-Gilford progeria syndrome. J Med Genet. 2004;41:715–717. doi: 10.1136/jmg.2004.019323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eriksson M, Brown WT, Gordon LB, Glynn MW, Singer J, Scott L, Erdos MR, Robbins CM, Moses TY, Berglund P, Dutra A, Pak E, Durkin S, Csoka AB, Boehnke M, Glover TW, Collins FS. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scaffidi P, Misteli T. Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nat Med. 2005;11:440–445. doi: 10.1038/nm1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scaffidi P, Misteli T. Lamin A-dependent nuclear defects in human aging. Science. 2006;312:1059–1063. doi: 10.1126/science.1127168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez S, Coppedè F, Sagelius H, Eriksson M. Increased expression of the Hutchinson-Gilford progeria syndrome truncated lamin A transcript during cell aging. Eur J Hum Genet. 2009;17:928–937. doi: 10.1038/ejhg.2008.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper ST, Lo HP, North KN. Single section Western blot: improving the molecular diagnosis of the muscular dystrophies. Neurology. 2003;61:93–97. doi: 10.1212/01.wnl.0000069460.53438.38. [DOI] [PubMed] [Google Scholar]
- 13.Liu GH, Barkho BZ, Ruiz S, Diep D, Qu J, Yang SL, Panopoulos AD, Suzuki K, Kurian L, Walsh C, Thompson J, Boue S, Fung HL, Sancho-Martinez I, Zhang K, Yates J 3rd, Izpisua Belmonte JC. Recapitulation of premature ageing with iPSCs from Hutchinson-Gilford progeria syndrome. Nature. 2011;472:221–225. doi: 10.1038/nature09879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McClintock D, Ratner D, Lokuge M, Owens DM, Gordon L, Collins FS, Djabali K. The mutant form of lamin A that causes Hutchinson-Gilford progeria is a biomarker of cellular aging in human skin. PLoS One. 2007;2:e1269. doi: 10.1371/journal.pone.0001269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies BS, Fong LG, Yang SH, Coffinier C, Young SG. The posttranslational processing of prelamin A and disease. Annu Rev Genomics Hum Genet. 2009;10:153–174. doi: 10.1146/annurev-genom-082908-150150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang SH. Blocking protein farnesyltransferase improves nuclear blebbing in mouse fibroblasts with a targeted Hutchinson-Gilford progeria syndrome mutation. Proc Natl Acad Sci U S A. 2005;102:10291–10296. doi: 10.1073/pnas.0504641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerace L, Blobel G. The nuclear envelope lamina is reversibly depolymerized during mitosis. Cell. 1980;19:277–287. doi: 10.1016/0092-8674(80)90409-2. [DOI] [PubMed] [Google Scholar]
- 18.Cao K, Capell BC, Erdos MR, Djabali K, Collins FS. A lamin A protein isoform overexpressed in Hutchinson-Gilford progeria syndrome interferes with mitosis in progeria and normal cells. Proc Natl Acad Sci U S A. 2007;104:4949–4954. doi: 10.1073/pnas.0611640104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scaffidi P, Misteli T. Lamin A-dependent misregulation of adult stem cells associated with accelerated ageing. Nat Cell Biol. 2008;10:452–459. doi: 10.1038/ncb1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varela I, Cadiñanos J, Pendás AM, Gutiérrez-Fernández A, Folgueras AR, Sánchez LM, Zhou Z, Rodríguez FJ, Stewart CL, Vega JA, Tryggvason K, Freije JM, López-Otín C. Accelerated ageing in mice deficient in Zmpste24 protease is linked to p53 signalling activation. Nature. 2005;437:564–568. doi: 10.1038/nature04019. [DOI] [PubMed] [Google Scholar]
- 21.Merideth MA, Gordon LB, Clauss S, Sachdev V, Smith AC, Perry MB, Brewer CC, Zalewski C, Kim HJ, Solomon B, Brooks BP, Gerber LH, Turner ML, Domingo DL, Hart TC, Graf J, Reynolds JC, Gropman A, Yanovski JA, Gerhard-Herman M, Collins FS, Nabel EG, Cannon RO 3rd, Gahl WA, Introne WJ. Phenotype and course of Hutchinson-Gilford progeria syndrome. N Engl J Med. 2008;358:592–604. doi: 10.1056/NEJMoa0706898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greising SM, Call JA, Lund TC, Blazar BR, Tolar J, Lowe DA. Skeletal muscle contractile function and neuromuscular performance in Zmpste24 (-/-) mice, a murine model of human progeria. Age (Dordr) 2012;34:805–19. doi: 10.1007/s11357-011-9281-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Csoka AB, English SB, Simkevich CP, Ginzinger DG, Butte AJ, Schatten GP, Rothman FG, Sedivy JM. Genome-scale expression profiling of Hutchinson-Gilford progeria syndrome reveals widespread transcriptional misregulation leading to mesodermal/mesenchymal defects and accelerated atherosclerosis. Aging Cell. 2004;3:235–243. doi: 10.1111/j.1474-9728.2004.00105.x. [DOI] [PubMed] [Google Scholar]
- 24.Wegner L, Anthonsen S, Bork-Jensen J, Dalgaard L, Hansen T, Pedersen O, Poulsen P, Vaag A. LMNA rs4641 and the muscle lamin A and C isoforms in twins--metabolic implications and transcriptional regulation. J Clin Endocrinol Metab. 2010;95:3884–3892. doi: 10.1210/jc.2009-2675. [DOI] [PubMed] [Google Scholar]
- 25.Haithcock E, Dayani Y, Neufeld E, Zahand AJ, Feinstein N, Mattout A, Gruenbaum Y, Liu J. Age-related changes of nuclear architecture in Caenorhabditis elegans . Proc Natl Acad Sci U S A. 2005;102:16690–16695. doi: 10.1073/pnas.0506955102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Porensky PN, Mitrpant C, McGovern VL, Bevan AK, Foust KD, Kaspar BK, Wilton SD, Burghes AH. A single administration of morpholino antisense oligomer rescues spinal muscular atrophy in mouse. Hum Mol Genet. 2012;21:1625–1638. doi: 10.1093/hmg/ddr600. [DOI] [PMC free article] [PubMed] [Google Scholar]


