Abstract
CHD proteins are members of the chromo domain family, a class of proteins involved in transcription, DNA degradation and chromatin structure. In higher eukaryotes, there are two distinct subfamilies of CHD proteins: CHD1 and CHD3/4. Analyses carried out in vitro indicate that the CHD3/4 proteins may regulate transcription via alteration of chromatin structure. However, little is known about the role of CHD proteins in vivo, particularly the CHD1 subfamily. To understand better the cellular function of CHD proteins, we initiated a study on the Chd1p protein from budding yeast. Using genomic DNA arrays, we identified genes whose expression is affected by the absence of Chd1p. A synthetic-lethal screen uncovered genetic interactions between SWI/SNF genes and CHD1. Biochemical experiments using Chd1p purified from yeast showed that it reconfigures the structure of nucleosome core particles in a manner distinct from the SWI–SNF complex. Taken together, these results suggest that Chd1p functions as a nucleosome remodeling factor, and that Chd1p may share overlapping roles with the SWI–SNF complex to regulate transcription.
Keywords: chromo domain/genomic array/nucleosome remodeling/synthetic lethality
Introduction
Chromo domain proteins are found in all eukaryotes (Koonin et al., 1995; Cavalli and Paro, 1998). The chromo domain was originally recognized as a 37 amino acid segment in the Drosophila Polycomb protein that shared close sequence similarity with a portion of the heterochromatin-associated protein HP1 (Paro and Hogness, 1991). Polycomb and HP1 were believed to act as regulators of transcription via formation of a higher order chromatin structure, hence the name ‘chromo domain’. Other chromo domain proteins, defined as having sequence similarity to this 37 amino acid segment, include Pdd1p, a Tetrahymena protein involved in programmed DNA degradation (Madireddi et al., 1996), and SWI6p, a protein required for the maintenance of heterochromatin-like regions in Schizosaccharomyces pombe (Ekwall et al., 1995). Although chromo domain proteins have been studied for over a decade, little was known about their molecular mode of action until recently.
A subset of chromo domain proteins, the CHD family, consists of proteins sharing three sequence features: a chromo domain, an ATPase/helicase and a DNA binding segment, although not all proteins termed CHD have this last domain (Delmas et al., 1993; Woodage et al., 1997). CHD proteins are well conserved, with members found from yeast to plants to mammals. In higher eukaryotes, up to four distinct CHD genes are present in the genome; for example, the human genome includes CHD1, CHD2, CHD3 and CHD4 (Woodage et al., 1997). The human CHD3 and CHD4 proteins have recently been shown to co-purify with each other and with the human histone deacetylase complex (HDAC) (Tong et al., 1998; Xue et al., 1998; Zhang et al., 1998). A Xenopus CHD protein related to the human CHD3/4 proteins has also been purified with a deacetylase complex (Wade et al., 1998). Additionally, the human CHD3/4 complex was shown to have nucleosome remodeling activity. These results suggest that at least some CHD proteins may be involved in altering the chromatin environment around genes. A model was proposed in which the activity of CHD3/4 increased access to the histones, allowing the deacetylase to modify the histone tails and subsequently leading to greater compaction of nucleosomal structure and inhibition of transcription (Zhang et al., 1998). Further support for a role of CHD3/4 proteins in transcriptional repression comes from the study of the Drosophila dMi-2 protein, a relative of the human CHD3 and CHD4 proteins. dMi-2 was found to interact in a two-hybrid screen with Hunchback, a protein required for the repression of homeotic genes, and mutations in dMi-2 affect both Hunchback- and Polycomb-mediated repression (Kehle et al., 1998).
In contrast, work on the Drosophila DmCHD1 suggests that it is involved in gene activation. Null mutations of DmCHD1 have not been reported, but, using immunofluorescence, the DmCHD1 protein was localized to puffs and interband regions on polytene chromosomes, areas generally associated with active transcription (Stokes et al., 1996). One possible explanation for the dichotomy of these results is that the CHD genes have diverged, such that different classes of CHD gene have different cellular roles. In fact, phylogenetic analysis of the CHD family revealed that the CHD3/4 class of genes is in a subfamily distinct from that of the CHD1 members (Woodage et al., 1997).
The genome of the budding yeast, Saccharomyces cerevisiae, encodes a single CHD protein, Chd1p, which most closely resembles the CHD1 subgroup from more complex eukaryotes (Woodage et al., 1997). CHD1-null mutations in yeast are viable but have subtle phenotypes when grown under special conditions (Woodage et al., 1997; Jin et al., 1998; Tsukiyama et al., 1999). For example, a chd1Δ strain is resistant to high concentrations of 6-azauracil (6AU), a pyrimidine analog that is toxic to wild-type cells. Since RNA polymerase II and elongation factor SII mutants are sensitive to 6AU, it was proposed that CHD1 may act as a negative regulator of transcription (Woodage et al., 1997). Taken together, the work from mammals, flies and yeast does not yet provide a clear understanding of the genetic roles of the CHD1 subgroup of proteins or how these proteins function biochemically.
To increase our understanding of the CHD proteins, and of the function of the CHD1 subfamily in particular, we have chosen to study the CHD1 gene of S.cerevisiae. Since biochemical and genetic experimentation in this organism are feasible, we applied both approaches simultaneously to this problem. Among other features, our investigation has revealed that Chd1p is a nucleosome remodeling factor that is biochemically distinct from the well characterized SWI–SNF complex, but may share overlapping functions with SWI–SNF in vivo.
Results
Genome-wide analysis of CHD1 function
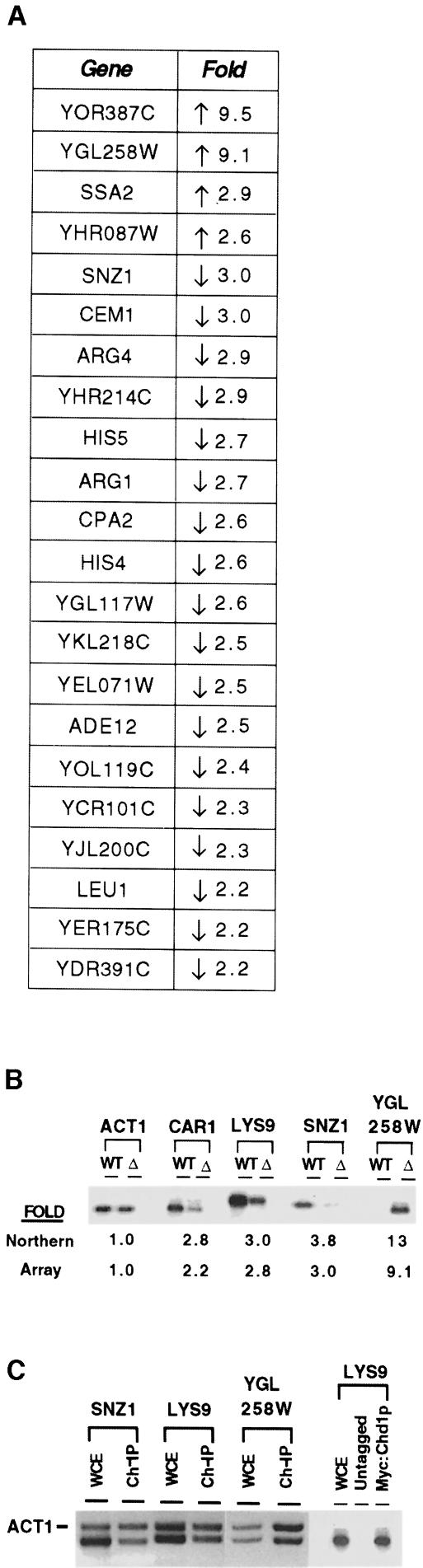
Since preliminary work in Drosophila and yeast suggests that CHD1 may be a transcriptional regulator, we carried out a genome-wide analysis of transcription, using DNA microarray technology (DeRisi et al., 1997), to examine the possible involvement of CHD1 in this process. We purified mRNA from wild-type and CHD1-deletion strains grown in minimal medium. Fluorescent probes from the two mRNA samples were synthesized and hybridized to a DNA microarray representing the yeast genome, and differential effects on transcript levels of ∼6000 genes were monitored. In two independent experiments, the expression of ∼2–4% of the genes in the genome was affected by the absence of CHD1 by a factor of ≥2.0 (complete data set available upon request). Twenty-three genes were consistently affected by the loss of CHD1 in both experiments (Figure 1A). The differences between the two array experiments are most likely due to minor differences in the growth conditions of the cells and in the quality of the RNA. Northern blot analyses verified that a selected subset of genes are differentially expressed in the wild-type and deletion strains (Figure 1B). Northern blots (using RNA from a third independent preparation) also indicate that genes affected in only one of the two array hybridizations are indeed affected by the absence of CHD1 (CAR1 and LYS9 in Figure 1B). The results from the DNA array experiment are consistent with the idea that Chd1p is a regulator of transcription.
Fig. 1. Analysis of CHD1’s effect on genome-wide expression. (A) Table of genes whose expression is affected by ≥2-fold in both microarrays. The ‘Fold’ column indicates the average between experiments. Upward-pointing arrows signify that gene expression increased in the deletion strain, while downward-pointing arrows indicate that gene expression decreased in the deletion strain. (B) Northern blot of genes identified from genomic DNA array analysis. Four genes affected by the absence of CHD1, as determined by the array experiment, show similar expression differences when analyzed by RNA Northern blots. (C) PCR analysis of DNA cross-linked to Chd1p. PCRs were carried out using either total DNA from whole cell extracts (WCE) or chromatin immunoprecipitated DNA (Ch-IP), and primers targeted against genes affected by the absence of CHD1. Primers for the ACT1 gene, which is unaffected by the loss of CHD1, were also added to the reactions. The right panel shows a control reaction with DNA from an untagged strain.
In an effort to determine which genes may be directly regulated by Chd1p, we performed a series of chromatin immunoprecipitation experiments. Briefly, Myc::Chd1p was cross-linked to DNA in vivo by treating cells with formaldehyde. A variety of cross-linking times, ranging from 1 to 15 min, were used in an attempt to eliminate non-specific cross-linking. The cells were harvested and lysed, and the chromatin was extracted and sheared by sonication to produce DNA fragments with an average size of ∼400 bp. Anti-Myc antibodies coupled to beads were added to the sheared chromatin to precipitate DNA that had been cross-linked to Chd1p. After removing cross-linked proteins, the DNA was extracted and analyzed using the polymerase chain reaction (PCR). Primers directed against the promoter region of Chd1p-regulated genes as well as primers for a gene (ACT1) not affected by Chd1p were added together in the PCRs.
Chromatin immunoprecipitations carried out in extracts derived from a MYC::CHD1 strain precipitated at least 500-fold more DNA than a control extract in which Chd1p was not tagged with Myc (our unpublished data; see Figure 1C), showing that Chd1p can be very effectively cross-linked to DNA. However, the immunoprecipitation step did not preferentially precipitate DNA from genes regulated by Chd1p compared with the control gene (Figure 1C). The ACT1 gene was detected along with SNZ1, LYS9 and YGL258W genes in the Chd1p-precipitated DNA pool. The most likely interpretation of this result is that Chd1p is an abundant protein that is bound to chromatin throughout the genome, and whose absence affects the expression of only certain genes.
The results from the chromatin immunoprecipitation experiment prompted us to estimate the concentration of Chd1p in cells. The Western blot signal of Chd1p in titrations of whole cell extract was compared with the signal of Chd1p in titrations of purified Chd1p (data not shown; see below). Using this technique, we estimate that there are ∼1000 molecules of Chd1p per cell. This amount corresponds to approximately one Chd1p molecule for every 12 500 bp (one every six genes) or about one molecule for every 50 nucleosomes.
Genetic interactions with SWI/SNF genes
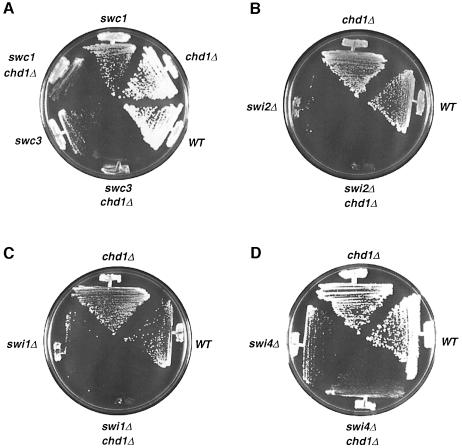
Given that the DNA array results support a role for CHD1 in transcription, we attempted to uncover genetically the cellular function of Chd1p responsible for its effect on gene expression. A synthetic-lethal screen was undertaken to identify genes that may share redundant or overlapping functions with CHD1. Since a CHD1-deleted strain is viable and has only subtle phenotypes, it is possible that other genes may be able to substitute for the loss of CHD1 function. Synthetic-lethal screening is a common method used to uncover mutations in a second gene that will require the cell to maintain a wild-type copy of the gene being studied in order to survive (Bender and Pringle, 1991). A colony sectoring assay was implemented to identify mutants of interest by their inability to lose a wild-type copy of CHD1.
The starting strain was deleted for the genomic copy of CHD1 and carried a plasmid containing a wild-type copy of CHD1 and ADE3. When grown on low adenine plates, cells form red colonies when they maintain the CHD1, ADE3 plasmid. Because CHD1 is not essential, cells can lose the plasmid and form red colonies with white sectors. After UV mutagenesis, 75 000 cells were plated on low adenine plates and colonies that stayed completely red, indicating that these cells cannot lose the CHD1 plasmid, were chosen for further analysis. Three mutants remained after subsequent testing; these were named swc1, swc2 and swc3 for synthetically sick with chd1. We observed that the swc mutants are not synthetically lethal with chd1; rather, they grow very slowly without a functional copy of CHD1, and hence have a growth advantage when they maintain the CHD1 plasmid (Figure 2A).
Fig. 2. A chd1 mutation is synthetically sick/lethal with swi/snf mutations. (A) A chd1Δ strain is severely sick when combined with a swc1/2 (swi2) or swc3 (alr1) mutation. (B and C) A chd1Δ swi2Δ double mutant (B) and a chd1Δ swi1Δ double mutant (C) are both unable to survive on 5-FOA when forced to lose a wild-type copy of CHD1 on a plasmid containing the URA3 gene (pAJ741). All strains in (B) and (C) possess the pAJ741 plasmid before being streaked on 5-FOA plates. (D) A swi4Δ chd1Δ strain is viable.
swc1 and swc2 form one complementation group and swc3 another. SWC3 was determined to be ALR1 by transformation of the swc3 mutant with a genomic library and isolation of plasmid sequences that complemented the slow growth defect of swc3. A disruption of the ALR1 gene and construction of an alr1Δ chd1Δ double mutant verified the synthetic sickness phenotype (our unpublished data). ALR1 codes for a magnesium transporter (MacDiarmid and Gardner, 1998) and is not an essential gene in the W303 strain (our unpublished data). We speculate that Chd1p may regulate another component of the magnesium uptake/metabolism pathway that, when eliminated, leads to cell death in an alr1 background. The data from the DNA microarray experiment cannot be used to identify this gene because of strain differences. We used a W303 strain in our screen and an S288C strain for the array experiment; there are significant differences in the phenotype of an alr1 deletion between the two strains.
swc1 and swc2 were complemented by transformation with an SWI2 plasmid. A chd1Δ swi2Δ double mutant was constructed and found to be inviable (Figure 2B). The inviability of the double mutant, in contrast to the synthetic sickness phenotype of the mutants generated from the screen, could be due to the strain differences or to the possibility that the swc1 and swc2 mutations are reduction-of-function rather than null alleles of SWI2. swc1, swc2 and swc3 were all transformed with a SWI2 plasmid, but only swc1 and swc2 were complemented by the plasmid. SWC1(2) was confirmed to be an allele of SWI2 by an allelism test (see Materials and methods for details).
The genetic interaction between CHD1 and SWI2 suggests that their gene products may have similar and redundant functions as regulators of transcription via chromatin alteration (Workman and Kingston, 1998). Swi2p is the ATPase ‘engine’ of the SWI–SNF remodeling complex and is required for the nucleosome remodeling activity of the complex in vitro (reviewed in Travers, 1999). It is possible that Chd1p’s biochemical activity may overlap with that of the entire SWI–SNF complex, and not simply with Swi2p. To test this idea, we checked for a genetic interaction between CHD1 and a gene coding for a different component of the SWI–SNF complex: SWI1 (Cairns et al., 1994; Peterson et al., 1994). A chd1Δ swi1Δ double mutant is also synthetically lethal (Figure 2B), implying that Chd1p shares functions with the SWI–SNF complex as a whole. As a control, we also looked for interactions with another SWI gene, SWI4, which does not code for a protein that is part of the SWI–SNF complex. A swi4Δ chd1Δ double mutant is viable (Figure 2D), demonstrating that CHD1 interacts specifically with genes encoding subunits of the SWI–SNF remodeling complex.
To investigate further the genetic interaction between CHD1 and SWI2, we compared the genome-wide expression profile of a chd1Δ strain with that of an snf2Δ (swi2Δ) strain (Sudersanam et al., 2000). The expression of ∼35 genes was similarly affected (>1.8-fold difference) by the absence of CHD1 and SNF2. Because the growth conditions and strain background were different for the chd1 and snf2 strains used in the microarray experiments, the comparison between the two data sets is not ideal, and this estimate of the overlap may not be completely accurate. Still, this result is consistent with the possibility that there is a small subset of genes coordinately regulated by both CHD1 and SWI2. It is possible that one, or more, essential gene requires both CHD1 and SWI2 for proper expression and the synthetic lethality between the two mutants is due to the lack of expression of this vital gene(s). In any case, the evidence indicates that CHD1 and SWI2 have overlapping, but not superimposed, functions in the cell.
Purification of Chd1p
Because chd1 is synthetically lethal with swi2, and because the two proteins show sequence similarity (Eisen et al., 1995), we tested whether Chd1p possesses biochemical activities resembling those of Swi2p. To this end, Chd1p was purified from yeast. The purification was facilitated by tagging Chd1p with a Myc6::His6 fragment at the N-terminus. We know that tagged Chd1p is functional in the cell because the hybrid CHD1 gene fully complemented a chd1Δ mutation (monitored by the ability of MYC::HIS::CHD1 to complement a chd1 swi2 mutant) when integrated into the yeast genome and transcribed from its own promoter. The expression level of the tagged protein is similar to that of the wild-type protein (our unpublished data).
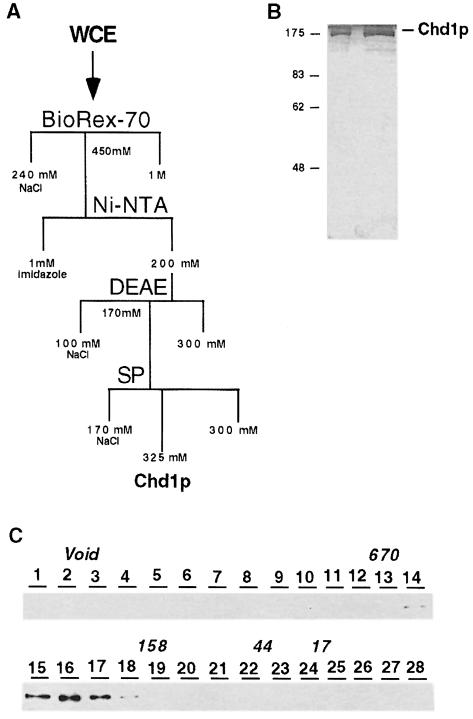
Using column chromatography to fractionate yeast whole cell extract and Western analysis to identify Chd1p-containing fractions, Myc-tagged Chd1p was purified to near homogeneity (Figure 3). The silver-stained gel shows that Chd1p is the predominant protein in the purest fractions and that no other proteins co-purified with Chd1p in stoichiometric amounts (Figure 3B). We believe that the faint bands in these fractions correspond to proteins that are contaminants, because these same proteins elute off the SP column at distinct peaks several fractions after the peak Chd1p fractions. Chd1p obtained from the first column eluted from a Superose-6 gel filtration column with a nominal size of ∼340 kDa (Figure 3C). Chd1p from the SP-Sepharose fractions eluted from the Superose-6 column with a similar apparent size (our unpublished data). These results suggest that the purified Chd1p is either a monomer with a large Stokes’ radius or a dimer. Also, during our purification, Western blots of early columns showed that there was no heterogeneity in the elution profile of Chd1p, suggesting that there was only one major form of Chd1p in our extracts. However, we cannot rule out the possibility that native Chd1p exists within the cell as a larger protein complex that disassembles during our cell lysis procedure.
Fig. 3. Purification of Chd1p. (A) Schematic flowchart of the chromatographic steps used to purify Chd1p. (B) Silver-stained gel of the purest fraction of Chd1p from the SP-Sepharose column. Lane 1 contains 1 µl (∼15 ng) and lane 2 contains 5 µl of protein. (C) Western blot showing the elution profile of Chd1p collected from a Superose 6 sizing column. The elution profile of molecular weight standards is indicated as 670 (kDa), 158, 44 and 17.
Chd1p alters the structure of nucleosome core particles in an ATP-dependent manner distinct from the SWI–SNF complex
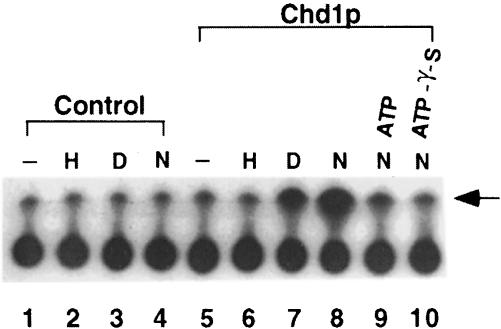
To begin a functional analysis of purified Chd1p, ATPase assays were conducted in the presence of a range of potential substrates. As shown in Figure 4, the ability of Chd1p to hydrolyze ATP is stimulated by free DNA and nucleosomal DNA, but not by core histones. In this respect, the ATPase activity resembles that of Swi2p-related subunits, which are also stimulated by free DNA and nucleosomal DNA (Laurent et al., 1993; Cairns et al., 1996).
Fig. 4. Chd1p possesses DNA- and nucleosome-stimulated ATPase activity. ATPase assays contained purified Chd1p (2 µl) and 1 pmol of nucleosomes (N) or the equivalent amount of DNA (D) or core histones (H). As competitors, unlabeled ATP and ATP-γ-S were added to the indicated reactions at a 5-fold molar excess over the standard ATP concentration. The arrow points to the signal derived from free phosphate.
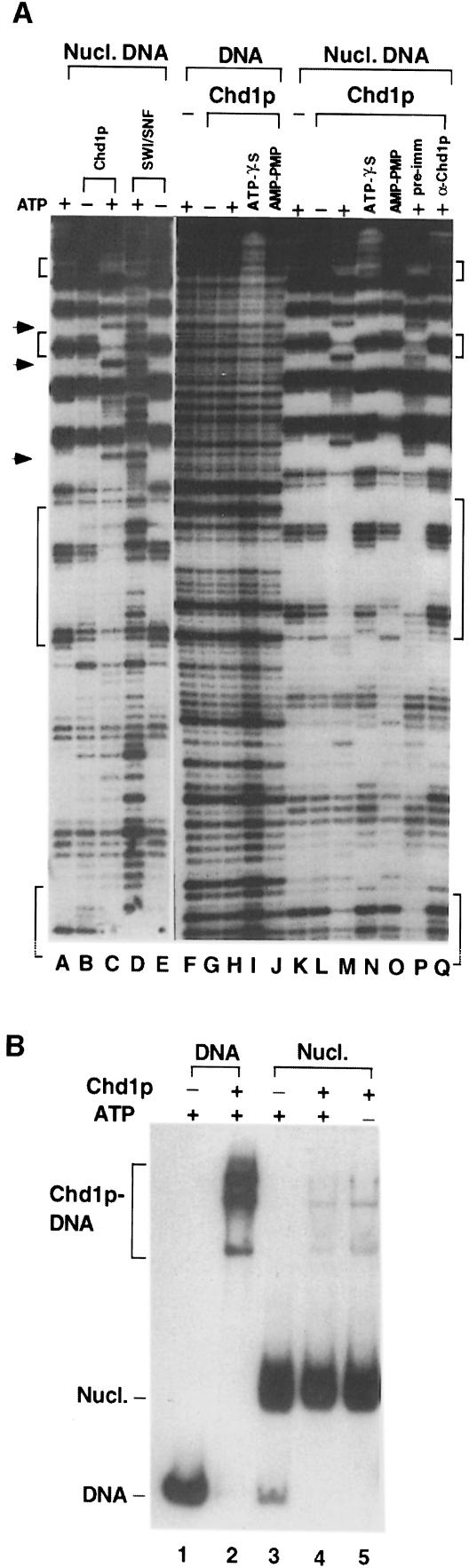
Complexes containing Swi2p-related subunits have been demonstrated to disrupt histone–DNA contacts within nucleosome core particles assembled with a sea urchin 5S rRNA gene fragment (for examples, see Cairns et al., 1996; Owen-Hughes et al., 1996). Disruption of the core particles is detected by a change in accessibility to DNase I digestion, and requires the energy released from ATP hydrolysis. To address whether Chd1p can remodel nucleosomes, we added purified Chd1p to 5S nucleosome core particles and examined the resulting DNase I cleavage pattern. Because the 5S rDNA sequence used for this analysis adopts a specific rotational position relative to the underlying octamer, DNase I predominantly cuts 5S mononucleosomes at sites ∼10 bp apart (Figure 5A, lane A). In the presence of ATP, but not in its absence, treatment with Chd1p generated protection from DNase I cleavage at several regions of the DNA and produced sites of hypersensitivity flanking these regions (Figure 5A, compare lane B with C, and lane L with M). In contrast, purified SWI–SNF increased the number of sites attacked by DNase I to produce a more uniform cleavage pattern throughout the DNA (Figure 5A, compare lane C with D). These data reveal that the ATP-dependent changes conferred by Chd1p are distinct from those of SWI–SNF.
Fig. 5. Chd1p alters nucleosome core particle structure. (A) Purified Chd1p (0.5 µl) or purified SWI–SNF was incubated with mock-reconstituted 5S DNA (DNA) or nucleosome-assembled 5S DNA (Nucl. DNA) and subjected to DNase I protection analysis. Where indicated, ATP and its respective analogs were added to a final concentration of 1 mM. To challenge Chd1p activity, 1 µl of anti-Chd1p polyclonal serum (α-Chd1p) or pre-immune serum (pre-imm) was incubated with Chd1p for 5 min before addition of the 5S nucleosome core particles. Brackets and arrows indicate DNase I-protected regions and DNase I-hypersensitive sites, respectively, which are generated by Chd1p activity on mononucleosomes. (B) Reactions identical to those in (A) were subjected to EMSA.
We also determined that ATP-γ-S and AMP-PMP, which are two non-hydrolyzable forms of ATP, did not enable purified Chd1p to alter core particle structure (Figure 5A, lanes N and O). Because Chd1p is able to bind ATP-γ-S, as demonstrated by the observation that ATP-γ-S can competitively inhibit Chd1p ATPase activity (Figure 4, lane 10), these results indicate that the remodeling requires energy released from ATP hydrolysis. To confirm that Chd1p was responsible for the ATP-dependent alterations, rather than other proteins in the purified fraction, we challenged the remodeling activity with antibodies raised against Chd1p purified from Escherichia coli. Changes to 5S mononucleosome structure were completely inhibited by the addition of anti-Chd1p polyclonal serum but not by pre-immune serum (Figure 5A, compare lane P with Q). Moreover, fractions immunodepleted of Chd1p produced little or no change to 5S core particle structure (data not shown). These results firmly establish that Chd1p alters the structure of 5S nucleosome core particles through interactions requiring ATP hydrolysis.
There are several possible explanations for the extended regions of DNase I protection observed in Chd1p-remodeled nucleosomes. First, Chd1p could be a sequence-specific DNA-binding protein and be responsible for the protected regions. Secondly, Chd1p could recognize a specific feature of the remodeled nucleosome and remain bound following ATP hydrolysis. Thirdly, the protected regions could result from changes in the path of the DNA around the histone core. Although Chd1p can bind naked DNA (Figure 5B, lane 2), it shows no specific protection of naked 5S rDNA (Figure 5A, lanes F–J). These results support the conclusion of earlier studies that Chd1p can bind DNA non-specifically (Delmas et al., 1993). Furthermore, this binding does not require ATP (see below; Figure 5B, lanes 4 and 5). To explore whether Chd1p remains bound to the nucleosome following remodeling, we monitored nucleosome remodeling assays by electrophoretic mobility shift analysis (EMSA) rather than DNase I digestion. Little or no shift in the mobility of the 5S nucleosome core particles was detected upon addition of Chd1p (Figure 5B, lanes 4 and 5). The shifted bands in these lanes migrate similarly to Chd1p–DNA complexes, and probably represent Chd1p–DNA complexes formed from the small amount of free DNA in the 5S nucleosome preparation. The simplest explanation for these results is that Chd1p does not remain stably bound to the 5S nucleosome following remodeling. Alternatively, it is possible that Chd1p does remain bound to the nucleosome but the interaction is not stable to electrophoresis or that Chd1p remains bound but does not produce an observable shift in the mobility of the nucleosome. In any case, because neither the amount nor the mobility of nucleosome core particles in the reaction with Chd1p and ATP changes, Chd1p must alter nucleosome structure without removing histones from the DNA.
Discussion
CHD proteins are highly conserved between yeast, flies, worms and mammals. Using genetic and biochemical approaches, we have uncovered the cellular function of Chd1p from budding yeast: it is a nucleosome remodeling factor with a likely role as a regulator of transcription.
CHD1 effects on genome-wide expression
The results of the DNA array experiments show that CHD1 has both positive and negative effects on transcription and that ∼2–4% of all genes are affected by the absence of CHD1. Northern blot analyses of a subset of affected genes confirm the expression differences seen in the microarray experiments. The SWI–SNF complex has recently been described as being both a positive and negative regulator of genes (Holstege et al., 1998; Murphy et al., 1999; Sudarsanam et al., 2000) and it is becoming more clear that nucleosome remodeling factors may not simply unmask promoter regions to help initiate transcription but may also alter chromatin structure to inhibit transcription.
Chd1p can be readily cross-linked to DNA in cells, but we were not able to detect preferential association of Chd1p to genes whose expression it affects. One interpretation of this result is that Chd1p is distributed throughout the genome, but, under the conditions observed, affects the expression of a subset of genes. Another possibility is that Chd1p is only transiently present at certain promoters, and once Chd1p has remodeled those promoters it is no longer needed in that region to maintain the altered state.
A connection to SWI–SNF function
Using a broad synthetic-lethal screen, we have determined that CHD1 genetically interacts with SWI2 and that cells require one or the other for viability. Our genetic analyses have also shown that a chd1Δ swi1Δ strain is inviable, revealing that CHD1 interacts with at least two genes encoding components of the SWI–SNF complex. [It has also been observed that chd1 is synthetically lethal with snf6, which encodes a third component of the SWI–SNF complex (E.Haswell, personal communication).] These results complement the in vitro remodeling data (see below) and strongly suggest that Chd1p functions in vivo to alter chromatin structure. The fact that Chd1p may have a similar role to the SWI–SNF complex would explain why neither is essential in cells.
Recently, CHD1 has also been shown to interact genetically with ISW genes. Under stress conditions, a chd1 isw1 isw2 strain is inviable (Tsukiyama et al., 1999). Because ISW proteins are also components of remodeling factors, CHD1’s genetic interaction with SWI/SNF and ISW genes suggests that all four of these chromatin remodeling components share some overlapping functions in yeast.
Purification of Chd1p
We have purified Chd1p to near homogeneity using four chromatographic steps. In our most purified fractions, no other proteins were present in stoichiometric amounts with Chd1p. In addition, no other proteins appeared to co-purify with Chd1p in the final column steps. The apparent size of Chd1p after the first purification step and after the last purification step remained approximately the same. In addition, an immunoprecipitation experiment performed on the BioRex-70 fraction did not reveal any proteins that specifically precipitated with Chd1p in stoichiometric amounts (our unpublished data). These results strongly suggest that the form of Chd1p purified was not associated with other proteins and argues against the possibility that a Chd1p complex disassembled between chromatographic steps. The human CHD1 protein is believed to exist in a large complex (Kelley et al., 1999), and the human CHD3 and CHD4 proteins have also been found to be associated with a multiprotein complex (Tong et al., 1998; Xue et al., 1998; Zhang et al., 1998). It is possible that the yeast Chd1p is a simpler version of its counterpart in more complex organisms; alternatively, the yeast Chd1p may exist in a complex that is unstable to lysis conditions used in our purification.
Chd1p is a remodeling factor
We have demonstrated that Chd1p, using the energy provided by ATP hydrolysis, alters the structure of reconstituted mononucleosomes. There is, however, a noticeable difference between Chd1p’s activity and that of other remodeling activities. SWI–SNF-like activities increase cutting by DNase I throughout the DNA template, whereas Chd1p allows increased accessibility in some locations but protects specific sites in other locations. Both the increased accessibility and the protection are dependent on Chd1p and require both nucleosomal DNA and ATP hydrolysis. It is possible that the protection arises from Chd1p remaining bound following the remodeling step. Alternatively, this protection may reflect repositioning of the underlying histones with the DNA (Hamiche et al., 1999; Langst et al., 1999; Whitehouse et al., 1999).
Active fractions of Chd1p from our purification are predominantly composed of the Chd1p polypeptide. Although other remodeling factors have been purified as multiprotein complexes, the SWI2-like catalytic subunit of these remodeling complexes on its own can alter nucleosomes similarly to that of the intact complexes (Corona et al., 1999; Phelan et al., 1999). Thus, although it is possible that Chd1p functions within a loose multiprotein complex in vivo, it may simply act as a single polypeptide as we observed in vitro.
The finding that Chd1p perturbs mononucleosomes assigns the first biochemical function for CHD1 proteins. Previous experiments, and other experiments reported in this paper, have provided clues that Chd1p may have a role in transcription and chromatin structure. Our results demonstrate that Chd1p can indeed remodel chromatin and that this activity is probably responsible for regulating gene expression.
Materials and methods
Strains, media and reagents
Strains yHT148 (S288C Matα ura3-52 lys2-801 ade2-101 leu2-Δ1 his3-Δ200 trp1-Δ1 + pRS314) and yHT147 (S288C Matα chd1Δ::TRP1 ura3-52 lys2-801 ade2-101 leu2-Δ1 his3-Δ200 trp1-Δ1) were used for the DNA array and for the RNA Northern blot procedures. Strain yHT153 (S288C Matα 6MYC::6HIS::CHD1::TRP1 ura3-52 lys2-801 ade2-101 leu2-Δ1 his3-Δ200 trp1-Δ1) was used for the chromatin immunoprecipitation experiment. All three strains were grown in SD-Trp medium (0.67% Bacto yeast nitrogen base without amino acids, 2% dextrose, 2% Bacto agar, supplemented amino acids without tryptophan) and harvested at an OD660 of 2.0.
Strains yHT40 (W303 Mata ura3 ade2-1 leu2-3 his3-11 trp1-1 chd1Δ::LEU2 + pAJ741) and yHT42 (W303 Matα ura3 ade2-1 leu2-3 his3-11 trp1-1 chd1Δ::TRP1 + pAJ741) were used for the synthetic-lethal screen. pAJ741 contains the CHD1, ADE3 and URA3 genes on a 2µ plasmid. Cells were grown on presporulation (prespo) plates (0.8% Bacto yeast extract, 0.3% Bacto peptone, 10% dextrose, 2% Bacto agar) or 5-fluoroorotic acid (5-FOA) plates (0.7% yeast nitrogen without amino acids, 2% agar, 0.001% uracil, 0.08% 5-FOA, 0.1% supplemented amino acids) during analysis of the mutants.
The strains used for analyses of swi1, swi2 and swi4 mutants were: CY258 (Matα swi1Δ::LEU2 ura3-Δ99 lys2-801 ade2-101 leu2-Δ1 his3-Δ200), CY521 (Matα swi4Δ::HIS3 ura3-52 lys2-801 ade2-101 leu2-Δ1 his3-Δ200) and CY26 derivatives (Mata chd1Δ::HIS3 or chd1Δ::LEU2 ura3-52 lys2-801 ade2-101 leu2-Δ1 his3-Δ200 trp1-Δ1), yHT2568 (Matα swi2Δ::HIS3 ura3-52 lys2-801 ade2-101 leu2-Δ1 his3-Δ200 HO::TRP1), yHT2540 (Mata chd1Δ::LEU2 ura3-52 lys2-801 ade2-101 leu2-Δ1 his3-Δ200 HO::TRP1).
Strain yHT149 (Mata 6MYC::6HIS::CHD1::TRP1 ura3-52 lys2-801 leu2-Δ1 his3-Δ200 trp1 pep4Δ::HIS3 prb1-Δ1.6R) was used for the purification of Chd1p and grown in 2× YEPD (2% Bacto yeast extract, 4% Bacto peptone, 4% dextrose).
pCHD1 plasmids all contain the CHD1 gene derived from a genomic CHD1 λ clone obtained from the American Type Culture Collection (ATCC) (Olson et al., 1986). The genomic library used to clone SWC3 was also obtained from the ATCC (Thrash et al., 1985; Rose et al., 1987).
Anti-Myc (9E10) antibodies and anti-Chd1p antibodies were obtained from Covance. Anti-Chd1p polyclonal antibodies were generated using an N-terminal fragment of Chd1p (amino acids 122–300) fused to glutathione S-transferase as an antigen.
DNA microarray
Poly(A)+ RNA was purified from total RNA using oligo(dT) resin (Invitrogen). Hybridizations and array analyses were carried out as described previously (DeRisi et al., 1997).
Northern blots
Poly(A)+ RNA (0.5 µg) was loaded per sample on an agarose–formaldehyde gel and electrophoresis was carried out in 1× MOPS buffer. The RNA was transferred to a GeneScreen membrane (Dupont) and radiolabeled DNA probes were hybridized to the membrane at 65°C for 12 h. The membrane was washed with detergent and transcript signals were quantified using a Molecular Dynamics PhosphorImager.
Chromatin immunoprecipitation
Cross-linking, cell lysis, sonication to shear chromatin, and immunoprecipitation were performed essentially as described previously (Aparicio et al., 1997) with the following modifications. Formaldehyde cross-linking was quenched after 1–15 min and chromatin was sonicated for 7 × 12 s to produce fragments ranging from 100 to 600 bp. For PCRs, an equal molar amount of primers for ACT1 and regulated genes was added in addition to either total DNA or immunoprecipitated DNA.
Synthetic-lethal screen
Strains yHT40 and yHT42 were grown in 5 ml of SD-URA to an OD660 of 0.5. The cells were harvested and washed twice with water, resuspended in water and sonicated. The cells were then mutagenized by UV irradiation at five different doses. A small aliquot of cells at each dose was plated (in the dark) to determine the percent viability of each pool. The remaining master stock was stored in the dark at 4°C. Cells from three pools with viability of 72, 21 and 14% were chosen for the screen. A total of ∼75 000 cells were plated on to prespo plates at a density of 300–400 cells per plate and grown at room temperature. Colonies that turned red and did not sector were chosen for restreaking. Those that remained red were restreaked again. Fifty mutants remained after the second restreak and were transformed with a pCHD1 plasmid (either TRP1 or LEU2 marked) and simultaneously streaked on to 5-FOA plates. Mutants that sectored after the introduction of a second pCHD1 plasmid and did not grow on 5-FOA when harboring only the original pCHD1, URA3 plasmid (pAJ741) were selected for further analysis (the three mutants that met this criterion, swc1, swc2 and swc3, all originated from the 21% viability pool). All three mutants were backcrossed three times and mated to each other to perform complementation tests. Diploids were tested for both wild-type growth and the ability to sector.
SWC1 and SWC2 were cloned via complementation of the slow growth and sectoring phenotypes of the swc1 and swc2 mutations by an SWI2 plasmid (pBD10, pBD3). To verify that SWC1 and SWC2 are indeed SWI2, an allelism test was carried out. The URA3 gene was integrated at the 3′ untranslated region (3′-UTR) (∼500 bp downstream of the termination codon) of the SWI2 locus of a chd1Δ haploid cell and mated to an swc1 chd1Δ mutant. After sporulation and tetrad dissection, the haploids were analyzed. swc1 (slow growing colonies) always segregated away from the URA3 gene in a 2:2 ratio (out of 21 tetrad dissections). In addition, a chd1Δ haploid (yHT2540) and an swi2Δ haploid (yHT2568) were mated, the diploid sporulated and spores analyzed for the double mutant phenotype. No double mutant spores were ever recovered (from 28 tetrad dissections). However, transformation of a CHD1 plasmid (pAJ741) into the diploid and subsequent sporulation and dissection showed that cells with genomic copies of CHD1 and SWI2 deleted were viable when harboring the pAJ741 plasmid, revealing that the synthetic interaction between CHD1 and SWI2 is indeed CHD1 dependent. The same dependence on pCHD1 for spore viability was seen with the chd1Δ swi1Δ strain. [Note: it has been observed that the TRP1 allele trp1-Δ1 makes swi/snf strains sicker than trp1-1 or TRP1 strains. The strains used in all of our manipulations of swi2Δ and swi2/swc1/swc2 were either trp1-1 or TRP1 (and not trp1-Δ1) and synthetic interactions between chd1Δ and swc1/2 as well as swi2Δ were observed for both trp1-1 and TRP1 strains.] To clone SWC3, a YCp50 genomic library was electroporated into an swc3 mutant and plasmids that complemented the slow growth defect were isolated and sequenced. By subcloning and retransformation of candidate SWC3 genes, it was determined that ALR1 was the only gene on the rescuing plasmids that complemented the swc3 mutation. SWC3 was confirmed to be ALR1 by the ability of high magnesium medium to complement the swc3 slow growth defect and by construction of an alr1Δ chd1Δ strain and demonstration that the double mutant was synthetically sick.
Purification
Twenty-four liters of strain yHT149 were grown in 2× YEPD and harvested at an OD660 of 6. The cell pellet (∼400 g) was washed with water and resuspended in 200 ml of 3× lysis buffer [750 mM NaCl, 150 mM HEPES pH 7.6, 30% glycerol, 0.3% Nonidet P-40 (NP-40), 30 mM Mg(OAc)2, 3 mM EDTA] and frozen in a dry ice–ethanol bath for storage. The cell paste was thawed and β-mercaptoethanol (BME) and protease inhibitors were added to the following final concentrations: 10 mM BME, 1 mM phenylmethylsulfonyl fluoride (PMSF), 2 mM benzamidine, 1 µg/ml each of pepstatin, leupeptin and bestatin. Biospec bead beating chambers (350 ml) were used to lyse cells. Cells were agitated for 30 s, with a 90 s cooling period, 20 times. The lysate was spun at 9000 r.p.m. in a Sorvall SS-34 rotor for 20 min and the supernatant was extracted with a 500 mM NaCl incubation step (30 min with stirring). The extract was clarified with a 35 000 r.p.m. spin using a 60 Ti rotor (Beckman) in an ultracentrifuge (Beckman L8-55M). The supernatant was then diluted to a final NaCl concentration of 240 mM.
The extract was loaded on to a 500 ml BioRex-70 column (Bio-Rad). The column was washed with 240 and 290 mM NaCl buffer A (20 mM HEPES pH 7.6, 10% glycerol, 0.1% NP-40, 0.1 mM EDTA, 1 mM PMSF, 2 mM benzamidine, 1 µg/ml each of pepstatin, leupeptin and bestatin, 10 mM BME). Chd1p was eluted from the column with 450 mM NaCl buffer A in 50 ml fractions. To one 50 ml peak fraction, 2.5 ml of Ni-nitrilotriacetic acid (Ni-NTA) (Qiagen) resin were added, in addition to imidazole to 1 mM final concentration. The slurry was nutated for 1.5 h and packed into a column. The Ni-NTA column was washed with 1, 2 and 4 mM imidazole buffer B (70 mM NaCl, 25 mM Tris pH 8.0, 10% glycerol, 0.1% NP-40, 0.1 mM EDTA, 1 mM PMSF, 2 mM benzamidine, 1 µg/ml each of pepstatin, leupeptin and bestatin, 10 mM BME). Imidazole buffer B (200 mM) was used to elute Chd1p from the column. Peak fractions were pooled and loaded on to a 0.75 ml DEAE–Sepharose column (Amersham–Pharmacia). The column was washed with 70 mM NaCl and 120 mM NaCl buffer C [50 mM Tris pH 8.0, 10% glycerol, 0.1% NP-40, 0.1 mM EDTA, 1 mM PMSF, 2 mM benzamidine, 1 µg/ml each of pepstatin, leupeptin and bestatin, 1 mM dithiothreitol (DTT)]. Chd1p was eluted with 170 mM NaCl buffer C. Peak fractions were then loaded on to a 1 ml HiTrap SP column (Amersham–Pharmacia). After washes with 170 mM NaCl and 325 mM NaCl buffer D (50 mM HEPES pH 7.6, 10% glycerol, 0.1% NP-40, 0.1 mM EDTA, 1 mM PMSF, 2 mM benzamidine, 1 µg/ml each of pepstatin, leupeptin and bestatin, 1 mM DTT), Chd1p was eluted from the column with 375 mM NaCl buffer D.
Nucleosome reconstitution, DNase I analysis and EMSA
An end-labeled ScaI–AvaII fragment of 172 bp from a sea urchin 5S rRNA gene was assembled into nucleosome core particles with purified HeLa nucleosomes as described previously (Steger and Workman, 1997). DNase I and EMSA reactions were performed in 20 µl with 10 mM HEPES pH 7.8, 50 mM KCl, 4 mM MgCl2, 5 mM DTT, 0.5 mM PMSF, 0.25 mg/ml bovine serum albumin (BSA) and 5% glycerol. Reactions were incubated at 30°C for 30 min. Samples with 5S nucleosome core particles contained ∼1–2 fmol of reconstituted probe and donor nucleosomes to give a total of 0.4 pmol of nucleosomes. Reactions with histone-free DNA contained the same amounts of mock-reconstituted probe DNA and nucleosomes as used for the mononucleosome reactions. The amount of Chd1p required to reconfigure all of the 5S nucleosome core particles was determined to be roughly stoichiometric with the total amount of nucleosomes in the reaction. DNase I digestion and gel electrophoresis were performed as described previously (Steger and Workman, 1997).
ATPase assay
Reactions were carried out under the same conditions used for the DNase I protection analysis with the addition of 0.5 mM ATP and 1 µCi of [γ-32P]ATP. After 30 min at 30°C, reactions were terminated by the addition of 1 µl of 0.5 M EDTA. A 1 µl sample from each reaction was spotted on to a polyethyleneimine cellulose plate for thin layer chromatography. Chromatography was carried out in 50 mM HCl to separate ATP and free phosphate.
Acknowledgments
Acknowledgements
We would like to thank F.Winston and P.Sudarsanam for sharing the SNF2 microarray data prior to publication and Craig Peterson for the CY26, CY258 and CY407 strains. We are grateful to Ahmed Hassan and Jerry Workman for providing purified SWI–SNF. We would also like to thank Anne Cassidy-Stone for assistance with chromatin immunoprecipitation experiments, Yi Wei Jiang and Danesh Moazed for technical advice, and the Johnson laboratory for comments on the manuscript.
References
- Aparicio O.M., Weinstein,D.M. and Bell,S.P. (1997) Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell, 91, 59–69. [DOI] [PubMed] [Google Scholar]
- Bender A. and Pringle,J.R. (1991) Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol., 11, 1295–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns B.R., Kim,Y.J., Sayre,M.H., Laurent,B.C. and Kornberg,R.D. (1994) A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc. Natl Acad. Sci. USA, 91, 1950–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns B.R. et al. (1996) RSC, an essential, abundant chromatin-remodeling complex. Cell, 87, 1249–1260. [DOI] [PubMed] [Google Scholar]
- Cavalli G. and Paro,R. (1998) Chromo-domain proteins: linking chromatin structure to epigenetic regulation. Curr. Opin. Cell Biol., 10, 354–360. [DOI] [PubMed] [Google Scholar]
- Corona D.F., Langst,G., Clapier,C.R., Bonte,E.J., Ferrari,S., Tamkun,J.W. and Becker,P.B. (1999) ISWI is an ATP-dependent nucleosome remodeling factor. Mol. Cell, 3, 239–245. [DOI] [PubMed] [Google Scholar]
- Delmas V., Stokes,D.G. and Perry,R.P. (1993) A mammalian DNA-binding protein that contains a chromodomain and an SNF2/SWI2-like helicase domain. Proc. Natl Acad. Sci. USA, 90, 2414–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi J.L., Iyer,V.R. and Brown,P.O. (1997) Exploring the metabolic and genetic control of gene expression on a genomic scale. Science, 278, 680–686. [DOI] [PubMed] [Google Scholar]
- Eisen J.A., Sweder,K.S. and Hanawalt,P.C. (1995) Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res., 23, 2715–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K., Javerzat,J.P., Lorentz,A., Schmidt,H., Cranston,G. and Allshire,R. (1995) The chromodomain protein Swi6: a key component at fission yeast centromeres. Science, 269, 1429–1431. [DOI] [PubMed] [Google Scholar]
- Hamiche A., Sandaltzopoulos,R., Gdula,D.A. and Wu,C. (1999) ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell, 97, 833–842. [DOI] [PubMed] [Google Scholar]
- Holstege F.C., Jennings,E.G., Wyrick,J.J., Lee,T.I., Hengartner,C.J., Green,M.R., Golub,T.R., Lander,E.S. and Young,R.A. (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell, 95, 717–728. [DOI] [PubMed] [Google Scholar]
- Jin Y.H. et al. (1998) Isolation and characterization of hrp1+, a new member of the SNF2/SWI2 gene family from the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet., 257, 319–329. [DOI] [PubMed] [Google Scholar]
- Kehle J., Beuchle,D., Treuheit,S., Christen,B., Kennison,J.A., Bienz,M. and Muller,J. (1998) dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science, 282, 1897–1900. [DOI] [PubMed] [Google Scholar]
- Kelley D.E., Stokes,D.G. and Perry,R.P. (1999) CHD1 interacts with SSRP1 and depends on both its chromodomain and its ATPase/helicase-like domain for proper association with chromatin. Chromosoma, 108, 10–25. [DOI] [PubMed] [Google Scholar]
- Koonin E.V., Zhou,S. and Lucchesi,J.C. (1995) The chromo superfamily: new members, duplication of the chromo domain and possible role in delivering transcription regulators to chromatin. Nucleic Acids Res., 23, 4229–4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langst G., Bonte,E.J., Corona,D.F. and Becker,P.B. (1999) Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell, 97, 843–852. [DOI] [PubMed] [Google Scholar]
- Laurent B.C., Treich,I. and Carlson,M. (1993) The yeast SNF2/SWI2 protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes Dev., 7, 583–591. [DOI] [PubMed] [Google Scholar]
- MacDiarmid C.W. and Gardner,R.C. (1998) Overexpression of the Saccharomyces cerevisiae magnesium transport system confers resistance to aluminum ion. J. Biol. Chem., 273, 1727–1732. [DOI] [PubMed] [Google Scholar]
- Madireddi M.T., Coyne,R.S., Smothers,J.F., Mickey,K.M., Yao,M.C. and Allis,C.D. (1996) Pdd1p, a novel chromodomain-containing protein, links heterochromatin assembly and DNA elimination in Tetrahymena. Cell, 87, 75–84. [DOI] [PubMed] [Google Scholar]
- Murphy D.J., Hardy,S. and Engel,D.A. (1999) Human SWI–SNF component BRG1 represses transcription of the c-fos gene. Mol. Cell. Biol., 19, 2724–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson M.V., Dutchik,J.E., Graham,M.Y., Brodeur,G.M., Helms,C., Frank,M., MacCollin,M., Scheinman,R. and Frank,T. (1986) Random-clone strategy for genomic restriction mapping in yeast. Proc. Natl Acad. Sci. USA, 83, 7826–7830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen-Hughes T., Utley,R.T., Cote,J., Peterson,C.L. and Workman,J.L. (1996) Persistent site-specific remodeling of a nucleosome array by transient action of the SWI/SNF complex. Science, 273, 513–516. [DOI] [PubMed] [Google Scholar]
- Paro R. and Hogness,D.S. (1991) The Polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc. Natl Acad. Sci. USA, 88, 263–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson C.L., Dingwall,A. and Scott,M.P. (1994) Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc. Natl Acad. Sci. USA, 91, 2905–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan M.L., Sif,S., Narlikar,G.J. and Kingston,R.E. (1999) Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell, 3, 247–253. [DOI] [PubMed] [Google Scholar]
- Rose M.D., Novick,P., Thomas,J.H., Botstein,D. and Fink,G.R. (1987) A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene, 60, 237–243. [DOI] [PubMed] [Google Scholar]
- Steger D.J. and Workman,J.L. (1997) Stable co-occupancy of transcription factors and histones at the HIV-1 enhancer. EMBO J., 16, 2463–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes D.G., Tartof,K.D. and Perry,R.P. (1996) CHD1 is concentrated in interbands and puffed regions of Drosophila polytene chromosomes. Proc. Natl Acad. Sci. USA, 93, 7137–7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsanam P., Iyer,V.R., Brown,P.O. and Winston,F. (2000) Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 97, 3364–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrash C., Bankier,A.T., Barrell,B.G. and Sternglanz,R. (1985) Cloning, characterization, and sequence of the yeast DNA topoisomerase I gene. Proc. Natl Acad. Sci. USA, 82, 4374–4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong J.K., Hassig,C.A., Schnitzler,G.R., Kingston,R.E. and Schreiber, S.L. (1998) Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature, 395, 917–921. [DOI] [PubMed] [Google Scholar]
- Travers A. (1999) An engine for nucleosome remodeling. Cell, 96, 311–314. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T., Palmer,J., Landel,C.C., Shiloach,J. and Wu,C. (1999) Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev., 13, 686–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade P.A., Jones,P.L., Vermaak,D. and Wolffe,A.P. (1998) A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr. Biol., 8, 843–846. [DOI] [PubMed] [Google Scholar]
- Whitehouse I., Flaus,A., Cairns,B.R., White,M.F., Workman,J.L. and Owen-Hughes,T. (1999) Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature, 400, 784–787. [DOI] [PubMed] [Google Scholar]
- Woodage T., Basrai,M.A., Baxevanis,A.D., Hieter,P. and Collins,F.S. (1997) Characterization of the CHD family of proteins. Proc. Natl Acad. Sci. USA, 94, 11472–11477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman J.L. and Kingston,R.E. (1998) Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem., 67, 545–579. [DOI] [PubMed] [Google Scholar]
- Xue Y., Wong,J., Moreno,G.T., Young,M.K., Cote,J. and Wang,W. (1998) NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell, 2, 851–861. [DOI] [PubMed] [Google Scholar]
- Zhang Y., LeRoy,G., Seelig,H.P., Lane,W.S. and Reinberg,D. (1998) The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell, 95, 279–289. [DOI] [PubMed] [Google Scholar]