Abstract
The remarkable ability of many parasites to evade host immunity is the key to their success and pervasiveness. The immune evasion is directly linked to the silencing of the members of extended families of genes that encode for major parasite antigens. At any time only one of these genes is active. Infrequent switches to other members of the gene family help the parasites elude the immune system and cause prolonged maladies. For most pathogens, the detailed mechanisms of gene silencing and switching are poorly understood. On the other hand, studies in the budding yeast Saccharomyces cerevisiae have revealed similar mechanisms of gene repression and switching and have provided significant insights into the molecular basis of these phenomena. This information is becoming increasingly relevant to the genetics of the parasites. Here we summarize recent advances in parasite epigenetics and emphasize the similarities between S. cerevisiae and pathogens such as Plasmodium, Trypanosoma, Candida, and Pneumocystis. We also outline current challenges in the control and the treatment of the diseases caused by these parasites and link them to epigenetics and the wealth of knowledge acquired from budding yeast.
Keywords: Antigenic variation, Allelic exclusion, Telomere position effect, Gene silencing, Epigenetic switch
Review
Mechanisms of antigenic variation and immune evasion
Many protozoan parasites and pathogenic fungi use antigenic variation as the major strategy to evade the host immune defenses [1-3]. The genomes of these species harbor extended families of genes that encode closely related surface proteins (Table 1). In any given cell, all but one gene of these families are repressed by compact chromatin structures. These structures are refractory to transcription and are epigenetically transmitted to daughter cells. Occasional and reversible switches to a different active gene confer antigenic variation. These ever-changing ‘cloaks of invisibility’ enable the pathogens to persist in the hosts with devastating efficiency [1,4].
Table 1.
Varying genes and known mechanisms of variation in different pathogens
| Species | Varying genes (number of genes given in parentheses) | Mechanisms involved in gene variation | Reference(s) |
|---|---|---|---|
|
Trypanosoma brucei |
VSG (>1,000) |
Epigenetic switches, DNA recombination |
[3] |
|
Pneumocystis carinii |
MSG (160) |
DNA recombination |
[5,6] |
|
Plasmodium falciparum |
VAR (60), rifin (150 to 200), and stevor (30 to 35) |
Epigenetic switches |
[7-10] |
|
Candida glabrata |
EPA (23) |
Epigenetic switches |
[2,11] |
|
Giardia lamblia |
VSP (220) |
RNA interference |
[12-14] |
|
Saccharomyces cerevisiae |
Various subtelomeric genes |
Epigenetic switches |
[15,16] |
| S. cerevisiae | Mating type loci (HMRα and HMLα) | DNA recombination | [17] |
Silent ‘donor’ genes in Trypanosoma and Pneumocystis
Trypanosomes are bloodstream parasites with a most remarkable ability to evade the immune system and to cause severe diseases such as nagana (Trypanosoma vivax, T. congolense) and sleeping sickness (T. brucei) [3]. These maladies are characterized by extreme fatigue and sleepiness. T. brucei is the prevalent pathogen in humans and has become a prototype for antigenic variation. It harbors a massive family of more than 1,000 mostly subtelomeric variant surface glycoprotein (VSG) genes and pseudogenes.
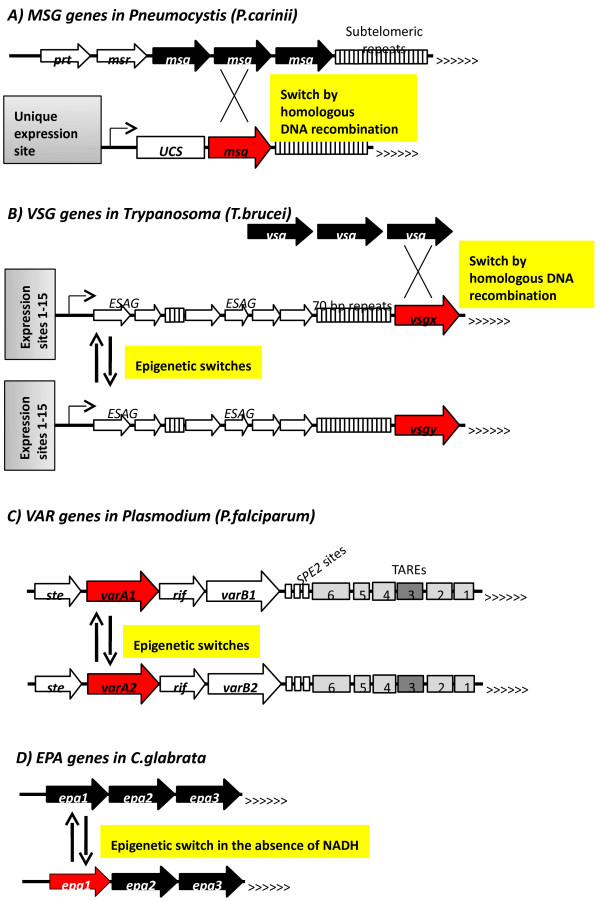
The fungi of the Pneumocystis family reside within the mammalian lungs and normally cause no symptoms, but can lead to serious infections in immunocompromised individuals and in HIV-infected patients [5,6]. Antigenic variation in Pneumocystis is produced by about 160 major surface glycoprotein (MSG) genes. Many of these are the last protein encoding genes at the telomeres of the 17 chromosomes [18,19] (Figure 1A). Interestingly, the interchromosomal MSG genes or pseudogenes are also surrounded by telomeric repeats [18].
Figure 1.
Schematics of varying genes and major mechanisms of variation in Pneumocystis carinii, Trypanosoma brucei, Plasmodium falciparum, and Candida glabrata. (A) In P. carinii, one to three MSG gene arrays (frequently flanked by MSR or PRT genes) are positioned next to a variable number of subtelomeric repeats and the telomeres (depicted by >>>). These genes (black arrows) lack promoters. To be expressed, a single MSG (red arrow) is transferred by homologous recombination to a unique expression site that contains an upstream conserved sequence (UCS). (B) In T. brucei, over 1,000 VSG gene donor sequences (black arrows) are exchanged by homologous recombination to one of fifteen expression sites. The VSG genes at these sites (red arrows) are adjacent to the telomeres and flanked by multiple 70 bp repeats. Several expression site-associated genes (ESAGs, open arrows) are distal to the telomere. All these and VSG are expressed in the direction of the telomere by one promoter (angled arrow). Only one of the fifteen expression sites is active at a time. Infrequent epigenetic switches of this site confers allelic exclusion and antigenic variation. (C) In P. falciparum, 60 VAR genes are positioned in tandem close to the telomeres or at interchromosomal locations (not shown). VARA (red arrow) point towards the telomere, while VARB (white arrow) point away. Six telomere-associated repeat elements (TARE) and several 12-base SPE sites (bind P. falciparum SPE2 interacting protein 2, PfSIP2) are located between VAR genes and the telomere. RIF and STE genes are frequently found in the vicinity of VAR genes. Only one VAR gene is expressed at a time. Switches between expressed VAR genes confer allelic exclusion and antigenic variation. (D) In C. glabrata, EPA genes are organized in arrays and are the last protein encoding genes before the telomere. In the example shown in the figure, EPA3 and EPA2 are mostly silenced, while EPA1 is expressed. ESAG, expression site-associated gene; PfSIP2, P. falciparum SPE2 interacting protein 2.
The only active VSG in T. brucei is expressed from one of 15 dedicated expression sites, while the active MSG in P. carinii is expressed from one unique expression site [4,5] (Figure 1A,B). These expression sites are adjacent to a telomere and contain gene promoters plus other regulatory elements. The pools of silent intact VSG and MSG genes plus pseudogenes and other VSG and MSG homologous sequences serve as a depot of donor elements that are translocated to the expression sites via DNA recombination [5,20] (Figure 1A,B). It is not known how the frequency of these recombination events is controlled [3]. However, it seems apparent that the silencing of the VSGs in T. brucei is accomplished by epigenetic means [21,22]. Strong support for this idea is offered by the observations that the knockdowns of key heterochromatin regulators (SIR2, RAP1, DOT1A) leads to their derepression [21,23,24]. No genetic evidence is available from Pneumocystis. It is noteworthy that a similar constitutive repression of ‘donor’ genes and their translocation to an active site governs the switching of the mating type in S. cerevisiae[17]. These parallels are extensively covered [3-5] and will not be reviewed here.
Epigenetic switching of subtelomeric genes in Trypanosoma, Plasmodium, and Candida
Of the 15 VSG expression sites in T. brucei, only one is active at a time. Early during the infection, the VSG switching is conducted mostly by rapid epigenetic on-off transitions between these expression sites (Figure 1B). Later on, the switching involves both epigenetic and DNA recombination events [3,4]. It is not known how the expression from a single site is achieved.
Different species of Plasmodium cause malaria by invading red blood cells in a wide variety of organisms. The pathogens undergo a complex life cycle that involves transmission by mosquitoes and a latency period in the livers of the hosts [7,8]. P. falciparum, one of the most extensively studied malaria pathogens, is chosen as a paradigm for gene variation by epigenetic switches. During blood-stage infection this pathogen expresses alternative forms of the immunodominant antigen P. falciparum erythrocyte membrane protein 1 (PfEMP1). The expressed PfEMP1 is trafficked by specialized vesicular structures and then displayed on the surface of the infected erythrocytes [7]. Using PfEMP1 as an adherent, the infected erythrocyte is sequestered to the vascular walls to contribute to the severe symptoms of malaria. PfEMP1 is encoded by a limited family of 60 VAR genes, which are positioned in the subtelomeric regions of the chromosomes and in several interchromosomal clusters. An elaborate and poorly understood mechanism of coordinated silencing of the VAR genes is combined with rare epigenetic switches to other variants to confer an ever-shifting antigenic makeup (Figure 1C). This strategy is sufficient to minimize recognition of PfEMP1 by the immune response [7] and, at the same time, prevents the exhaustion of the reservoir of VAR genes [25-27]. Other families of surface proteins (rifin, stevor, PfMC-2TM) also contribute to antigenic variation, but PfEMP1 is believed to be the critical driving force of the immune evasion [7,9]. Besides epigenetic switches, there is solid evidence for frequent gene conversions between VAR genes [9]. While these contribute to the diversification of the gene family, such events do not directly contribute to the switching of the PfEMP1 surface antigens.
The coordinated silencing of all but one VAR gene is the crux of prolonged malaria infections and the slowly developing and incomplete immunity to the pathogen. Consequently, the factors that contribute to antigenic variation in P. falciparum have been extensively studied [7,9,25,28]. While a significant body of information has been acquired, the mechanisms of VAR silencing remain unknown [9,26,27,29-31]. For example, it is not clear what kind of cis-elements serve as VAR silencers. The introns [32] and a conserved region upstream of the VAR promoters [30,33,34] have been proposed to act as silencing elements, but later on the significance of the intron has been debated [7,30]. The pairing of VAR genes has also been proposed to contribute to repression, but the mechanistic details are yet to be elucidated [30,34]. However, it has been conclusively shown that histone acetylation, histone methylation, and the propagation of heterochromatin away from the telomeres control the VAR genes [35-39]. Another line of evidence suggests that the tethering of VAR genes to poorly characterized subdomains in the nuclear periphery could be critical for both their repression and switching [31,40,41]. In addition, there is widespread expression of long non-coding RNAs in blood-stage P. falciparum[42-45], but no conclusive evidence for the regulation of VAR genes by such RNAs has been obtained [46-48].
Candida glabrata is an opportunistic parasite that causes prolonged urinal tract infections [6]. The key event in these infections is the adhesion of C. glabrata to host epithelial cells via epithelial adhesin (EPA) genes. Antigenic variation in this species is conferred by 23 EPA genes, which are positioned in the subtelomeric regions (Figure 1D) and are repressed by the NADH-dependent histone deacetylase SIR2[49,50]. Interestingly, C. glabrata is a nicotinic acid auxotroph. It is believed that the repression of EPA genes relies on the NADH provided by the host. Once the parasite moves to the urinary tract (there is very little nicotinic acid in the urine) the activity of Sir2p diminishes and the EPAs, and EPA1 in particular, are expressed [2,11].
RNA interference in Giardia
Giardia lamblia is a human intestinal pathogen that causes mild to severe diarrhea. Infection is transmitted by ingestion of cysts followed by the attachment of Giardia to the intestinal cells via variant-specific surface protein (VSP) genes. G. lamblia displays distinctive antigen variation through changes in the expression of about 220 VSP genes [51]. However, there are notable distinctions in the control of variation between Giardia and the previously described species. First, VSP genes are not clustered in the subtelomeric regions. Second, it has been suggested that multiple VSP genes are expressed and then all but one of the VSP mRNAs are repressed by an elaborate endogenous RNAi system that remains to be fully characterized [12-14]. No DNA methylation has been demonstrated in this organism [12].
Telomere position effect (TPE) in S. cerevisiae: similarities in pathogens
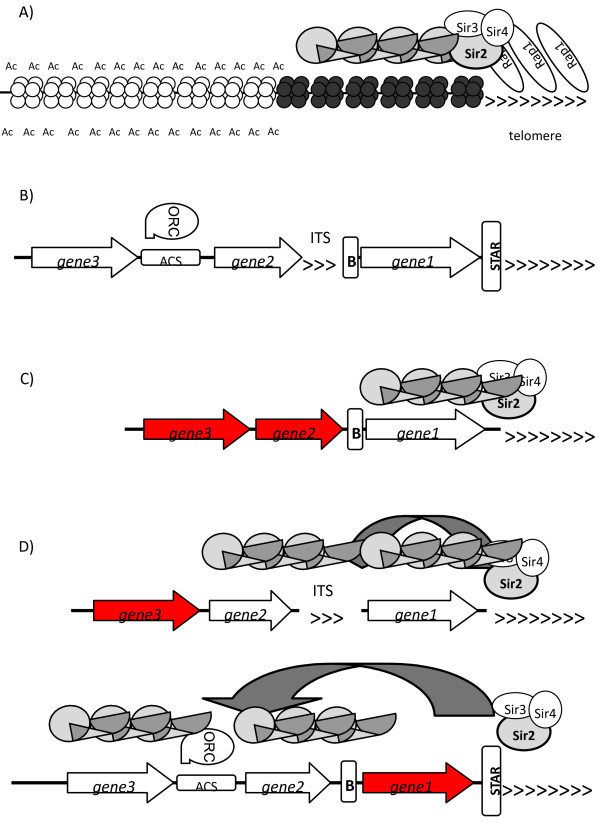
Allelic exclusion and variation is the crux in the infections by the above pathogens. However, the mechanisms of gene silencing and switching are not so well understood. On the other hand, a similar (but not identical) phenomenon (referred to as telomere position effect, TPE) in the innocent budding yeasts is significantly better characterized [15,52]. Briefly, silencing is mediated by compact heterochromatin, which is re-established after each cell division. Rare conversions between the silent and active states confer a quasi-stable pattern of epigenetically controlled gene expression in the vicinity of the telomeres [53]. TPE has been extensively reviewed [15,53,54] and is schematically presented in Figure 2A.
Figure 2.
Subtelomeric gene silencing in Saccharomyces cerevisiae. (A) Spreading of histone deacetylation away from the telomere. Rap1 proteins associate with the telomere repeats and recruit Sir2/Sir3/Sir4 proteins. Sir2p is an enzyme that deacetylates the histones in the adjacent nucleosome. More Sir2/Sir3/Sir4 proteins are recruited by the now deacetylated nucleosome (dark octamer) to eventually spread histone deacetylation to the next nucleosome (depicted by the curved arrow above the nucleosomes). Histone deacetylation and silent information regulator (SIR) proteins can spread several kilobases away from the telomeres. (B) Subtelomeric cis-elements in S. cerevisiae. Repetitive core X and Y’ elements contain dormant origins of DNA replication (ACS, it binds origin recognition complex, ORC), internal telomeric sequences (ITS, they bind Rap1 proteins), chromatin boundaries (depicted by B, and subtelomeric anti-silencing regions (STARs). (C) Chromatin boundaries restrict the spreading of histone deacetylation and prevent the silencing of telomere-distal genes (red arrows). (D)ITS and ACS are protosilencers, which extend the spreading of SIR proteins or confer telomere-dependent silencing of genes (white arrows) beyond an active subtelomeric gene (red arrow). A hypothetical STAR and a chromatin boundary contribute to the maintenance of the active gene. ORC, origin recognition complex; SIR, silent information regulator.
Unlike many higher eukaryotes, DNA methylation plays no role in the repression of subtelomeric genes in S. cerevisiae. In addition, while long non-coding RNA (called telomeric repeat-containing RNA, TERRA) is produced by the yeast subtelomeric DNA, a role of this RNA in gene silencing and switching has not been established [55]. These peculiarities have rendered this model organism somewhat irrelevant to epigenetics in higher eukaryotes. However, they make it quite relevant to the parasites that were discussed earlier. While budding yeasts cannot represent the whole variety of mechanisms for antigenic variation, some noteworthy similarities do exist. In Plasmodium, Candida, Trypanosoma, and Pneumocystis, the gene families that contribute to antigenic variation are mostly or exclusively located in the subtelomeric regions of the chromosomes. The repression of these subtelomeric genes is highly dependent on histone modifications [23,24,39,49,51,56-58], whereas in other eukaryotes the formation and maintenance of heterochromatin is more complex and involves additional levels of regulation. For example, similarly to S. cerevisiae and in contrast to higher eukaryotes, DNA methylation and RNAi do not seem to contribute to the silencing of the variance genes [7,15].
In S. cerevisiae and P. falciparum the silenced variation loci cluster in the nuclear periphery [8,15,59]. In S. cerevisiae, the relocation of these loci leads to derepression [59]. In P. falciparum, relocation to another ‘active’ domain, still in the nuclear periphery, is believed to contribute to the switching and the activation of VAR genes [7,33].
In S. cerevisiae, P. falciparum, and possibly other pathogens, the epigenetic silencing of genes needs a passage through the S phase [32,53], but it is not clear if DNA replication itself is the required process [60].
Shared strategies of antigenic variation and the means to combat them
In order to implement antigenic variation, the parasites must execute three distinct tasks. First, they need to selectively and exclusively activate one gene of the family at a time. Second, they need to effectively repress all but one of the genes of the family. Third, they need to switch the active gene at a frequency that runs ahead of the building immune response but does not exhaust the repertoire of the gene family.
Selective expression of one gene
The key question, how to selectively express one and only one gene of the extended family, remains unanswered. Ostensibly, the means must be coupled to the repression of all other genes, but how a gene is singled out is a persisting mystery. A common theme in the studies in Pneumocystis and Trypanosoma is the existence of expression sites [3,7]. In Pneumocystis the single expression site determines the expressed variant. This situation calls for regulated DNA recombination events at a frequency that will not jeopardize the pool of MSG genes. The same applies to the VSG genes in trypanosomes except that the situation there is complicated by the multiplicity of expression sites and their exchange through bona fide epigenetic means. A clue from S. cerevisiae suggests that the expressed site in trypanosomes could be related to the position of the active locus in the nucleus. In S. cerevisiae the telomeres cluster in several compartments in the nuclear periphery. Upon translocation to the nucleoplasm the telomeric genes lose repression [59]. A few proteins, including the Ku antigen and the nuclear pore components, contribute to this peripheral clustering [59,61]. A similar clustering of the inactive VAR genes in the nuclear periphery is apparent in Plasmodium, but the active VAR gene remains in the nuclear periphery slightly away from the repressed cluster [7,8]. These observations are consistent with the idea that the sub-nuclear localization of the VAR or VSG loci is linked to the expression of these genes. However, it is not clear if this differential localization is the cause or the consequence of the switch from silenced to active state.
Another clue from budding yeasts points to the fine architecture of subtelomeric DNA as combined with the balance of transcriptional activators and repressors. In S. cerevisiae, subtelomeric DNA consists of conserved core X and Y’ elements and harbors degenerate internal telomeric repeats (ITS), silent origins of DNA replication (ACS), and isolated binding sites for Rap1p (Figure 2B). These act as protosilencers and relay the spreading of SIR proteins away from the chromosome ends (Figure 2D) [62]. Additional complexity is provided by subtelomeric anti-silencing regions (STARs) and chromatin boundaries (Figure 2C) [63-65]. Likewise, the folding of the telomere and the establishment of t-loops and G-quadruplexes also contributes to complexity [63,66,67]. In this vein, G-quadruplex structures have been recently characterized in P. falciparum[68], while protosilencers have been conclusively identified in C. glabrata[69,70]. The SPE sites and the subtelomeric TARE3 in P. falciparum (Figure 1C) have also displayed properties consistent with protosilencing or boundary activities [45,71]. It is therefore tempting to speculate that assemblies of cis-elements similar to these in S. cerevisiae also exist in parasites.
It has been shown that in S. cerevisiae, the protosilencers, STARs, and chromatin boundaries can confer isolated expression of a gene imbedded in a heterochromatic region (Figure 2D) [64,66]. At the same time, studies in S. cerevisiae and D. melanogaster have shown that the abundance/strength of transcriptional activators counteract the silencing of target genes [72-74]. In S. cerevisiae, it has been demonstrated that overexpression of the trans-activator Ppr1p antagonizes the silencing of a telomeric URA3 reporter and that progression through S phase was necessary for the establishment of the active state [73]. It is conceivable that in parasites a similar mechanism of silent to active transition could exist. For example, an increase of variance gene-specific transcriptional activators and/or STAR binding factors accompanied by a passage through S phase could destabilize the repression of all genes in the family and predispose them to a conversion (see model in Additional file 1). Although the currently expressed gene has the advantage to remain active through epigenetic heritance [75], another gene could compete via the engagement of the chromatin boundaries and the gradual sequestration of limiting gene-specific activators. Reversion to a lower abundance of such activators would reinstate the robust repression of the other variance genes and uphold the conversion. Hence, the interplay between weak cis-elements and subtle changes in the abundance of transcription factors could significantly contribute to the elusive mechanism of epigenetic switches. In support, such temporary destabilization and expression of multiple VAR genes before a single VAR gene is selected has already been observed in P. falciparum[30,31,75]. Interestingly, there was an increase in the rate of switching at subtelomeric VAR loci as compared to switching at the internal loci. By all means, a closer look at the subtelomeric DNA of these parasites and a search for protosilencers, boundary, and/or anti-silencing elements and factors is warranted.
Repression of the varying genes
Lessons from budding yeasts have provided a basic framework for the understanding of this process in parasites. The central mechanism of the spreading of deacetylation from the telomeres operates in these and many other eukaryotes (Figure 2A) [15,52]; however, some exceptions need to be mentioned. As in budding yeast, Sir2p is a critical factor for the silencing of the varying genes in P. falciparum and C. glabrata, but it is not essential for the VSGs in T. brucei and its role in P. carinii is unknown [11,35,40,49,56-58,76]. Similarly, the telomere-binding protein Rap1 is essential for telomeric silencing in T. brucei and C. glabrata[56], but no evidence for its role in Plasmodium or Pneumocystis is available. Another feature that appears conserved between parasites and yeasts is the existence of histone variants that are specific to silent and active chromatin. In S. cerevisiae, H2A.Z antagonizes telomeric silencing and is enriched at chromatin boundaries [77]. A similar but more complex exchange of histones functions in P. falciparum where the unusual H2A.Z/H2B.Z double-variant nucleosomes are prevalent at active genes, but are excluded from silenced VAR genes [78]. Histone variants have also been described in T. brucei, but their relevance to gene silencing, if any, is not clear [79,80].
The methylation of histones poses even greater uncertainty. Methylation at specific K/R residues is associated with both gene activation and gene repression and is catalyzed by two classes of histone methyltransferases, SET and DOT1 [81]. In S. cerevisiae, the trimethylation of H3K79 by Dot1p has long been considered a key event in telomeric silencing [81], while the methylation of H3K36 by Set2p has been continuously linked to active transcription [82]. It was surprising to learn that the methylation of H3K36 by P. falciparum variant-silencing SET (PfSETvs) was critical for the repression of VAR genes in P. falciparum[39]. In T. brucei, the deletion of DOT1B does not lead to a general derepression, but increases the duration of the epigenetic switch [24]. In summary, although histone methylation at specific residues certainly contributes to gene silencing, significant variations between different parasites and S. cerevisiae could be expected.
Where does this notion lead us? It is feasible that the disruption of histone acetylation and methylation will preclude gene silencing and will cause the expression of many if not all of the varying genes. Indeed, it has been shown that the deletion of the homologues of SIR2, DOT1, or SET2 can produce pathogens that display multiple antigens [24,39,58]. These mutants, when properly attenuated, can be used for the successful generation of vaccines. Currently, the lack of vaccines is one of the most haunting issues in malaria and sleeping sickness [7,39,83-85]. In this respect, the gained knowledge of gene silencing can deliver a major breakthrough in the prevention of these devastating maladies. It is also conceivable that the drug targeting of the parasite homologues of the Sir2, Rap1, Set2, or Dot1 proteins can be used to combat the infections. To date, inhibitors of histone deacetylases or methyltransferases have shown promise under laboratory conditions [38,86]. However, this approach certainly needs fine-tuning. Because the varying surface antigens are directly linked to morbidity, their potential overexpression could produce ‘super-pathogens’ in the patients and will offset any gain in immunity. The risk of such a possibility has been demonstrated in vitro in C. glabrata[87].
Reversible epigenetic switches
An alternative to the risky and harmful overexpression of surface antigens is the reduction in the frequency of epigenetic switches. The rationale is that the immune system would gain ample opportunity to combat and clear the parasite. Unfortunately, the actual mechanisms of epigenetic conversions in S. cerevisiae or in the pathogens are not known [52]. While many regulators of TPE have been identified, the majority of them expand or contract the subtelomeric heterochromatin domain [15,88]. Hence, they do not necessarily alter the frequency of switching. Mutations in other regulators produce higher levels of the expression of otherwise silenced reporters [89,90], but it is hard to tell if modest loss of repression or frequent epigenetic conversions have yielded these results.
A decrease in the rate of switching is a reliable criterion for true deregulation, but it is rarely observed. To our knowledge, only one study in parasites (T. brucei) has shown such an effect. As mentioned above, in this species the deletion of DOT1B retards the epigenetic switch to a point where the cells express two VSGs for weeks [24]. Interestingly, in S. cerevisiae the trimethylation of H3K79 (it is catalyzed by Dot1p), but not DOT1 itself, increases the rate of the silencing establishment [91]. Five other studies in S. cerevisiae have reported the so-called ‘enhanced memory for heritable transmission’. Two of them have characterized mutations in histone H4, which increase the stability of both the repressed and the active states of subtelomeric reporters [92,93]. Two other papers [94,95] have pointed out that SIR1 alters the frequency of conversion at the mating type loci. Sir1p binds to dormant origins of DNA replication (these act as silencers of the mating loci) and recruits Sir2p [53]. Recently, we reported that the deletion of CAC1 reduces the frequency of epigenetic conversions of subtelomeric reporters [96]. Cac1p is a component of chromatin assembly factor I (CAF-I), which travels along with the replication forks and reassembles H3/H4 into nucleosomes on newly synthesized DNA [97]. It seems that both local silencers and the passage of replication forks act to occasionally change the epigenetic state of genes.
It is of particular interest that a replication fork factor contributes to epigenetic conversions. It is well known that the passage of the fork disperses the existing nucleosomes [52]. The subsequent reassembly combined with subtle variations in the abundance of variant gene-specific factors could both bestow the opportunity for a switch, as depicted in Additional file 1. This notion is in tune with the observations in S. cerevisiae and P. falciparum that the establishment of silencing requires a passage through S phase [7,52]. In a similar vein, a recent study in P. falciparum has demonstrated that the repositioning of a gene (P. falciparum reticulocyte binding protein-like homologue 4, PfRh4) to an active site in the nuclear periphery is associated with more frequent active to silent epigenetic switching [98]. It is attractive to speculate that these conversions are promoted by the open chromatin environment and that both replication forks and the state of existing nucleosomes determine the frequency of epigenetic switches. At present, CAF-I is the only candidate that could potentially confer reduced switching and non-varying phenotype in the parasites. However, other histone chaperones such as the homologues of the yeast ASF1, Rtt106, FACT, or HST should be considered. The exploration of this possibility may generate new drug targets and a truly new class of anti-pathogen drugs.
Conclusions
Parasites like Plasmodium and Trypanosoma cause devastating maladies in millions of people and are a leading cause of death in many developing countries. Others such as Pneumocystis could be deadly opportunistic agents. They all share a common powerful weapon: antigenic variation. The remote yeast S. cerevisiae has provided a paradigm and a framework to study positional effects, which are very relevant to the underlying mechanisms of antigenic variation. In the opinion of the authors, researchers need to turn more often to yeasts for clues on how to disarm such pathogens.
Abbreviations
ACS: ARS consensus sequence; ARS: Autonomously replicating sequence; CAF-I: Chromatin assembly factor I; DOT1: Disruptor of telomeric silencing 1; EPA: Epithelial adhesin; ESAG: Expression site-associated gene; ITS: Internal telomeric sequences; MSG: Major surface glycoprotein; ORC: Origin recognition complex; PfEMP1: P. falciparum erythrocyte membrane protein 1; PfRh4: P. falciparum reticulocyte binding protein-like homologue 4; PfSETvs: P. falciparum variant-silencing SET; PfSIP2: P. falciparum SPE2 interacting protein 2; RAP1: Repressor activator protein-1; RNAi: RNA interference; SIR: Silent information regulator; STAR: Subtelomeric anti-silencing region; TARE: Telomere-associated repeat element; TERRA: Telomeric repeat-containing RNA; TPE: Telomere position effect; UCS: Upstream conserved sequence; VSG: Variant surface glycoprotein; VSP: Variant-specific surface protein.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
BW, RO, and DJ participated in the review of the literature, in the writing of the manuscript, and in the preparation of the figures. KY conceived and drafted the review and wrote the final version of the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Model for epigenetic conversions driven by subtle fluctuations of activators and/or silencing factors. (1) All but one of a family of hypothetical varying genes (VG1 to VGN) are maintained in a silenced state. These genes are flanked by a subtelomeric anti-silencing region (STAR) and a chromatin boundary (B). (2) A subtle increase in the abundance of gene activators (green circle) and/or factors that engage STAR (purple circle) and the concomitant passage of replication forks would allow the activators access to the promoters of the VG genes and (3) would predispose all VG loci to derepression. (4) Consequently, during the next stage (re-establishment of silencing), the derepressed VG genes (VG2 to VGN, pink) will compete with the currently active gene (VG1, red). (5) During this stage, a decline in the abundance of activators and STAR-acting factors would aid in the formation of heterochromatin and the limiting activators would be gradually sequestered to a single locus. There is a high probability that the currently active gene (VG1) will be reinstated as the active locus. However, switches to another gene are possible (VGN). The likelihood of such switches, depicted by the width of the arrow, represents the frequency of epigenetic conversions.
Contributor Information
Brandon A Wyse, Email: bwyse@uoguelph.ca.
Roxanne Oshidari, Email: roshidar@uoguelph.ca.
Daniel CB Jeffery, Email: djeffery@uoguelph.ca.
Krassimir Y Yankulov, Email: yankulov@uoguelph.ca.
Acknowledgements
The writing of this review is in part supported by a grant from the National Science and Engineering Research Council of Canada (NSERC) (#217548-2010) to KY and an NSERC graduate studentship to DJ.
References
- Deitsch KW, Lukehart SA, Stringer JR. Common strategies for antigenic variation by bacterial, fungal and protozoan pathogens. Nat Rev Microbiol. 2009;7(7):493–503. doi: 10.1038/nrmicro2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filler SG. Candida-host cell receptor-ligand interactions. Curr Opin Microbiol. 2006;9(4):333–339. doi: 10.1016/j.mib.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Morrison LJ, Marcello L, McCulloch R. Antigenic variation in the African trypanosome: molecular mechanisms and phenotypic complexity. Cell Microbiol. 2009;11(12):1724–1734. doi: 10.1111/j.1462-5822.2009.01383.x. [DOI] [PubMed] [Google Scholar]
- Vink C, Rudenko G, Seifert HS. Microbial antigenic variation mediated by homologous DNA recombination. FEMS Microbiol Rev. 2011. doi: 10.1111/j.1574-6976.2011.00321.x. [DOI] [PMC free article] [PubMed]
- Cushion MT, Stringer JR. Stealth and opportunism: alternative lifestyles of species in the fungal genus Pneumocystis. Annu Rev Microbiol. 2010;64:431–452. doi: 10.1146/annurev.micro.112408.134335. [DOI] [PubMed] [Google Scholar]
- Jain N, Fries BC. Antigenic and phenotypic variations in fungi. Cell Microbiol. 2009;11(12):1716–1723. doi: 10.1111/j.1462-5822.2009.01384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherf A, Lopez-Rubio JJ, Riviere L. Antigenic variation in Plasmodium falciparum. Annu Rev Microbiol. 2008;62:445–470. doi: 10.1146/annurev.micro.61.080706.093134. [DOI] [PubMed] [Google Scholar]
- Scherf A, Riviere L, Lopez-Rubio JJ. SnapShot: var gene expression in the malaria parasite. Cell. 2008;134(1):190. doi: 10.1016/j.cell.2008.06.042. [DOI] [PubMed] [Google Scholar]
- Kirkman LA, Deitsch KW. Antigenic variation and the generation of diversity in malaria parasites. Curr Opin Microbiol. 2012;15(4):456–462. doi: 10.1016/j.mib.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwakalinga SB, Wang CW, Bengtsson DC, Turner L, Dinko B, Lusingu JP, Arnot DE, Sutherland CJ, Theander TG, Lavstsen T. Expression of a type B RIFIN in Plasmodium falciparum merozoites and gametes. Malar J. 2012;11:429. doi: 10.1186/1475-2875-11-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domergue R, Castano I, De Las PA, Zupancic M, Lockatell V, Hebel JR, Johnson D, Cormack BP. Nicotinic acid limitation regulates silencing of Candida adhesins during UTI. Science. 2005;308(5723):866–870. doi: 10.1126/science.1108640. [DOI] [PubMed] [Google Scholar]
- Prucca CG, Slavin I, Quiroga R, Elias EV, Rivero FD, Saura A, Carranza PG, Lujan HD. Antigenic variation in Giardia lamblia is regulated by RNA interference. Nature. 2008;456(7223):750–754. doi: 10.1038/nature07585. [DOI] [PubMed] [Google Scholar]
- Prucca CG, Lujan HD. Antigenic variation in Giardia lamblia. Cell Microbiol. 2009;11(12):1706–1715. doi: 10.1111/j.1462-5822.2009.01367.x. [DOI] [PubMed] [Google Scholar]
- Nash TE, Banks SM, Alling DW, Merritt JW Jr, Conrad JT. Frequency of variant antigens in Giardia lamblia. Exp Parasitol. 1990;71(4):415–421. doi: 10.1016/0014-4894(90)90067-M. [DOI] [PubMed] [Google Scholar]
- Ottaviani A, Gilson E, Magdinier F. Telomeric position effect: from the yeast paradigm to human pathologies? Biochimie. 2008;90(1):93–107. doi: 10.1016/j.biochi.2007.07.022. [DOI] [PubMed] [Google Scholar]
- Yankulov K. Dare to challenge the silence? Telomeric gene silencing revisited. Nucleus. 2011;2(6):513–516. doi: 10.4161/nucl.2.6.17710. [DOI] [PubMed] [Google Scholar]
- Rusche LN, Rine J. Switching the mechanism of mating type switching: a domesticated transposase supplants a domesticated homing endonuclease. Genes Dev. 2010;24(1):10–14. doi: 10.1101/gad.1886310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keely SP, Renauld H, Wakefield AE, Cushion MT, Smulian AG, Fosker N, Fraser A, Harris D, Murphy L, Price C, Quail MA, Seeger K, Sharp S, Tindal CJ, Warren T, Zuiderwijk E, Barrell BG, Stringer JR, Hall N. Gene arrays at Pneumocystis carinii telomeres. Genetics. 2005;170(4):1589–1600. doi: 10.1534/genetics.105.040733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkin SM, Stringer JR. Translocation of surface antigen genes to a unique telomeric expression site in Pneumocystis carinii. Mol Microbiol. 1996;19(2):283–295. doi: 10.1046/j.1365-2958.1996.375905.x. [DOI] [PubMed] [Google Scholar]
- Robinson NP, Burman N, Melville SE, Barry JD. Predominance of duplicative VSG gene conversion in antigenic variation in African trypanosomes. Mol Cell Biol. 1999;19(9):5839–5846. doi: 10.1128/mcb.19.9.5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo LM, Cross GA, Janzen CJ. Epigenetic regulation in African trypanosomes: a new kid on the block. Nat Rev Microbiol. 2009;7(7):504–513. doi: 10.1038/nrmicro2149. [DOI] [PubMed] [Google Scholar]
- Rudenko G. Epigenetics and transcriptional control in African trypanosomes. Essays Biochem. 2010;48(1):201–219. doi: 10.1042/bse0480201. [DOI] [PubMed] [Google Scholar]
- Hughes K, Wand M, Foulston L, Young R, Harley K, Terry S, Ersfeld K, Rudenko G. A novel ISWI is involved in VSG expression site downregulation in African trypanosomes. EMBO J. 2007;26(9):2400–2410. doi: 10.1038/sj.emboj.7601678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo LM, Janzen CJ, Cross GA. A histone methyltransferase modulates antigenic variation in African trypanosomes. PLoS Biol. 2008;6(7):e161. doi: 10.1371/journal.pbio.0060161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portugal S, Carret C, Recker M, Armitage AE, Gonçalves LA, Epiphanio S, Sullivan D, Roy C, Newbold CI, Drakesmith H, Mota MM. Host-mediated regulation of superinfection in malaria. Nat Med. 2011;17(6):732–737. doi: 10.1038/nm.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recker M, Buckee CO, Serazin A, Kyes S, Pinches R, Christodoulou Z, Springer AL, Gupta S, Newbold CI. Antigenic variation in Plasmodium falciparum malaria involves a highly structured switching pattern. PLoS Pathog. 2011;7(3):e1001306. doi: 10.1371/journal.ppat.1001306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recker M, Nee S, Bull PC, Kinyanjui S, Marsh K, Newbold C, Gupta S. Transient cross-reactive immune responses can orchestrate antigenic variation in malaria. Nature. 2004;429(6991):555–558. doi: 10.1038/nature02486. [DOI] [PubMed] [Google Scholar]
- Scherf A, Hernandez-Rivas R, Buffet P, Bottius E, Benatar C, Pouvelle B, Gysin J, Lanzer M. Antigenic variation in malaria: in situ switching, relaxed and mutually exclusive transcription of var genes during intra-erythrocytic development in Plasmodium falciparum. EMBO J. 1998;17(18):5418–5426. doi: 10.1093/emboj/17.18.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horrocks P, Pinches R, Christodoulou Z, Kyes SA, Newbold CI. Variable var transition rates underlie antigenic variation in malaria. Proc Natl Acad Sci USA. 2004;101(30):11129–11134. doi: 10.1073/pnas.0402347101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzikowski R, Li F, Amulic B, Eisberg A, Frank M, Patel S, Wellems TE, Deitsch KW. Mechanisms underlying mutually exclusive expression of virulence genes by malaria parasites. EMBO Rep. 2007;8(10):959–965. doi: 10.1038/sj.embor.7401063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M, Dzikowski R, Amulic B, Deitsch K. Variable switching rates of malaria virulence genes are associated with chromosomal position. Mol Microbiol. 2007;64(6):1486–1498. doi: 10.1111/j.1365-2958.2007.05736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitsch KW, Calderwood MS, Wellems TE. Malaria: cooperative silencing elements in var genes. Nature. 2001;412(6850):875–876. doi: 10.1038/35091146. [DOI] [PubMed] [Google Scholar]
- Voss TS, Healer J, Marty AJ, Duffy MF, Thompson JK, Beeson JG, Reeder JC, Crabb BS, Cowman AF. A var gene promoter controls allelic exclusion of virulence genes in Plasmodium falciparum malaria. Nature. 2006;439(7079):1004–1008. doi: 10.1038/nature04407. [DOI] [PubMed] [Google Scholar]
- Voss TS, Tonkin CJ, Marty AJ, Thompson JK, Healer J, Crabb BS, Cowman AF. Alterations in local chromatin environment are involved in silencing and activation of subtelomericvar genes in Plasmodium falciparum. Mol Microbiol. 2007;66(1):139–150. doi: 10.1111/j.1365-2958.2007.05899.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas-Junior LH, Hernandez-Rivas R, Ralph SA, Montiel-Condado D, Ruvalcaba-Salazar OK, Rojas-Meza AP, Mancio-Silva L, Leal-Silvestre RJ, Gontijo AM, Shorte S, Scherf A. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell. 2005;121(1):25–36. doi: 10.1016/j.cell.2005.01.037. [DOI] [PubMed] [Google Scholar]
- Lopez-Rubio JJ, Gontijo AM, Nunes MC, Issar N, Hernandez Rivas R, Scherf A. 5′ flanking region of var genes nucleate histone modification patterns linked to phenotypic inheritance of virulence traits in malaria parasites. Mol Microbiol. 2007;66(6):1296–1305. doi: 10.1111/j.1365-2958.2007.06009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chookajorn T, Dzikowski R, Frank M, Li F, Jiwani AZ, Hartl DL, Deitsch KW. Epigenetic memory at malaria virulence genes. Proc Natl Acad Sci USA. 2007;104(3):899–902. doi: 10.1073/pnas.0609084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmquist NA, Moss TA, Mecheri S, Scherf A, Fuchter MJ. Small-molecule histone methyltransferase inhibitors display rapid antimalarial activity against all blood stage forms in Plasmodium falciparum. Proc Natl Acad Sci USA. 2012;109(41):16708–16713. doi: 10.1073/pnas.1205414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Mu J, Zhang Q, Ni T, Srinivasan P, Rayavara K, Yang W, Turner L, Lavstsen T, Theander TG, Peng W, Wei G, Jing Q, Wakabayashi Y, Bansal A, Luo Y, Ribeiro JM, Scherf A, Aravind L, Zhu J, Zhao K, Miller LH. PfSETvs methylation of histone H3K36 represses virulence genes in Plasmodium falciparum. Nature. 2013;499(7457):223–227. doi: 10.1038/nature12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duraisingh MT, Voss TS, Marty AJ, Duffy MF, Good RT, Thompson JK, Freitas-Junior LH, Scherf A, Crabb BS, Cowman AF. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell. 2005;121(1):13–24. doi: 10.1016/j.cell.2005.01.036. [DOI] [PubMed] [Google Scholar]
- Ralph SA, Scheidig-Benatar C, Scherf A. Antigenic variation in Plasmodium falciparum is associated with movement of var loci between subnuclear locations. Proc Natl Acad Sci USA. 2005;102(15):5414–5419. doi: 10.1073/pnas.0408883102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epp C, Li F, Howitt CA, Chookajorn T, Deitsch KW. Chromatin associated sense and antisense noncoding RNAs are transcribed from the var gene family of virulence genes of the malaria parasite Plasmodium falciparum. RNA. 2009;15(1):116–127. doi: 10.1261/rna.1080109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekera AM, Patankar S, Schug J, Eisen G, Kissinger J, Roos D, Wirth DF. Widespread distribution of antisense transcripts in the Plasmodium falciparum genome. Mol Biochem Parasitol. 2004;136(1):35–42. doi: 10.1016/j.molbiopara.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Mourier T, Carret C, Kyes S, Christodoulou Z, Gardner PP, Jeffares DC, Pinches R, Barrell B, Berriman M, Griffiths-Jones S, Ivens A, Newbold C, Pain A. Genome-wide discovery and verification of novel structured RNAs in Plasmodium falciparum. Genome Res. 2008;18(2):281–292. doi: 10.1101/gr.6836108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadbent KM, Park D, Wolf AR, Van Tyne D, Sims JS, Ribacke U, Volkman S, Duraisingh M, Wirth D, Sabeti PC, Rinn JL. A global transcriptional analysis of Plasmodium falciparum malaria reveals a novel family of telomere-associated lncRNAs. Genome Biol. 2011;12(6):R56. doi: 10.1186/gb-2011-12-6-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum J, Papenfuss AT, Mair GR, Janse CJ, Vlachou D, Waters AP, Cowman AF, Crabb BS, de Koning-Ward TF. Molecular genetics and comparative genomics reveal RNAi is not functional in malaria parasites. Nucleic Acids Res. 2009;37(11):3788–3798. doi: 10.1093/nar/gkp239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raabe CA, Sanchez CP, Randau G, Robeck T, Skryabin BV, Chinni SV, Kube M, Reinhardt R, Ng GH, Manickam R, Kuryshev VY, Lanzer M, Brosius J, Tang TH, Rozhdestvensky TS. A global view of the nonprotein-coding transcriptome in Plasmodium falciparum. Nucleic Acids Res. 2009;38(2):608–617. doi: 10.1093/nar/gkp895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph SA, Bischoff E, Mattei D, Sismeiro O, Dillies MA, Guigon G, Coppee JY, David PH, Scherf A. Transcriptome analysis of antigenic variation in Plasmodium falciparum–var silencing is not dependent on antisense RNA. Genome Biol. 2005;6(11):R93. doi: 10.1186/gb-2005-6-11-r93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Las PA, Pan SJ, Castano I, Alder J, Cregg R, Cormack BP. Virulence-related surface glycoproteins in the yeast pathogen Candida glabrata are encoded in subtelomeric clusters and subject to RAP1- and SIR-dependent transcriptional silencing. Genes Dev. 2003;17(18):2245–2258. doi: 10.1101/gad.1121003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas-Hernandez LL, Juarez-Reyes A, Arroyo-Helguera OE, De Las PA, Pan SJ, Cormack BP, Castano I. yKu70/yKu80 and Rif1 regulate silencing differentially at telomeres in Candida glabrata. Eukaryot Cell. 2008;7(12):2168–2178. doi: 10.1128/EC.00228-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulakova L, Singer SM, Conrad J, Nash TE. Epigenetic mechanisms are involved in the control of Giardia lamblia antigenic variation. Mol Microbiol. 2006;61(6):1533–1542. doi: 10.1111/j.1365-2958.2006.05345.x. [DOI] [PubMed] [Google Scholar]
- Yankulov K. Dynamics and stability: epigenetic conversions in position effect variegation. Biochem Cell Biol. 2013;91(1):6–13. doi: 10.1139/bcb-2012-0048. [DOI] [PubMed] [Google Scholar]
- Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- Rehman MA, Yankulov K. The dual role of autonomously replicating sequences as origins of replication and as silencers. Curr Genet. 2009;55(4):357–363. doi: 10.1007/s00294-009-0265-7. [DOI] [PubMed] [Google Scholar]
- Feuerhahn S, Iglesias N, Panza A, Porro A, Lingner J. TERRA biogenesis, turnover and implications for function. FEBS Lett. 2010;584(17):3812–3818. doi: 10.1016/j.febslet.2010.07.032. [DOI] [PubMed] [Google Scholar]
- Yang X, Figueiredo LM, Espinal A, Okubo E, Li B. RAP1 is essential for silencing telomeric variant surface glycoprotein genes in Trypanosoma brucei. Cell. 2009;137(1):99–109. doi: 10.1016/j.cell.2009.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsford S, Kawahara T, Isamah C, Horn D. A sirtuin in the African trypanosome is involved in both DNA repair and telomeric gene silencing but is not required for antigenic variation. Mol Microbiol. 2007;63(3):724–736. doi: 10.1111/j.1365-2958.2006.05553.x. [DOI] [PubMed] [Google Scholar]
- Tonkin CJ, Carret CK, Duraisingh MT, Voss TS, Ralph SA, Hommel M, Duffy MF, Silva LM, Scherf A, Ivens A, Speed TP, Beeson JG, Cowman AF. Sir2 paralogues cooperate to regulate virulence genes and antigenic variation in Plasmodium falciparum. PLoS Biol. 2009;7(4):e84. doi: 10.1371/journal.pbio.1000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A, Gasser SM. Structure and function in the budding yeast nucleus. Genetics. 2012;192(1):107–129. doi: 10.1534/genetics.112.140608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchmaier AL, Rine J. DNA replication-independent silencing in S. cerevisiae. Science. 2001;291(5504):646–650. doi: 10.1126/science.291.5504.646. [DOI] [PubMed] [Google Scholar]
- Maillet L, Gaden F, Brevet V, Fourel G, Martin SG, Dubrana K, Gasser SM, Gilson E. Ku-deficient yeast strains exhibit alternative states of silencing competence. EMBO Rep. 2001;2(3):203–210. doi: 10.1093/embo-reports/kve044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourel G, Lebrun E, Gilson E. Protosilencers as building blocks for heterochromatin. Bioessays. 2002;24(9):828–835. doi: 10.1002/bies.10139. [DOI] [PubMed] [Google Scholar]
- Fourel G, Magdinier F, Gilson E. Insulator dynamics and the setting of chromatin domains. Bioessays. 2004;26(5):523–532. doi: 10.1002/bies.20028. [DOI] [PubMed] [Google Scholar]
- Fourel G, Revardel E, Koering CE, Gilson E. Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J. 1999;18(9):2522–2537. doi: 10.1093/emboj/18.9.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power P, Jeffery D, Rehman MA, Chatterji A, Yankulov K. Sub-telomeric core X and Y’ elements in S. cerevisiae suppress extreme variations in gene silencing. PLoS One. 2011;6(3):e17523. doi: 10.1371/journal.pone.0017523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryde FE, Louis EJ. Limitations of silencing at native yeast telomeres. EMBO J. 1999;18(9):2538–2550. doi: 10.1093/emboj/18.9.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E, Geli V. How telomeres are replicated. Nat Rev Mol Cell Biol. 2007;8(10):825–838. doi: 10.1038/nrm2259. [DOI] [PubMed] [Google Scholar]
- Smargiasso N, Gabelica V, Damblon C, Rosu F, De Pauw E, Teulade-Fichou MP, Rowe JA, Claessens A. Putative DNA G-quadruplex formation within the promoters of Plasmodium falciparum var genes. BMC Genomics. 2009;10:362. doi: 10.1186/1471-2164-10-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallegos-Garcia V, Pan SJ, Juarez-Cepeda J, Ramirez-Zavaleta CY, Martin-del-Campo MB, Martinez-Jimenez V, Castano I, Cormack B, De Las PA. A novel downstream regulatory element cooperates with the silencing machinery to repress EPA1 expression in Candida glabrata. Genetics. 2012;190(4):1285–1297. doi: 10.1534/genetics.111.138099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez-Reyes A, Ramirez-Zavaleta CY, Medina-Sanchez L, De Las PA, Castano I. A protosilencer of subtelomeric gene expression in Candida glabrata with unique properties. Genetics. 2012;190(1):101–111. doi: 10.1534/genetics.111.135251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flueck C, Bartfai R, Niederwieser I, Witmer K, Alako BT, Moes S, Bozdech Z, Jenoe P, Stunnenberg HG, Voss TS. A major role for the Plasmodium falciparum ApiAP2 protein PfSIP2 in chromosome end biology. PLoS Pathog. 2010;6(2):e1000784. doi: 10.1371/journal.ppat.1000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renauld H, Aparicio OM, Zierath PD, Billington BL, Chhablani SK, Gottschling DE. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 1993;7(7A):1133–1145. doi: 10.1101/gad.7.7a.1133. [DOI] [PubMed] [Google Scholar]
- Aparicio OM, Gottschling DE. Overcoming telomeric silencing: a trans-activator competes to establish gene expression in a cell cycle-dependent way. Genes Dev. 1994;8(10):1133–1146. doi: 10.1101/gad.8.10.1133. [DOI] [PubMed] [Google Scholar]
- Ahmad K, Henikoff S. Modulation of a transcription factor counteracts heterochromatic gene silencing in Drosophila. Cell. 2001;104(6):839–847. doi: 10.1016/S0092-8674(01)00281-1. [DOI] [PubMed] [Google Scholar]
- Fastman Y, Noble R, Recker M, Dzikowski R. Erasing the epigenetic memory and beginning to switch–the onset of antigenic switching of var genes in Plasmodium falciparum. PLoS One. 2012;7(3):e34168. doi: 10.1371/journal.pone.0034168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B, Pan SJ, Domergue R, Rigby T, Whiteway M, Johnson D, Cormack BP. High-affinity transporters for NAD + precursors in Candida glabrata are regulated by Hst1 and induced in response to niacin limitation. Mol Cell Biol. 2009;29(15):4067–4079. doi: 10.1128/MCB.01461-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneghini MD, Wu M, Madhani HD. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112(5):725–736. doi: 10.1016/S0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers WA, Salcedo-Amaya AM, Smits AH, Francoijs KJ, Treeck M, Gilberger TW, Stunnenberg HG, Bartfai R. H2A.Z/H2B.Z double-variant nucleosomes inhabit the AT-rich promoter regions of the Plasmodium falciparum genome. Mol Microbiol. 2013;87(5):1061–1073. doi: 10.1111/mmi.12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S. Chromatin-based transcriptional punctuation. Genes Dev. 2009;23(9):1037–1041. doi: 10.1101/gad.1806409. [DOI] [PubMed] [Google Scholar]
- Siegel TN, Hekstra DR, Kemp LE, Figueiredo LM, Lowell JE, Fenyo D, Wang X, Dewell S, Cross GA. Four histone variants mark the boundaries of polycistronic transcription units in Trypanosoma brucei. Genes Dev. 2009;23(9):1063–1076. doi: 10.1101/gad.1790409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AT, Zhang Y. The diverse functions of Dot1 and H3K79 methylation. Genes Dev. 2011;25(13):1345–1358. doi: 10.1101/gad.2057811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol. 2012;13(2):115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz L, Brown GV, Genton B, Moorthy VS. A review of malaria vaccine clinical projects based on the WHO rainbow table. Malar J. 2012;11:11. doi: 10.1186/1475-2875-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Greca F, Magez S. Vaccination against trypanosomiasis: can it be done or is the trypanosome truly the ultimate immune destroyer and escape artist? Hum Vaccin. 2011;7(11):1225–1233. doi: 10.4161/hv.7.11.18203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seder RA, Chang LJ, Enama ME, Zephir KL, Sarwar UN, Gordon IJ, Holman LA, James ER, Billingsley PF, Gunasekera A, Richman A, Chakravarty S, Manoj A, Velmurugan S, Li M, Ruben AJ, Li T, Eappen AG, Stafford RE, Plummer SH, Hendel CS, Novik L, Costner PJ, Mendoza FH, Saunders JG, Nason MC, Richardson JH, Murphy J, Davidson SA, Richie TL. et al. Protection against malaria by intravenous immunization with a nonreplicating sporozoite vaccine. Science. 2013;341(6152):1359–1365. doi: 10.1126/science.1241800. [DOI] [PubMed] [Google Scholar]
- Prusty D, Mehra P, Srivastava S, Shivange AV, Gupta A, Roy N, Dhar SK. Nicotinamide inhibits Plasmodium falciparum Sir2 activity in vitro and parasite growth. FEMS Microbiol Lett. 2008;282(2):266–272. doi: 10.1111/j.1574-6968.2008.01135.x. [DOI] [PubMed] [Google Scholar]
- Castano I, Pan SJ, Zupancic M, Hennequin C, Dujon B, Cormack BP. Telomere length control and transcriptional regulation of subtelomericadhesins in Candida glabrata. Mol Microbiol. 2005;55(4):1246–1258. doi: 10.1111/j.1365-2958.2004.04465.x. [DOI] [PubMed] [Google Scholar]
- Doheny JG, Mottus R, Grigliatti TA. Telomeric position effect–a third silencing mechanism in eukaryotes. PLoS One. 2008;3(12):e3864. doi: 10.1371/journal.pone.0003864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace JA, Orr-Weaver TL. Replication of heterochromatin: insights into mechanisms of epigenetic inheritance. Chromosoma. 2005;114(6):389–402. doi: 10.1007/s00412-005-0024-6. [DOI] [PubMed] [Google Scholar]
- Tabancay AP Jr, Forsburg SL. Eukaryotic DNA replication in a chromatin context. Curr Top Dev Biol. 2006;76:129–184. doi: 10.1016/S0070-2153(06)76005-7. [DOI] [PubMed] [Google Scholar]
- Osborne EA, Hiraoka Y, Rine J. Symmetry, asymmetry, and kinetics of silencing establishment in Saccharomyces cerevisiae revealed by single-cell optical assays. Proc Natl Acad Sci USA. 2011;108(4):1209–1216. doi: 10.1073/pnas.1018742108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu EY, Bi X, Holland MJ, Gottschling DE, Broach JR. Mutations in the nucleosome core enhance transcriptional silencing. Mol Cell Biol. 2005;25(5):1846–1859. doi: 10.1128/MCB.25.5.1846-1859.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CM, Haimberger ZW, Johnson CO, Wolf AJ, Gafken PR, Zhang Z, Parthun MR, Gottschling DE. Heritable chromatin structure: mapping “memory” in histones H3 and H4. Proc Natl Acad Sci USA. 2002;99(4):16454–16461. doi: 10.1073/pnas.182424999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillus L, Rine J. Epigenetic inheritance of transcriptional states in S. cerevisiae. Cell. 1989;59(4):637–647. doi: 10.1016/0092-8674(89)90009-3. [DOI] [PubMed] [Google Scholar]
- Xu EY, Zawadzki KA, Broach JR. Single-cell observations reveal intermediate transcriptional silencing states. Mol Cell. 2006;23(2):219–229. doi: 10.1016/j.molcel.2006.05.035. [DOI] [PubMed] [Google Scholar]
- Jeffery DC, Wyse BA, Rehman MA, Brown GW, You Z, Oshidari R, Masai H, Yankulov KY. Analysis of epigenetic stability and conversions in Saccharomyces cerevisiae reveals a novel role of CAF-I in position-effect variegation. Nucleic Acids Res. 2013;41(18):8475–8488. doi: 10.1093/nar/gkt623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabert C, Groth A. Chromatin replication and epigenome maintenance. Nat Rev Mol Cell Biol. 2012;13(3):153–167. doi: 10.1038/nrm3288. [DOI] [PubMed] [Google Scholar]
- Coleman BI, Ribacke U, Manary M, Bei AK, Winzeler EA, Wirth DF, Duraisingh MT. Nuclear repositioning precedes promoter accessibility and is linked to the switching frequency of a Plasmodium falciparum invasion gene. Cell Host Microbe. 2012;12(6):739–750. doi: 10.1016/j.chom.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Model for epigenetic conversions driven by subtle fluctuations of activators and/or silencing factors. (1) All but one of a family of hypothetical varying genes (VG1 to VGN) are maintained in a silenced state. These genes are flanked by a subtelomeric anti-silencing region (STAR) and a chromatin boundary (B). (2) A subtle increase in the abundance of gene activators (green circle) and/or factors that engage STAR (purple circle) and the concomitant passage of replication forks would allow the activators access to the promoters of the VG genes and (3) would predispose all VG loci to derepression. (4) Consequently, during the next stage (re-establishment of silencing), the derepressed VG genes (VG2 to VGN, pink) will compete with the currently active gene (VG1, red). (5) During this stage, a decline in the abundance of activators and STAR-acting factors would aid in the formation of heterochromatin and the limiting activators would be gradually sequestered to a single locus. There is a high probability that the currently active gene (VG1) will be reinstated as the active locus. However, switches to another gene are possible (VGN). The likelihood of such switches, depicted by the width of the arrow, represents the frequency of epigenetic conversions.