Abstract
Cytoplasmic aspartyl-tRNA synthetase (AspRS) from Saccharomyces cerevisiae is a homodimer of 64 kDa subunits. Previous studies have emphasized the high sensitivity of the N-terminal region to proteolytic cleavage, leading to truncated species that have lost the first 20–70 residues but that retain enzymatic activity and dimeric structure. In this work, we demonstrate that the N-terminal extension in yeast AspRS participates in tRNA binding and we generalize this finding to eukaryotic class IIb aminoacyl-tRNA synthetases. By gel retardation studies and footprinting experiments on yeast tRNAAsp, we show that the extension, connected to the anticodon-binding module of the synthetase, contacts tRNA on the minor groove side of its anticodon stem. Sequence comparison of eukaryotic class IIb synthetases identifies a lysine-rich 11 residue sequence (29LSKKALKKLQK39 in yeast AspRS with the consensus xSKxxLKKxxK in class IIb synthetases) that is important for this binding. Direct proof of the role of this sequence comes from a mutagenesis analysis and from binding studies using the isolated peptide.
Keywords: aminoacyl-tRNA synthetase/aspartyl-tRNA synthetase/RNA-binding motif/synthetase extension
Introduction
The 20 aminoacyl-tRNA synthetases (aaRSs) have been classified into two classes, based on the structural organization of their catalytic site (Eriani et al., 1990). In addition to the class-specific catalytic domains, they contain idiosyncratic modules involved in the recognition and binding of the homologous tRNA molecules (Delarue and Moras, 1993; Moras, 1993), this recognition being limited in most cases to the acceptor and/or the anticodon branches (Schimmel, 1987; Giegé et al., 1993). Differences between prokaryotic and eukaryotic synthetases lie in the presence of eukaryote-specific extra-domains at the N- or the C-terminus of the protein, outside the structural consensus defining the two classes of synthetases. These extra-domains can be removed by controlled proteolysis and are not essential for aminoacylation activity (e.g. Lorber et al., 1988; Mirande, 1991; Agou et al., 1996). In higher eukaryotes, they are believed to participate in the multisynthetase complex formation composed of nine synthetases and three auxiliary proteins (Mirande, 1991). In lower eukaryotes, however, such complexes have not been found and no functional role could be ascribed explicitly to these appendices.
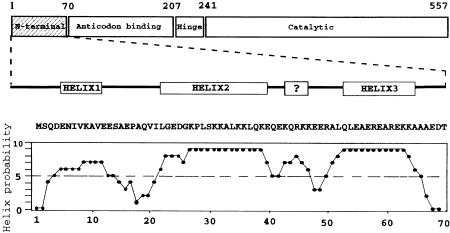
N-terminal extensions are found in eukaryotic class IIb synthetases. In yeast aspartyl-tRNA synthetase (AspRS), a representative class IIb homodimeric protein of 128 kDa, the extension reaches a length of 70 amino acids and is localized next to the anticodon module of the synthetase (Figure 1). When AspRS is purified from yeast cells, the extension is partly proteolyzed, leading to a heterogeneous mixture of molecules lacking their first 13–32 amino acids (Lorber et al., 1987). This heterogeneity hampers crystallization of the free enzyme (Lorber et al., 1987; Sauter et al., 1999) but not that of the complex with tRNA (Giegé et al., 1980; Lorber et al., 1983). In the structure of the complex (Ruff et al., 1991), however, the extension is not visible, probably as the consequence of crystalline disorder due to the heterogeneous AspRS. Since absence of the complete 70 amino acid extension does not affect the in vitro aminoacylation efficiency of the enzyme (Lorber et al., 1988) and does not inhibit yeast growth (Eriani et al., 1991), it can be concluded that the structure of the enzyme in the complex (Ruff et al., 1991) and that recently solved for the free enzyme truncated of its first 70 amino acids (Sauter et al., 2000) corresponds to the functional core of the AspRS molecule required in tRNA aminoacylation. This does not exclude, however, an ancillary role for the extension in the aminoacylation of tRNAAsp, a view supported by a wealth of indirect evidence. Indeed, the extra-domain is characterized by a large number of positively charged amino acids probably organized in helical domains (Figure 1). Circular dichroism and NMR experiments confirmed a polyanion-induced helicity of peptides derived from the extension (Agou et al., 1995). Moreover, the amphiphilic nature of the helices, with the presentation of the charged residues on one side, explains the binding of yeast AspRS to heparin and the absence of binding of the truncated enzyme to this sulfonated polysugar (Lorber et al., 1988). Since negatively charged heparin is known to mimic nucleic acids, these data indicate a strong probability of at least some of these 70 amino acids being involved in nucleic acid recognition in general, and in tRNAAsp binding in particular.
Fig. 1. Schematic representation of the modular structure of yeast AspRS and organization of its N-terminal extension. The anticodon recognition module (71–207) is linked to the catalytic domain (242–557) by a small hinge region (207–241). The N-terminal extension (1–70), specific to eukaryotic AspRSs, is linked to the anticodon recognition module. It comprises three putative helical domains predicted with high probability, and perhaps a fourth helix of lower probability. The sequence of the extension (Sellami et al., 1986) and the helix probability plot computed according to the network procedure (Rost and Sanders, 1993, 1994; Rost et al., 1994) are shown. All published work on AspRS prepared from yeast cells concerns a protein heterogeneously cleaved in the median part of the extension (see text for details).
Here we demonstrate that the extension present at the N-terminus of yeast AspRS plays a role in tRNAAsp binding. The necessity to bind tRNA tightly explains conservation of the extension in eukaryotic AspRSs and in other class IIb synthetases. Our demonstration is supported by an extensive comparison between the native form and a number of deleted forms of yeast AspRS. We show the capacity of a 70 amino acid peptide to bind to tRNAAsp with a strong affinity and a specificity dependent on structural features. The presence of this extension considerably increases the stability of the complex between AspRS and its homologous tRNA. Finally, we determine the tRNA domain and the AspRS residues involved in this recognition and we define a new amino acid motif conserved in the N-terminal extensions of class IIb synthetases in eukaryotes responsible for RNA binding. The biological implications of these findings are discussed.
Results and discussion
The N-terminal extension of yeast AspRS is involved in tRNAAsp binding
Considering previous work carried out on the N-terminal extension of yeast AspRS, we took into account the potential of this peptide to structure itself when binding to polyanions (Agou et al., 1995) and thus we investigated its involvement in tRNA binding. We determined the dissociation constants (Kd) of tRNAAsp for both forms of AspRS, namely the native enzyme (1–557) and its deleted version (71–557) deprived of the N-terminal extension. Interestingly, these two proteins diverge largely in their tRNA-binding capacities, as estimated by band shift experiments (Figure 2A, left and middle), having Kds differing by two orders of magnitude (Table I). This result was confirmed by an in vitro determination of the catalytic constants (kcat and Km) for tRNAAsp aminoacylation: Km is increased 100 times when the N-terminal extension of the protein is missing, while the catalytic rate constant kcat for tRNA aminoacylation remains the same for both enzymes (Table I).
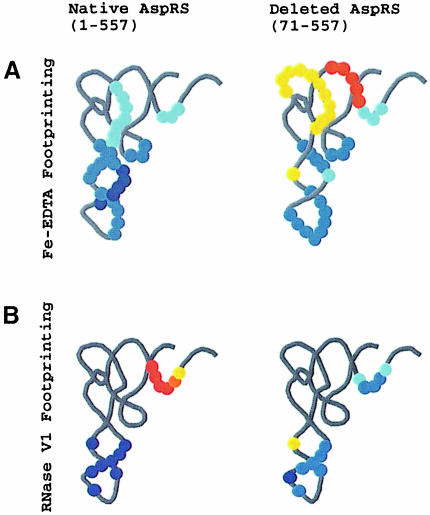
Fig. 2. Autoradiograms of gel retardation and footprint experiments. (A) Gel retardation experiments obtained in the presence of 3′-end-labeled native yeast tRNAAsp complexed to native AspRS (1–557) (1.95–250 nM) (left), deleted AspRS (71–557) (312 nM to 40 µM) (middle) and the N-terminal extension of AspRS (1–70) (right). Note the presence of two types of tRNA–AspRS (1–557) complexes: a fast migrating complex (middle band) corresponding to two tRNA molecules per dimeric AspRS (doubling the negative charges is dependent on increasing the global molecular weight of the complex by 15%) and a slow complex with one tRNA per enzyme. (B) Fe-EDTA and (C) RNase V1 mapping of 3′-end-labeled tRNAAsp in the absence (0) or presence of native or deleted (–70) AspRSs. (D) RNase V1 mapping of 3′-end-labeled tRNAAsp in the absence (0) or presence of increasing concentrations of N-terminal extension. Lanes T1 and AH correspond to RNase T1 and alkaline hydrolysis ladders, respectively. The control lane C contains tRNA without treatment; controls of incubation with proteins are not shown. Numbering of bands is according to G residues. In (B), protected regions are indicated by solid lines and intensifications by dashed lines. In (C) and (D), the major cleavage point at position 42 in the anticodon stem is emphasized.
Table I. Binding and tRNAAsp aminoacylation properties of full-length and truncated yeast AspRS.
| AspRS | Kd (nM) | Km (nM) | kcat (/s) |
|---|---|---|---|
| Full-length (1–557) | 30 | 5–10 | 0.10–0.15 |
| Truncated (71–557) | 5000 | 500–1000 | 0.10–0.15 |
Obvious questions arising from these observations are the way in which the extra-domain of AspRS recognizes tRNA and the possibility that the increase in tRNA affinity for the full-length enzyme triggers an increase in specificity.
Search for the region in tRNA that binds to the N-terminal extension of AspRS
Previous solution studies of the yeast AspRS–tRNAAsp complex have pointed out contact regions between the protein and the tRNA that lie on the side of the variable region of the tRNA, including the anticodon loop and stem (e.g. Romby et al., 1985; Garcia et al., 1990; Rudinger et al., 1992). Regions of tRNAAsp in close proximity to AspRS comprise nucleotides of the D-stem (10–14), the anticodon stem and loop (24–39) and the acceptor stem (66–76), in agreement with the contacts observed on the crystal structure (Ruff et al., 1991; Cavarelli et al., 1993).
To monitor variations in the protection patterns on tRNAAsp obtained in the presence of the native yeast AspRS (1–557) and its deleted version (71–557), and to determine the region recognized by the extension (1–70), we undertook extensive footprinting experiments. A first set of footprints with Fe-EDTA as the structural probe (Latham and Cech, 1989; Giegé et al., 1999) allowed systematic scanning of the RNA riboses for their proximity to AspRS. Clear differences in the protection profiles were found (Figure 2B). When tRNAAsp is complexed to the deleted protein, nucleotides 22–26 in the D- and anticodon stems, 31–39 in the anticodon loop and 68–70 in the acceptor stem are protected. In contrast, nucleotides 43 in the anticodon stem, 47–57 in the variable region and the T-stem and loop, and 63–67 in the T- and acceptor stems are more accessible to the probe. Taken together, these results lead to an interaction scheme where nucleotides inside the tRNA L-shaped structure are in close proximity to the core of the synthetase, whereas the other side of the tRNA is more accessible, indicating a conformational change during complex formation (Figure 3A). A similar conclusion was obtained with other structural probes, such as iodine (Rudinger et al., 1992), and agrees with the crystal structure of the complex. We recall, however, that all these previous results were obtained with heterogeneous yeast AspRS samples deprived of part of the N-terminal extension.
Fig. 3. Residues protected by native and deleted AspRS against hydrolysis by (A) Fe-EDTA or (B) RNase V1. Protections and intensifications are displayed on L-shaped structures of tRNAAsp (data were taken from Figure 2B and C), the differences in strength, determined on a Bio-imager, are indicated by different intensities of blue dots for protected positions (weak, 20–40%; moderate, 40–60%; and strong, 60–100%) and of orange dots for intensifications (weak, 10–40%; moderate, 40–60%; and strong, >60%).
The protection profile obtained with the native enzyme (1–557) superimposes well on the pattern obtained with the deleted AspRS (71–557) in the tRNA acceptor stem and partly in the anticodon domain (i.e. at nucleotides 21–29 and 35–40), although clear variations in the intensities of the footprints are observed (Figures 2B and 3A). Outside these discrete regions, the tRNA is the target of additional protection due to the presence of the N-terminal extension. This occurs at positions 27–29, 35–39 and 42–53 in the anticodon domain and the variable region. In contrast, residues 32, 33 and 34 in the anticodon loop are no longer protected, and the signals of conformational changes (cleavage enhancements) observed with AspRS (71–557) are missing (Figure 3A).
Additional footprints using cobra venom RNase V1, a probe directed toward double-stranded RNA regions (Favorova et al., 1981), confirm the conclusions from Fe-EDTA mapping. Indeed, both strands of the tRNAAsp anticodon stem are strongly and specifically cut, leading to an easy monitoring of the cleavage signals at nucleotides 26–30 and 32 in the 5′ strand and nucleotide 42 in the 3′ strand of the anticodon stem (Figure 2C). Incubation with native AspRS (1–557) enhances the protection observed with AspRS (71–557) on residues 26–30 and induces a specific protection on residue 42 that does not exist with AspRS (71–557) (Figure 3B). The percentages of protection at position 42 are reported in Table II and were used to follow the contact between different AspRS constructs and tRNAAsp (see below). Concerning the native yeast AspRS molecule, our results indicate that its N-terminal extension makes direct contacts in the minor groove of the tRNAAsp anticodon stem and leads to a closer proximity between the major groove and the rest of the synthetase (Figure 3). The main outcome is the formation of a stronger complex, catalytically more efficient, as reflected by the retardation gel and aminoacylation experiments (Table I).
Table II. Binding of tRNAAsp to yeast AspRS constructs with variations in the N-terminal extensions and extent of protection of tRNAAsp by these synthetase variants against hydrolysis by RNase V1 at position 42 in the anticodon stem of the tRNA.
| AspRS variants | Kd (nM) | RNase V1 protection at position 42 of tRNAAsp (%) |
|---|---|---|
| Native AspRS (1–557) | 20 | 92 |
| Truncated AspRSs | ||
| –17 (18–557) | 20 | 98 |
| –25 (26–557) | 20 | 97 |
| –33 (34–557) | 100 | 53 |
| –40 (41–557) | 2500 | 25 |
| –46 (47–557) | 10 000 | 2 |
| –70 (71–557) | 5000 | 9 |
| N-terminal domain (1–70) | 500 | 78 |
| Mutated AspRSsa | ||
| L29A | 20 | 69 |
| S30A | 20 | 70 |
| K31A | 20 | 86 |
| K32A | 100 | 69 |
| L34A | 150 | 11 |
| K35A | 500 | 0 |
| K36A | 100 | 43 |
| Chimeric AspRSsb | ||
| human AspRS #1 | 5000 | 0 |
| human AspRS #2 | 500 | 31 |
| yeast LysRS #1 | 500 | 40 |
| yeast LysRS #2 | 20 | 76 |
| yeast AsnRS #1 | 20 | 70 |
| yeast AsnRS #2 | 100 | 56 |
aMutations are in the truncated –25 form (26–557) of yeast AspRS.
bThe N-terminal extensions are from human AspRS and from yeast LysRS and AsnRS and are appended to the core structure of yeast AspRS as schematized in Figure 5B. The accuracy of measurements of Kd values and of percentage protection is ∼20%.
Interestingly, when high concentrations of total tRNA are used (5–40 µM), the same aminoacylation plateaus are reached with both native and truncated AspRS, but the aminoacylation rate is reduced two to three times with the truncated enzyme (data not shown). This means that non-cognate tRNAs bind to AspRS lacking the N-terminus but are not (or only marginally) aspartylated. Thus the extension participates in the global efficiency of the aspartylation process by hindering the correct binding of non-cognate tRNAs.
Specificity of the recognition between tRNAAsp and the AspRS extension
Several transcripts of tRNAAsp mutated in the anticodon stem were used in footprinting experiments to test their effect on the protection pattern by full-length AspRS (data not shown). Replacements in tRNAAsp of the two base pairs G27–C43 and G28–C42 by C27–G43/C28–G42 (variant probed by RNase V1 and Fe-EDTA), of G30–U40 by G30–C40, and of C31–G39 by G31–C39 (probed by RNase V1) have produced no significant modification in the protection patterns. This implies that the sequence of the anticodon stem is of no importance for its recognition by the N-terminal extension.
In a second series of experiments, we demonstrate that the extension indeed does not participate significantly in the specific recognition of the homologous tRNA. To reach this conclusion, yeast total tRNA was aminoacylated either by the native (1–557) or the truncated (71–557) AspRS. After each aminoacylation reaction, the charged tRNA molecules were isolated from bulk RNA on an EF-Tu-substituted affinity column where only aminoacylated tRNAs are retained (Sprinzl, 1994; Becker et al., 1997). Sequencing of the two isolated families of tRNAs did not detect any non-aspartate sequences. Therefore, no significant tRNA mischarging occurs with truncated AspRS, indicating that the specific recruitment of tRNAAsp by AspRS is not fundamentally dependent on the presence of the N-terminal extension.
The AspRS extension binds to the anticodon stem of tRNAAsp
The 70 amino acid peptide corresponding to the N-terminal extension of yeast AspRS was cloned, overexpressed and subjected to gel retardation and footprinting experiments in order to determine its own potential to bind tRNAAsp. The peptide by itself is able to shift with the same efficiency either tRNAAsp (Figure 2A, right) or total tRNA (from yeast or Escherichia coli, data not shown) with a Kd of 500 nM (Table II). This confirms its participation in tRNA binding and shows that it is not able to distinguish tRNAAsp from other tRNAs. In addition, the footprints indicate clearly that, even when isolated, the peptide still binds to the 3′ strand of the tRNA anticodon stem, recognizing only the minor groove of the helix even at high concentrations (Figure 2D). This leads to the general conclusion that even if this peptide cannot recognize a given sequence, it binds strongly and specifically to the minor groove of double-stranded RNAs.
To determine the region in the extension encompassing such a strong RNA-binding domain, progressive deletants of the AspRS N-terminal extremity were designed and tested in binding and footprinting experiments. Deleted AspRSs correspond to the native molecule deprived of its 17, 25, 33, 40 or 46 N-terminal amino acids (called –17, –25, –33, –40 and –46 AspRSs, respectively) (Figure 4A). Footprinting (Figure 4B) and gel retardation experiments (not shown) indicate a perfect correlation between the loss of contact with the tRNAAsp anticodon stem (at position 42) and the decrease in affinity. Quantification of these protections is given in Table II. The contact with tRNA residue 42 disappears abruptly when amino acids between positions 25 and 40 are removed (Figure 4B), with a consequent decrease in affinity as reflected by an ∼100-fold increase of the Kd of the complex (Table II). Thus, the peptidic region 25–40 contains the information for improved tRNA binding.
Fig. 4. (A) Amino acid sequence of the yeast AspRS N-terminal extension. (B–D) Footprinting patterns obtained with (B) progressively deleted AspRSs, (C) mutated AspRSs and (D) chimeric synthetases. In (A), the arrows indicate the sites chosen for progressive deletions performed after residues –17, –25, –33, –40, –46 and –70, respectively. On autoradiograms (B–D), C indicates control, and T1 and AH indicate RNase T1 and alkaline hydrolysis ladders; incubation controls with proteins are not shown.
Surprisingly, loosening the strong contact between the anticodon stem and the N-terminal extension of the protein also correlates with a closer proximity of the end of the acceptor stem to the catalytic domain of the enzyme. Indeed, nucleotides 69–73 become protected from RNase V1 hydrolysis (Figure 4B), implying the existence of a compensation effect between the two extremities of the L-shaped tRNA molecule during complex formation. Thus, when the anticodon extremity is strongly anchored to full-length AspRS, additional contacts at the level of the acceptor stem do not seem necessary to direct the CCA end correctly in the active site, whereas these contacts become more important when the anticodon domain is not tightly bound to the synthetase, as is the case with the truncated AspRS.
A peptidic motif responsible for improving tRNA binding
Sequence alignments of AspRSs with N-terminal extra-domains allowed the identification in the S.cerevisiae synthetase of the motif 29LSKKxLKKxxK39 strongly conserved in the enzymes from lower eukaryotes (Candida albicans and Schizosaccharomyces pombe) and partially conserved in those from higher eukaryotes (Drosophila melanogaster, Arabidopsis thaliana, Plasmodium falciparum and Caenorhabditis elegans) (Figure 5A). This motif is virtually absent in the short extensions of human and rat AspRSs, with only the two lysine residues homologous to K32 and K36 conserved (Jacobo-Molina et al., 1989; Mirande and Waller, 1989).
Fig. 5. (A) Sequence alignments of the N-terminal extension of eukaryotic aminoacyl-tRNA synthetases from subclass IIb and (B) design of the different chimeric synthetases. (A) Sequences of AsnRSs are on top, of AspRSs in the middle and of LysRSs on the bottom. The N-terminal extensions are followed by part of the core structure (after position 161). The conserved RNA-binding motif in the N-terminal extensions is highlighted in gray and pink for the most conserved residues. Amino acids conserved in families of synthetases of the same identity are indicated in red for AsnRSs, in purple for AspRSs and in yellow for LysRSs. Note the conservation at position 160 at the junction of the extension with the anticodon-binding module of the synthetases. (B) The chimeras were obtained by fusing the core of yeast AspRS with the N-terminal extensions of human AspRS, and yeast LysRS and AsnRS. Sequences originating from yeast AspRS are represented in yellow for the active core of the synthetase and in blue for the extension. Those from yeast LysRS, AsnRS and human AspRS are in gray. The conserved RNA-binding motif is in pink. In the chimera, the N-terminal sequences from non-homologous synthetases were ligated to residue A65 in yeast AspRS (note that the peptide stretch 65–69 belongs to the extension of yeast AspRS). They correspond to polypeptides 1–51 from yeast LysRS, 1–96 from yeast AsnRS and 1–18 from human AspRS. In yeast AspRS, 30 amino acids separate the conserved motif and the anticodon-binding domain. In chimeric proteins #1, this distance is 15 amino acids for LysRS, 30 amino acids for AsnRS and six amino acids for human AspRS. Variations were introduced in chimeric proteins #2 by fusing the N-terminal domains from yeast LysRS, yeast AsnRS and human AspRS to yeast AspRS residues L52, A51 and K43, respectively. In doing so, 30, 45 and 30 amino acid linkers (between the conserved motif and the anticodon-binding domain) were obtained. Sequences of yeast AspRS, LysRS and AsnRS are according to Sellami et al. (1986), Martinez et al. (1991) and Johnston et al. (1994), respectively; that of human AspRS is according to Jacobo-Molina et al. (1989).
Each of the conserved amino acids between positions 29 and 36 was substituted individually by alanine (mutations were performed on AspRS-25), and the corresponding mutated AspRSs were tested for their capacity to contact the minor groove of the tRNAAsp anticodon stem (Figure 4C). All of the conserved amino acids were found to be involved in tRNA binding, since each mutation leads to decreased protection of position 42, the strongest effects being observed for residues L34 and K35 (Figure 4C and Table II). In previous studies, secondary structure predictions (Lorber et al., 1988; Agou et al., 1995) suggested that the peptide encompassing the consensus motif adopts a helical fold, with the four lysine residues (K31, K32, K35 and K36) located on the same side of the helix. This makes them good candidates for non-specific interactions with the anticodon stem of the tRNA. The other three amino acids of the motif (L39, S32 and L34), and especially the hydrophobic leucines, are probably important for the stability of the helix and the correct presentation of the lysine residues towards RNA.
Sequence comparison of N-terminal extensions of synthetases from class IIb
A lysine-rich amino acid motif is also present in the N-terminal extensions of LysRSs and AsnRSs from eukaryotes (Figure 5A). When compared with the motif identified in AspRSs (29LSKKxLKKxxK39), a general consensus sequence xSKxxLKKxxK appears for the three families of eukaryotic class IIb synthetases. This consensus, although less conserved in eukaryotic LysRSs, retains residues homologous to S30, K31, L34, K35, K36 and K39 in yeast AspRS. Degeneracy is increased in AsnRSs where only the second part of the motif (L34, K35, K36 and K39) is present.
The ability of the N-terminal extensions of yeast LysRS and AsnRS to form contacts with the anticodon stem of yeast tRNAAsp was tested by fusing them to the core of yeast AspRS. As a negative control, we also fused the short extension of human AspRS (Jacobo-Molina et al., 1989), which shows only a marginal amino acid conservation with the yeast motif. Two sets of constructs were designed for each chimera. In the first set (chimera #1), the extra-domain was fused directly to the anticodon domain of AspRS, whereas, in the second one (chimera #2), the fusions were done in order to modulate the distance between the AspRS anticodon domain and the conserved peptidic motif (Figure 5B). All constructs were tested in footprinting experiments to visualize the protection of the 3′ strand of the tRNAAsp anticodon stem (Figure 4D and Table II).
With all the chimeric proteins, the protection is, at least partially, recovered at position 42, even in the case of the human extension. However, the extent of protection is strongly dependent upon the distance between the RNA-binding motif and its connection to the anticodon-binding module of AspRS. Only if this distance is the same as that found in the yeast AspRS (30 residues) is the rescue by the foreign extension optimal. In other words, optimal binding of the peptidic motif to its tRNAAsp target occurs with yeast LysRS chimera #2 and yeast AsnRS chimera #1. In the case of human AspRS chimera #2, the motif is too degenerated to recover good protection of nucleotide 42. If this distance of 30 amino acids is not maintained, as in human AspRS chimera #1, or in yeast LysRS chimera #1 and yeast AsnRS chimera #2 (Figure 5B), the binding to tRNA is strongly affected and the protection is decreased, or even abolished in the case of human AspRS chimera #1 (Table II).
General conclusions and perspectives
Based on an explicit demonstration with yeast AspRS, this work shows that a short lysine-rich peptide sequence in the N-terminal extension of this class IIb synthetase is involved in tRNAAsp binding by sequence-independent contacts. In Figure 6, we have modeled the three helices predicted in this extension on the crystallographic structure of the AspRS–tRNAAsp complex. While the exact location of helices 1 and 3 is rather poorly defined, especially that of helix 3, we are confident about the location of helix 2 containing the RNA-binding motif that lies in the minor groove of the tRNA anticodon stem. This assumption is supported by, amongst other evidence, the strong interaction of the isolated N-terminal (1–70) peptide in the minor groove of tRNAAsp. Taken together, the proposed model accounts well for the improved anchoring of tRNAAsp on the native synthetase, especially for its anticodon domain. As a consequence, the stability of the complex is increased, with the net result that aspartate identity is globally better expressed in the native enzyme (the catalytic efficiency kcat/Km is 100-fold higher, see Table I). Noticeably, the additional contacts that the extension makes with the anticodon stem are accompanied by a long-distance adaptation between the acceptor stem and the catalytic site of the enzyme.
Fig. 6. Three-dimensional structure of a full monomer of the AspRS–tRNAAsp complex of yeast. The structure of the core enzyme (in yellow) complexed to the tRNA is that determined by crystallography (Ruff et al., 1991); that of the N-terminal extension (in blue) was modeled taking into account the data from this work. Note the location of the three helices in proximity to the tRNA, and particularly that of helix 2 (with the RNA-binding motif in pink) lying in the minor groove of the anticodon stem helix. The figure was generated using the program SETOR (Evans, 1993).
In the case of other eukaryotic AspRSs, as well as in the two other representatives of eukaryotic class IIb synthetases, AsnRSs and LysRSs, this peptidic sequence is also present in a more or less conserved version (xSKxxLKKxxK), but is located at different distances from the branching point with the anticodon binding modules of these proteins (see Figure 5). It is likely that this variability reflects faint architectural differences in class IIb synthetases and that, within their own synthetase context, this motif would also bind to tRNA as it does in the context of yeast AspRS. Other lysine-rich sequences were identified in extensions of other eukaryotic synthetases, i.e. in HisRS, IleRS, MetRS, GlnRS, SerRS, ThrRS and ValRS from yeast (Lorber et al., 1988; Mirande, 1991). Only in the case of yeast MetRS and GlnRS were these extensions shown to increase the affinity of the synthetase for the cognate tRNA (Glasfeld and Schimmel, 1997; Whelihan and Schimmel, 1997; Wang and Schimmel, 1999). Thus, it appears that the strategy used by nature for the recognition of tRNA by synthetase in the eukaryotic world combines both specific (identity elements) and non-specific (structural features) components distributed in distinct tRNA and protein domains. Interestingly, the property of binding tRNA non-specifically can be compared with what has been shown for Arc-1p. Indeed, this protein, when associated with yeast MetRS, leads to increased affinity of the tRNA for MetRS (Simos et al., 1996).
Lysine-rich sequences closely related to the consensus sequence of the RNA-binding motif present in class IIb eukaryotic synthetases were also found in a series of double-stranded RNA-recognizing proteins (Krovat and Jantsch, 1996), and their helical nature was visualized in the crystal structure of such a protein from Xenopus laevis complexed to double-stranded RNA (Ryter and Schultz, 1998).
In conclusion, this work demonstrates the participation of the N-terminal extensions of class IIb synthetases in optimizing specific tRNA recognition by synthetases. At this stage, we cannot exclude additional physiological functions of these extensions, and more generally of the extra-domains of eukaryotic synthetases. In higher eukaryotes, their participation in multisynthetase complex formation has experimental support (Quevillon et al., 1999; Rho et al., 1999). In lower eukaryotes, however, where such complexes have not been detected, other reasons have to be invoked to account for their conservation, such as subcellular localization or trafficking of synthetases (Melki et al., 1991; Schimmel and Wang, 1999) and/or synthetase regulation. We are exploring these possibilities.
Materials and methods
Construction and purification of native, truncated, mutated and fusion proteins
All DNA sequences were cloned into pQE-70 (Qiagen) between SphI and BglII restriction sites and encode proteins derived from yeast AspRS with a C-terminal His6 tag sequence under the control of an inducible T5 promoter. Native and N-terminal truncated proteins were amplified by PCR from pRS314-AspRS (Eriani et al., 1991). PCR mutagenesis was used to produce the point mutations on the –25 truncated form of AspRS. N-terminal domains from yeast AsnRS and LysRS were PCR amplified from yeast total DNA and used as primer in a second PCR amplification reaction on AspRS DNA fusing heterologous domains to the core of AspRS (amino acids 71–557). Each construct was checked by sequencing both extremities.
The His6-tagged protein variants were purified by nickel affinity chromatography, as described by Qiagen, and conserved at –20°C in 50 mM phosphate buffer pH 7.2, 150 mM KCl, 50% glycerol, 1 mM EDTA and 1 mM dithiothreitol (DTT). An additional purification step on a UnoS column (Bio-Rad) was necessary for point-mutated AspRSs containing heterogeneities at their N-termini after the first purification step. Proteins were quantified by Bradford assays (Bio-Rad).
tRNA 3 ′ end labeling
Yeast tRNAAsp and E.coli (ATP,CTP):tRNA nucleotidyltransferase or CCase were a kind gift from A.Théobald-Dietrich and were purified according to Ruff et al. (1988) and Cudny and Deutscher (1986), respectively. The 3′ end labeling of yeast tRNAAsp was achieved by [α-32P]ATP exchange in the presence of CCase (G.Keith, personal communication). Labeled tRNA was purified by electrophoresis on a 12% polyacrylamide gel followed by passive elution in 0.5 M NH4OAc pH 6.0, 0.1 mM EDTA, 0.1% SDS, 10 mM Mg(OAc)2, and ethanol precipitated. The samples were then redissolved in water and renatured by heating for 2 min at 65°C and cooling down for 10 min at room temperature.
Gel retardation experiments
For each complex formation, ∼10 000 Cerenkov counts of tRNA were incubated with variable protein concentrations (from 2 nM to 40 µM) in a total volume of 20 µl containing 25 mM phosphate buffer pH 7.2, 75 mM KCl, 10 mM MgCl2 and 25% glycerol for 15 min on ice and loaded on native 6% (29:1) acrylamide:bisacrylamide gels (200 × 200 × 1.5 mm3, 140 V for 45 min) in TBE at 4°C when performed with full-length synthetases, and on 10% (29:1) acrylamide:bisacrylamide gels when performed with the N-terminal domain peptide alone.
Nuclease mapping and footprints
Digestions with nuclease V1 were done in 25 mM phosphate buffer pH 7.2, 75 mM KCl, 10% glycerol and 10 mM MgCl2 for 10 min on ice. Each reaction was performed in a total volume of 25 µl in the presence of 10 ng/µl RNA, 3′-end-labeled tRNA (∼50 000 Cerenkov counts) and 1.2 × 10–2 U of RNase V1. tRNA and 10 µM AspRS were incubated for 5 min on ice prior to enzymatic digestion. Reactions were stopped by addition of 1 vol. of ‘Stop Mix’ (0.6 M NaOAc, 4 mM EDTA, 0.1 µg/µl total tRNA), followed by a phenol/chloroform extraction and ethanol precipitation. An alkaline ladder was obtained by incubation of 2 µg of tRNA in 50 mM NaHCO3 pH 9.0 at 95°C for 5 min and a guanine ladder by denaturing RNase T1 digestion: 2 µg of RNA were first denatured by heating for 5 min at 55°C followed by rapid cooling on ice, and digested in 7 M urea, 1 mM EDTA, 10 mM sodium citrate pH 4.5 in the presence of 1 U of RNase T1 for 10 min at 55°C.
Radioactive samples were washed with 70% ethanol, dried and quantified (Cerenkov) before being solubilized in loading buffer (90% v/v formamide, 20 mM EDTA and 0.5% w/v dyes), heated for 3 min at 85°C and electrophoresed on 15% polyacrylamide gels. Signal analysis and quantification were done on a FUJIX Bio-Imaging Analyser BAS 2000 with Work Station Software (v1.1).
Fe-EDTA footprints
Mapping with Fe-EDTA was performed in a 20 µl reaction volume on ice containing the pre-incubted complexes between tRNAAsp (0.5 µM) and the different synthetases (10 µM) in 25 mM phosphate buffer pH 7.2, 75 mM KCl, 10 mM MgCl2. A 1 µl aliquot of each of the following listed solutions was added: 40 mg/ml ammonium iron (II) sulfate hexahydrate (Aldrich Chemicals), 250 mM EDTA, 100 mM DTT and 0.5% (v/v) hydrogen peroxide. The reaction was run for 10 min on ice, stopped and analyzed as described above. Since identical protection patterns were obtained in the presence of 10 and 20 µM AspRS (71–557), we decided to conduct our experiments with the lowest (10 µM) synthetase concentration. Under such conditions, the protection patterns do not reflect a partial dissociation of the complexes.
Determination of catalytic constants
Aminoacylation of yeast tRNAAsp was performed at 30°C in 100 mM HEPES–KOH pH 7.5, 10 mM MgCl2, 5 mM ATP, 30 mM KCl, 5 mM DTT and 25 µM 3H-labeled l-aspartic acid. Samples were quenched and treated in the conventional way described by Perret et al. (1990). tRNA concentrations varied between 5 and 50 nM in the presence of 10 nM native AspRS, and 125 and 1000 nM in that of 20 nM N-terminal deleted AspRS (71–557). Kinetic constants were derived from Lineweaver–Burk plots and represent an average of at least two independent experiments.
EF-Tu selection
A 120 µg aliquot of yeast total tRNA in a total reaction volume of 500 µl was aminoacylated with 1 µM native AspRS or AspRS (71–557), using the same buffer conditions as listed above, for 20 min at 30°C. Aminoacylated tRNAs were purified on an EF-Tu affinity column as described by Becker et al. (1997). After deacylation, eluted tRNAs were 3′-end labeled, purified on polyacrylamide gel and each band was sequenced enzymatically (Donis-Keller et al., 1977).
Sequence alignments and modeling
Extensions of synthetases first were aligned independently using ClustalX (Thompson et al., 1997), then grouped together and adjusted manually. Modeling of the N-terminal extension was based on the crystallographic structure of the yeast AspRS–tRNAAsp complex (PDB ID code 1ASZ). The first 66 residues were added and the helices built using O (Jones et al., 1991). The final model was refined to ensure good stereochemical geometry.
Acknowledgments
Acknowledgements
We thank J.Gangloff, O.Poch and C.Sauter for advice and stimulating discussions. We are also indebted to M.Mirande for many discussions on eukaryotic synthetases. This work was supported by grants from the Centre National de la Recherche Scientifique (CNRS), the Ministère de l’Education Nationale, de la Recherche et de la Technologie (MENRT), Université Louis Pasteur, Strasbourg and the European Community (BIO4 CT98 0189).
References
- Agou F., Yang,Y., Gesquière,J.-C., Waller,J.-P. and Guittet,E. (1995) Polyanion-induced α-helical structure of a synthetic 23-residue peptide representing the lysine-rich segment of the N-terminal extension of yeast cytoplasmic aspartyl-tRNA synthetase. Biochemistry, 34, 569–576. [DOI] [PubMed] [Google Scholar]
- Agou F., Waller,J.-P. and Mirande,M. (1996) Expression of rat aspartyl-tRNA synthetase in Saccharomyces cerevisiae. Role of the NH2-terminal polypeptide extension on enzyme activity and stability. J. Biol. Chem., 271, 29295–29303. [DOI] [PubMed] [Google Scholar]
- Becker H.D., Reinbolt,J., Kreutzer,R., Giegé,R. and Kern,D. (1997) Existence of two distinct aspartyl-tRNA synthetases in Thermus thermophilus. Structural and biochemical properties of the two enzymes. Biochemistry, 36, 8785–8797. [DOI] [PubMed] [Google Scholar]
- Cavarelli J., Rees,B., Ruff,M., Thierry,J.-C. and Moras,D. (1993) Yeast tRNAAsp recognition by its cognate class II aminoacyl-tRNA synthetase. Nature, 362, 181–184. [DOI] [PubMed] [Google Scholar]
- Cudny H. and Deutscher,M.P. (1986) High-level overexpression, rapid purification, and properties of Escherichia coli tRNA nucleo tidyltransferase. J. Biol. Chem., 261, 6450–6453. [PubMed] [Google Scholar]
- Delarue M. and Moras,D. (1993) The aminoacyl-tRNA synthetases family: modules at work. BioEssays, 15, 675–687. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam,A.M. and Gilbert,W. (1977) Mapping adenines, guanines and pyrimidines in RNA. Nucleic Acids Res., 4, 2527–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriani G., Delarue,M., Poch,O., Gangloff,J. and Moras,D. (1990) Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature, 347, 203–206. [DOI] [PubMed] [Google Scholar]
- Eriani G., Prevost,G., Kern,D., Vincendon,P., Dirheimer,G. and Gangloff,J. (1991) Cytoplasmic aspartyl-tRNA synthetase from Saccharomyces cerevisiae. Study of its functional organization by deletion analysis. Eur. J. Biochem., 200, 337–343. [DOI] [PubMed] [Google Scholar]
- Evans S. (1993) SETOR: hardware lighted three-dimensional model representations of macromolecules. J. Mol. Graph., 11, 134–138. [DOI] [PubMed] [Google Scholar]
- Favorova O.O., Fasiolo,F., Keith,G., Vassilenko,S.K. and Ebel,J.-P. (1981) Partial digestion of tRNA-aminoacyl-tRNA synthetase complexes with cobra venom ribonuclease. Biochemistry, 20, 1006–1011. [DOI] [PubMed] [Google Scholar]
- Garcia A., Giegé,R. and Behr,J.-P. (1990) New photoactivatable structural and affinity probes of RNA: specific features and applications for mapping of spermine binding sites in yeast tRNAAsp and interaction of this tRNA with yeast aspartyl-tRNA synthetase. Nucleic Acids Res., 18, 89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegé R., Lorber,B., Ebel,J.-P., Moras,D. and Thierry,J.-C. (1980) Cristallisation du complexe formé entre l’aspartate-tRNA de levure et son aminoacyl-tRNA synthétase spécifique. C.R. Acad. Sci. Paris D-2, 291, 393–396. [PubMed] [Google Scholar]
- Giegé R., Puglisi,J.D. and Florentz,C. (1993) tRNA structure and aminoacylation efficiency. Prog. Nucleic Acid Res. Mol. Biol., 45, 129–206. [DOI] [PubMed] [Google Scholar]
- Giegé R., Helm,M. and Florentz,C. (1999) Chemical and enzymatic probing of RNA structure. In Barton,D.H.R. and Nakanishi,K. (eds), Comprehensive Natural Product Chemistry. Pergamon, Oxford, UK, Vol. 6, pp. 63–80. [Google Scholar]
- Glasfeld E. and Schimmel,P. (1997) Zinc-dependent tRNA binding by a peptide element within a tRNA synthetase. Biochemistry, 36, 6739–6744. [DOI] [PubMed] [Google Scholar]
- Jacobo-Molina A., Peterson,R. and Yang,D.C.H. (1989) cDNA sequence, predicted primary structure, and evolving amphiphilic helix of human aspartyl-tRNA synthetase. J. Biol. Chem., 264, 16608–16612. [PubMed] [Google Scholar]
- Johnston M. et al. (1994) Complete nucleotide sequence of Saccharomyces cerevisiae chromosome VIII. Science, 265, 2077–2082. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for the building of protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Krovat B.C. and Jantsch,M.F. (1996) Comparative mutational analysis of the double-stranded RNA binding domains of Xenopus laevis RNA-binding protein A. J. Biol. Chem., 271, 28112–28119. [DOI] [PubMed] [Google Scholar]
- Latham J.A. and Cech,T.R. (1989) Defining the inside and outside of a catalytic RNA molecule. Science, 245, 276–282. [DOI] [PubMed] [Google Scholar]
- Lorber B., Giegé,R., Ebel,J.-P., Berthet,C., Thierry,J.-C. and Moras,D. (1983) Crystallization of a tRNA–aminoacyl-tRNA synthetase complex. Characterization and first crystallographic data. J. Biol. Chem., 258, 8429–8435. [PubMed] [Google Scholar]
- Lorber B., Kern,D., Mejdoub,H., Boulanger,Y., Reinbolt,J. and Giegé,R. (1987) The microheterogeneity of the crystallizable yeast cytoplasmic aspartyl-tRNA synthetase. Eur. J. Biochem., 165, 409–417. [DOI] [PubMed] [Google Scholar]
- Lorber B., Mejdoub,H., Reinbolt,J., Boulanger,Y. and Giegé,R. (1988) Properties of N-terminal truncated yeast aspartyl-tRNA synthetase and structural characteristics of the cleaved domain. Eur. J. Biochem., 174, 155–161. [DOI] [PubMed] [Google Scholar]
- Martinez R., Latreille,M.T. and Mirande,M. (1991) A PMR2 tandem repeat with a modified C-terminus is located downstream from the KRS1 gene encoding lysyl-tRNA synthetase in Saccharomyces cerevisiae. Mol. Gen. Genet., 227, 149–154. [DOI] [PubMed] [Google Scholar]
- Melki R., Kerjan,P., Waller,J.-P., Carlier,M.-F. and Pantaloni,D. (1991) Interaction of microtubule-associated proteins with microtubules: yeast lysyl- and valyl-tRNA synthetases and tau 218–235 synthetic peptide as model systems. Biochemistry, 30, 11536–11545. [DOI] [PubMed] [Google Scholar]
- Mirande M. (1991) Aminoacyl-tRNA synthetase family from prokaryotes and eukaryotes: structural domains and their implications. Prog. Nucleic Acid Res. Mol. Biol., 40, 95–142. [DOI] [PubMed] [Google Scholar]
- Mirande M. and Waller,J.-P. (1989) Molecular cloning and primary structure of cDNA encoding the catalytic domain of rat liver aspartyl-tRNA synthetase. J. Biol. Chem., 264, 842–847. [PubMed] [Google Scholar]
- Moras D. (1993) Structural aspects and evolutionary implications of the recognition between tRNAs and aminoacyl-tRNA synthetases. Biochimie, 75, 651–657. [DOI] [PubMed] [Google Scholar]
- Perret V., Garcia,A., Grosjean,H., Ebel,J.-P., Florentz,C. and Giegé,R. (1990) Relaxation of transfer RNA specificity by removal of modified nucleotides. Nature, 344, 787–789. [DOI] [PubMed] [Google Scholar]
- Quevillon S., Robinson,J.-C., Berthonneau,E. Siatecka,M. and Mirande,M. (1999) Macromolecular assemblage of aminoacyl-tRNA synthetases: identification of protein–protein interactions and characterization of a core protein. J. Mol. Biol., 285, 183–195. [DOI] [PubMed] [Google Scholar]
- Rho S.B., Kim,M.J., Lee,J.S., Seol,W., Motegi,H., Kim,S. and Shiba,K. (1999) Genetic dissection of protein–protein interactions in multi-tRNA synthetase complex. Proc. Natl Acad. Sci. USA, 96, 4488–4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romby P., Moras,D., Bergdoll,M., Dumas,P., Vlassov,V.V., Westhof,E., Ebel,J.-P. and Giegé,R. (1985) Yeast tRNAAsp tertiary structure in solution and areas of interaction of the tRNA with aspartyl-tRNA synthetase. A comparative study of the yeast phenylalanine system by phosphate alkylation experiments with ethylnitrosourea. J. Mol. Biol., 184, 455–471. [DOI] [PubMed] [Google Scholar]
- Rost B. and Sanders,C. (1993) Prediction of protein structure at better than 70% accuracy. J. Mol. Biol., 232, 584–599. [DOI] [PubMed] [Google Scholar]
- Rost B. and Sanders,C. (1994) Combining evolutionary information and neural networks to predict protein secondary structure. Proteins, 19, 55–72. [DOI] [PubMed] [Google Scholar]
- Rost B., Sanders,C. and Schneider,R. (1994) PHD—an automatic mail server for protein secondary structure prediction. Comput. Appl. Biosci., 10, 53–60. [DOI] [PubMed] [Google Scholar]
- Rudinger J., Puglisi,J.D., Pütz,J., Schatz,D., Eckstein,F., Florentz,C. and Giegé,R. (1992) Determinant nucleotides of yeast tRNAAsp interact directly with aspartyl-tRNA synthetase. Proc. Natl Acad. Sci. USA, 89, 5882–5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff M., Cavarelli,J., Mikol,V., Lorber,B., Mitschler,A., Giegé,R., Thierry,J.-C. and Moras,D. (1988) A high resolution diffracting crystal form of the complex between yeast tRNAAsp and aspartyl-tRNA synthetase. J. Mol. Biol., 201, 235–236. [DOI] [PubMed] [Google Scholar]
- Ruff M., Krishnaswamy,S., Boeglin,M., Poterszman,A., Mitschler,A., Podjarny,A., Rees,B., Thierry,J.-C. and Moras,D. (1991) Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNAAsp. Science, 252, 1682–1689. [DOI] [PubMed] [Google Scholar]
- Ryter J.M. and Schultz,S.C. (1998) Molecular basis of double-stranded RNA–protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J., 17, 7505–7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter C., Lorber,B., Kern,D., Cavarelli,J., Moras,D. and Giegé,R. (1999) Crystallogenesis studies on aspartyl-tRNA synthetase: use of phase diagram to improve crystal quality. Acta Crystallogr. D, 55, 149–156. [DOI] [PubMed] [Google Scholar]
- Schimmel P. (1987) Aminoacyl-tRNA synthetases: general scheme of structure–function relationships in the polypeptides and recognition of transfer RNAs. Annu. Rev. Biochem., 56, 125–158. [DOI] [PubMed] [Google Scholar]
- Schimmel P. and Wang,C.C. (1999) Getting tRNA synthetases into the nucleus. Trends Biochem. Sci., 24, 127–128. [DOI] [PubMed] [Google Scholar]
- Sellami M., Fasiolo,F., Dirheimer,G., Ebel,J.-P. and Gangloff,J. (1986) Nucleotide sequence of the gene coding for yeast cytoplasmic aspartyl-tRNA synthetase (APS); mapping of the 5′ and 3′ termini of AspRS mRNA. Nucleic Acids Res., 14, 1657–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simos G., Segref,A., Fasiolo,F., Hellmuth,K., Shevchenko,A., Mann,M. and Hurt,E.C. (1996) The yeast protein Arc 1p binds to tRNA and functions as a cofactor for the methionyl- and glutamyl-tRNA synthetases. EMBO J., 15, 5437–5448. [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M. (1994) Elongation factor Tu: a regulatory GTPase with an integrated effector. Trends Biochem. Sci., 19, 245–250. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.-C. and Schimmel,P. (1999) Species barrier to tRNA recognition overcome with nonspecific RNA binding domains. J. Biol. Chem., 274, 16508–16512. [DOI] [PubMed] [Google Scholar]
- Whelihan E.F. and Schimmel,P. (1997) Rescuing an essential enzyme–RNA complex with a non-essential appended domain. EMBO J., 16, 2968–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]