Abstract
Despite the strong progress has been made in the field of melanoma epigenetics, the importance of genome-wide demethylation or hypomethylation remains underestimated. However, this phenomenon might also reflect important epigenetic alterations due to its ability to cause genetic instability. Furthermore, no methylation-based distinction has been drawn among the diverse primary melanoma subtypes. To assess global methylation we measured the methylation level on the 6 CpG sites of LINE1 sequences in 46 primary melanomas in association with patients’ survivals and the clinicopathological characteristics of specimens. We demonstrate that LINE1 hypomethylation is accompanied by the shortened relapse-free survival of melanoma patients; however, Cox regression analysis shows a direct relationship between the overall loss of 5-methylcytosine and metastatic potential of primary melanomas, which is confirmed by Kruskal-Wallis tests with Dunn’s Multiple Comparison Post-test showing that not only the presence but the number of metastases during the 5-year follow-up period is associated with the transposon demethylation. In this study, we demonstrate the strong influence of global DNA demethylation in the metastatic formation of primary melanomas during the follow-up period.
Keywords: Melanoma progression, epigenetics, DNA methylation, global hypomethylation, LINE1
Introduction
Global DNA hypomethylation is an important component of epigenetic modifications causing genetic instability [1-3]. The global hypomethylation is related to the overall loss of 5-methylcytosine which is believed to correspond to the methylcytosine of repetitive transposable elements (LINE and SINE sequences) having integrated into the human genome during evolution and gained protection from transcription due to their higher levels of methylcytosine [4,5]. As these elements are reactivated by hypomethylation, they can recombine with each other, causing karyotypic instability [3]. The LINE1 sequences are 5000 to 6000 bp long, and 104 copies of these elements are spread throughout the genome [2,6]. Considering that LINE1 elements along with Alu (SINE) sequences constitute 40% of the human genome, it is clear why the literature generalises the transposable demethylation as global hypomethylation [2].
In melanoma, the most aggressive form of skin neoplasms, the presence of genome-wide hypomethylation has been reported by several groups [7-9]. The degree of global demethylation has been proven sufficient in distinguishing a benign naevus from primary and metastatic melanomas [7]; however, no demethylation-based distinction has been drawn among the diverse primary melanoma subtypes with different clinical behaviours. Nonetheless, the relationship between transposable hypomethylation and patients’ survival is still a topic of doubt and discussion, as a single group has reported the prolonged survival of stage III melanoma patients with hypomethylation [5]. However, this finding stands in sharp contrast to data from other types of cancers, which exhibit global demethylation [10,11].
Our aim was to investigate the extent of LINE1 methylation level in primary malignant melanoma samples and to determine that how transposable methylation is associated with melanoma progression.
Materials and methods
Melanoma samples; genomic DNA extraction
Forty-six primary melanomas were used for LINE1 methylation analysis; the clinicopathological data of the melanoma patients are summarised in Table 1.
Table 1.
Clinical-pathological parameters of primary melanomas
| Variables | No. of tumors analyzed by LINE1 pyrosequencing |
|---|---|
| All patients | 46 |
| Histological subtype | |
| SSM1 | 30 |
| NM2 | 16 |
| Gender | |
| Female | 24 |
| Male | 22 |
| Age (years) | |
| 20-50 | 14 |
| ≥50 | 32 |
| Breslow thickness (mm)3 | |
| ≤2 | 18 |
| 2-4 | 12 |
| >4 | 16 |
| Clark’s stage | |
| I-III (early) | 27 |
| IV-V (late) | 19 |
| Location of primary tumor | |
| Extremity | 25 |
| Trunk | 20 |
| Head | 1 |
| Metastasis formation4 | |
| Absent | 24 |
| Present | 22 |
| Patient’s survival5 | |
| Alive | 24 |
| Exitus | 22 |
| Ulceration | |
| Absent | 24 |
| Present | 22 |
superficial spreading melanoma;
nodular melanoma;
thickness categories are based on the current staging system;
metastasis of the examined primary tumours;
patients with at least 5-year follow-up period were included.
The tumour tissues were obtained from the Department of Dermatology, University of Debrecen, Hungary. All human studies were conducted in accordance with the principles outlined in the Declaration of Helsinki, were approved by the Regional and Institutional Ethics Committee of the University of Debrecen Medical and Health Science Centre and were conducted according to regulations (Protocol #2836-2008). Written informed consent was obtained from each patient.
After surgical excision, the G-spin™ Genomic DNA Extraction Kit (Intron, Korea) was used to isolate high-molecular-weight DNA from 46 primary melanomas according to the manufacturer’s protocol. To determine the quantity of DNA obtained, we used a NanoDrop ND-1000 UV-Vis Spectrophotometer. The DNA integrity was verified via 1.2% agarose gel electrophoresis.
Bisulphite pyrosequencing of LINE1
Global hypomethylation was measured via 6 CpG sites located on the transposable LINE1 sequence. The genomic DNA specimens (n=46) were treated by applying sodium bisulphite using the EZ DNA Methylation-Gold Kit (Zymo Research, Orange, CA, USA) to 500 ng of each DNA sample following the manufacturer’s recommendations. LINE1 pyrosequencing analysis was performed on the PSQ96 (Biotage) using the following primers: sense, 5’-BIOTIN-TAGGGAGTGTTAGATAGTGG, antisense, 5’-AACTCCCTAACCCCTTAC, and sequencing primer, 5’-AACTCCCTAACCCCTTAC. Hot-start PCR was performed using the HotStarTaq Master Mix Kit (Qiagen), and the pyrosequencing was carried out in accordance with the manufacturer’s protocol (Biotage). The target CpGs were evaluated by converting the resulting pyrograms to numerical values for peak heights. The following sequence containing six unique CpG sites was analysed: 5’-RCCCTACTTCRACTCRCRCACRATACR.
Statistical analyses
To estimate the effect of the LINE1 methylation status on patients’ survival, the LINE1 methylation level represented by continuous variables was dichotomised at its cut-off value determined for each CpG site by the ROC curve analysis in which the true positive rate (Sensitivity) is plotted in function of the false positive rate (100-Specificity) for different cut-off points of a parameter. Each point on the ROC curve represents a sensitivity/specificity pair corresponding to a particular decision threshold. The area under the ROC curve (AUC) is a measure of how well a parameter can distinguish between the hypomethylated and non-hypomethylated groups. Kaplan-Meier survival probability curve was drawn for the groups using the log-rank test to examine the difference in survival by LINE1 methylation level. The clinical parameters including patient age and gender, Breslow thickness, Clark level, histological subtype, metastasis formation during the 5-year follow up period and tumour surface ulceration were examined by the Cox proportional hazards regression model, and the effect of hypomethylation on the survival was presented as hazard ratios (HR) with corresponding 95% confidence intervals (CI). To control for the above-noted additional confounders, a stepwise regression was applied to select variables.
As the initial D’Agostino and Pearson omnibus normality test revealed that LINE1 methylation values do not follow the Gaussian distribution, we used the nonparametric Mann Whitney and Kruskal-Wallis tests with Dunn’s Multiple Comparison Post-test to verify the direct relationship between the LINE1 methylation status and clinical parameters.
Results
Relationship of patients’ survival and LINE1 hypomethylation
Our experimental design included 46 primary malignant melanoma samples (Table 1). Six CpG sites on the LINE1 transposable sequence were measured for their global methylation pattern. Not having demonstrated a Gaussian distribution, the continuous variables of the LINE1 pyrosequencing results were dichotomised, and the simplicity groups were constructed using the threshold values determined by the ROC curve analysis. The threshold levels were the following: CpG_1 (55.5), CpG_2 (68.1), CpG_3 (46.8), CpG_4 (56.8), CpG_5 (83.6) and CpG_6 (61.6).
The sample groups were defined as LINE1 hypomethylated (specimens with a LINE1 methylation < threshold value) and non-hypomethylated (samples with a LINE1 methylation ≥ threshold value).
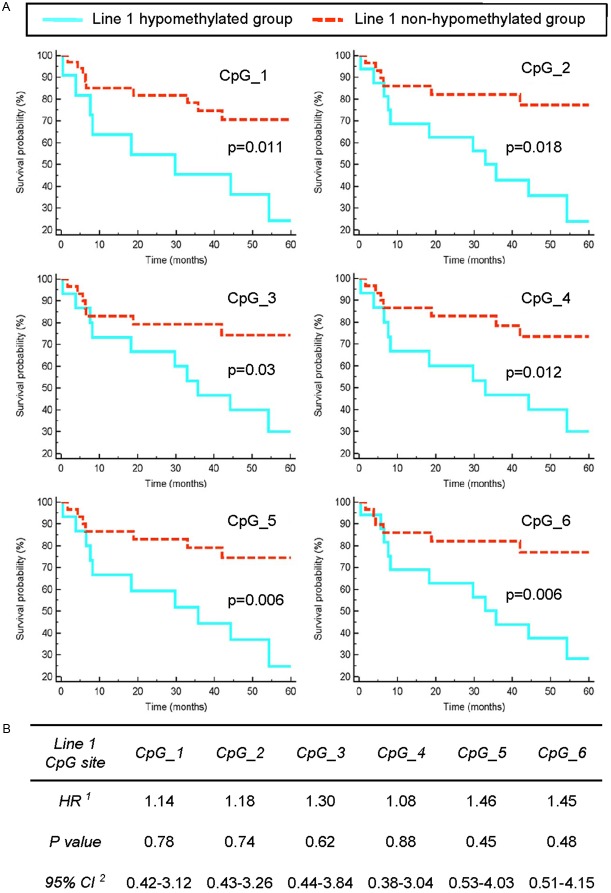
As shown in Figure 1A, the Kaplan-Meier curve analysis revealed significant differences between the hypomethylated and non-hypomethylated patients, with a decreased overall survival (OS) rate for the hypomethylated group for each CpG site (CpG_1: p=0.011; CpG_2: p=0.018; CpG_3: p=0.03; CpG_4: p=0.012; CpG_5: p=0.006; and CpG_6: p=0.006). Notably, the hazard ratios (HR) were higher than 3.0 in each hypomethylated group for all six CpG sites. The Cox proportional hazard regression model allowed for the control of additional clinical covariates as well as for the patient gender and age. As the stepwise regression method indicated metastatic potential (when primary melanomas develop metastasis during the five-year follow up period) to influence patient survival, this potential was included in the model, and the adjusted HR values were calculated. As demonstrated in Figure 1B, the adjusted HR values did not remain higher than 1.46 nor did the adjusted p-values remain significant.
Figure 1.
Relationship between survival and LINE1 hypomethylation in primary melanomas. A: Kaplan-Meier analysis of primary melanoma (n=46) patients survival according to LINE1 methylation levels which were measured by pyrosequencing at 6 CpG sites. Kaplan- Meyer function for Overall Survival rate (OS) was calculated for CM patients according to cut-off values (determined by ROC curves) of methylation of CpG_1, CpG_2, CpG_3, CpG_4, CpG_5 and CpG_6 sites of LINE1, respectively. Dashed red line refers for patients with LINE1 methylation above, whereas solid green line depicts LINE1 methylation below the cut-off value, respectively. Cumulative survival by LINE1 methylation level was evaluated using the Log-Rank test and reported P values are two sided. B: Clinical cofounders including age and gender of patients, Breslow thickness, Clark level, histological subtype, metastasis formation during the 5-year follow up period and tumour surface ulceration was examined by Cox proportional hazards regression model whereas the effect of hypomethylation on survival was presented as Hazard Ratios (HR) with corresponding 95% Confidence Intervals (CI).
LINE1 hypomethylation is related to metastatic capacity of melanomas
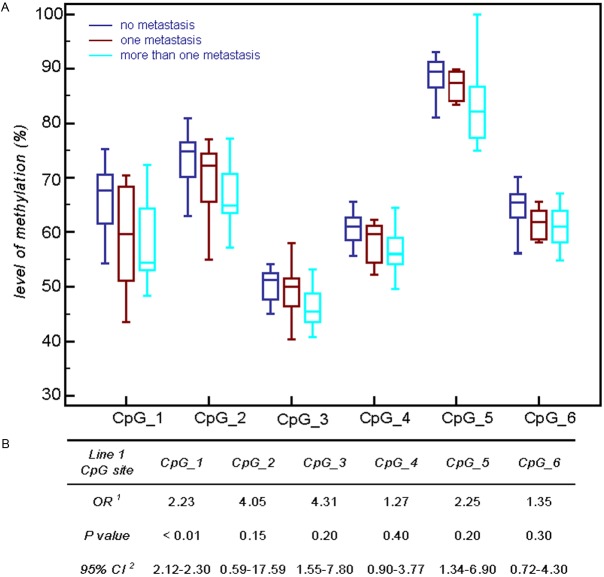
As the stepwise method of Cox proportional hazard regression model suggested the direct relationship of the overall loss of 5-methylcytosine with metastatic potential, we built a logistic regression model with a stepwise selection for LINE1 hypomethylation and for the clinical parameters. Figure 2 demonstrates the association of metastatic capacity and global demethylation in primary melanomas. For each CpG site, we found high odds ratios (OR) for the hypomethylated sample groups; however, the OR remained significant for CpG1 (Figure 2B). The Mann-Whitney test confirmed that global hypomethylation of all six CpGs was significantly associated with metastasis formation. To increase the statistical power of the univariate test, primary melanomas with metastasis were divided into subgroups according to the number of metastases developed during the follow-up period, and the Kruskal-Wallis test with Dunn’s Multiple Comparison Post-test was applied to measure the strength of the relationship. Figure 2A depicts significantly different levels of global methylation for most CpGs, except for CpG4, among tumours without metastasis and melanomas with more than one metastasis over 5 years.
Figure 2.
Association between the number of metastasis and LINE1 hypomethylation. A: Primary melanomas that developed metastasis were divided into subgroups according to the number of metastasis formed during the follow-up period and Kruskall Wallis test with Dunn’s Multiple Comparison Post-test was applied to study association of global hypomethylation and metastatic capacity which reached significant level in the majority of CpGs. B: Logistic regression model for LINE1 hypomethylation and the clinical cofounders summarize the association of metastatic capacity and global demethylation in primary melanomas. For each CpGs high Odds Ratios (OR) were evaluated in the hypomethylated sample groups with corresponding 95% Confidence Intervals (CI). Association reached significance for CpG_1.
Discussion
The so-called ‘genome-wide hypomethylation’ is believed to correspond to the loss of methylcytosine, which belong to repetitive transposable elements and can cause karyotypic instability [3,12], as the genome contains approximately 5000 full-length LINE1 elements, 60-100 of which are still capable of retrotransposition [4,13]. It was indicated that LINE1 hypomethylation can offer a new facility that emphasises early detection in the case of applying a technical approach that may make rapid, systematic screening possible [14].
To date, genome-wide hypomethylation has been demonstrated in 16 melanoma cell lines compared to melanocytes via repetitive elements [7]. Regrettably, we were unable to determine the actual clinical effects of genome-wide methylation, as the above-mentioned study focused only on comparing cell lines to controls instead of drawing a demethylation-based distinction between cell lines with different characteristics.
Notably, data from other types of neoplasms, such as ovarian and colon cancer [6], have established a relationship between shortened survival and global demethylation [15]. Additionally, Hoshimoto et al. described the shortened relapse-free survival of demethylated melanomas [9]. Sigalotti et al. conducted a valuable study aimed at estimating the LINE1 methylation status of 42 short-term cultures from surgically removed IIIC melanomas. Surprisingly, among the three LINE1 CpG sites, the hypomethylation of two CpG sites was associated with prolonged patient survival [5].
Our study comprised 46 primary melanomas measured at 6 LINE1 CpG sites using quantitative pyrosequencing. In contrast to Sigalotti’s research group, we demonstrated a shortened relapse-free survival in LINE1 hypomethylation patients in all six CpG sites using the log-rank test. More importantly, using the same statistical approach as Sigalotti’s group, the metastatic capacity was significantly associated with global hypomethylation for any of the 6 CpGs. When the primary melanomas with metastasis were divided into subgroups based on the number of metastases formed during the 5-year follow-up period, significantly different levels of global methylation were observed between the groups for the majority of CpGs. Our results contrast with the findings of Sigalotti’s group [5]. However, their study comprised cell cultures derived from homogeneous staged specimens, whereas our experiments were performed using primary melanomas characteristic of distinct clinical behaviour.
In conclusion, we demonstrated that LINE1 hypomethylation status strongly predicts the metastatic capacity of primary melanomas suggesting the role of transposons’ hypomethylation in the progression of primary melanomas.
Acknowledgements
This research was supported by the Hungarian National Research Fund (OTKA K75191), the Hungarian Academy of Sciences (grant number 2011 TKI 473), the UD Faculty of Medicine Research Fund (Bridging Fund 2012), the TÁMOP-4.2.2/B-10/1-2010-0024 and TÁMOP-4.2.2.A-11/1/KONV-2012-0031 projects; the TÁMOP projects are co-financed by the European Union and the European Social Fund. The authors have no connection with any of the companies or products mentioned in this article.
Disclosure of conflict of interest
None.
References
- 1.van den Hurk K, Niessen HE, Veeck J, van den Oord JJ, van Steensel MA, Zur Hausen A, van Engeland M, Winnepenninckx VJ. Genetics and epigenetics of cutaneous malignant melanoma: a concert out of tune. Biochim Biophys Acta. 2012;1826:89–102. doi: 10.1016/j.bbcan.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 2.Heyn H, Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nat Rev Genet. 2012;13:679–692. doi: 10.1038/nrg3270. [DOI] [PubMed] [Google Scholar]
- 3.Balazs M, Ecsedi S, Vizkeleti L, Begany A. Genomics of human malignant melanoma. In: Tanaka Y, editor. Breakthroughs in melanoma research. Rijeka: InTech; 2011. pp. 237–263. [Google Scholar]
- 4.Wild L, Flanagan JM. Genome-wide hypomethylation in cancer may be a passive consequence of transformation. Biochim Biophys Acta. 2010;1806:50–57. doi: 10.1016/j.bbcan.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Sigalotti L, Fratta E, Bidoli E, Covre A, Parisi G, Colizzi F, Coral S, Massarut S, Kirkwood JM, Maio M. Methylation levels of the “long interspersed nucleotide element-1” repetitive sequences predict survival of melanoma patients. J Transl Med. 2011;9:78. doi: 10.1186/1479-5876-9-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sunami E, de Maat M, Vu A, Turner RR, Hoon DS. LINE-1 hypomethylation during primary colon cancer progression. PLoS One. 2011;6:e18884. doi: 10.1371/journal.pone.0018884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tellez CS, Shen L, Estecio MR, Jelinek J, Gershenwald JE, Issa JP. CpG island methylation profiling in human melanoma cell lines. Melanoma Res. 2009;19:146–155. doi: 10.1097/cmr.0b013e32832b274e. [DOI] [PubMed] [Google Scholar]
- 8.Haqq C, Nosrati M, Sudilovsky D, Crothers J, Khodabakhsh D, Pulliam BL, Federman S, Miller JR 3rd, Allen RE, Singer MI, Leong SP, Ljung BM, Sagebiel RW, Kashani-Sabet M. The gene expression signatures of melanoma progression. Proc Natl Acad Sci U S A. 2005;102:6092–6097. doi: 10.1073/pnas.0501564102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoshimoto S, Kuo CT, Chong KK, Takeshima TL, Takei Y, Li MW, Huang SK, Sim MS, Morton DL, Hoon DS. AIM1 and LINE-1 epigenetic aberrations in tumor and serum relate to melanoma progression and disease outcome. J Invest Dermatol. 2012;132:1689–1697. doi: 10.1038/jid.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daskalos A, Nikolaidis G, Xinarianos G, Savvari P, Cassidy A, Zakopoulou R, Kotsinas A, Gorgoulis V, Field JK, Liloglou T. Hypomethylation of retrotransposable elements correlates with genomic instability in non-small cell lung cancer. Int J Cancer. 2009;124:81–87. doi: 10.1002/ijc.23849. [DOI] [PubMed] [Google Scholar]
- 11.Lee HS, Kim BH, Cho NY, Yoo EJ, Choi M, Shin SH, Jang JJ, Suh KS, Kim YS, Kang GH. Prognostic implications of and relationship between CpG island hypermethylation and repetitive DNA hypomethylation in hepatocellular carcinoma. Clin Cancer Res. 2009;15:812–820. doi: 10.1158/1078-0432.CCR-08-0266. [DOI] [PubMed] [Google Scholar]
- 12.de Maat MF, van de Velde CJ, Benard A, Putter H, Morreau H, van Krieken JH, Meershoek Klein-Kranenbarg E, de Graaf EJ, Tollenaar RA, Hoon DS. Identification of a quantitative MINT locus methylation profile predicting local regional recurrence of rectal cancer. Clin Cancer Res. 2010;16:2811–2818. doi: 10.1158/1078-0432.CCR-09-2717. [DOI] [PubMed] [Google Scholar]
- 13.Sunami E, Vu AT, Nguyen SL, Hoon DS. Analysis of methylated circulating DNA in cancer patients’ blood. Methods Mol Biol. 2009;507:349–356. doi: 10.1007/978-1-59745-522-0_25. [DOI] [PubMed] [Google Scholar]
- 14.Szekvolgyi L, Imre L, Minh DX, Hegedus E, Bacso Z, Szabo G. Flow cytometric and laser scanning microscopic approaches in epigenetics research. Methods Mol Biol. 2009;567:99–111. doi: 10.1007/978-1-60327-414-2_7. [DOI] [PubMed] [Google Scholar]
- 15.Pattamadilok J, Huapai N, Rattanatanyong P, Vasurattana A, Triratanachat S, Tresukosol D, Mutirangura A. LINE-1 hypomethylation level as a potential prognostic factor for epithelial ovarian cancer. Int J Gynecol Cancer. 2008;18:711–717. doi: 10.1111/j.1525-1438.2007.01117.x. [DOI] [PubMed] [Google Scholar]