Abstract
Active targeted delivery of nanoparticle-encapsulated agents to tumor cells in vivo is expected to enhance therapeutic effect with significantly less non-specific toxicity. Active targeting is based on surface modification of nanoparticles with ligands that bind with extracellular targets and enhance payload delivery in the cells. In this study, we have used label-free Raman micro-spectral analysis and kinetic modeling to study cellular interactions and intracellular delivery of C6-ceramide using a non-targeted and an epidermal growth factor receptor (EGFR) targeted biodegradable polymeric nano-delivery systems, in EGFR-expressing human ovarian adenocarcinoma (SKOV3) cells. The results show that EGFR peptide-modified nanoparticles were rapidly internalized in SKOV3 cells leading to significant intracellular accumulation as compared to non-specific uptake by the non-targeted nanoparticles. Raman micro-spectral analysis enables visualization and quantification of the carrier system, drug-load, and responses of the biological systems interrogated, without exogenous staining and labeling procedures.
Keywords: Biodegradable polymeric nanoparticle, targeted drug delivery, epidermal growth factor receptor, Raman micro-spectroscopy
1. INTRODUCTION
Pharmaceutical nanotechnology offers a number of advantages in systemic delivery of therapeutic agents targeted towards specific diseases, such as cancer 1–3. Polymer and lipid nanosystems can encapsulate diverse types of payloads and protect these in the systemic circulation for efficient availability at the target site and in the cells of interest.2,4–6 The opportunity to develop nanoparticle carriers that respond to physiological stimuli, feasibility for delivery of multiple therapeutic agents in a single formulation, a combination of imaging and drug therapy to monitor effects in real time, and the opportunity to combine drugs with energy physical stimuli (heat, light, and sound) delivery for synergistic therapeutic effects are additional benefits from multifunctional pharmaceutical nanosystems. Regardless of the inherent properties of the drug candidates, the pharmacokinetics and distribution pattern upon systemic administration will be dictated by the properties of the nanocarrier systems.7
For systemic therapy, passive and active targeting strategies are utilized in order to accumulate the nanoparticle-encapsulated agent to the disease site and in the cell of interest.8–10 Passive targeting relies on the properties of the delivery system and the disease pathology in order to preferentially accumulate the drug at the site of interest and avoid nonspecific distribution.11 Active targeting, on the other hand, relies on the coupling of a specific ligand on the nanoparticle surface that will be recognized by the tissue or cells present at the disease site.9 In tumor targeting, for example, there are several strategies that can be adopted for the surface modification of nanocarrier systems for effective targeted delivery to the tumor cells or to endothelial cells of the tumor blood vessels. Since tumor cells are rapidly proliferating, they overexpress receptors to enhance the uptake of nutrients and growth factors. The phage display method has been used to identify specific peptide sequences that can be used for targeting to tumors and other disease areas in the body.12
Raman microspectral imaging is a label-free approach for high resolution non-destructive chemical analysis of biological samples, including living cells.13–17 The technique relies on detection and characterization of the intrinsic chemical bond vibrations within all biomolecules of any system. Coupled with optical microscopy, Raman micro-spectral method of analysis allows for optical sectioning with diffraction-limited spatial resolution. Subsequent analysis of the obtained hyper-spectral data sets with multivariate statistical algorithms yields a biochemical snapshot of the cellular sample.18,19 One such statistical analysis employed is the Vertex Component Analysis (VCA), which renders information about biochemically distinct micro-environments within a cell.17 The spectral data evaluated has specific information regarding the chemical components within the system, which for cellular analysis refers to the nucleus and the cell body, membrane-rich organelles such as the mitochondria, as well as inherent and exogenous intra-cellular inclusions.17 Hence, Raman micro-spectroscopy coupled with statistical methods of analysis allows us to assess the sub-cellular architecture with synergistic characterization of dynamics of an internalized nanoparticle system.
In a previous study, we have utilized Raman micro-spectral imaging to characterize and evaluate the intracellular delivery of biodegradable polymeric nanoparticle-encapsulated therapeutics.17 Poly(ethylene oxide) (PEO)-modified poly(epsilon-caprolactone) (PCL) and PEO-modified poly(D,L-lactide-co-glycolide) (PLGA) nanoparticles were evaluated for intracellular delivery and degradation in HeLa cells. The results established faster uptake kinetics and intracellular dynamics for the non-loaded PLGA system. Also, intracellular translocation patterns differed for the two systems: PCL nanoparticles demonstrated a haphazard intracellular distribution, while PLGA particulates showed an affinity toward the perinuclear region.
In the present study we have interrogated the uptake kinetics and intracellular dynamics of the non-targeted polymeric nanoparticles (PLGA-PEG/PCL) with a modified system, specifically targeting epidermal growth factor receptor (EGFR). EGFR is overexpressed in a variety of highly aggressive tumors, including ovarian cancer.20,21 Time-dependent Raman and fluorescence analyses were carried to characterize the intracellular availability of nanoparticles. We have utilized Raman micro-spectral imaging to understand the uptake and cellular distribution of non-targeted and EGFR-targeted PEG-PLGA and PCL blend nanoparticles in SKOV-3 human ovarian adenocarcinoma cells.
Upon verifying uptake kinetics of the polymeric nanostructures, we encapsulated a therapeutic payload (C6-ceramide) into the nano-formulations. C6-ceramide is known to mimic the endogenous ceramide within the cellular architecture, decreasing the apoptotic threshold.22,23 We have implemented the drug in the nanoparticle formulation as a model system for characterization of its biochemical patterns of dissociation from the delivery systems as well as interrogate efficiency of EGFR-targeting capabilities in speed and efficiency of delivery. The signal pertaining to the C6-ceramide was observed to co-localize with the sub-cellular signal rich in endoplasmic reticulum and mitochondria. We developed an algorithm for quantifying drug levels using Raman micro-spectral data and used kinetic modeling to compare uptake kinetics in targeted and non-targeted systems. We demonstrate that Raman micro-spectral imaging has the capability to monitor a number of variable systems simultaneously, without extensive labeling methods.
2. MATERIALS AND METHODS
2.1 Nanoparticle Preparation
Non-targeted and EGFR targeted polymer blend nanoparticles were prepared using a solvent displacement method as reported previously.24 Briefly, an 11-amino acid EGFR targeting peptide, termed G11, was synthesized with four additional glycine and a terminal cysteine residue (i.e., YHWYGYTPQNVI-GGGGC) at the Tufts University’s Peptide Synthesis Laboratory (Medford, MA). The peptide was conjugated with PEG-PLGA diblock copolymer, where the terminal maleimide functionality of PEG reacted with the SH group of cysteine residue. Following purification and characterization of the peptide-PEG-PLGA conjugate, the blend nanoparticles were prepared with PCL at a 20:80 PEG-PLGA to PCL w/w ratio. The peptide-PEG-PLGA conjugate (or the PEG-PLGA conjugate for non-targeted particles) and PCL were dissolved in 2 mL 50/50 acetonitrile/DMF, and placed in a 37°C water bath for 10 minutes to facilitate dissolution. This polymer solution was added drop-wise to 20 mL deionized distilled water while stirring. The preparation was covered with aerated parafilm, allowed to stir overnight, centrifuged at 10,000g for 30 minutes, and then re-suspended in deionized distilled water. To synthesize EGFR-targeted nanoparticles, the peptide-PEG-PLGA conjugate was added to the nanoparticle formulation at 20% w/w total polymer, with an additional 10% w/w of PEG-PLGA conjugate. Likewise, the PEG-PLGA conjugate was added at a concentration of 20% w/w total polymer such that the total blend composition remains 20:80 as with non-targeted nanoparticles. In order to obtain fluorescing images, nanoparticles were loaded with 1% (w/w) Rhodamine 123 fluorescent dye. Following preparation and purification, both control and EGFR-targeted nanoparticles were characterized for size and surface charge using dynamic light scattering.
EGFR targeted and non-targeted polymer blend nanoparticles containing C6-ceramide were synthesized using a similar solvent displacement method.24,25 Briefly, the PLGA-PEG-peptide conjugate (or the PLGA-PEG conjugate for non-targeted particles), PCL, and D11-C6-ceramide (20% w/w, or 4mg) were dissolved in 2 mL 50/50 acetonitrile/DMF, and placed in a 37°C water bath for 10 minutes to facilitate dissolution. Fluorophore, namely NBD-ceramide, was labeled at a concentration of 0.5% w/w to ceramide. This polymer/drug solution was added dropwise to 20 mL distilled, deionized water while stirring. The preparation was covered with aerated parafilm, allowed to stir overnight, centrifuged at 10,000g for 30 minutes, and then re-suspended in phosphate-buffered saline (pH 7.4)
2.2 Cell Culture Conditions
Human SKOV-3 cells (ATCC, Manassas, VA) were grown in 75 cm2 culture flasks (Fisher Scientific) with 7 mL of Dulbecco’s Modified Eagle’s medium (DMEM, ATCC) and 10% fetal bovine serum (FBS, ATCC). Cells were incubated at 37 °C and 5% CO2. Cells were grown on CaF2 windows to avoid background signals from the regular glass slides.
Prior to Raman biochemical analysis, cells were fixed in 10% formalin (Sigma-Aldrich, St. Louis, MO) and subsequently submerged into phosphate buffered saline solution.
2.3 Raman Data Acquisition
The cellular images presented in this paper were recorded using a CRM200 confocal Raman microscope (WITec GmbH, Ulm, Germany) employing 488 nm (ca. 40 mW out of output fiber, with ~5 mW at sample) excitation wavelength from a solid state laser (Spectral Physics model “cyan” laser). The exciting laser radiation is coupled into a Zeiss microscope through a wavelength-specific single-mode optical fiber. The incident laser beam is collimated via an achromatic lens and passes a holographic band-pass filter before it is focused onto the sample through the water immersion objective (60×/NA 1.00, WD 2.0 mm).
The sample is located on a piezo-electrically driven microscope scanning stage with an x,y resolution of ca. 3 nm and a repeatability of ±5 nm and z resolution of ca. 0.3 nm and ±2 nm repeatability. The sample is scanned through the laser focus in a raster pattern at a constant stage speed of fractions of a micrometer per second. The continuous motion prevents sample degradation at the focal point of the laser beam. Raman spectra are collected with a resolution or step size of 500 nm at 250 ms per pixel
Raman backscattered radiation was collected through the microscope objective and passed through a holographic edge filter to block Rayleigh scattering and reflected laser light before being focused into a multimode optical fiber. The 50 µm diameter single-mode input fiber and the 50 µm diameter multimode output fiber provided the optical apertures for the confocal measurement. The light emerging from the output optical fiber was dispersed by a 30 cm focal length, f/4 CzernyTurner monochromator, incorporating interchangeable gratings (1800/mm, blazed at 500 nm, and 600/mm, blazed at 500 nm). The light was finally detected by a back-illuminated deep-depletion, 1024 × 128 pixel charge-coupled device camera operating at −82 °C.
2.4 Multivariate Data Analysis: Vertex Component Analysis (VCA)
All raw Raman data sets were imported into a software package developed in-house using the MATLAB (The Mathworks, Natick, MA) environment. All spectra in the dataset were corrected for cosmic rays, and interpolated to constant wavenumber increments between intensity data points. Subsequently, multivariate color decomposition was performed using Vertex Component Analysis (VCA, see above). VCA images were produced by overlaying three or four endmember abundance images to result in pseudo-color images depicting the abundance (concentration) of cellular components.
Briefly, VCA decomposes the spectra from the cellular hyper-spectral dataset into contributions from the most dissimilar spectra, which are considered the “pure component” or “end-member” spectra. Each end-member spectrum has a corresponding abundance or intensity variation associated with a region within the cellular architecture. The abundance values for a given end-member are scaled and converted to a monochrome intensity map of the imaged object. The intensities depict the extent of spectral contribution of the end-member spectrum to each of the pixels within the dataset. An overlay of these RGB color maps yields a pseudo-color map with mixed colors indicating the intensity values of each of three end-members.
2.5 Quantification of Drug Content and Uptake Kinetics
Quantification of drug internalization upon delivery of drug alone or via targeted or non-targeted nano-formulations was analyzed at each of 6 time points. Since a pixel represents a 3-dimensional volume of 500 × 500 × ~1500 nm3, stretching intensities within the Raman spectra correlate with the abundances of the functional groups within the chemical environment. Thus, the number of pixels containing evident spectral contributions of C–D stretching intensities of the drug was recorded and tabulated from each cell and for each time point. The threshold for determination of the drug presence within the cell was set by the user, which included spectral C–D stretching contributions exceeding >5 CCD intensity counts (compare to the spectral contributions of protein Amide I intensities of 80–100 CCD counts). Such information was taken after the VCA analysis, where the intensity map of the cell was reconstructed based on the abundance spectra of the drug and the nucleus. We have annotated the intensities and corresponding pixel count of the nucleus and taken that as a constant used to normalize pixel count corresponding to drug, since nuclear size of a cell within the same culture varies only slightly. Thus, from each of the cells, pixel number was summed over all the voxels corresponding to the presence of the drug, and all the pixels corresponding to the nucleus.
3. RESULTS AND DISCUSSION
A number of studies have shown that biodegradable polymeric nanoparticles can serve a useful purpose in enhancing tissue-specific and intracellular drug delivery upon systemic administration.26–29 Using PLGA and PCL blend nanoparticles, we have compared cellular uptake and intracellular availability of non-targeted and EGFR-targeted nanoparticles in SKOV3 human ovarian adenocarcinoma cells. Our previous studies26 have demonstrated the encapsulation capabilities a hydrophobic chemotherapeutic within the polymeric nanoparticles, when administered to both breast and ovarian cancer cells both in vitro and in vivo in tumor xenograft models.27 Figure 1 shows the schematic representation of the synthesis of peptide-modified PEG-PLGA di-block copolymer and subsequent fabrication of the nanoparticles by blending with PCL. The final peptide-modified PEG-PLGA/PCL weight ratio was optimized to be 20:80 for encapsulation of the hydrophobic drug ceramide.27 Non-targeted nanoparticles were formulated similarly except using PEG-PLGA/PCL blend. The sizes of the control and EGFR-targeted, loaded and non-loaded nanoparticles and the surface charges are enumerated in Figure 1.
Figure 1.
Schematic representation of preparation of non-targeted and epidermal growth factor receptor (EGFR)-targeted nanoparticles. The drug load utilized for the experiments is D11-C-6-ceramide, which is depicted in the schematic as well. The Table below is enumerating the size and zeta potentials of the two systems utilized.
Subsequent to establishing a consistent protocol for targeted and non-targeted nanoparticle formulations, we have encapsulated a hydrophobic drug – C6-ceramide – within the particle core. The physicochemical characteristics of the loaded nano-formulation are also annotated in Figure 1. In lieu of attaching a fluorophore onto the C6-ceramide side-chain, or implementing an NBD-labeled ceramide, we have modified C6-ceramide via isotopic labeling, deuterating the drug’s side chain (D11-C6-ceramide). This does not significantly alter the physicochemical properties of the drug as long as the deuterium is covalently bonded, nor does it affect encapsulation or intracellular biodistribution. This modification, however, enhances dramatically the sensitivity of Raman micro-spectral analysis to the payload.
Chemical signatures of organic systems fall into two distinct spectral regions in Raman spectroscopy, namely the fingerprint region (750 – 1800 cm−1) as well as the C–H stretching region (2800 – 3100 cm−1). Thus, the loaded and non-loaded polymeric nanoparticles analyzed exhibit their distinct vibrational fingerprints within the aforementioned spectral ranges. In order to establish the chemical signatures of the nanoparticulates, the nanoparticle suspensions were dried on CaF2 slides and subsequently analyzed using Raman micro-spectroscopy. The annotated spectra for the two characterized formulations are shown in Figure 2. The spectra for the non-targeted PEG-PLGA/PCL nanoparticles as well as for the EGFR-targeted PEG-PLGA/PCL nanoparticles show very similar signatures. This is due to their variability solely in the minute concentrations of the EGFR-targeting peptide, which is a 17-mer peptide used only in a 20% (w/w) concentration. Spectral signatures of the major components of the nanoparticles can be seen in Supplemental Figure 1.
Figure 2.
Raman spectra of the non-targeted and EGFR-targeted PLGA-PEG-PCL nanoparticles along with their D-11-C6-loaded counterparts. Major peaks regarding the nanoparticle components are identified: 1730 cm−1 – ester stretching peaks due to PCL, which is the dominant polymeric component; peaks between 2000 and 2300 cm−1 are all due to stretching frequency of C–D, specific to the D11-C6-ceramide; 2620 and 2950 cm−1 – peaks within the C–H stretching region pertaining to PCL and PLGA, respectively.
As is mentioned above, the encapsulated C6-ceramide had to be slightly modified for spectroscopic analysis in order to easily distinguish the signatures of the drug within the biochemical environment of the cell spectroscopically. The spectra collected of the D11-C6-ceramide encapsulated in each of the polymeric systems show a similar spectral profile within the fingerprint and the C–H stretching region (Figure 2). In particular, the carbon-deuterium (C–D) stretching vibrations are easily distinguished within the spectral region of 2000 – 2300 cm−1, which is barren of any inherently biological signal.
PCL is the major polymer within the nanoparticle formulation [at 80% (w/w)] and thus, its spectral signatures dominate those of the polymer grafts (PLGA-PEG and PLGA-PEG-EGFR). Thus, nanoparticle-characteristic peaks are observed at 1730 cm−1 (due to the C=O stretching vibrations, specific to the ester linkages of PCL) as well as the 2920 cm−1 (due to the C–H stretching vibrations), and are seen in Figure 2. As can be seen from the mentioned figure, however, biocompatible and biodegradable polymers for nanoparticle preparations exhibit characteristic peak intensities within the same spectral range as biological systems. This results in an overlap of the vibrational frequencies of the biological material with the signal coming from exogenous polymeric systems. Hence, biochemical contrast must be achieved in order to easily distinguish the exogenous systems within any biological specimen. We utilize a multivariate statistical method of analysis, namely Vertex Component Analysis (VCA), in order to discriminate the spectral signatures of the cellular components and the nanoparticles. Utility of VCA lies in visualization of distribution of chemical components within cells.
VCA is a mathematical algorithm, which “unmixes” the spectra within the dataset, based on the principle that biochemical spectra are comprised of “pure” component spectra. Hence, \VCA is a multivariate color decomposition method, which decomposes the spectrum of each pixel into contributions from the most dissimilar spectra within the dataset, which are ultimately considered the “pure component” or “end-member” spectra. Since Raman spectral datasets of a biological sample may be comprised of upward of 10,000 individual pixel-spectra, VCA also renders the hyper-spectral datasets more manageable, reducing their dimensionality. The output of the VCA algorithm is an abundance map for each of the end-members; the number of end-members is typically between three and six. An RGB(Y) overlay of these monochrome images yields a pseudo-color map with mixed colors indicating the intensity values of each of the three (or four) biochemical components; the resulting images contain enormous amounts of biochemical information regarding the sample.
For the investigations of uptake patterns of targeted and non-targeted nanoparticles, we have utilized SKOV-3 ovarian cancer cell line. The SKOV-3 cells were cultured directly on CaF2 slides, which is the preferred substrate for Raman imaging because it avoids Raman signals associated with glass slides. The cells were incubated with nanoparticle-containing media for various time intervals, in order to establish dynamics of the internalization patterns of the two nano-assemblies - PLGA-PEG/PCL and EGFR-targeted PLGA-PEG/PCL assemblies. The resulting hyper-spectral datasets were subsequently analyzed using VCA.
Six distinct time points (15 min, 30 min, 1, 2, 3 and 6 hrs) were utilized in order to verify via fluorescence imaging the penetration kinetics of the two polymeric nanoparticle systems (data not shown). We have previously published data demonstrating that the EGFR-targeted nanoparticles accumulate in greater quantities within the cytosol, versus the non-targeted nanoparticles.17 The uptake kinetics of the two systems we also found to be very distinct, where the fluorescence signal of the EGFR-targeted nanoparticles was seen after 15 min of incubation, surpassing that of non-targeted particulates.
However, the major disadvantage, of fluorescence imaging is the limitation of functional information regarding the biological samples. In order to view any specimen, it must be labeled with a specific fluorophore, which implies that some basic information regarding the biochemical properties of systems must be known prior to labeling. This poses a challenge to collecting biochemical data regarding penetration kinetics and dynamics of various drugcarriers, along with their payload dissociation patterns. The most widely utilized experimental procedure involves encapsulation of fluorophores into the nano-assembly. However, localization of the fluorophore’s intracellular compartmentalization poses a great challenge. Fluorescence from rhodamine, a commonly utilized fluorophore that is loaded into the nanoparticle core, is easily detected within the cell, however, its signal is registered from voxels throughout the cytosol. This implies the possibility of Rhodamine molecules leaching out of the polymeric nano-structure. Hence integrity of intracellular localization of distinct nanoparticles may be compromised.
Thus, we report Raman micro-spectral imaging data analysis of cells incubated cultures with targeted and non-targeted nanoparticles to complement the fluorescence data. Raman micro-spectral image analysis rendered four most distinct spectral features within the biological sample: signatures characteristic of nuclear and cytoplasmic proteins and DNA/RNA signal are shown in blue; membrane-rich organelles such as the mitochondria and the Golgi apparatus are shown in green; intracellular structures rich in lipids and phospholipids, inherent to the SKOV-3 cell line are depicted in yellow; while the nanoparticle-characteristic signal is seen in red.
Cells at the earliest time points, namely the 15, 30 min and 1 hr incubation periods, did not exhibit any distinguishable nanoparticle-characteristic peak intensities within the hyper-spectral dataset. This is seen in Figure 3, from the lack of red intracellular inclusions, which depict the signal of the internalized nanostructures. This small discrepancy with the fluorescence data, however, may be due to the higher sensitivity of fluorescence detection due to generated high intensity signal of the fluorophore utilized. Although, it is not clear whether or not the fluorescent signal is from free fluorophores released from nanoparticles detected within the cytosol, or rather from distinct internalized nanoparticles, depicting its localization.
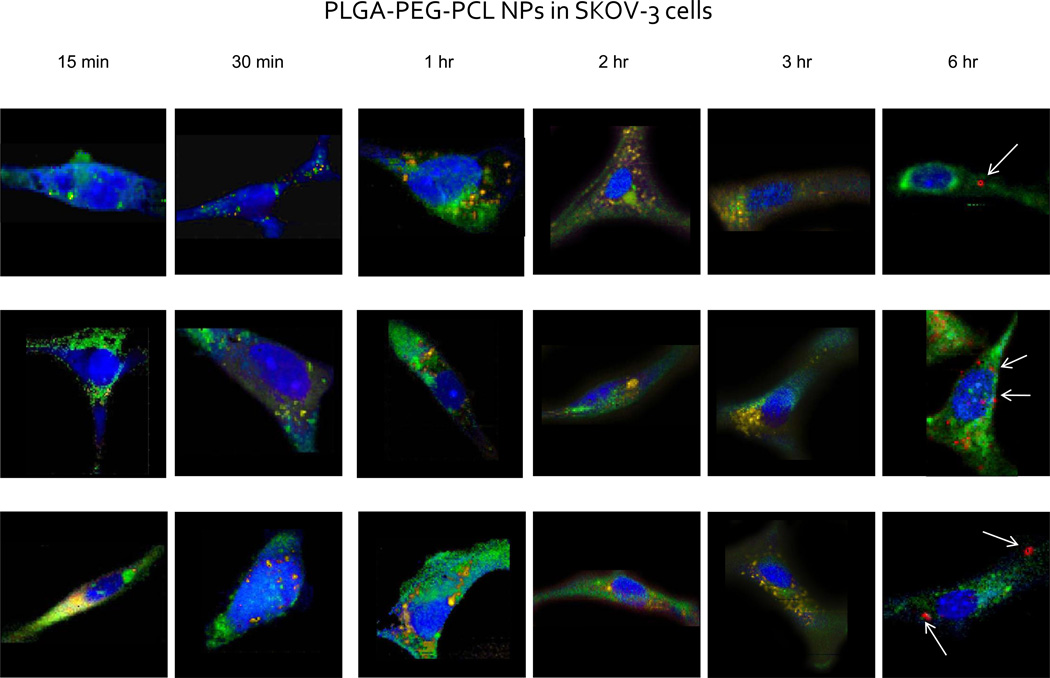
Figure 3.
Raman images of single SKOV-3 cells incubated with non-targeted PLGA-PEG-PCL nanoparticles. Single cells were imaged using a confocal Raman micro-spectral system and analyzed using VCA. Pseudo-colored images represent: cell body and nuclei – blue; membrane-rich organelles – green; early endocytic vesicles – yellow; nanoparticles – red. Arrows point out points of intracellular aggregation of the polymeric system.
After an incubation period of three hours, however, a small percentage of the cell population demonstrates spectral changes within the C–H stretching region as well as shifting of the ester peak within the fingerprint region of the spectrum (spectra not shown). However, the changes are too minute for their visual detection within any one spectrum alone, within the overwhelming amount of biochemical information. Hence, upon implementation of the VCA algorithm, onset of chemical discrimination patterns in a form of cellular response by nanoparticle accumulation sets a threshold for the onset of nanoparticle internalization.
Longer time points of nanoparticle incubation (e.g., around 6 hours), resulted in visible biochemical changes within the cell, as can be seen in Figure 3. Green and yellow intensity variations within the image still delineate the mitochondria-rich regions as well as phospholipid inclusions, respectively. However, some of the phospholipid-rich clusters begin to carry a signal due to a small amount of internalized nanoparticles therein. The red intensity clusters, on the other hand, exhibit strong signal unique to the PLGA-PEG-PCL nanoparticles. This is corroborated with the biochemical analysis of the pure PCL and PLGA nanoparticles where the internalization, translocation, and degradation dynamics of the two polymers varied drastically.24,28,30
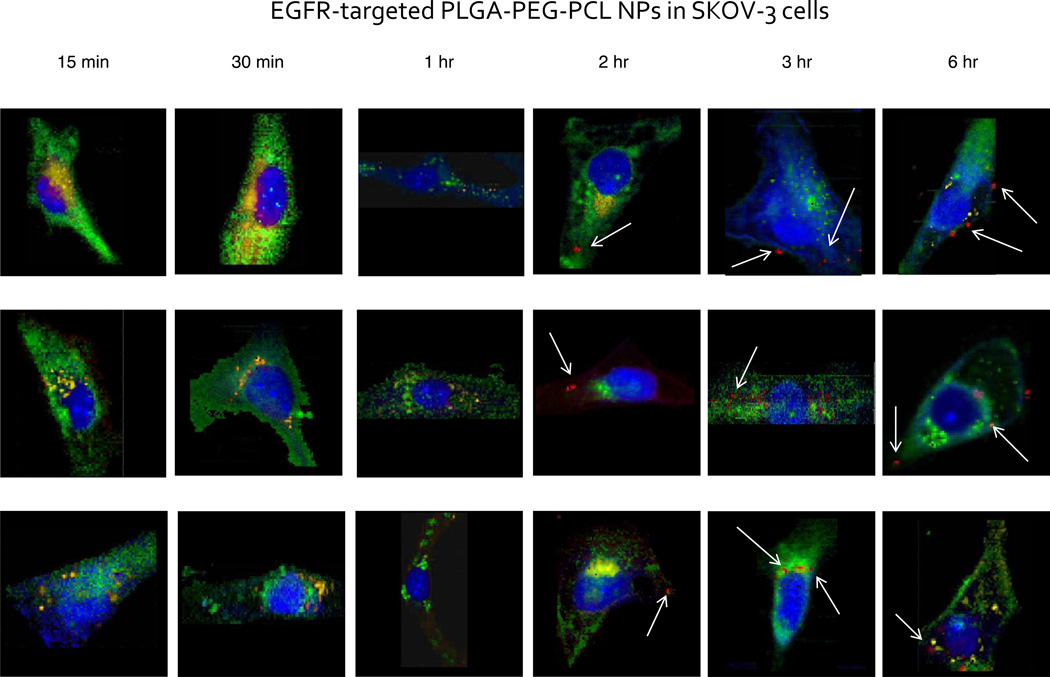
Subsequent EGFR targeting of the PEG-PLGA/PCL blend nanoparticles to the epidermal growth factor receptor via a short peptide chain linked to PLGA-PEG polymer yields dramatic improvement to internalization kinetics of the nanoparticle, as can be seen in Figure 4. Earliest time points do not show an observable accumulation of the targeted nanoparticles within the cell body. However, after 1 hour incubation period, some cells exhibit biochemical signal within the vesicular compartments of the cell body related to a small concentration of internalized targeted nanoparticles (spectral data not shown). This may be visualized by the individual spectrum taken from a particular intracellular inclusion.
Figure 4.
Raman images of single SKOV-3 cells incubated with EGFR-targeted PLGA-PEG-PCL nanoparticles over the specified time periods. Single cells were imaged using a confocal Raman micro-spectral system and analyzed using VCA. Pseudo-colored images represent: cell body and nuclei – blue; membrane-rich organelles – green; early endocytic vesicles – yellow; nanoparticles – red. Arrows point out points of intracellular aggregation of the targeted polymeric system.
After 2 hour incubation, saturation of cells by the EGFR-targeted nanoparticles is reached, since the number of nanoparticles within the cytosol reaches a constant level. This can be deduced from the constant number of red inclusions present within the cell body as is further seen in Figure 4. At a 6 hour incubation time point, the nanoparticle-characteristic signal is still distinguishable within the sub-cellular architecture. The constant level of the sequestered nanoparticles within the cell shows evidence of a threshold of tolerable amounts of intracellular nanoparticle accumulation. From 1 hour to 6 hours incubation of the cell culture with the targeted nanoparticles, it is also evident that they tend to aggregate within the perinuclear region more so than the non-targeted nanoparticles.
Distinct kinetics of the two formulations of nanoparticles inadvertently demonstrates discrete rates for cellular penetration, as has been previously noted in the literature.31,32 Non-targeted PLGA-PEG, PCL polymer blend has been previously established to be internalized into cells via non-specific endocytosis. The EGFR-targeted nanoparticles, on the other hand, tend to follow cellular uptake similar to the endogenous EGFR substrate. Previous studies conducted on the EGFR-internalization pathway have demonstrated that upon binding of the endogenous substrate to the EGFR complex following external activation (i.e. radiation), EGFR-bound substrate is internalized via processes inherent to protein tyrosine kinase. This phenomenon is quite rapid, occurring within 10–20 minutes upon stimulation. Thus, it has been speculated, as the EGFR-targeted nanoparticles harbor in essence an extension to the EGFR peptide, the mechanism of their internalization must follow the similar patterns to the receptor/substrate internalization pathway.
Spectroscopic detection of the polymeric nanostructures this early within the cellular milieu does present a difficulty, solely due to the densely packed intra-cellular matrix, concentrations of which overwhelm the backscattering information collected by the CCD. This is due to the proximity of the spectral fingerprint of the nanoparticles. However, as the ultimate goal is to implement this nanoparticle in a clinical setting, we have utilized polymeric nanostructures precisely due to their biodegradable and biocompatible nature. Raman spectroscopy has the potential to detect populated regions of sequestered nanostructures, which occurs after an hour of incubation with the nanoparticle-rich media. The algorithm presented here shows great promise in decomposing the data enough to get a glimpse into the dynamics of the nanostructures without the use of various bulky labels. Hence this technique is the one of the few, which directly probes the chemistry of the system-of-interest without indirect or surrogate measurements. The following studies conducted with loaded nanoparticulates would demonstrate the detection sensitivity of our instrumentation to the dissociated therapeutic cargo, due to its distinct spectral signatures.
Incubation of cell cultures with D11-C6-ceramide-loaded nanoparticles underscores the previously established kinetics of the two formulations. Furthermore, C6-ceramide is known to have an affinity for accumulation within the mitochondria, which we have analyzed using label-free Raman imaging, corroborating the findings with conventional fluorescence staining. We have thus counterstained the nanoparticle-incubated cell cultures with a Mitotracker dye, in order to verify the accumulation propensity of the drug therein. Figures 5 and 6 demonstrate the acquired results for both the targeted and the non-targeted nano-formulations. For ease of visualization, the cell body and nucleus are pseudo-colored blue, mitochondrial aggregations as well as intracellular inclusions are delineated in green, while the red intensities pertain to the intracellular D11-C6-ceramide.
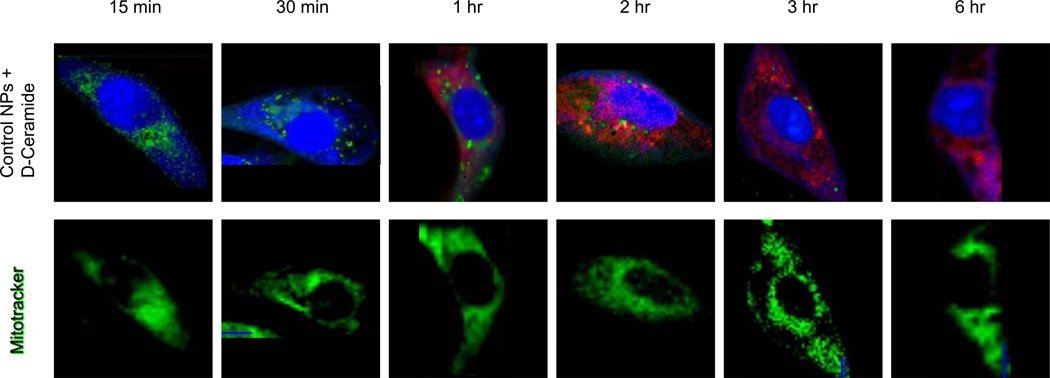
Figure 5.
Top row: Raman images of single SKOV-3 cells incubated with non-targeted PLGA-PEG-PCL nanoparticles loaded with D11-C6-ceramide, over enumerated time periods. Single cells were imaged using a confocal Raman micro-spectral system and analyzed using VCA. Pseudo-colored images represent: cell body and nuclei – blue; membrane-rich organelles and vesicles – green; D11-C6-ceramide – red. Bottom row: corresponding cells counterstained after Raman data acquisition with Mitotracker Green. Same microscope setup was used to analyze fluorescence stain, at the same focal plane.
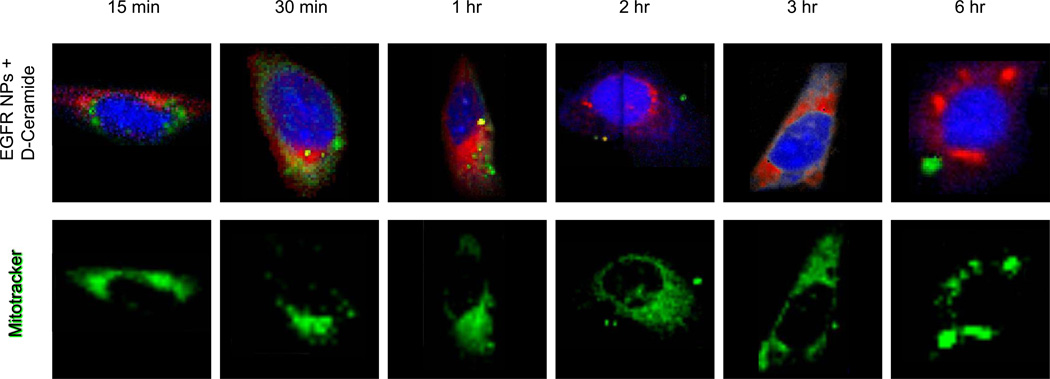
Figure 6.
Top row: Raman images of single SKOV-3 cells incubated with EGFR-targeted PLGA-PEG-PCL nanoparticles loaded with D11-C6-ceramide, over enumerated time periods. Single cells were imaged using a confocal Raman micro-spectral system and analyzed using VCA. Pseudo-colored images represent: cell body and nuclei – blue; membrane-rich organelles and vesicles – green; D11-C6-ceramide – red. Bottom row: corresponding cells counterstained after Raman data acquisition with Mitotracker Green. Same microscope setup was used to analyze fluorescence stain, at the same focal plane.
Although single polymeric nanoparticles are not easily distinguished within the sub-cellular matrix, as their signal is masked and overwhelmed by the stronger and more abundant intensities of various cellular components, sensitivity of Raman micro-spectral imaging to isotopically labeled D11-C6-cearmide is evident. Non-targeted polymeric nanoparticles are not easily detected within the sub-cellular environment before at least a 3 hour incubation period (Figure 3), however, the drug is seen to permeate the cell within a 1 hour incubation (Figure 5, top row, red intensities). This phenomenon is suggestive of almost single nanoparticle penetration into the cellular matrix and delivery of the cargo therein, since Raman is very sensitive to the C–D stretching intensities, which are evidently present at a 1 hour time point. The concentration of the drug within a single nanoparticle, although minute when compared to the signal of the cellular components, is still detectable within the sea of sub-cellular constituents. This also points to the fast kinetics of nanoparticle internalization and subsequent release of the cargo within the cellular milieu, within minutes of penetration. Drug release can be visualized from its translocation into the mitochondria-rich cellular regions, as opposed to tightly packed clusters of sequestered nanoparticles.
As we analyze the affinity of the drug to the sub-cellular compartments it becomes evident that C6-ceramide accumulates generally within the mitochondria. This is seen from the Mitotracker dye, with which we have counterstained the same cells. Figure 5 clearly demonstrates that the drug-specific intracellular regions (red intensities, top row) are superimposable with the mitochondria (green intensities, bottom row). Intracellular distribution profile of the drug is also noteworthy throughout the time-course experiment, where mitochondrial aggregations become detectable. Following the increasing intracellular accumulation of C6-ceramide, mitochondria begin to migrate and aggregate within the cytosol, as is seen from the clusters of green intensities of the Mitotracker dye from a 15 min to a 6 hr incubation period. Intensities due to the drug concentration within the cell are following a synergistic pattern, further underscoring their direct association with the mitochondria.
Incubation of the cell culture with the D11-C6-ceramide-loaded EGFR-targeted nanoparticles reveals a similar distribution pattern, except with much faster kinetics. After a mere 15 minutes the signal of the drug is already seen dispersed within the intracellular matrix (Figure 6, bright red intracellular regions in top row). Moreover, it is seen to co-localize with the mitochondria, as can be observed from the bright red and green intensities of the drug and mitochondria, respectively.
Spectral intensities render information regarding the chemical environment probed as well as a concentration profile of the sample. Intracellular concentration of D11-C6-ceramide was quantitatively examined via the spectral data generated using Raman micro-spectral imaging accompanied by statistical algorithm – VCA. Three systems were compared in their efficiency in drug delivery, namely the non-targeted polymeric nano-formulation as well as the EGFR-targeted system, with no delivery vehicle whatsoever. The cell culture was incubated with a free D11-C6-ceramide in the same concentration as included for the loading of the nano-carrier, and incubated for the same time periods as with the delivery systems for comparison.
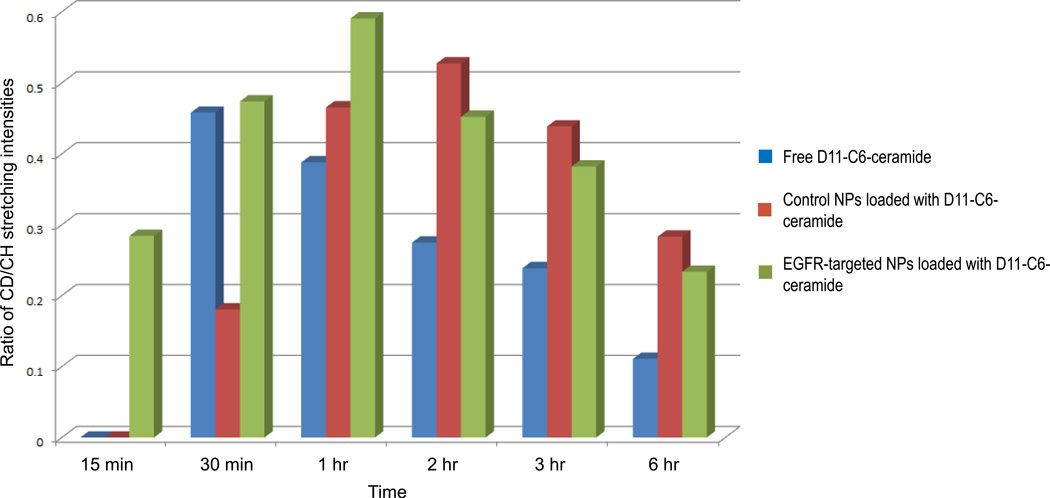
We have thus analyzed the concentration intensities of the drug within the cell, comparing them to a consistent standard, namely the nucleus of each cell. C–D stretching intensities of the drug from each cell were analyzed at every time point of the experiment, after which the number of pixels carrying information regarding presence of the drug was tabulated. Alongside this data we have quantified the consequent pixel count of the nucleus and taken that as a constant, since nuclear size of a cell within the same culture varies only slightly. The tabulated results are displayed in Figure 7.
Figure 7.
Ratios of pixel (each pixel is 500 × 500 × ~1500 nm3) intensity counts due to C–H stretching intensities of the nucleus versus pixel counts of C–D stretching intensities due to internalized D11-C6-ceramide within the mitochondria. Average of 3 cells per time point.
EGFR-targeted polymeric nano-formulation system clearly surpasses in speed and sustained delivery of the drug into the cytosol over the non-targeted and free drug administrations. Within 15 minutes of incubation, D11-C6-ceramide is seen to be associated with the mitochondria, as is seen from Figures 6 and 7, and further corroborated by the super-imposable image of the stained mitochondria (Figure 6). Intracellular accumulation of the drug via a non-targeted method of delivery mimics the accumulation patters of ceramide introduced via a targeted system. However, the internal concentration as well as the latency of its intracellular accumulation is lower than those introduced using an EGFR-targeted carrier. Administration of the free drug into the cell culture, on the other hand, does not seem to follow a Gaussian bell-curve spread of internal drug concentration over time. D11-C6-ceramide is seen to rapidly accumulate within the intracellular matrix within 30 min of incubation, at a concentration similar to the drug introduced via a targeted method of delivery. However, from that time point, ceramide concentration drastically decreases. Analysis of the data, in fact, reveals that all groups followed a similar interesting trend with respect to D11-C6-ceramide amount in cells over time. Cells display an overall increase in drug concentration over a period of 1–3 hours (depending on the carrier) followed by a gradual decrease; however, the rate of decrease is lower for nanoparticle delivery systems than for free drug. This demonstrates the sustainable delivery of the drug using a nano-delivery system, while also demonstrating faster kinetics for a targeted delivery.
Since Raman is a weak effect, larger concentrations of a weak photon scatterer are required, which incidentally in our case is the polymeric nano-formulation, rather than the dosed drug. Decreasing the administered dose would yield a more refined image in terms of uptake and distribution speed and dynamics of the free drug versus the dissociated ceramide from the delivery system.
Drug uptake kinetic constants were calculated for each carrier based on the time period during which the drug concentration in cells increases in an effort to quantify differences in uptake kinetics of the different systems. Absorption was expressed as a first order process, with a rate proportional to drug concentration in the solution in contact with the cells:
where I is the normalized pixel number, kuptake is the drug uptake kinetic constant in normalized pixel intensity/(min*µM), and C0 is the initial drug concentration (100 µM) in the solution dosed to the cells. This expression is based on the assumption that the concentration of drug in the solution dosed to the cells is approximately constant. Area under the uptake curve over 6 hours for each carrier was also estimated based on the trapezoidal rule as an indication of total exposure to D-ceramide (Table 1).
Table 1.
Calculated drug uptake parameters
| Control NPs + D-ceramide | EGFR NPs + D-ceramide | Free D-ceramide | |
|---|---|---|---|
| Uptake kinetic constant (intensity/(min*µM)) | 0.051 | 0.114 | 0.153 |
| Max drug amount (intensity) | 0.527 | 0.591 | 0.458 |
| AUC (intensity*min) | 128.89 | 141.40 | 100.74 |
The parameters of three representative dosing groups, namely the solution form of D11-C6-ceramide dosing of the cell cultures, non-targeted, and EGFR-targeted PLGA/PCL blend nanoparticles loaded with the drug. Uptake kinetic constant, maximum intracellular drug amount, as well as area-under-the-curve (AUC) for each dosing type were calculated based on the intracellular drug concentrations during the 6 distinct incubation periods.
Cells incubated with free D-ceramide showed a higher D-ceramide uptake rate compared to both EGFR-targeted and control nanoparticles (Table 1). However, maximum D-ceramide concentration achieved and AUC values were higher for nanoparticles compared to free D-ceramide, EGFR-nanoparticle being the highest. These results highlight the importance of considering kinetics of uptake processes for understanding effects of carrier systems. Ceramide is an intermediate molecule in the synthesis and degradation of sphingomyelin, gangliosides and other glycolipids in cells.33,34 Therefore, ceramide may be metabolized and degrade inside cells,35 which may explain the decrease in ceramide amount in cells over time. Higher drug concentration and AUC achieved in cells with nanoparticles compared to free drug may be due to the protection of the nanoparticles of D-ceramide against degradation in cells.
Raman micro-spectral analysis is able to distinguish not only the carrier system, but also the drug-load, as well as the responses of the biological system interrogated, without exogenous staining and labeling procedures. One such cellular response to the presence of D11-C6-ceramide is noticeable in the clustering of mitochondria throughout the cytosol. Onset of mitochondrial aggregations becomes visible following a 6 hour incubation of the cell culture with the free drug as well as with the dissociated drug from the non-targeted delivery system. The drug delivered via the EGFR-targeted nano-formulation, on the other hand, showed a more dramatic packaging and clumping of mitochondria throughout the cytosol, commencement of which is observed as early as 3 hours post incubation (Figure 6). This may be due to a much earlier presence of the drug within the mitochondria, which is known to reduce the apoptotic threshold, acting as a cytotoxic agent.
4. CONCLUSIONS
We have demonstrated the capability of spectral imaging to detect and localize polymeric nano-structures as well as their cargo in cells without the use of various fluorophores, which may leach out of the carrier system or influence their subsequent uptake. Raman micro-spectral imaging method was able to detect and corroborate the distinct mechanisms of internalization pathways of the non-targeted polymer blend nanoparticles (PLGA-PEG, PCL) with those of EGFR-targeted polymer blend (PLGA-PEG-EGFR, PCL). The EGF receptor targeted nanoparticles tend to get internalized much faster into the cell body and sequestered within large vesicular compartments in the perinuclear region or the cell periphery. This renders the delivered drug cargo to be release much earlier into the sub-cellular matrix, where it may influence cellular responses and various biochemical processes. All these distinct events may ultimately be monitored and characterized simultaneously and non-invasively using Raman micro-spectral method of sample interrogation.
Supplementary Material
Supplemental Figure 1. Spectral signatures of the main components of the nanoparticle are shown. PCL and PLGA are the main components of the nanostructure. A graft copolymer spectrum is also shown to depict the intensities due to the polymers as well as that of the peptide (1665 cm−1). The spectra of the final nanostructure formulations are also shown. Distinctions of spectral features within the C-H stretching region are shown for each polymer, as well as the ester stretching region (1735–1765 cm−1), which shows shifting of peak height and ratios of the two main polymers utilized.
ACKNOWLEDGEMENTS
This study was supported by the National Institutes of Health, National Cancer Institute’s grants R01-CA119617, R01-CA119617S1 (ARRA Supplement), and R21-CA135594 to Dr. Amiji and R01-CA090346 to Dr. Diem.
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest
REFERENCES
- 1.Amiji M. Improving targeted delivery. Drug Deliv. 2007:1–4. [Google Scholar]
- 2.Park J, Ye M, Park K. Biodegradable polymers for microencapsulation. Molecules. 2005;10:146–161. doi: 10.3390/10010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Torchilin V. Recent Approaches to Intracellular Delivery of Drugs and DNA and Organelle Targeting. Annu. Rev. Biomed. Eng. 2006;8:343–375. doi: 10.1146/annurev.bioeng.8.061505.095735. [DOI] [PubMed] [Google Scholar]
- 4.Allen T, Cullis P. Drug Delivery Systems: Entering the Mainstream. Science. 2004;303:1818–1822. doi: 10.1126/science.1095833. [DOI] [PubMed] [Google Scholar]
- 5.Uhrich K, Cannizzaro S, Langer R, Shakesheff K. Polymeric Systems for Controlled Drug Release. Chem. Rev. 1999;99:3181–3198. doi: 10.1021/cr940351u. [DOI] [PubMed] [Google Scholar]
- 6.Torchilin VP. Micellar Nanocarriers: Pharmaceutical Perspectives. Pharm Res. 2006;24:1–16. doi: 10.1007/s11095-006-9132-0. [DOI] [PubMed] [Google Scholar]
- 7.Derycke A. Liposomes for photodynamic therapy. Adv Drug Deliv. Rev. 2004;56:17–30. doi: 10.1016/j.addr.2003.07.014. [DOI] [PubMed] [Google Scholar]
- 8.Lasic D, Templeton N. Liposomes in gene therapy. Adv. Drug Deliv. Rev. 1996;20:221–266. [Google Scholar]
- 9.Torchilin V. Structure and design of polymeric delivery systems. J. Control Rel. 2001;73:137–172. doi: 10.1016/s0168-3659(01)00299-1. [DOI] [PubMed] [Google Scholar]
- 10.Kell D, Dobson P. The Cellular Uptake of Pharmaceutical Drugs is Mainly Carrier-mediated and is thus an Issue not so Much of Biophysics but of Systems Biology. Systems Chem. 2008:149–168. [Google Scholar]
- 11.Faraji A, Wipf P. Nanoparticles in cellular drug delivery. Bioorganic & Medicinal Chem. 2009;17:2950–2962. doi: 10.1016/j.bmc.2009.02.043. [DOI] [PubMed] [Google Scholar]
- 12.Singhal S, Nie S, Wang M. Nanotechnology Applications in Surgical Oncology. Annu. Rev. Med. 2010;61:359–373. doi: 10.1146/annurev.med.60.052907.094936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dobson P, Kell D. Carrier-mediated cellular uptake of pharmaceutical drugs: an exception or the rule? Nature Rev. 2008;7:205–220. doi: 10.1038/nrd2438. [DOI] [PubMed] [Google Scholar]
- 14.Wu J, Sawa T, Matsumura Y, Hori K, Maeda H. Tumor vascular permeability and the EPR effect in macromolecular therapeutics: a review. J Control Rel. 2000;65:271–284. doi: 10.1016/s0168-3659(99)00248-5. [DOI] [PubMed] [Google Scholar]
- 15.Muro S, Koval M, Muzykantov V. Endothelial Endocytic Pathways: Gates for Vascular Drug Delivery. Current Vascular Pharmacology. 2004;2:281–299. doi: 10.2174/1570161043385736. [DOI] [PubMed] [Google Scholar]
- 16.Smith G, Petrenko V. Phage display. Chemical Reviews. 1997;97:391–410. doi: 10.1021/cr960065d. [DOI] [PubMed] [Google Scholar]
- 17.Naumann D. FT-infrared and FT-Raman spectroscopy in biomedical research. Applied Spectroscopy Reviews. 2001;36:239–298. [Google Scholar]
- 18.Wartewig S, Neubert R. Pharmaceutical applications of Mid-IR and Raman spectroscopy. Adv. Drug Deliv. Rev. 2005;57:1144–1170. doi: 10.1016/j.addr.2005.01.022. [DOI] [PubMed] [Google Scholar]
- 19.Nijssen A, Koljenović S, Schut TCB, Caspers PJ, Puppels GJ. Towards oncological application of Raman spectroscopy. J Biophoton. 2009;2:29–36. doi: 10.1002/jbio.200810055. [DOI] [PubMed] [Google Scholar]
- 20.Pysz M, Gambhir S, Willmann J. Molecular imaging: current status and emerging strategies. Clinical Radiology. 2010;65:500–516. doi: 10.1016/j.crad.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chernenko T, Matthäus C, Milane L, Quintero L, Amiji M, Diem M. Label-Free Raman Spectral Imaging of Intracellular Delivery and Degradation of Polymeric Nanoparticle Systems. ACS Nano. 2009;3:3552–3559. doi: 10.1021/nn9010973. [DOI] [PubMed] [Google Scholar]
- 22.Krafft C, Dietzek B, Popp J. Raman and CARS microspectroscopy of cells and tissues. Analyst. 2009;134:1046. doi: 10.1039/b822354h. [DOI] [PubMed] [Google Scholar]
- 23.Miljković M, Chernenko T, Romeo M, Bird B, Matthäus C. Diem., Label-free imaging of human cells: algorithms for image reconstruction of Raman hyperspectral datasets. Analyst. 2010;135:2002. doi: 10.1039/c0an00042f. [DOI] [PubMed] [Google Scholar]
- 24.Chernenko T, Matthäus C, Milane L, Quintero L, Amiji M, Diem M. Label-Free Raman Spectral Imaging of Intracellular Delivery and Degradation of Polymeric Nanoparticle Systems. ACS Nano. 2009;3:3552–3559. doi: 10.1021/nn9010973. [DOI] [PubMed] [Google Scholar]
- 25.Nascimento J, Dias J. Vertex component analysis: a fast algorithm to unmix hyperspectral data. IEEE Trans. Geosci. Remote Sensing. 2005;43:898–910. [Google Scholar]
- 26.Baselga J. Critical Update and Emerging Trends in Epidermal Growth Factor Receptor Targeting in Cancer. Journal of Clinical Oncology. 2005;23:2445–2459. doi: 10.1200/JCO.2005.11.890. [DOI] [PubMed] [Google Scholar]
- 27.Ono M, Kuwano M. Molecular Mechanisms of Epidermal Growth Factor Receptor (EGFR) Activation and Response to Gefitinib and Other EGFR-Targeting Drugs. Clinical Cancer Research. 2006;12:7242–7251. doi: 10.1158/1078-0432.CCR-06-0646. [DOI] [PubMed] [Google Scholar]
- 28.Chawla J, Amiji M. Biodegradable poly(epsilon-caprolactone) nanoparticles for tumor-targeted delivery of tamoxifen. 2007;249:127–138. doi: 10.1016/s0378-5173(02)00483-0. [DOI] [PubMed] [Google Scholar]
- 29.Jabr-Milane L, van Vlerken L, Yadav S, Amiji M. Multi-functional nanocarriers for targeted delivery of drugs and genes. Journal of Controlled Release. 2008;130:121–128. doi: 10.1016/j.jconrel.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 30.Milane L, Duan Z, Amiji M. Development of EGFR-Targeted Polymer Blend Nanocarriers for Combination Paclitaxel/Lonidamine Delivery To Treat Multi-Drug Resistance in Human Breast and Ovarian Tumor. Cells. Mol. Pharmaceutics. 2011;8:185–203. doi: 10.1021/mp1002653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bei J, Wang S, Chen D. Polycaprolactone microparticles and their biodegradation. Polymer Degradation and Stability. 2000;67:455–459. [Google Scholar]
- 32.Oh K, Yin H, Lee E, Bae Y. Polymeric nanovehicles for anticancer drugs with triggering release mechanisms. J. Mater. Chem. 2007;17:3987. [Google Scholar]
- 33.van Apeldoorn A, van Manen H, Bezemer J, de Bruijn J, van Blitterswijk C, Otto C. Raman Imaging of PLGA Microsphere Degradation Inside Macrophages. J. Am. Chem. Soc. 2004;126:13226–13227. doi: 10.1021/ja0459936. [DOI] [PubMed] [Google Scholar]
- 34.Nascimento J, Dias J. Vertex component analysis: a fast algorithm to unmix hyperspectral data. IEEE Transactions on Geoscience and Remote Sensing. 2005;43:898–910. [Google Scholar]
- 35.Li S. Hydrolytic Degradation Characteristics of Aliphatic Polyesters Derived from Lactic Glycolic Acids. J. Biomed. Mater. Res. Part B: Applied Biomaterials. 1999;48:342–353. doi: 10.1002/(sici)1097-4636(1999)48:3<342::aid-jbm20>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 36.Orth J. A Novel Endocytic Mechanism of Epidermal Growth Factor Receptor Sequestration and Internalization. Cancer Research. 2006;66:3603–3610. doi: 10.1158/0008-5472.CAN-05-2916. [DOI] [PubMed] [Google Scholar]
- 37.Dittmann K, Mayer C, Kehlbach R, Rodemann H. Radiation-induced caveolin-1 associated EGFR internalization is linked with nuclear EGFR transport and activation of DNA-PK. Mol Cancer. 2008;7:69. doi: 10.1186/1476-4598-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Spectral signatures of the main components of the nanoparticle are shown. PCL and PLGA are the main components of the nanostructure. A graft copolymer spectrum is also shown to depict the intensities due to the polymers as well as that of the peptide (1665 cm−1). The spectra of the final nanostructure formulations are also shown. Distinctions of spectral features within the C-H stretching region are shown for each polymer, as well as the ester stretching region (1735–1765 cm−1), which shows shifting of peak height and ratios of the two main polymers utilized.