Abstract
Hsp47 is a heat-shock protein that interacts transiently with procollagen during its folding, assembly and transport from the endoplasmic reticulum (ER) of mammalian cells. It has been suggested to carry out a diverse range of functions, such as acting as a molecular chaperone facilitating the folding and assembly of procollagen molecules, retaining unfolded molecules within the ER, and assisting the transport of correctly folded molecules from the ER to the Golgi apparatus. Here we define the substrate recognition of Hsp47, demonstrating that it interacts preferentially with triple-helical procollagen molecules. The association of Hsp47 with procollagen coincides with the formation of a collagen triple helix. This demonstrates that Hsp47’s role in procollagen folding and assembly is distinct from that of prolyl 4-hydroxylase. These results indicate that Hsp47 acts as a novel molecular chaperone, potentially stabilizing the correctly folded collagen helix from heat denaturation before its transport from the ER.
Keywords: Hsp47/molecular chaperone/procollagen/protein folding
Introduction
The lumen of the endoplasmic reticulum (ER) contains a number of proteins that interact with polypeptide chains during their maturation and transport through the ER to the Golgi apparatus (for review see Leitzgen and Haas, 1998). These proteins include enzymes that catalyse folding events, such as disulfide bond formation and peptidyl-prolyl cis–trans isomerization, and molecular chaperones that interact with folding polypeptide chains to prevent premature aggregation or non-specific interactions. One of the general features of these proteins is that they interact with the unfolded or unassembled polypeptide chains and do not interact with the transport competent native structure. Hence, interaction of unfolded polypeptides with these ER-resident proteins leads to their retention within this organelle, providing a mechanism for quality control of proteins entering the secretory pathway (Hammond and Helenius, 1995).
Procollagen is an excellent example of a molecule that interacts with a number of ER-resident enzymes and molecular chaperones during its folding and assembly (McLaughlin and Bulleid, 1998). The fibrillar procollagen molecule is a trimer consisting of three distinct regions, two trimeric regions (the C- and N-propeptides), separated by a long triple-helical forming region (Kadler, 1994). Once the individual chains have been translocated into the lumen of the ER, the C-propeptide globular domain folds. If this initial folding event is prevented, the unfolded C-propeptides associate with immunoglobulin heavy chain binding protein (BiP), which eventually leads to proteosomal-mediated degradation (Chessler and Byers, 1993; Lamandé et al., 1995; Fitzgerald et al., 1999). Once folded, the C-propeptides either assemble to form trimers immediately or associate with protein disulfide isomerase (PDI) until interacting chains are synthesized (Wilson et al., 1998). After trimerization, the triple-helical domain is folded; this process occurs in a C- to N-direction and is dependent on the hydroxylation of proline residues within the triple-helical forming region (Engel and Prockop, 1991; Bulleid et al., 1997). Trimeric non-triple-helical molecules interact with prolyl 4-hydroxylase (P4H), an association that depends on the folding status of the protein rather than hydroxylation state (Walmsley et al., 1999). Once the triple helix has formed, the protein is transported to the Golgi apparatus where it forms higher-order aggregates resulting in characteristic distensions of the Golgi cisternae (Bonfanti et al., 1998).
Procollagen chains have also been reported to interact with Hsp47 throughout this process but it remains unclear exactly what role this protein plays during procollagen folding, assembly and transport (Nagata, 1996). Using a variety of techniques, Hsp47 has been shown to associate with either nascent polypeptide chains (Sauk et al., 1994), monomeric chains, unhydroxylated non-triple-helical trimers and hydroxylated triple-helical molecules (Nakai et al., 1992; Satoh et al., 1996). Such promiscuity in binding is difficult to reconcile with a role as a molecular chaperone as these proteins would classically interact with specific intermediates in the folding pathway. More recently, evidence has been presented for interaction of Hsp47 with synthetic peptides with the sequence (Gly-Pro-Pro)n (Koide et al., 1999). These results suggest that the binding of Hsp47 to these peptides is dependent on the length of the peptides and the hydroxylation state of the proline in the Y position of the Gly-X-Y triplet. Thus Hsp47 has a preference for longer peptides containing unhydroxylated proline residues and a low affinity for peptides with hydroxylated proline residues. It remains to be established whether the binding of Hsp47 to these peptides is representative of the binding of Hsp47 to procollagen chains within the lumen of the ER.
One characteristic of procollagen molecules that separates them from most proteins synthesized by cells is that their triple-helical domain is not thermostable, being denatured at temperatures a few degrees above physiological temperature. Thermal stability depends on the content of hydroxyproline residues within the triple-helical domain (Kivirikko et al., 1992). Thus a triple-helical domain containing unhydroxylated proline residues unfolds at temperatures above 25°C. It has been known for some time that procollagen within the cell is more thermostable than the isolated protein (Bruckner and Eikenberry, 1984), which would suggest that the intracellular environment could protect the collagen helix from heat denaturation. The fact that Hsp47 binds to procollagen during its biosynthesis and that it is a heat shock protein would make it a prime candidate for a chaperone that stabilizes the thermally unstable collagen helix. To investigate this possibility and to determine the substrate specificity of Hsp47 during procollagen folding and assembly, we determined its ability to interact with a variety of different procollagen chains during different stages of their folding and assembly in a well characterized semi-permeabilized (SP) cell system (Wilson et al., 1995). We have used such a system previously to highlight the role of PDI and P4H in procollagen assembly and to identify key amino acid sequences required to ensure subunit assembly and nucleation of the triple helix (Bulleid et al., 1997; Lees et al., 1997; Wilson et al., 1998; Walmsley et al., 1999).
Results
We have previously demonstrated that the folding and assembly of procollagen can be divided into three distinct phases, each of which can be reconstituted in an SP cell-free translation system (Bulleid, 1996). By carefully selecting procollagen chains and the conditions during translation, we can synthesize polypeptides that either remain monomeric (Lees and Bulleid, 1994), assemble to form trimers but do not form triple helices or fold and assemble completely to form triple-helical molecules that are stable at physiological temperature (Bulleid et al., 1996). The procollagen constructs used in this study and an indication of the endpoints in their folding pathway are outlined in Table I. Thus, full-length proα1(III) chains, proα1(III)Δ1 chains (which contain a shortened triple-helical domain) and proα2(1):(III)BGR chains (which contain a modified C-propeptide) fold and assemble to form triple-helical molecules (Lees et al., 1997). If any of these chains is synthesized in the presence of inhibitors of proline hydroxylation, they associate to form trimers but the triple-helical domain does not form. The proα1(III)Δ1 chain with the first cysteine within the C-propeptide mutated to serine does not assemble to form trimers and remains monomeric, as do the proα1(I), proα2(I) and proα2(I)Δ1 chains (Lees and Bulleid, 1994).
Table I. Description of constructs used in this study.
| Name | Description | Folding end point |
|---|---|---|
| proα1(III) | full-length type III procollagen chain | triple-helical trimer |
| proα1(I) | full-length type I procollagen chain | monomeric, can potentially form trimers (Uitto, 1979) |
| proα2(I) | full-length type I procollagen chain | monomeric |
| proα1(III)Δ1 | type III procollagen chain with internal deletion in triple-helical domain | triple-helical trimer |
| proα1(III)Δ1cys1 | type III procollagen chain with internal deletion in triple-helical domain with the first cysteine in C-propeptide mutated to serine | monomer |
| proα2(I)Δ1 | the proα2 chain of type I procollagen with an internal deletion in the triple-helical domain | monomer |
| proα2(I):(III)BGR | the proα2 chain of type I procollagen with an internal deletion in the triple-helical domain, the C-propeptide is modified to allow homotrimer formation | triple-helical trimer with reduced thermal stability |
Does Hsp47 bind to monomeric or trimeric procollagen chains?
A key question to address when considering the role of Hsp47 in the biosynthesis of procollagen is what forms of the protein it interacts with. To address this question we studied the interaction of Hsp47 with newly synthesized proα1(III) chains. When mRNA encoding this protein was translated in the presence of SP cells for 90 min, >80% of the translocated translation product formed correctly-folded triple-helical molecules, as judged by resistance of the triple helical domain to digestion with proteases (Figure 1A). Potential interactions between translation products and endogenous ER proteins were stabilized with the thiol-cleavable cross-linking agent dithio-bis(propionic acid) N-hydroxy succinimide ester (DSP), which cross-links Hsp47 to various forms of type I procollagen (Satoh et al., 1996). Immunoprecipitation of any cross-links were then carried out with antibodies raised against PDI (identical to the β-subunit of P4H), Hsp47, the α-subunit of P4H or a control antibody (Figure 1B). Little or no cross-linked product was precipitated with antibodies to either the α-subunit of P4H, PDI or the control antibody (Figure 1B, lanes 2, 4 and 5). However, a cross-linked product was precipitated with antibodies to Hsp47 (Figure 1B, lane 3). These results confirm that, although the α- and β-subunits of P4H have previously been shown to interact with non-triple helical procollagen (Bulleid et al., 1996; Walmsley et al., 1999), this interaction is lost upon folding of the triple helix. Conversely, Hsp47 would seem to interact with correctly folded and assembled proα1(III).
Fig. 1. Cross-linking of procollagen chains to molecular chaperones. Transcripts encoding proα1(III), proα2(I) and proα1(I) were translated in a rabbit reticulocyte lysate in the presence of SP cells. Translation products were chemically cross-linked using the thiol-cleavable cross-linker DSP. Cross-linked samples were immunoprecipitated using the indicated antibodies before separation by SDS–PAGE. (A) Products of translation of proα1(III) RNA either untreated (lane 1) or treated with a combination of proteases (chymotrypsin, trypsin and pepsin) (lane 2). (B–D) The products of translation from (B) proα1(III), (C) proα2(I) and (D) proα1(I) RNA were cross-linked with DSP and separated by SDS–PAGE before (lane 1) or after immunoprecipitation with antibodies to PDI (lane 2), Hsp47 (lane 3), the α-subunit of P4H (lane 4) or anti-Myc antibody (9E-10) (lane 5).
To assess further the interaction of Hsp47 with procollagen chains at different stages of assembly, we investigated its interaction with chains that remain monomeric. For these experiments we translated individually the two type I procollagen chains, proα1(I) and proα2(I), which would normally form heterotrimers. When translated individually, neither of these chains folded to form triple-helical molecules as judged by resistance to digestion with proteases (results not shown). After cross-linking, products were immunoprecipitated by antibodies to PDI and the α-subunit of P4H (Figure 1C and D, lanes 2 and 4) but little or no product was precipitated by antibodies to Hsp47 (Figure 1C and D, lane 3). These results suggest that, if folding and assembly are prevented and the procollagen chains remain monomeric, then they interact with the subunits of P4H but not with Hsp47. Clearly we cannot determine whether PDI is interacting with these chains as a subunit of P4H or on its own; we can conclude that there is a definite distinction between the substrate specificity of Hsp47 and P4H.
As we are using different procollagen chains in these experiments, the different specificities could reflect the differences in amino acid sequences rather than folding status. To address this point we made use of (i) a proα1(III) chain with a point mutation, in the C-propeptide, that prevents self-assembly and (ii) a proα2(I) chain with an altered C-propeptide that allows the formation of homotrimeric molecules. Both these constructs have shortened triple-helical regions that have previously been shown to have the ability to form triple-helical structures (Lees et al., 1997). We initially determined whether these smaller molecules shared the same Hsp47- and P4H-binding characteristics as the full-length proteins. When the proα1(III)Δ1 chain was translated in the presence of SP cells under conditions that allow folding and assembly of the triple-helical domain, cross-links were found to Hsp47, but not to the subunits of P4H (Figure 2A). Conversely, when the proα2(I) Δ1 chain was translated in the presence of SP cells, no cross-linked products were immunoprecipitated with antibodies to Hsp47 but were seen with antibodies to the subunits of P4H (Figure 2C). The binding specificity of both these chains was altered when they were modified either to prevent or allow trimerization and triple helix formation. Thus the proα1(III)Δ1Cys chain, which remains monomeric, now binds P4H subunits and not Hsp47 (Figure 2B), whereas the proα2(I): (III)BGR, which forms trimers, now binds Hsp47 (Figure 2D, lane 3). This chain also binds the subunits of P4H (Figure 2D, lanes 2 and 4); this could reflect the fact that the alterations in the C-propeptide were only partially effective and there was, as shown previously, a mixture of trimeric triple-helical and monomeric molecules (Lees et al., 1997). These results demonstrate that the different specificity of Hsp47 and P4H for procollagen chains is probably a reflection of the different folding and assembly states rather than differences in amino acid compositions of the triple-helical forming regions.
Fig. 2. Comparison of cross-linked products to procollagen chains that form either monomers or trimers. Transcripts encoding proα1(III)Δ1, proα1(III)Δ1Cys-Ser, proα2(I)Δ1 and proα2(I):(III)BGR were translated in rabbit reticulocyte lysate in the presence of SP cells. Products of translation were isolated and chemically cross-linked using DSP. Cross-linked samples were immunoprecipitated using the indicated antibodies and separated by SDS–PAGE under reducing conditions. The (A) proα1(III)Δ1, (B) proα1(III)Δ1Cys-Ser, (C) proα2(I)Δ1 and (D) proα2(I):(III)BGR translation products were cross-linked with DSP and separated by SDS–PAGE either before (lane 1) or after immunoprecipitation with antibodies to PDI (lane 2), Hsp47 (lane 3), the α-subunit of P4H (lane 4) or control antibody 9E-10 (lane 5).
Does Hsp47 bind to hydroxylated or unhydroxylated procollagen chains?
The experiments described above indicate that Hsp47 binds to chains that form trimers but not to chains that remain monomeric. All the procollagen chains translated under the conditions so far described will be hydroxylated even if they remain monomeric, so it would appear that the binding of Hsp47 to the procollagen chains is not regulated by the hydroxylation state of the protein. To ascertain if this is indeed the case, we translated proα1(III)Δ1 in the presence of α,α′-dipyridyl, a reagent that chelates Fe2+ and, therefore, inhibits both proline and lysine hydroxylation (Kivirikko et al., 1992). Under these conditions, the C-propeptides associate and a trimeric structure is formed but the triple helix does not fold (Bulleid et al., 1996). After translation and cross-linking, association of procollagen chains was only observed to PDI and the α-subunit of P4H (Figure 3A, lanes 2 and 4) and not to Hsp47 (Figure 3A, lane 3).
Fig. 3. Interaction of Hsp47 with unhydroxylated procollagen trimers. (A) Transcripts encoding proα1(III)Δ1 were translated in a rabbit reticulocyte lysate in the presence of SP cells. Products of translation were isolated and cross-linked using DSP. Cross-linked samples were separated by SDS–PAGE either before (lane 1) or after immunoprecipitation with antibodies to PDI (lane 2), Hsp47 (lane 3), P4H (lane 4) and control antibody 9E-10 (lane 5). (B) Proα1(III)Δ1 RNA was translated in the presence of SP cells for 60 min before inhibiting further synthesis by adding cycloheximide to 5 mM. A sample was removed and incubated for a further 60 min at 30°C, cross-linked with DSP and immunoprecipitated with anti-Hsp47 antibody (lane 6). Ferrous sulfate was added to 5 mM to the remaining translation mix, which was then divided into five aliquots and incubated at 30°C for the indicated time period. Following this incubation, each sample was cross-linked with DSP and immunoprecipitated with anti-Hsp47 antibody (lanes 1–5).
To investigate this binding specificity further, we translated proα1(III)Δ1 in the presence of α,α′-dipyridyl, added cycloheximide to prevent further protein synthesis, and then added an excess of Fe2+ to quench the chelator. At various time points after addition of iron, samples were cross-linked and immunoprecipitated. Our previous results showed that, while both subunits of P4H associate with the procollagen chains before the addition of iron, they dissociated after iron addition; this dissociation coincides with both hydroxylation and triple helix formation (Bulleid et al., 1996). Here we show that the reverse was true for the association of Hsp47 with the procollagen chain. Hence when proα1(III)Δ1 was translated in the presence of α,α′-dipyridyl, no association was found, yet after iron addition a steady increase in association of Hsp47 with the procollagen chain was seen (Figure 3B, lanes 1–5). If no iron was added and the samples were incubated for 60 min, no association with Hsp47 occurred (Figure 3B, lane 6). These results indicate that either Hsp47 binding requires hydroxylation for binding or, as hydroxylation is required to allow triple helix formation, Hsp47 only binds to the triple-helical structure.
Is Hsp47 binding regulated by triple helix formation or proline hydroxylation?
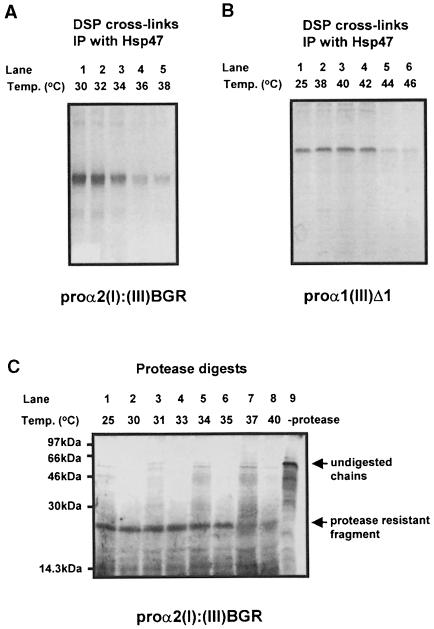
To resolve the question of whether Hsp47 recognizes the triple-helical structure or hydroxylated procollagen chains, we studied the binding of Hsp47 during thermal denaturation of the triple-helical domain of either proα2(I): (III)BGR or proα1(III)Δ1. As described in the previous experiments both these chains form triple-helical structures and interact with Hsp47. Following translation the cells containing these procollagen chains were heated to varying temperatures, then translation products were cross-linked with DSP and cross-linked products were immunoprecipitated with an antibody to Hsp47. As the temperature increased, Hsp47 dissociated from both proα2(I):(III)BGR (Figure 4A) and proα1(III)Δ1 chains (Figure 4B). Most, but not all, of the Hsp47 dissociated between 34 and 36°C for proα2(I):(III)BGR, which coincided with the melting temperature for this chain as judged by protease sensitivity (Figure 4C). The temperature of dissociation of Hsp47 from proα1(III)Δ1 was between 42 and 44°C, which is slightly higher than the previously reported melting temperature (39–40°C) (Bulleid et al., 1996). This discrepancy could be explained by the fact that this experiment was carried out with intact cells whereas the melting temperature reported previously was carried out after solubilization. It should be noted that the melting temperature of procollagen chains within intact cells is generally higher than after solubilization (Bruckner and Eickenberry, 1984). As the temperature of dissociation of Hsp47 from these two procollagen chains is different, the dissociation is unlikely to be due to thermal denaturation of Hsp47 itself. It is more likely to be due to melting of the triple-helical domain. These results also provide convincing evidence that high-affinity binding of Hsp47 to procollagen chains is regulated by formation of the triple-helical domain rather than by the state of hydroxylation of the proline or lysine residues.
Fig. 4. Stability of Hsp47 interaction to heat denaturation. Products of translation in SP cells of proα2(I):(III)BGR (A and C) and proα1(III)Δ1 RNA (B) were heated at indicated temperatures before cross-linking with DSP and immunoprecipitation with anti-Hsp47 antibody (A and B). Alternatively the proα2(I):(III)BGR translation product in SP cells was heated to various temperatures for 10 min and separated by SDS–PAGE either untreated (lane 9) or treated with proteases as described in Figure 1A (lanes 1–8).
Further evidence to support this assumption came from an experiment where we first heat-denatured the procollagen chains and then allowed the triple helix to refold after reducing the temperature. For this experiment we used the proα2(I):(III)BGR chain. Similar results were obtained with the proα1(III)Δ1 chain (results not shown). This treatment resulted in a rapid reformation of the collagen triple helix as judged by resistance to digestion with chymotrypsin and trypsin (Figure 5A, lanes 3–7). After heat denaturation only trace amounts of translation product were resistant to digestion (Figure 5A, compare lanes 2 and 3). After only 5 min on ice, a significant fraction of the translation products regained resistance to proteolytic digestion (Figure 5A, lane 4). When the translation products were assayed for association with Hsp47 after heat denaturation, no association was detected (Figure 5B, lane 1). However, after only 5 min on ice, Hsp47 re-associated with the procollagen chain (Figure 5B, lane 2). These results clearly show that Hsp47 dissociates from procollagen chains after heat denaturation but reassociates upon refolding of the triple helix.
Fig. 5. Refolding of procollagen after heat denaturation and reassociation with Hsp47. (A) After translation of proα2(I):(III), SP cells were isolated and resuspended in KHM before heating at 47°C for 15 min to denature the triple-helical domain. Aliquots of heat-denatured SP cells were separated by SDS–PAGE either before (lane 1) or after incubation on ice for the times indicated before solubilization proteolysis using chymotrypsin, trypsin and pepsin (lanes 3–7). A control sample, which was not heat-denatured, was also subjected to proteolysis (lane 2). (B) Aliquots of heat-denatured SP cells were incubated on ice for the times indicated before cross-linking using DSP and immunoprecipitation with anti-Hsp47 antibody (lanes 1–5).
Discussion
The main objective of this study was to determine the specificity of Hsp47 binding to a variety of procollagen chains during their synthesis, folding and assembly in functionally intact ER. Our results demonstrate that, within the experimental system used here, high-affinity Hsp47 binding to procollagen depends upon the formation of the collagen triple helix. Hsp47 was dissociated from the procollagen chain after heat denaturation of the triple helix; re-association occurred when the triple-helical domain refolded. In the absence of Hsp47 binding, P4H bound to the triple-helical region, confirming previous studies that have demonstrated a preference for binding of this protein to the non-triple helical structure (Walmsley et al., 1999). The binding of Hsp47 to a variety of triple-helical sequences, from full-length α1(III) to the shortened α1(III)Δ1 and α2(I)Δ1 chains, would suggest that this protein does not recognize a specific amino acid sequence but recognizes a structure that these sequences have in common. This preference for structural characteristics has also been suggested by the fact that Hsp47 can bind to the synthetic peptide sequences (Pro-Pro-Gly)n (Koide et al., 1999), an observation that would be difficult to explain if Hsp47 recognized specific amino acids within the triple-helical domain.
Previous work has suggested that Hsp47 binds to procollagen chains during various stages in its folding and assembly (Nakai et al., 1992; Sauk et al., 1994; Satoh et al., 1996). Using the SP cell approach adopted in this report, we are synthesizing relatively low concentrations of procollagen chains into ER that contains relatively high concentrations of P4H and Hsp47. Under these conditions the binding will be determined by the relative affinity of these proteins for the procollagen intermediates. P4H has a high affinity for (X-Y-Gly)n sequences that are non-triple-helical and adopt a polyproline type II helix, whereas it has a relatively low affinity for the triple-helical structure (Juva and Prockop, 1969; Kivirikko et al., 1992). This high affinity for non-triple-helical chains would ensure that P4H preferentially binds to this particular intermediate within our system. However, if the concentration of procollagen chains within the ER is increased significantly, then the amount of P4H present would be saturated, potentially allowing binding of Hsp47 to non-helical chains. This assumes that the affinity of non-helical chains for P4H is higher than that for Hsp47, an assumption that is verified by in vitro binding experiments (Juva and Prockop, 1969, Natsume et al., 1994). The experimental approach adopted previously to study Hsp47 binding involved either over-expression of procollagen chains by transfection, or use of chick tendon fibroblasts with a very high level of procollagen expression (Nakai et al., 1992; Satoh et al., 1996; Hosakawa and Nagata, 2000). Thus the discrepancy between previous results and those described here can be explained by the relative concentration of procollagen chains within the ER and the different affinities of P4H and Hsp47 for non-helical molecules. Indeed, in cell lines that synthesize a lower amount of procollagen, either endogenously (RD cells; Hosakawa and Nagata, 2000) or after transfection (Walmsley et al., 1999), little or no Hsp47 association with non-helical molecules could be detected.
If the primary role of Hsp47 is to bind to triple-helical structures, then what is the physiological consequence of this binding? Most chaperones that interact with their substrates do so in order to stabilize intermediates in the folding pathway, so why would Hsp47 bind to the fully folded triple-helical domain? These questions are difficult to address but some clues could come from the intrinsic thermal instability of regions of the triple-helical domain (Bächinger and Davis, 1990) and the potential requirement for a chaperone to prevent denaturation. Stabilizing amino acids, such as hydroxyproline, are not uniformly distributed along the collagen triple helix. This gives rise to well characterized regions within the helix that undergo temporary relaxation of conformation (Ryhänen et al., 1983). We hypothesize that Hsp47 binds to and stabilizes these regions of instability and that, during heat shock, expression of Hsp47 is induced to prevent thermal denaturation. If this hypothesis is correct then these regions of instability would need to be stabilized by a different mechanism once the procollagen chains exit the ER, as Hsp47 dissociates before ER–Golgi transport.
One mechanism that could account for this stabilization would be the lateral association of procollagen molecules, an event that occurs either before exit from the ER or within the intermediate compartment between the ER and the Golgi (Bonfanti et al., 1998). Alternatively, Hsp47 could bind to the regions of instability within the triple helix until a further stabilization event, such as hydration, occurs. Evidence to support this latter hypothesis comes from studies of the binding of Hsp47 to synthetic peptides (Koide et al., 1999). Here binding was shown to be dependent upon the length of the peptides (requiring more than seven Pro-Pro-Gly triplets) and upon whether the proline residues in the Y position of the X-Y-Gly repeats were hydroxylated (binding decreased as the number of hydroxyprolines increased). These results could be explained by the relative stabilities of triple helices formed by the synthetic peptides. The dependence upon peptide length could be explained by the fact that peptides with fewer than seven Pro-Pro-Gly triplets do not form a triple helix whereas longer peptides do (Bruckner et al., 1975). The decrease in binding following progressive substitution of proline for hydroxyproline could be a result of increasing the thermal stability of the helix formed. The more hydroxyproline residues present the greater the stability of the helix. Thus if Hsp47 recognizes regions of instability of the triple helix, one would predict that it would bind to peptides with a low thermal stability but not to peptides with high thermal stability. Hsp47 could therefore, recognize a conformation that is in transition between triple helix and polyproline type II helix. Further experimentation will be required to ascertain if this hypothesis is correct.
In summary, we have expressed a variety of procollagen chains that form clearly defined intermediates in the folding and assembly pathway in functionally intact ER in order to determine the binding specificity of Hsp47 and P4H. We suggest that there is competition between P4H and Hsp47 for binding to the non-triple-helical molecules with P4H preferentially binding to this intermediate. Hsp47 binds to procollagen molecules that have formed triple helices; we hypothesize that this binding is required to stabilize regions of the helix with low thermal stability. The consequence of this association is that procollagen chains are prevented from further transport through the secretory pathway until a further maturation event, such as lateral association of the procollagen molecules or further stabilization of the triple helical domain, occurs.
Materials and methods
Construction of recombinant plasmids
Construction of plasmids pα1(III), pα1(III)Δ1, pα1(III)α1Cys-Ser, pα2(I)Δ1 and pα2(I):(III)BGR has been described previously (Lees and Bulleid, 1994; Lees et al., 1997). The full-length cDNA clones encoding proα1(I) and proα2(I) were a kind gift from Dr D.J.Prockop (MCP Hahnemann University, Philadelphia, PA, USA).
Transcription in vitro
Transcription reactions were carried out as described by Gurevich et al. (1987). Recombinant plasmids pα1(III), pα1(III)Δ1, pα1(III)Δ1Cys, or plasmids pα2(I), pα2(I)Δ1, or pα2(I):(III)BGR were transcribed using T3 RNA polymerase or T7 RNA polymerase (Promega, Southampton, UK) for 24 h at 37°C. Following purification over RNeasy columns (Qiagen, Dorking, UK), RNA was resuspended in 100 µl RNase-free water containing 1 mM dithiothreitol (DTT) and 40 units RNasin (Promega).
Translation in vitro
RNA was translated using a rabbit reticulocyte lysate (FlexiLysate, Promega) for 90 min at 30°C unless otherwise indicated. The translation reaction (25 µl) contained 16.5 µl reticulocyte lysate, 0.5 µl 1 mM amino acids (minus methionine), 0.4 µl 100 mM KCl, 0.25 µl ascorbic acid (5 mg/ml), 15 µCi l-[35S]methionine (Amersham International, Amersham, UK), 0.5 µl transcribed RNA and 5 µl (∼2 × 105) semi-permeabilized HT-1080 cells (SP cells) prepared as described by Wilson et al. (1995). After translation, N-ethylmaleimide was added to a final concentration of 20 mM. SP cells were isolated by centrifugation in a microcentrifuge at 10 000 g for 5 min and the pellet was resuspended in an appropriate buffer for subsequent enzymic digestion or gel electrophoresis.
Proteolytic digestion
Isolated SP cells were resuspended in 20 µl CTT buffer [50 mM Tris–HCl pH 7.4, 0.15 M NaCl, 10 mM EDTA with 1% (v/v) Triton X-100]. Samples were then digested with a combination of chymotrypsin (250 µg/ml) and trypsin (100 µg/ml) (Sigma, Poole, UK) for 5 min at room temperature. Reactions were stopped by adding soybean trypsin inhibitor (Sigma) to a final concentration of 500 µg/ml. Pepsin digests were carried in the presence of 0.1 M HCl. Acidified samples were incubated with pepsin (100 µg/ml) for 2 h at 30°C or 16h at 4°C. Reactions were stopped by neutralization with Tris base (100 mM) and boiling SDS–PAGE loading buffer.
Thermal denaturation experiments
Translation products in SP cells were resuspended in KHM (110 mM potassium acetate, 20 mM HEPES, 2 mM magnesium acetate, pH 7.2) and aliquots were placed in a thermal cycler. A stepwise temperature gradient was set up over an appropriate temperature range; each temperature step was held for 10 min. After each incubation the sample was centrifuged at 10 000 g for 60 s, resuspended in CTT buffer and treated with a combination of chymotrypsin and trypsin, as described above.
A similar procedure was applied during experiments where DSP cross-linking was carried out following heating of samples at different temperatures. In this case samples removed from the thermal cycler were cross-linked using 1 mM DSP (Pierce and Warriner, Chester, UK) after each heating step as outlined below.
Chemical cross-linking
Translation products in SP cells were resuspended in 50 µl KHM. Cross-linking was carried out using 1 mM DSP [from a 50 mM stock solution in dimethylsulfoxide (DMSO)] for 10 min at 20°C followed by a further 10 min incubation after addition of 100 mM glycine to quench the DSP reaction. Cross-linked products were immunoprecipitated using the appropriate antibody.
Immunoprecipitation
Detailed protocols have been described previously (Wilson et al., 1998). Briefly SP cells, after cross-linking, were isolated and solubilized in denaturing buffer (50 mM Tris–HCl pH 7.4, 1% SDS, 1% Nonidet P-40). Insoluble material was removed by centrifugation at 13 000 g for 10 min. Supernatants were diluted in IP buffer [50 mM Tris–HCl, 0.15 M NaCl, 2 mM EDTA, 1% (v/v) Triton X-100] to 1 ml. Samples were pre-incubated with 50 µl of protein A–Sepharose [10% (w/v) in PBS] (Zymed Laboratories, San Francisco, CA, USA) for 30 min at 4°C to remove protein A-binding components. Cross-linked products were immunoprecipitated using the appropriate antibody and 50 µl protein A–Sepharose, overnight at 4°C. Antibodies against PDI were as described previously (John et al., 1993). Antibodies against α-subunit P4H and Myc were from Calbiochem (Nottingham, UK) and antibodies against Hsp47 were from Stressgen Biotechnologies Corporation (Victoria, BC, Canada). Immunoprecipitated complexes were retrieved by centrifugation at 13 000 g for 30 s and washed three times in IP buffer.
Refolding of denatured procollagen molecules
SP cells containing translated material were resuspended in 50 µl aliquots of KHM and heated at 47°C for 15 min to denature any triple-helical material. Samples were then incubated on ice for specific time periods before subjecting them to proteolysis as outlined above.
Refolding of triple-helix after inhibition of hydroxylation by α,α ′-dipyridyl
After 60 min translation in the presence of 0.5 mM α,α′-dipyridyl, cycloheximide was added to 5 mM, and samples were incubated for further time periods up to 60 min at 30°C in the presence or absence of 5 mM iron sulfate to allow hydroxylation to occur post-translationally. Aliquots were removed at fixed intervals and centrifuged to isolate SP cells. Isolated SP cells were cross-linked with DSP and immunoprecipitated with the anti-Hsp47 antibody.
SDS–PAGE
Samples were resuspended in SDS–PAGE loading buffer [0.0625 M Tris–HCl pH 6.8, 2% (w/v) SDS, 10% (v/v) glycerol and Bromophenol Blue] in the presence of 50 mM DTT and boiled for 5 min. After electrophoresis, gels were processed for autoradiography and exposed to Kodak X-Omat AR film, or images were quantified by phosphoimage analysis.
Acknowledgments
Acknowledgements
This work was supported by The Royal Society and grants from The Wellcome Trust (reference: 48356 and 50600). We thank Dr Jordi Bella for useful comments and suggestions on the manuscript.
References
- Bächinger H.-P. and Davis,J.M. (1990) Sequence specific thermal stability of the collagen triple helix. Int. J. Biol. Macromol., 13, 152–156. [DOI] [PubMed] [Google Scholar]
- Bonfanti L. et al. (1998) Procollagen traverses the Golgi stack without leaving the lumen of the cisternae: evidence for cisternal maturation. Cell, 95, 993–1003. [DOI] [PubMed] [Google Scholar]
- Bruckner P. and Eikenberry,E.F. (1984) Procollagen is more stable in cellulo than in vitro. Eur. J. Biochem., 140, 397–399. [DOI] [PubMed] [Google Scholar]
- Bruckner P., Rutschmann,B., Engel,J. and Rothe,M. (1975) A classical synthesis of the collagen-like peptides with the sequence 2(GlyProPro)nObut and their characterisation with circular dichroism and ultracentrifugation. Helv. Chim. Acta, 58, 1276–1287. [DOI] [PubMed] [Google Scholar]
- Bulleid N.J. (1996) Novel approach to study the initial events in the folding and assembly of procollagen. Sem. Cell Dev. Biol., 7, 667–672. [Google Scholar]
- Bulleid N.J., Wilson,R. and Lees,J.F. (1996) Type III procollagen assembly in semi-intact cells: chain association, nucleation and triple helix folding do not require formation of inter-chain disulfide bonds but triple helix nucleation does require hydroxylation. Biochem. J., 317, 195–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulleid N.J., Dalley,J.A. and Lees,J.F. (1997) The C-propeptide domain of procollagen can be replaced with a transmembrane domain without affecting trimer formation or collagen triple helix folding during biosynthesis. EMBO J., 16, 6694–6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chessler S.D. and Byers,P.H. (1993) BiP binds type I procollagen proα chains with mutations in the carboxy-terminal propeptide synthesized by cells from patients with osteogenesis imperfecta. J. Biol. Chem., 268, 18226–18233. [PubMed] [Google Scholar]
- Engel J. and Prockop,D.J. (1991) The zipper-like folding of collagen triple helices and the effects of mutations that disrupt the zipper. Annu. Rev. Biophys. Chem., 20, 137–152. [DOI] [PubMed] [Google Scholar]
- Fitzgerald J., Lamandé,S.R. and Bateman,J.F. (1999) Proteosomal degradation of unassembled mutant type I collagen proα1(I) chains. J. Biol. Chem., 274, 27392–27398. [DOI] [PubMed] [Google Scholar]
- Gurevich V.V., Pokrovskaya,I.D., Obukhova,T.A. and Zozulya,S.A. (1987) Preparative in vitro mRNA synthesis using SP6 and T7 polymerases. Anal. Biochem., 195, 207–213. [DOI] [PubMed] [Google Scholar]
- Hammond C. and Helenius,A. (1995) Quality control in the secretory pathway. Curr. Opin. Cell Biol., 7, 523–529. [DOI] [PubMed] [Google Scholar]
- Hosakawa N. and Nagata,K. (2000) Procollagen binds to both prolyl 4-hydroxylase/protein disulphide isomerase and Hsp47 within the endoplasmic reticulum in the absence of ascorbate. FEBS Lett., 466, 19–25. [DOI] [PubMed] [Google Scholar]
- John D.C.A., Grant,M.E.G. and Bulleid,N.J. (1993) Cell-free synthesis and assembly of prolyl-4-hydroxylase: Association of the α-subunit with the β-subunit (PDI) prevents its misfolding and aggregation. EMBO J., 12, 1587–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juva K. and Prockop,D.J. (1969) Formation of enzyme–substrate complexes with protocollagen proline hydroxylase and large polypeptide substrates. J. Biol. Chem., 23, 6486–6492. [PubMed] [Google Scholar]
- Kadler K. (1994) Fibril forming collagens. In Sheterline,P., (ed.), Protein Profiles: Extracellular Matrix 1; Academic Press, Oxford, UK. [Google Scholar]
- Kivirikko K.I., Myllylä,R. and Pihlajaniemi,T. (1992) Hydroxylation of proline and lysine residues in collagens and other animal and plant proteins. In Harding,J.J. and Crabbe,M.J.C. (eds), Post-translational Modification of Proteins. CRC Press, Boca Raton, FL, USA, pp. 1–51. [Google Scholar]
- Koide T., Asada,S. and Nagata,K. (1999) Substrate recognition of collagen-specific molecular chaperone HSP47. J. Biol. Chem., 274, 34523–34526. [DOI] [PubMed] [Google Scholar]
- Lamandé S.H., Chessler,S.D., Golub,S.B., Byers,P.H., Chan,D., Cole,W.G., Sillence,D.O. and Bateman,J.F. (1995) Endoplasmic reticulum-mediated quality control of type I collagen production by cells from osteogenesis imperfecta patients with mutations in the proα1(I) chain carboxy-terminal propeptide which impair subunit assembly. J. Biol. Chem., 270, 8642–8649. [DOI] [PubMed] [Google Scholar]
- Lees J.F. and Bulleid,N.J. (1994) The role of cysteine residues in the folding and association of the COOH-terminal propeptide of types I and III procollagen. J. Biol. Chem., 269, 24354–24360. [PubMed] [Google Scholar]
- Lees J.F., Tasab,M. and Bulleid,N.J. (1997) Identification of the molecular recognition sequence which determines the type-specific assembly of procollagen. EMBO J., 16, 908–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitzgen K. and Haas,I.G. (1998) Protein maturation in the endoplasmic reticulum. Chemtracts Biochem. Mol. Biol., 11, 423–445. [Google Scholar]
- McLaughlin S.H. and Bulleid,N.J. (1998) Molecular recognition in procollagen chain assembly. Matrix Biol., 16, 369–377. [DOI] [PubMed] [Google Scholar]
- Nagata K. (1996) Hsp47: a collagen-specific molecular chaperone. Trends Biochem. Sci., 21, 23–26. [DOI] [PubMed] [Google Scholar]
- Nakai A., Satoh,M., Hirayoshi,K. and Nagata,K. (1992) Involvement of the stress protein HSP47 in procollagen processing in the endoplasmic reticulum. J. Cell Biol., 117, 903–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsume T., Koide,T., Yokota,S., Hirayoshi,K. and Nagata,K. (1994) Interactions between collagen-binding stress protein Hsp47 and collagen: analysis of kinetic parameters by surface plasmon resonance biosensor. J. Biol. Chem., 269, 31224–31228. [PubMed] [Google Scholar]
- Ryhänen L., Zaragoza,E.J. and Uitto,J. (1983) Conformational stability of type I collagen triple helix: evidence for temporary and local relaxation of the protein conformation using a proteolytic probe. Arch. Biochem. Biophys., 223, 562–571. [DOI] [PubMed] [Google Scholar]
- Satoh M., Hirayoshi,K., Yokota,S.-I, Hosokawa,N and Nagata,K. (1996) Intracelluar interaction of collagen-specific stress protein HSP47 with newly synthesised procollagen. J. Cell Biol., 133, 469–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauk J.J., Smith,T., Norris,K. and Ferreira,L. (1994) Hsp47 and the translocation–translocation machinery co-operate in the production of αI(I) chains of procollagen. J. Biol. Chem., 269, 3941–3946. [PubMed] [Google Scholar]
- Uitto J. (1979) Collagen polymorphism: isolation and partial characterisation of α1(I)-trimer molecules in normal human skin. Arch. Biochem. Biophys., 192, 371–379. [DOI] [PubMed] [Google Scholar]
- Walmsley A.R., Batten,M.R., Lad,U. and Bulleid,N.J. (1999) Intra-cellular retention of procollagen within the endoplasmic reticulum is mediated by prolyl 4-hydroxylase. J. Biol. Chem., 274, 14884–14892. [DOI] [PubMed] [Google Scholar]
- Wilson R., Allen,A.J., Oliver,J., Brookman,J.L., High,S. and Bulleid,N.J. (1995) Development of a semi-permeabilised cell system to study the translocation, folding, assembly and transport of secretory proteins. Biochem. J., 307, 679–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R., Lees,J.F. and Bulleid,N.J. (1998) Protein disulphide isomerase acts as a molecular chaperone during the assembly of procollagen. J. Biol. Chem., 273, 9637–9643. [DOI] [PubMed] [Google Scholar]