Summary
Contrary to the standard of eliminating antimicrobial hits that collapse bacterial proton motive force (PMF), in this issue of Chemistry and Biology, Farha et al., describe the value of screens to identify molecules that dissipate PMF, yet are non-bacteriolytic and selectively toxic.
The urgent need to discover and develop novel antimicrobial agents is underscored by the rapid spread of bacterial pathogens exhibiting resistance to most classes of clinically used antibiotics. Yet the indispensable bacterial membrane that houses several essential proteins and the prokaryotic respiratory chain remain underexploited for discovering new antibiotics(Hurdle et al., 2011). It is well accepted that newly discovered antimicrobial chemotypes should be efficacious against multi-drug resistant organisms, whilst avoiding cytotoxicity or adverse toxicity in animals to warrant progression as clinical candidates(O’Neill and Chopra, 2004). In this regard, chemical libraries of natural and synthetic origins have been extensively screened by academic groups and pharmaceutical companies to find new antibiotic chemotypes. Such screens routinely identify hits with potent antibacterial activity, but often result in the disturbance of the functional integrity and associated bioenergetics of the prokaryotic membrane(Payne et al., 2007). These hits include molecules that inhibit the bacterial respiratory chain, lyse or induce curvature of the cytoplasmic membrane or possess a mode of action that is challenging to elucidate, such as molecules that co-interact with the membrane and peptidoglycan components or membrane embedded proteins (Figure 1)(Hurdle et al., 2011; Pogliano et al., 2012). Many of these hits contain lipophilic moieties and also cause damage to mammalian membranes or display cytotoxicity, making them undesirable for progression. Therefore, an ear-consensus has emerged in the antibiotic discovery field that such molecules are nuisances that should be removed early in the drug discovery process, to avoid downstream toxicity problems in mammals(O’Neill and Chopra, 2004; Payne et al., 2007). Hence counter-screening assays have been exploited to eliminate hits disrupting the functional properties of the membrane. Whilst it is certainly pertinent to eliminate molecules that are cytotoxic and cause leakage of the prokaryotic cytosol, that could induce septicemia, it is now apparent that not all molecules targeting the functional properties of the bacterial membrane are undesirable or even lack a therapeutic index for safe use in humans(Hurdle et al., 2011). Indeed, within the last decade, we have seen: the clinical development of drugs daptomycin and telavancin whose potency in part arise from membrane interaction; the ongoing clinical development of membrane-active agents such as HT61 and XF73 for nasal decolonization of methicillin-resistant Staphylococcus aureus; and the emergence of the respiratory chain as a leading drug target in Mycobacterium tuberculosis(Hurdle et al., 2011; Koul et al., 2011).As demonstrated with daptomycin, some antimicrobial peptides and other membrane-active molecules, selective killing of bacteria can be achieved by exploiting the major differences in the lipid compositions between prokaryotic and mammalian membranes (Hurdle et al., 2011; Verkleij et al., 1973). These examples therefore challenge the traditional view that the membrane is an unsuitable target. Thus, in this issue, the study by Farha et al.,(2013) is valiant, and against the mainstream dogma, in that it seeks to prioritize rather than eliminate membrane-active hit molecules. This may well prompt revisiting such molecules that have been discarded because they affect the bacterial membrane properties.
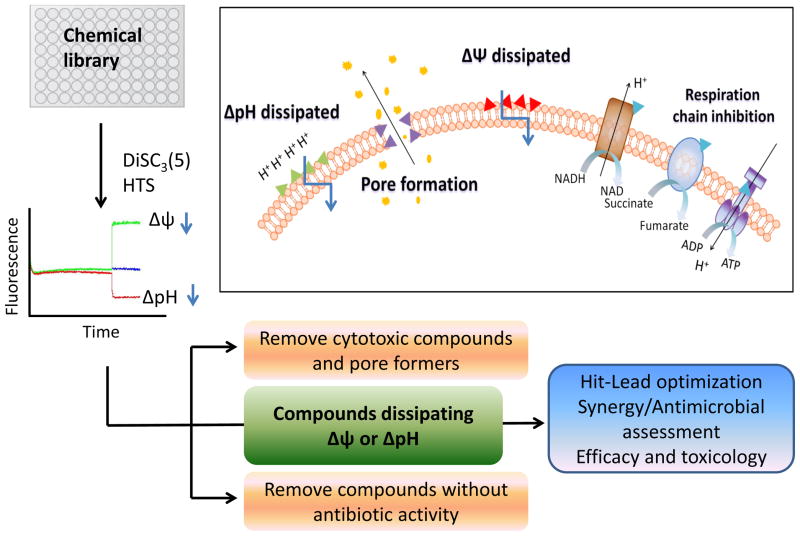
Figure 1.
Screening and preclinical path for hits dissipating the membrane’s electrical potential (Δψ) or proton gradient (ΔpH). The inset simplifies the membrane and respiratory chain as drug target sites; not shown is the peptidoglycan and unknown membrane-embedded proteins that could interact with membrane-active agents. The DiSC3(5) florescence data in the graph was derived from Farha et al., (2013).
In their strategy Farha et al., (2013) optimized the standard assay that adopts the membrane-potential sensitive dye 3,5-dipropylthiacarbocyanine-DISC3(5)- to screen for molecules that either dissipates the electrical potential (Δψ) or the proton gradient (ΔpH) of the S. aureus membrane (Figure 1). Paradoxically, DiSC3(5) assays have been exploited by others to remove membrane-active molecules(Gentry et al., 2010). Both Δψ and ΔpH are the two components of the cellular proton motive force (PMF) that is intricate to bioenergetics, solute transport, motility and maintaining the intracellular pH for protein function(Krulwich et al., 2011). By screening a modest bioactive library (30,000 compounds) the authors recovered 72 compounds appearing as dissipators of the Δψ and 272 appearing as dissipators of the ΔpH. Importantly, follow-up dose response assays and control assays were performed to remove false hits that mimic the florescence of DiSC3(5) or quenchits florescence in a manner that is independent of changes in the PMF. Consequently, three hits were identified and confirmed as specific dissipators of the Δψ and three as ΔpH dissipators. Most of these hits only induced cytotoxicity at concentrations above their minimum inhibitory concentration. Also, as expected, combinations of Δψ dissipators with ΔpH dissipators were synergistic against S. aureus, as this causes the complete collapse of the PMF. Therefore, the authors point to synergistic combinations of Δψ and ΔpH dissipators as a means of dose sparing that lowers the cytotoxicity of individual compounds, whilst retaining potency against bacteria(Farha et al., 2013). Although this is a comprehensive concept, its application to systemic infections is challenged by the need to match the pharmacology of the compounds to active drug in sufficient concentrations at the site of infection. Probably, it may more readily apply to treating topical diseases such as staphylococcal skin infections. It is also worth noting thatDiSC3(5)response assays does not provide adequate insight to the biochemical or molecular mechanism responsible for perturbation of the membrane’s properties. Hence suitable hits will need to undergo detailed studies to understand their action at the membrane target site. For example, we only now understand that the interaction of daptomycin with the membrane does not only dissipate Δψ, but it dramatically affects peptidoglycan biosynthesis and assembly of cell division proteins(Pogliano et al., 2012).
With the challenges of identifying antibiotic leads, the propensity of bacteria to develop resistance and the difficulty of treating dormant bacterial infections, membrane-active that are bactericidal and less prone to resistance selection may become more attractive. The work by Farha et al., (2013) therefore encourages further debate on the utility of membrane-active hits. As this area expands a number a pertinent questions will need to be addressed to harness the benefits of these types of molecules. Certainly, it will be vital to address structure-activity relationships required to attain selectivity and whether interaction with prokaryote specific components in the cell envelope could render compounds more selective. A need also exist to extend this concept to Gram-negative pathogens for which there is a desperate need for new antibiotics; we speculate this will be most challenging for lipophilic species that are precluded from reaching the cytoplasmic membrane by the lipopolysaccharide layer. In closing, although chemical libraries contain several nuisance compounds, they also contain membrane-active molecules, with some selectivity, which could be polished by medicinal chemistry to produce clinical candidates that could be considered diamonds recovered from the rough.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Farha MA, Verschoor CP, Bowdish D, Brown ED. Collapsing the proton motive force to identify synergistic combinations against Staphylococcusaureus. Chem Biol. 2013;20:1–11. doi: 10.1016/j.chembiol.2013.07.006. [DOI] [PubMed] [Google Scholar]
- Gentry DR, Wilding I, Johnson JM, Chen D, Remlinger K, Richards C, Neill S, Zalacain M, Rittenhouse SF, Gwynn MN. A rapid microtiter plate assay for measuring the effect of compounds on Staphylococcusaureus membrane potential. J Microbiol Methods. 2010;83:254–256. doi: 10.1016/j.mimet.2010.08.012. [DOI] [PubMed] [Google Scholar]
- Hurdle JG, O’Neill AJ, Chopra I, Lee RE. Targeting bacterial membrane function: an underexploited mechanism for treating persistent infections. Nat Rev Microbiol. 2011;9:62–75. doi: 10.1038/nrmicro2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koul A, Arnoult E, Lounis N, Guillemont J, Andries K. The challenge of new drug discovery for tuberculosis. Nature. 2011;469:483–490. doi: 10.1038/nature09657. [DOI] [PubMed] [Google Scholar]
- Krulwich TA, Sachs G, Padan E. Molecular aspects of bacterial pH sensing and homeostasis. Nat Rev Microbiol. 2011;9:330–343. doi: 10.1038/nrmicro2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill AJ, Chopra I. Preclinical evaluation of novel antibacterial agents by microbiological and molecular techniques. Expert opinion on investigational drugs. 2004;13:1045–1063. doi: 10.1517/13543784.13.8.1045. [DOI] [PubMed] [Google Scholar]
- Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Discov. 2007;6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- Pogliano J, Pogliano N, Silverman JA. Daptomycin-mediated reorganization of membrane architecture causes mislocalization of essential cell division proteins. J Bacteriol. 2012;194:4494–4504. doi: 10.1128/JB.00011-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkleij AJ, Zwaal RF, Roelofsen B, Comfurius P, Kastelijn D, van Deenen LL. The asymmetric distribution of phospholipids in the human red cell membrane. A combined study using phospholipases and freeze-etch electron microscopy. Biochim Biophys Acta. 1973;323:178–193. doi: 10.1016/0005-2736(73)90143-0. [DOI] [PubMed] [Google Scholar]