Abstract
Genomic instability is a hallmark of cancer that leads to an increase in genetic alterations, thus enabling the acquisition of additional capabilities required for tumorigenesis and progression. Substantial heterogeneity in the amount and type of instability (nucleotide, microsatellite, or chromosomal) exists both within and between cancer types, with epithelial tumors typically displaying a greater degree of instability than hematological cancers. While high-throughput sequencing studies offer a comprehensive record of the genetic alterations within a tumor, detecting the rate of instability or cell-to-cell viability using this and most other available methods remains a challenge. Here, we discuss the different levels of genomic instability occurring in human cancers and touch on the current methods and limitations of detecting instability. We have applied one such approach to the surveying of public tumor data to provide a cursory view of genome instability across numerous tumor types.
Keywords: Genomic instability, Cancer, CIN, MSI, Nucleotide instability
Introduction
Cancer is a disease characterized and fuelled by dynamic genomic changes. The vast number of structural abnormalities present in cancer genomes is largely attributed to genomic instability, a transient or persistent state that increases the spontaneous mutation rate, leading to gross genetic alterations such as rearrangements and changes in chromosome number (aneuploidy). Genomic instability is therefore a driving force of tumorigenesis in that continuous modification of tumor cell genomes promotes the acquisition of further DNA alterations, clonal evolution, and tumor heterogeneity [1]. It is a feature of almost all cancers and has been observed in a range of malignant stages, from pre-neoplastic lesions prior to acquired TP53 mutations to advanced cases [2–4]. Numerous theories regarding the source of genome instability have been proposed. These theories, which include the mutator phenotype, DNA damage-induced replication stress, telomere dysfunction, and mitotic checkpoint failure [5–11], vary principally in their supposition of how early in tumorigenesis instability occurs, mechanisms leading to sequence level alteration, and whether instability initiates tumorigenesis or is merely a consequence of malignant transformation. While these mechanisms may all contribute to instability phenotypes to some extent in cancer in general, their prevalence varies across tumors derived from distinct cell types or in response to different carcinogens or selective pressures.
Genomic instability refers to a variety of DNA alterations, encompassing single nucleotide to whole chromosome changes, and is typically subdivided into three categories based on the level of genetic disruption. Nucleotide instability (NIN) is characterized by an increased frequency of base substitutions, deletions, and insertions of one or a few nucleotides; microsatellite instability (MIN or MSI) is the result of defects in mismatch repair genes which leads to the expansion and contraction of short nucleotide repeats called microsatellites; chromosomal instability (CIN) is the most prevalent form of genomic instability and leads to changes in both chromosome number and structure [12]. While instability is a characteristic of almost all human cancers, cancer genomes vary considerably in both the amount and type of genomic instability they harbor. Importantly, the instability phenotype has implications in patient prognosis as well as patient management, specifically with the choice of therapeutic agents [13–15].
Currently, detection of genome instability can be achieved using a variety of technologies, ranging from single-cell approaches to high-throughput multicellular techniques, each capable of detecting different levels of genomic changes. However, at present, no assay is capable of reliably measuring the rate (cell-to-cell variability) of small chromosomal changes such as deletions, amplification, and inversions within a population of cells. There is therefore a great need for sensitive, high-resolution techniques capable of detecting genomic instability over time as this would afford critical insights into the mechanisms that underlie genomic instability and the role of instability in tumorigenesis. In this review, we discuss the different levels of genomic instability and various methods of and limitations to detecting instability and describe global trends in genome instability across numerous tumor types.
Levels of genomic instability
Nucleotide instability
NIN typically develops due to replication errors and impairment of the base excision repair and nucleotide excision repair pathways, leading to subtle sequence changes involving only one or a few nucleotides (substitutions, deletions, insertions, etc.) which can affect gene structure and/or expression (Fig. 1a). While less common than the other forms of genomic instability, when present, single nucleotide alterations can cause dramatic phenotypes. For example, inherited defects in these repair pathways (germline mutations in XPC, ERCC2, DDB2, and MYH) lead to disorders such as xeroderma pigmentosum and MYH-associated polyposis, which result in genomic instability and the accumulation of DNA mutations, consequently predisposing these individuals to skin and colon cancers, respectively [16, 17]. Similar to the nuclear genome, the mitochondrial genome also displays NIN, and coupling of the high rate of reactive oxygen species generation with inefficient DNA repair can result in a rate of mtDNA mutations that is substantially higher than that of nuclear DNA [18, 19].
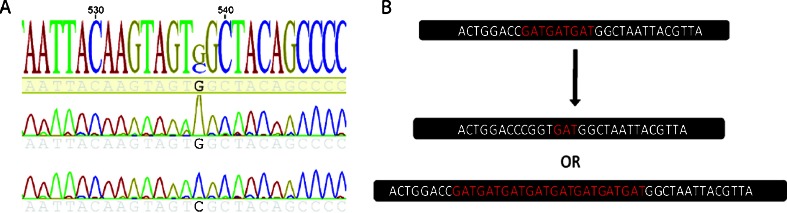
Fig. 1.
Nucleotide and microsatellite instability. a Detection of a G>C variant encoding a Gly>Arg amino acid change by Sanger sequencing in two lung cancer cell lines. b Defects in MMR lead to the expansion or contraction of microsatellites throughout the genome
Microsatellite instability
Microsatellites are repetitive DNA sequences comprising 1–6 bp located throughout the genome [20–22]. Within the population, microsatellite size is highly variable; however, each individual possesses unique microsatellites of a set length. MSI results from defects in DNA mismatch repair (MMR), specifically alterations of the MLH1, MSH2, MSH6, and PMS2 genes, which causes deletions or random insertion and expansion of microsatellites and a hypermutable phenotype (Fig. 1b). MSI is a characteristic feature of a number of cancers, including gastric, endometrial, ovarian, lung, and colorectal cancer (CRC), where it was first described and has been studied most extensively [23–28]. MSI occurs in approximately 15 % of CRC, which typically arise in the proximal colon, posses a normal karyotype, and are associated with a better prognosis than non-MSI tumors.
MSI occurs in both hereditary (Lynch syndrome) and sporadic forms of colon cancer, although via distinct mechanisms [13]. Hereditary non-polyposis colorectal cancer (Lynch syndrome) is characterized by inactivating germline mutations to MSH2, MSH6, PMS2, or MLH1, whereas sporadic CRC with MSI is associated with hypermethylation and loss of expression of MLH1 [27–32]. The majority of sporadic CRC with MSI arise in a background of extensive aberrant promoter methylation—referred to as the CpG island methylator phenotype (CIMP) [33–35]. CIMP tumors develop and progress by methylating the promoters of tumor suppressor genes such as p16, IGF-2, and MLH1 and possess clinical features distinct from non-CIMP tumors [33, 36–38].
Chromosomal instability
CIN is an increase in the rate of gain or loss of segmental and whole chromosomes during cell division and is the most prominent form of genomic instability in solid tumors, with roughly 90 % of human cancers exhibiting chromosomal abnormalities and aneuploidy [3, 39]. CIN tumors are characterized by global aneuploidy, amplifications, deletions, loss of heterozygosity (LOH), homozygous deletions, translocations, and inversions (Fig. 2). These alterations lead to karyotypic instability and the simultaneous growth of diverse tumor subpopulations, resulting in genomic inter- and intra-tumor heterogeneity [39]. CIN develops early in tumorigenesis (detectable in premalignant lesions) and is associated with intrinsic multidrug resistance [40] and poor prognosis [15, 41], making its detection clinically relevant.
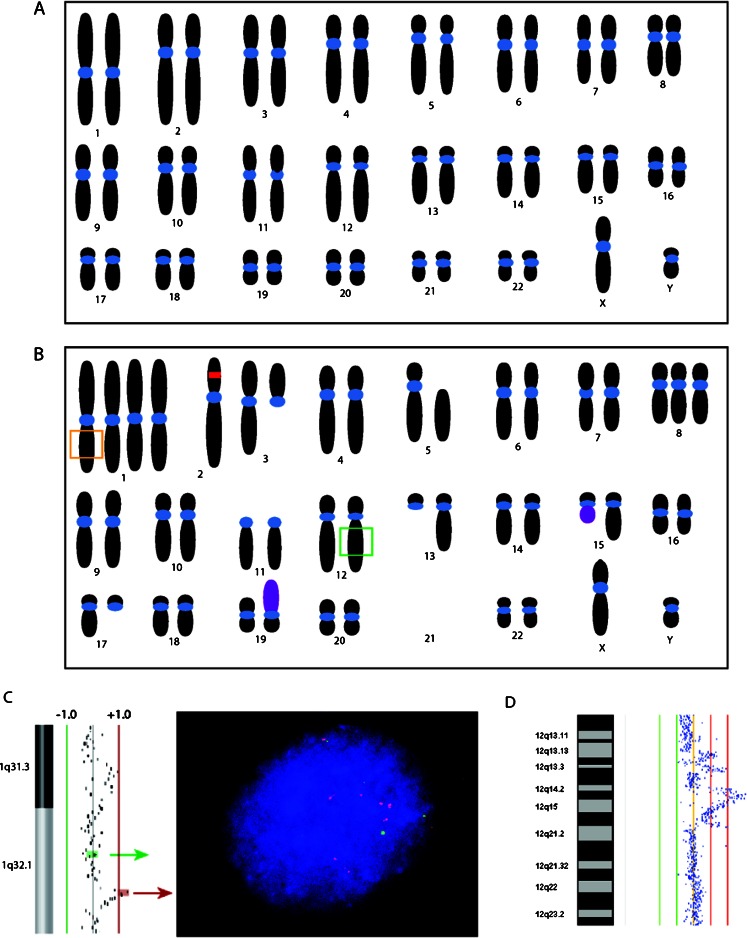
Fig. 2.
Chromosomal instability. a Normal karyotype. b Example of a potential karyotype of a cell with chromosomal instability and aneuploidy. The red box indicates an inversion and the purple chromosomes represent translocations. The orange and green boxes indicate the chromosomal regions depicted in (c) and (d) which harbor amplifications and can be visualized by FISH (c) and array-CGH (d)
Despite the prominence and fundamental importance of CIN to cancer biology, the molecular mechanisms underlying CIN in sporadic cancers remain poorly understood. This is due primarily to the fact that disruption of countless genes can give rise to CIN, including, but not limited to, those involved in chromosome condensation and segregation (STAG2) [42], telomere dysfunction (TRF1 and Tankyrase) [43], as well as DNA damage (ATM) [44, 45] and spindle checkpoint genes (BUB1, Mad2) [46–48], highlighting the heterogeneous nature of CIN in sporadic cancers. Attempts to explain the presence and molecular basis of CIN in sporadic cancers have led to the development of three prevailing theories: the mutator hypothesis, the oncogene-induced DNA damage model, and instability due to telomere erosion, which are reviewed in [5, 10, 49].
The advent of sequencing technologies has led to the recent discovery of an intriguing form of genome chaos and CIN, whereby only one or a few distinct chromosomes in a cancer cell are characterized by the presence of upwards of hundreds of complex genomic rearrangements [11]. These distinct chromosomal rearrangements were proposed by Stephens et al. to have developed through chromosome shattering (“thripsis” in Greek) or incomplete fragmentation and the inaccurate stitching together of chromosomes in a single stochastic event in a process termed “chromothripsis,” an event in contrast to the widely accepted notion of gradual accumulation of cancer genome rearrangements. Chromothripsis has been proposed to occur in ∼2–3 % of a wide spectrum of cancers (with a higher incidence in bone cancers), where chromosome-specific massive rearrangements have been described [11, 50]. The mechanisms underlying chromothripsis, and its clinical implications, have been recently reviewed by Forment et al. [51].
Interplay between instability types
While all levels of instability can co-occur within the same cell, and work in concert to disrupt a single gene, protein complex, or pathway, in colorectal and endometrial cancers, an inverse relationship between CIN and MIN has been observed [52, 53]. Although both types of instability appear to occur early in tumor development and increase with tumor progression, cancers with an MMR deficiency tend to be diploid and exhibit normal rates of gross chromosomal changes, whereas MMR-proficient tumors are typically aneuploid and display increased rates of chromosomal alterations [12]. Moreover, the fusion of MIN and CIN cells results in CIN, but not MIN, suggesting that CIN is a dominant phenotype that may result from gain-of-function alterations rather than gene inactivation [3, 46].
Methods for the detection and analysis of genome instability
A number of established strategies exist to detect genomic instability in cancer. However, it is important to keep in mind that genomic instability is a matter of rate of chromosomal alterations and is therefore a gauge of variability in chromosomal state between individual cells within a tumor [54]. To accurately assess instability, repeated measurements across cell populations throughout tumor evolution or, ideally, measurements in individual cancer cells are required to define the actual rate or variability in genomic changes for a particular tumor [54]. Although these measurements are more easily obtainable for cancer cell lines, measuring genome stability accurately in clinical tumor specimens where material is often limited and substantial cellular heterogeneity exists is considerably more difficult. As a result, few studies have determined the actual rate of chromosomal alterations in different cancer types and, thus, characterized true genomic instability [54]. Because of the difficulty in measuring actual genomic instability, various methods to calculate the frequency and extent of genomic changes for static tumor cell populations have been used as a surrogate to describe genomic instability. Therefore, caution must be taken when interpreting claims about instability in cancer. Since genomic instability occurs across multiple genetic levels, any method capable of detecting chromosomal, microsatellite, or nucleotide changes is adequate to measure a component of genomic instability. Such methods include, but are not limited to, karyotyping, flow cytometry, single nucleotide polymorphism (SNP) arrays, genome sequencing, and polymerase chain reaction (PCR), which are summarized in Table 1.
Table 1.
Currently available methods of detecting genome instability
| Method | Cellularity | Alterations detected | Rate and state | |
|---|---|---|---|---|
| Karyotyping | Single cell | Whole and segmental CIN, aneuploidy | –a | –a |
| Single-cell sequencing | Single cell | Whole and segmental CIN, translocations, insertions, deletions, and mutations | –a | –c |
| Flow cytometry | Multi-cell | Cell ploidy/aneuploidy | –b | –b |
| Array-CGH | Multi-cell | Whole and segmental CIN | N/A | –a |
| SNP arrays | Multi-cell | Whole and segmental CIN, SNP, UPD, LOH | N/A | –a |
| Whole-genome sequencing | Multi-cell | Whole and segmental CIN, translocations, insertions, deletions, and mutations | N/A | –c |
| PCR | Multi-cell | MSI, mitochondrial instability | –b | –a |
SNP single nucleotide polymorphisms, UPD uniparental disomy, LOH loss of heterozygosity, CGH comparative genomic hybridization, N/A cannot detect
aBest approach for measuring rate and state
bUseful but not ideal at measuring rate and state
cNot very useful at measuring rate and state
Single-cell approaches
Karyotyping is the visualization of a cell’s entire complement of chromosomes, or karyotype. Assessment of a cell karyotype enables the identification of abnormalities in chromosome number (aneuploidy) and large structural rearrangements like inversions and translocations [55, 56]. Traditionally, metaphase chromosomes are stained with a DNA-binding dye, such as Giemsa stain, which is taken up readily by gene-poor A,T-rich genomic regions and results in a chromosome-specific banding pattern that can be used to differentiate chromosomes and identify abnormalities. The use of multicolored fluorescence in situ hybridization (FISH) probes has greatly facilitated the assessment of CIN and is referred to as spectral karyotyping (SKY). The SKY technique results in coloring, or painting, of each chromosome with a different colored fluorophore, readily enabling the identification of chromosomes and rearrangements [55, 57]. Although excellent for detecting global CIN changes, even the most advanced FISH strategies cannot accurately measure somatic mutations throughout the genome. While karyotyping is one of the few techniques available that enable the identification of alterations within a single cell, and the only one capable of profiling both clonal and non-clonal chromosomal alterations [58], like most other methods, it offers only a static picture of the state of chromosomal alterations with no information regarding the extent of variability between cells. Furthermore, it is labor-intensive and metaphase spreads from even short-term cultures can acquire culturing artifacts that induce additional genomic changes. Despite these limitations, karyotyping remains the most reliable method to detect non-clonal chromosomal aberrations and assess genomic variability among cells.
Advances in next-generation sequencing and whole-genome amplification technologies have enabled the advent of single-cell sequencing, which offers promising insight into understanding genomic instability as it provides not only a comprehensive look at the state of genomic alterations of a tumor cell but also cell-to-cell heterogeneity. Because single-cell sequencing relies on gene amplification, sequence bias and adequate genome coverage remain major challenges. However, new amplification methodologies such as multiple annealing and looping-based amplification cycles, which enable over 90 % genome coverage and can accurately detect mutations and copy number variations [59], are in development and have the potential to greatly improve single-cell sequencing. Although many obstacles remain before single-cell sequencing can be routinely implemented as a standard procedure for detecting genome instability, it has the ability to provide an unprecedented view of genomic instability.
Multicellular approaches
Flow cytometry, which measures cells in suspension as they pass through a laser, scatter light, and emit fluorescence, can be used to approximate cellular aneuploidy. This strategy estimates cell ploidy based on DNA content (which correlates to the intensity of fluorescence) and the stage of cells in the cell cycle. Comparison of the estimated ploidy in the G0/G1 fraction of malignant and normal cells allows a gross estimate of genome instability in cancer cells [60, 61]. While flow cytometry is extremely accurate in its ability to estimate ploidy, it provides no information regarding NIN, MSI, or the segmental or whole-chromosome aberration components of CIN.
Array comparative genomic hybridization (aCGH) offers the ability to quantitatively detect and visualize whole and segmental chromosomal alterations such as gains, losses, amplifications, and LOH [62, 63]. Briefly, reference genomic DNA and test DNA are differentially labeled, pooled, and hybridized onto arrays comprising BAC, cDNA, or oligonucleotides, and imbalances are visualized as differences in fluorescence intensity. The advent of SNP arrays offered improved resolution, enabling more precise mapping of copy number alterations and the detection of uniparental disomy (copy neutral loss of heterozygosity) as well as the ability to distinguish alleles at specific polymorphic sites [64–66]. However, neither aCGH nor SNP arrays are able to detect translocations, inversions, or somatic mutations.
PCR is the gold standard for detecting MSI. PCR is used to amplify known microsatellite regions, and the lengths of the short tandem repeats (PCR products) are compared in tumor and normal DNA to determine the state of MSI [37, 67, 68]. This approach is therefore limited to assessing MSI. PCR is also used routinely for the analysis of mitochondrial instability. The ability to isolate mtDNA from total DNA using mitochondrial-specific primers rather than through centrifugation not only reduced tissue requirements but also enabled the use of archival paraffin-embedded tissues, greatly expanding the number samples available for analysis [19]. Commonly used markers of mitochondrial genome instability detected by PCR and followed by direct sequencing include point mutations, insertions, deletions, and length changes in homopolymeric or dimeric nucleotide tracts. Competitive PCR, in which a competitor DNA fragment is added to the DNA sample, can be used to determine mitochondrial DNA copy number by determining the ratio between the intensities of the control and the sample PCR product band [18].
While not routinely performed at the single-cell level, whole-genome sequencing is arguably the most comprehensive and informative method of profiling the cancer genome. In a single experiment, sequencing is capable of identifying nucleotide substitutions, insertions or deletions, and larger genomic rearrangements such as copy number changes, inversions, and translocations, simultaneously capturing all levels of genomic instability for a given population of tumor cells (Fig. 3) [69, 70]. The detection and extent of somatic mutations is determined informatically using computational programs for variant calling. These programs compare the sequences of both the tumor and patient-matched normal sample to a reference genome to reveal somatic and germline alterations, providing confidence calls for each mutation [69, 71]. Copy number analysis by sequencing (both by high or low coverage) offers substantial benefits over array-based methods, including higher resolution (down to a single base) and precise delineation of breakpoints [72, 73]. Copy number ratios at each genomic locus are estimated by counting and comparing the number of reads in both tumor and normal samples. Furthermore, whole-genome sequencing provides data on non-coding regions (promoters, enhances, introns, and non-coding RNA) as well as un-annotated regions, requiring no a priori knowledge of genome sequence, facilitating the discovery of novel DNA sequences.
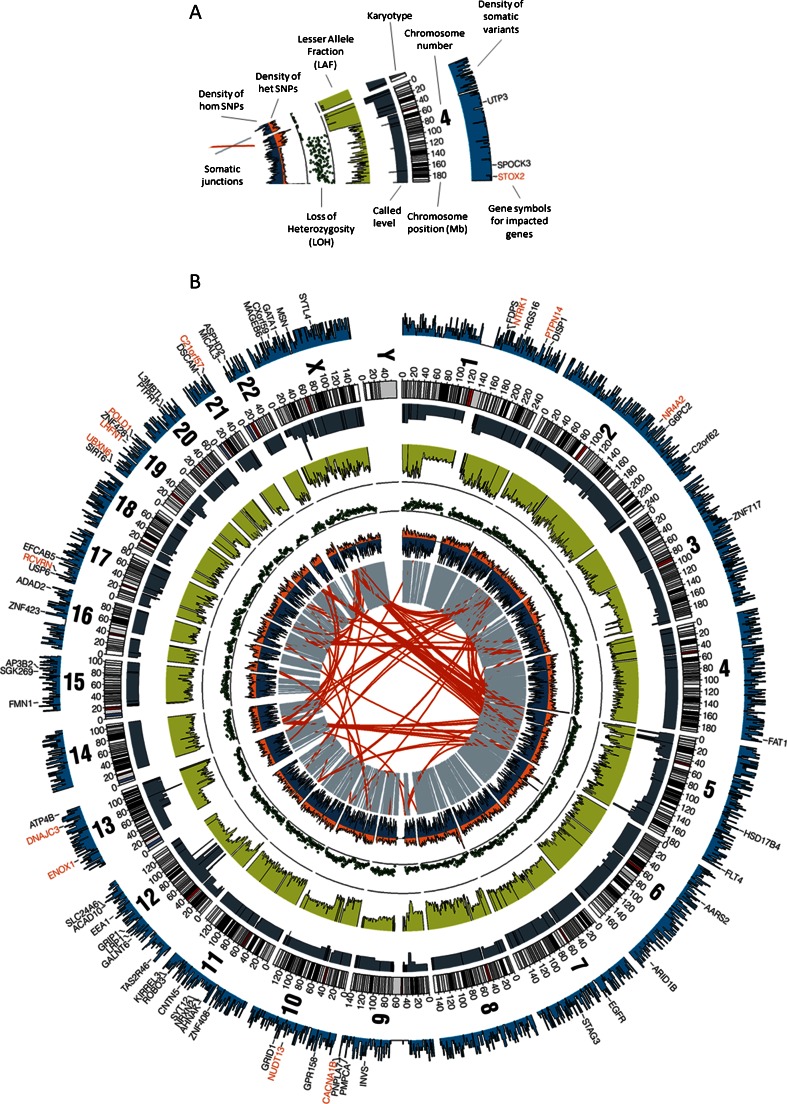
Fig. 3.
Information provided from whole-genome sequencing. a Legend depicting the genomic data (rearrangements, SNPs, LOH, lesser allele fraction, copy number, somatic mutations, and genes affected by these alterations) available following whole-genome sequencing. b Circos plot of a lung adenocarcinoma tumor from a never smoker referenced against the matched non-malignant tissue
Sequencing studies have provided massive amounts of data on cancer genomes, revealing great diversity in the mutation frequency across tumor types and identifying novel rearrangements in epithelial cancers. As data from sequencing studies continue to emerge in the public domain, a large-scale pan-cancer comparison of genomic instability in different cancers will be feasible. Such an analysis may shed more light on the mechanistic differences of cancer development in different tissues, which itself will improve our understanding of cancer biology and our ability to develop rationally designed therapies. The interpretation of whole-genome sequencing data in the context of heterogeneous tumors, however, remains a considerable challenge to the application of such data to patient care.
The fundamental limitation of these multicellular approaches is that they provide only a snapshot of the state of alterations in a tumor sample and are incapable of defining the rate of chromosomal changes within a tumor—two features that define genomic instability. While single-cell approaches such as karyotyping or single-cell array-CGH allow for unbiased comparisons of variability in chromosomal alterations between cells, they are not amenable to automation and are therefore time-consuming and labor-intensive. Collection of repeated tumor biopsy samples and advances in single-cell profiling technologies will help generate more accurate metrics of genomic instability.
Pan-cancer trends in CIN
It is well established that vast genome instability exists at different levels and to different extents in various tumor types. In the last decade, several large-scale sequencing studies have been undertaken in an attempt to characterize recurrent alterations in cancer genomes [74–79]. While thousands of mutations have been identified, these studies have shown that very few genes are recurrently mutated, deleted, or amplified at high frequencies within a tumor type. Of the handful of recurrently altered genes, TP53 is the most frequently altered gene in all tumor types, while the others (CDKN2A, PTEN, EGFR, and RAS) have roles in regulating growth and encode classical tumor suppressors and oncogenes [74, 76, 80, 81].
In general, epithelial tumors are thought to be more genomically unstable than hematologic and mesenchymal malignancies, in which a high proportion of cases are characterized by specific genetic rearrangements such as translocations [82]. Interestingly, certain cancer types display characteristic instability phenotypes. For instance, BRCA-associated breast and ovarian cancers demonstrate high levels of CIN, whereas lung cancer in smokers and never smokers differs in the extent of segmental alterations and subsequently, genome instability [83–86]. Moreover, specific subtypes of breast, ovarian, and lung cancers exhibit distinct patterns of alterations; the basal-like subtype of breast cancers (typically estrogen receptor-negative) have greater CIN than luminal subtypes, while type II high-grade serous ovarian carcinomas have greater CIN than type I serous ovarian cancers [87, 88]. In lung cancer, adenocarcinoma and squamous cell carcinoma demonstrate distinct patters of genomic alterations, and within lung adenocarcinoma, the magnoid subtype displays higher CIN than other adenocarcinoma subtypes [89, 90]. A review of genome sequencing studies revealed that epithelial-derived cancers such as breast, non-small cell lung, small-cell lung, melanoma, and prostate cancers have a greater number of somatic mutations than blood cancers including acute myeloid leukemia [91], which could suggest that epithelial cancers have greater nucleotide instability. However, specific environmental exposures, such as tobacco smoke, can have specific signatures in terms of epigenetic and genetic alterations in tumors, making it difficult to determine whether the mutations detected arose from nucleotide instability within a tumor or from carcinogen exposure [91]. As more cancer genome sequence data become publicly available, it will be interesting to determine whether specific cancer types exhibit a mutator phenotype and harbor greater nucleotide instability than others.
To compare CIN trends in a pan-cancer manner, we accessed and interrogated copy number data for a set of 2,201 tumor samples representing 24 cancer types made publically available by the Broad and Dana Farber Cancer Institutes (Table 2). Segmented tumor data were downloaded (http://www.broadinstitute.org/tumorscape/pages/portalHome.jsf); any segment with a log2 ratio exceeding ±0.1 was defined as a segmental alteration. Next, we calculated the fraction of each cancer genome encompassed by segmental alterations to determine the proportion of the genome altered (PGA) and summarized the PGA across the various malignancies (Table 2) [86]. Cancer cell lines were not included in our analysis.
Table 2.
Proportion of genome altered (log2 ratio ± 0.1) for various cancer types (n = 2,201)
| Type | Count | Median | Average | SD |
|---|---|---|---|---|
| Mesothelioma | 18 | 0.579305 | 0.539195 | 0.261045 |
| Lung SC | 17 | 0.574731 | 0.539328 | 0.265144 |
| Melanoma | 3 | 0.533935 | 0.63286 | 0.178692 |
| Breast | 193 | 0.361504 | 0.359875 | 0.228679 |
| Ovarian | 95 | 0.330162 | 0.348426 | 0.298813 |
| Lung NSC | 629 | 0.318028 | 0.324707 | 0.234076 |
| Esophageal squamous | 2 | 0.293933 | 0.293933 | 0.415683 |
| Hepatocellular | 110 | 0.242876 | 0.288188 | 0.222686 |
| Glioma | 28 | 0.198592 | 0.22249 | 0.143089 |
| Neuroblastoma | 25 | 0.196659 | 0.287776 | 0.24191 |
| Colorectal | 128 | 0.189425 | 0.261818 | 0.244976 |
| Renal | 99 | 0.130901 | 0.226044 | 0.224458 |
| Medulloblastoma | 119 | 0.110602 | 0.208003 | 0.250298 |
| Meningioma | 6 | 0.088144 | 0.228436 | 0.347694 |
| Endometrial | 1 | 0.07679 | 0.07679 | N/A |
| Prostate | 83 | 0.065822 | 0.180566 | 0.231366 |
| Synovial sarcoma | 2 | 0.030057 | 0.030057 | 0.042508 |
| Acute lymphoblastic leukemia | 378 | 0.02309 | 0.120554 | 0.256989 |
| Schwannoma | 5 | 0.011336 | 0.015021 | 0.012035 |
| Sarcoma NOS | 1 | 0.009686 | 0.009686 | N/A |
| Myelodysplasia | 19 | 0.000621 | 0.007157 | 0.009818 |
| Thyroid | 9 | 0.000445 | 0.083437 | 0.157782 |
| GIST | 16 | 0.000207 | 0.1654 | 0.341026 |
| Myeloproliferative disorder | 215 | 0.000135 | 0.011227 | 0.048053 |
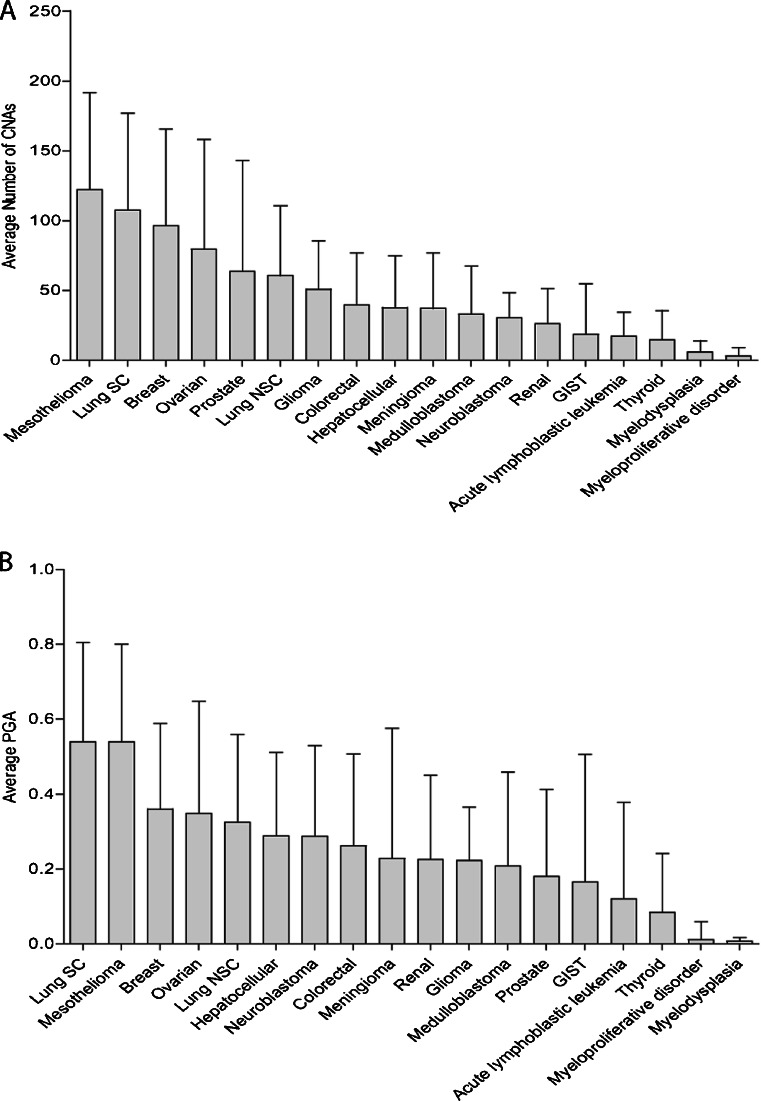
Of the 18 cancer types with at least six representative samples, mesothelioma and small-cell lung cancer had the greatest PGA and average number of copy number alterations (CNAs), suggesting that they may be the most genomically unstable in the context of CIN (Fig. 4). These two cancers were followed by breast, ovarian, non-small cell lung cancer, and liver, all epithelial cancers. The number of CNAs was highly correlated with PGA as greater PGAs were associated with a greater number of CNAs (Pearson’s correlation: r = 0.77) across all tumor samples. Of note is that hematological malignancies including acute lymphoblastic leukemia, myelodysplasia, and myeloproliferative disorder harbored some of the lowest PGAs. Thus, these results suggest that CNAs are highly correlated with PGA and that CIN may be greater in epithelial tumors than in hematological cancers, consistent with previously reported trends (Table 2 and Fig. 4).
Fig. 4.
Pan-cancer trends in genome instability. a Average number of copy number alterations for cell lines from each cancer type. Error bars represent standard deviation. b Average percent of the genome altered for all cell lines within each tumor type. Error bars represent standard deviation
Conclusion
Genomic instability occurs early in tumorigenesis, increasing the spontaneous mutation rate and enabling the acquisition of DNA alterations that promote the hallmarks of cancer, thereby driving tumor development. While the molecular basis of instability is well understood in hereditary cancers, where it is linked to mutations in DNA repair genes, the basis of instability in sporadic cancers remains poorly defined. This limited understanding is due both to the genomic heterogeneity in different tumor types as well as within individual tumors and a lack of methods capable of capturing both the state and rate of instability, which are required to determine the true measure of instability. Genome sequencing studies have provided a wealth of information regarding the state of instability in a variety of cancers, highlighting the diversity in both the types and amounts of instability observed in tumor genomes. As the amount of starting materials for whole-genome sequencing experiments continues to decrease, single-cell sequencing will become feasible for solid tumors; with this will come an expanded understanding of which mechanisms of genomic instability are selected for and precisely how specific patterns of instability support tumor growth in unique systems. In combination with repeat biopsies and sequencing of multiple areas in a single tumor, detailed maps of how genomic instability changes over time will emerge, which can then be interpreted in the context of unique selective pressures in the tumor microenvironment (e.g., the immune system, chemotherapy) or correlated to specific clinical features (e.g., tumor progression). Genomic instability remains an important, yet poorly defined, mechanism by which tumors accelerate their own evolution and survival. At the same time, once uncovered, these same mechanisms will undoubtedly present to the researcher a host of novel therapeutic opportunities.
Acknowledgments
This work was supported by funds from the Canadian Institutes for Health Research (CIHR; MOP 86731, MOP 94867, MOP-110949), Canadian Cancer Society Research Institute (no. 700809), U.S Department of Defence (CDMRP W81XWH-10-1-0634), NCI Early Detection Research Network, and the Canary Foundation. LAP and KLT are supported by Vanier Canada Graduate Scholarships. EAV is supported by Frederick Banting and Charles Best Canada Graduate Scholarship from CIHR.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Sieber OM, Heinimann K, Tomlinson IP. Genomic instability—the engine of tumorigenesis? Nature Reviews. Cancer. 2003;3(9):701–708. doi: 10.1038/nrc1170. [DOI] [PubMed] [Google Scholar]
- 2.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194(4260):23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 3.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386(6625):623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 4.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434(7035):907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 5.Loeb LA. A mutator phenotype in cancer. Cancer Research. 2001;61(8):3230–3239. [PubMed] [Google Scholar]
- 6.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434(7035):864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 7.Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444(7119):633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 8.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319(5868):1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 9.Janssen A, van der Burg M, Szuhai K, Kops GJ, Medema RH. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science. 2011;333(6051):1895–1898. doi: 10.1126/science.1210214. [DOI] [PubMed] [Google Scholar]
- 10.Hackett JA, Greider CW. Balancing instability: dual roles for telomerase and telomere dysfunction in tumorigenesis. Oncogene. 2002;21(4):619–626. doi: 10.1038/sj.onc.1205061. [DOI] [PubMed] [Google Scholar]
- 11.Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144(1):27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396(6712):643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 13.Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138(6):2073–2087. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Nicolantonio F, Martini M, Molinari F, Sartore-Bianchi A, Arena S, Saletti P, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 2008;26(35):5705–5712. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 15.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nature Genetics. 2006;38(9):1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 16.Dworaczek H, Xiao W. Xeroderma pigmentosum: a glimpse into nucleotide excision repair, genetic instability, and cancer. Critical Reviews in Oncogenesis. 2007;13(2):159–177. doi: 10.1615/CritRevOncog.v13.i2.20. [DOI] [PubMed] [Google Scholar]
- 17.Al-Tassan N, Chmiel NH, Maynard J, Fleming N, Livingston AL, Williams GT, et al. Inherited variants of MYH associated with somatic G:C→T:A mutations in colorectal tumors. Nature Genetics. 2002;30(2):227–232. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 18.Lee HC, Li SH, Lin JC, Wu CC, Yeh DC, Wei YH. Somatic mutations in the D-loop and decrease in the copy number of mitochondrial DNA in human hepatocellular carcinoma. Mutation Research. 2004;547(1–2):71–78. doi: 10.1016/j.mrfmmm.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 19.Lee HC, Yin PH, Lin JC, Wu CC, Chen CY, Wu CW, et al. Mitochondrial genome instability and mtDNA depletion in human cancers. Annals of the New York Academy of Sciences. 2005;1042:109–122. doi: 10.1196/annals.1338.011. [DOI] [PubMed] [Google Scholar]
- 20.Weissenbach J, Gyapay G, Dib C, Vignal A, Morissette J, Millasseau P, et al. A second-generation linkage map of the human genome. Nature. 1992;359(6398):794–801. doi: 10.1038/359794a0. [DOI] [PubMed] [Google Scholar]
- 21.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260(5109):816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 22.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363(6429):558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 23.Risinger JI, Berchuck A, Kohler MF, Watson P, Lynch HT, Boyd J. Genetic instability of microsatellites in endometrial carcinoma. Cancer Research. 1993;53(21):5100–5103. [PubMed] [Google Scholar]
- 24.Rhyu MG, Park WS, Meltzer SJ. Microsatellite instability occurs frequently in human gastric carcinoma. Oncogene. 1994;9(1):29–32. [PubMed] [Google Scholar]
- 25.Halling KC, Harper J, Moskaluk CA, Thibodeau SN, Petroni GR, Yustein AS, et al. Origin of microsatellite instability in gastric cancer. The American Journal of Pathology. 1999;155(1):205–211. doi: 10.1016/S0002-9440(10)65114-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim WS, Park C, Hong SK, Park BK, Kim HS, Park K. Microsatellite instability (MSI) in non-small cell lung cancer (NSCLC) is highly associated with transforming growth factor-beta type II receptor (TGF-beta RII) frameshift mutation. Anticancer Research. 2000;20(3A):1499–1502. [PubMed] [Google Scholar]
- 27.Fishel R, Lescoe MK, Rao MR, Copeland NG, Jenkins NA, Garber J, et al. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75(5):1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 28.Leach FS, Nicolaides NC, Papadopoulos N, Liu B, Jen J, Parsons R, et al. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75(6):1215–1225. doi: 10.1016/0092-8674(93)90330-S. [DOI] [PubMed] [Google Scholar]
- 29.Bronner CE, Baker SM, Morrison PT, Warren G, Smith LG, Lescoe MK, et al. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368(6468):258–261. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- 30.Nicolaides NC, Papadopoulos N, Liu B, Wei YF, Carter KC, Ruben SM, et al. Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature. 1994;371(6492):75–80. doi: 10.1038/371075a0. [DOI] [PubMed] [Google Scholar]
- 31.Palombo F, Gallinari P, Iaccarino I, Lettieri T, Hughes M, D'Arrigo A, et al. GTBP, a 160-kilodalton protein essential for mismatch-binding activity in human cells. Science. 1995;268(5219):1912–1914. doi: 10.1126/science.7604265. [DOI] [PubMed] [Google Scholar]
- 32.Papadopoulos N, Nicolaides NC, Wei YF, Ruben SM, Carter KC, Rosen CA, et al. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994;263(5153):1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- 33.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(15):8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Issa JP. CpG island methylator phenotype in cancer. Nature Reviews. Cancer. 2004;4(12):988–993. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 35.Goel A, Nagasaka T, Arnold CN, Inoue T, Hamilton C, Niedzwiecki D, et al. The CpG island methylator phenotype and chromosomal instability are inversely correlated in sporadic colorectal cancer. Gastroenterology. 2007;132(1):127–138. doi: 10.1053/j.gastro.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 36.Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Meyerhardt JA, Loda M, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58(1):90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vilar E, Gruber SB. Microsatellite instability in colorectal cancer—the stable evidence. Nature Reviews. Clinical Oncology. 2010;7(3):153–162. doi: 10.1038/nrclinonc.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ward R, Meagher A, Tomlinson I, O'Connor T, Norrie M, Wu R, et al. Microsatellite instability and the clinicopathological features of sporadic colorectal cancer. Gut. 2001;48(6):821–829. doi: 10.1136/gut.48.6.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gagos S, Irminger-Finger I. Chromosome instability in neoplasia: chaotic roots to continuous growth. The International Journal of Biochemistry & Cell Biology. 2005;37(5):1014–1033. doi: 10.1016/j.biocel.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 40.Lee AJ, Endesfelder D, Rowan AJ, Walther A, Birkbak NJ, Futreal PA, et al. Chromosomal instability confers intrinsic multidrug resistance. Cancer Research. 2011;71(5):1858–1870. doi: 10.1158/0008-5472.CAN-10-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walther A, Houlston R, Tomlinson I. Association between chromosomal instability and prognosis in colorectal cancer: a meta-analysis. Gut. 2008;57(7):941–950. doi: 10.1136/gut.2007.135004. [DOI] [PubMed] [Google Scholar]
- 42.Solomon DA, Kim T, Diaz-Martinez LA, Fair J, Elkahloun AG, Harris BT, et al. Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science. 2011;333(6045):1039–1043. doi: 10.1126/science.1203619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genome instability. Nature Reviews Molecular Cell Biology. 2010;11(3):171–181. doi: 10.1038/nrg2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levitt NC, Hickson ID. Caretaker tumour suppressor genes that defend genome integrity. Trends in Molecular Medicine. 2002;8(4):179–186. doi: 10.1016/S1471-4914(02)02298-0. [DOI] [PubMed] [Google Scholar]
- 45.Rotman G, Shiloh Y. ATM: from gene to function. Human Molecular Genetics. 1998;7(10):1555–1563. doi: 10.1093/hmg/7.10.1555. [DOI] [PubMed] [Google Scholar]
- 46.Cahill DP, Lengauer C, Yu J, Riggins GJ, Willson JK, Markowitz SD, et al. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392(6673):300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- 47.Hanks S, Coleman K, Reid S, Plaja A, Firth H, Fitzpatrick D, et al. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nature Genetics. 2004;36(11):1159–1161. doi: 10.1038/ng1449. [DOI] [PubMed] [Google Scholar]
- 48.Li Y, Benezra R. Identification of a human mitotic checkpoint gene: hsMAD2. Science. 1996;274(5285):246–248. doi: 10.1126/science.274.5285.246. [DOI] [PubMed] [Google Scholar]
- 49.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability—an evolving hallmark of cancer. Nature Reviews Molecular Cell Biology. 2010;11(3):220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 50.Maher CA, Wilson RK. Chromothripsis and human disease: piecing together the shattering process. Cell. 2012;148(1–2):29–32. doi: 10.1016/j.cell.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Forment JV, Kaidi A, Jackson SP. Chromothripsis and cancer: causes and consequences of chromosome shattering. Nature Reviews. Cancer. 2012;12(10):663–670. doi: 10.1038/nrc3352. [DOI] [PubMed] [Google Scholar]
- 52.Abdel-Rahman WM, Katsura K, Rens W, Gorman PA, Sheer D, Bicknell D, et al. Spectral karyotyping suggests additional subsets of colorectal cancers characterized by pattern of chromosome rearrangement. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(5):2538–2543. doi: 10.1073/pnas.041603298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muresu R, Sini MC, Cossu A, Tore S, Baldinu P, Manca A, et al. Chromosomal abnormalities and microsatellite instability in sporadic endometrial cancer. European Journal of Cancer. 2002;38(13):1802–1809. doi: 10.1016/S0959-8049(02)00152-1. [DOI] [PubMed] [Google Scholar]
- 54.Geigl JB, Obenauf AC, Schwarzbraun T, Speicher MR. Defining ‘chromosomal instability’. Trends in Genetics: TIG. 2008;24(2):64–69. doi: 10.1016/j.tig.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 55.Wan TS, Ma ES. Molecular cytogenetics: an indispensable tool for cancer diagnosis. Chang Gung Medical Journal. 2012;35(2):96–110. doi: 10.4103/2319-4170.106161. [DOI] [PubMed] [Google Scholar]
- 56.Beheshti B, Park PC, Sweet JM, Trachtenberg J, Jewett MA, Squire JA. Evidence of chromosomal instability in prostate cancer determined by spectral karyotyping (SKY) and interphase FISH analysis. Neoplasia. 2001;3(1):62–69. doi: 10.1038/sj.neo.7900125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bayani J, Squire JA. Advances in the detection of chromosomal aberrations using spectral karyotyping. Clinical Genetics. 2001;59(2):65–73. doi: 10.1034/j.1399-0004.2001.590201.x. [DOI] [PubMed] [Google Scholar]
- 58.Ye CJ, Stevens JB, Liu G, Bremer SW, Jaiswal AS, Ye KJ, et al. Genome based cell population heterogeneity promotes tumorigenicity: the evolutionary mechanism of cancer. Journal of Cellular Physiology. 2009;219(2):288–300. doi: 10.1002/jcp.21663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338(6114):1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Darzynkiewicz Z, Halicka HD, Zhao H. Analysis of cellular DNA content by flow and laser scanning cytometry. Advances in Experimental Medicine and Biology. 2010;676:137–147. doi: 10.1007/978-1-4419-6199-0_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.D'Urso V, Collodoro A, Mattioli E, Giordano A, Bagella L. Cytometry and DNA ploidy: clinical uses and molecular perspective in gastric and lung cancer. Journal of Cellular Physiology. 2010;222(3):532–539. doi: 10.1002/jcp.21991. [DOI] [PubMed] [Google Scholar]
- 62.Ishkanian AS, Malloff CA, Watson SK, DeLeeuw RJ, Chi B, Coe BP, et al. A tiling resolution DNA microarray with complete coverage of the human genome. Nature Genetics. 2004;36(3):299–303. doi: 10.1038/ng1307. [DOI] [PubMed] [Google Scholar]
- 63.Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nature Genetics. 1998;20(2):207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 64.Heinrichs S, Look AT. Identification of structural aberrations in cancer by SNP array analysis. Genome Biology. 2007;8(7):219. doi: 10.1186/gb-2007-8-7-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao X, Li C, Paez JG, Chin K, Janne PA, Chen TH, et al. An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer Research. 2004;64(9):3060–3071. doi: 10.1158/0008-5472.CAN-03-3308. [DOI] [PubMed] [Google Scholar]
- 66.Gondek LP, Tiu R, Haddad AS, O'Keefe CL, Sekeres MA, Theil KS, et al. Single nucleotide polymorphism arrays complement metaphase cytogenetics in detection of new chromosomal lesions in MDS. Leukemia: Official Journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2007;21(9):2058–2061. doi: 10.1038/sj.leu.2404745. [DOI] [PubMed] [Google Scholar]
- 67.Goel A, Nagasaka T, Hamelin R, Boland CR. An optimized pentaplex PCR for detecting DNA mismatch repair-deficient colorectal cancers. PloS One. 2010;5(2):e9393. doi: 10.1371/journal.pone.0009393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Janavicius R, Matiukaite D, Jakubauskas A, Griskevicius L. Microsatellite instability detection by high-resolution melting analysis. Clinical Chemistry. 2010;56(11):1750–1757. doi: 10.1373/clinchem.2010.150680. [DOI] [PubMed] [Google Scholar]
- 69.Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nature Reviews Genetics. 2010;11(10):685–696. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- 70.Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452(7189):872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 71.Ng PC, Kirkness EF. Whole genome sequencing. Methods in Molecular Biology. 2010;628:215–226. doi: 10.1007/978-1-60327-367-1_12. [DOI] [PubMed] [Google Scholar]
- 72.Campbell PJ, Stephens PJ, Pleasance ED, O'Meara S, Li H, Santarius T, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nature Genetics. 2008;40(6):722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chiang DY, Getz G, Jaffe DB, O'Kelly MJ, Zhao X, Carter SL, et al. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nature Methods. 2009;6(1):99–103. doi: 10.1038/nmeth.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455(7216):1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bell D. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McLendon R. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486(7403):395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Seo JS, Ju YS, Lee WC, Shin JY, Lee JK, Bleazard T, Lee J, Jung YJ, Kim JO, Yu SB et al. (2012). The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Research, 22, 2109–2119. [DOI] [PMC free article] [PubMed]
- 79.Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470(7333):214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321(5897):1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318(5853):1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 82.Mitelman F, Johansson B, Mertens F. Fusion genes and rearranged genes as a linear function of chromosome aberrations in cancer. Nature Genetics. 2004;36(4):331–334. doi: 10.1038/ng1335. [DOI] [PubMed] [Google Scholar]
- 83.Vollebergh MA, Jonkers J, Linn SC. Genomic instability in breast and ovarian cancers: translation into clinical predictive biomarkers. Cellular and Molecular Life Sciences: CMLS. 2012;69(2):223–245. doi: 10.1007/s00018-011-0809-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huang YT, Lin X, Liu Y, Chirieac LR, McGovern R, Wain J, et al. Cigarette smoking increases copy number alterations in nonsmall-cell lung cancer. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(39):16345–16350. doi: 10.1073/pnas.1102769108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Massion PP, Zou Y, Chen H, Jiang A, Coulson P, Amos CI, et al. Smoking-related genomic signatures in non-small cell lung cancer. American Journal of Respiratory and Critical Care Medicine. 2008;178(11):1164–1172. doi: 10.1164/rccm.200801-142OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thu KL, Vucic EA, Chari R, Zhang W, Lockwood WW, English JC, et al. Lung adenocarcinoma of never smokers and smokers harbor differential regions of genetic alteration and exhibit different levels of genomic instability. PloS One. 2012;7(3):e33003. doi: 10.1371/journal.pone.0033003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bowtell DD. The genesis and evolution of high-grade serous ovarian cancer. Nature Reviews. Cancer. 2010;10(11):803–808. doi: 10.1038/nrc2946. [DOI] [PubMed] [Google Scholar]
- 88.Fang M, Toher J, Morgan M, Davison J, Tannenbaum S, Claffey K. Genomic differences between estrogen receptor (ER)-positive and ER-negative human breast carcinoma identified by single nucleotide polymorphism array comparative genome hybridization analysis. Cancer. 2011;117(10):2024–2034. doi: 10.1002/cncr.25770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lockwood WW, Wilson IM, Coe BP, Chari R, Pikor LA, Thu KL, et al. Divergent genomic and epigenomic landscapes of lung cancer subtypes underscore the selection of different oncogenic pathways during tumor development. PloS One. 2012;7(5):e37775. doi: 10.1371/journal.pone.0037775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wilkerson MD, Yin X, Walter V, Zhao N, Cabanski CR, Hayward MC, et al. Differential pathogenesis of lung adenocarcinoma subtypes involving sequence mutations, copy number, chromosomal instability, and methylation. PloS One. 2012;7(5):e36530. doi: 10.1371/journal.pone.0036530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Loeb LA. Human cancers express mutator phenotypes: origin, consequences and targeting. Nature Reviews. Cancer. 2011;11(6):450–457. doi: 10.1038/nrc3063. [DOI] [PMC free article] [PubMed] [Google Scholar]