Abstract
We have determined the crystal structure of the Escherichia coli ribosome recycling factor (RRF), which catalyzes the disassembly of the termination complex in protein synthesis. The L-shaped molecule consists of two domains: a triple-stranded antiparallel coiled-coil and an α/β domain. The coil domain has a cylindrical shape and negatively charged surface, which are reminiscent of the anticodon arm of tRNA and domain IV of elongation factor EF-G. We suggest that RRF binds to the ribosomal A-site through its coil domain, which is a tRNA mimic. The relative position of the two domains is changed about an axis along the hydrophobic cleft in the hinge where the alkyl chain of a detergent molecule is bound. The tRNA mimicry and the domain movement observed in RRF provide a structural basis for understanding the role of RRF in protein synthesis.
Keywords: crystal structure/protein synthesis/ribosome recycling factor/translation/tRNA
Introduction
It is well established that cellular protein synthesis consists of three steps: initiation, elongation and termination. In the termination step, release factors (RF1 and RF2 in prokaryotes, eRF1 in eukaryotes) recognize the stop codon in the A-site of the ribosome and activate the hydrolysis of the polypeptide chain from tRNA in the P-site. Then, the dissociation of these factors from the ribosomal A-site is promoted by release factor 3 (RF3) in a GTP-dependent reaction (Freistroffer et al., 1997). For the next translation cycle, breakdown of the termination complex and recycling of the ribosome are required (Kaji et al., 1998). It was found that the ribosome recycling factor (RRF) and elongation factor G (EF-G) have a role in catalyzing ribosome recycling with GTP hydrolysis (Pavlov et al., 1997a; Kaji et al., 1998). In contrast, RF3 was reported to be able to substitute for EF-G in ribosome recycling by dissociating deacylated tRNA with RRF (Grentzmann et al., 1998). A second role for RRF is proposed to be in error reduction during peptide elongation (Janosi et al., 1996). The ribosome recycling step has been confirmed to be essential in protein synthesis since RRF inactivation was bacteriostatic in the growing phase and bactericidal during the transition between the stationary and growing phases (Janosi et al., 1998). The absence of RRF caused cell death in Escherichia coli (Janosi et al., 1994), which might be due to increased translation errors (Janosi et al., 1996) and unscheduled reinitiation (Ryoji et al., 1981; Janosi et al., 1998).
The molecular mechanism of RRF action in ribosome recycling and translation error prevention is still not clear. In the model that has been proposed based on the role of EF-G in translocating peptidyl-tRNA during translation elongation, EF-G translocates RRF from the A-site to the P-site and ejects deacylated tRNA from the ribosome (Janosi et al., 1996; Nakamura et al., 1996; Pavlov et al., 1997a). Then, the termination complex is dissociated into mRNA, tRNA and the ribosome for the next translation cycle in this model (Kaji et al., 1998). However, the proposal that tRNA is ejected from the ribosome by RRF and EF-G seems to contradict the recent results of Karimi et al. (1999), who showed that RRF and EF-G cannot accelerate the dissociation of deacylated tRNA from the ribosome. They proposed a new model in which RRF, EF-G and GTP only catalyze the dissociation of the 50S subunit from the termination complex, followed by tRNA removal from the 30S⋅tRNA⋅mRNA complex by initiation factor 3 (IF3).
The gene encoding RRF is widely distributed in prokaryotes with high sequence homology (Janosi et al., 1996; Figure 1). By sequence analysis, several genomic sequences from eukaryotes have been identified as the gene encoding an RRF homolog that appears to be present in organelles (Janosi et al., 1996). In spinach, the RRF homolog localized in the chloroplast exerted a competitive inhibitory action on the bacterial temperature-sensitive RRF (Rolland et al., 1999). However, it is known that the RRF homolog in yeast is necessary for protein synthesis in mitochondria (Kanai et al., 1998) but is not essential for cell growth (Kaji et al., 1998). Therefore, eukaryotic RRFs present in organelles might not be essential for cell viability since their inhibition could not influence the synthesis of cytoplasmic proteins. The absence of RRF homologs in archaea, which have protein synthesis machinery similar to that of eukaryotes, also supports the notion that RRF is unnecessary for eukaryotic protein synthesis. The bactericidal effect of removing RRF and its non-essential role in eukaryotes make RRF a potential target for developing a new antibacterial agent (Kaji et al., 1998).
Fig. 1. Multiple sequence alignment of RRFs from prokaryotes. The sequences of RRFs from E.coli (E.coli), Haemophilus influenzae (H.inf), Pseudomonas aeuruginosa (P.aeu) and Bacillus subtilis (B.sub) are used for multiple sequence alignment. The secondary structures of E.coli RRF are indicated by an arrow for a β-strand and by a cylinder for an α-helix. The amino acids that make contact with the alkyl chain of the detergent are marked with asterisks (*). The mutated residues identified in temperature-sensitive mutants of RRF (Janosi et al., 1998) are indicated by # marks. The heptad pattern (abc defg) of each residue in the coiled-coil is also shown. In four sequences, identical residues are boxed in black and homologous residues are boxed in gray.
It was proposed that RRF binds to the A-site of the ribosome and competes for that binding site with release factor 1 (RF1) in E.coli (Pavlov et al., 1997b). The binding affinity of RRF for the ribosome suggests that it may have a structural motif similar to other translation factors that show binding affinity for the A-site of the ribosome. The tRNA mimicry domain postulated in several translation factors might be the structural motif shared among them (Nakamura et al., 1996; Brock et al., 1998). In order to elucidate the structural relevance of RRF to other translation factors and to understand better its role in ribosome recycling, we have determined the crystal structure of E.coli RRF at 2.3 Å resolution.
Results
Structure determination
The crystals were grown as described in Materials and methods. The presence of a detergent, decyl-β-d-maltopyranoside, in the protein solution was essential for crystallization of E.coli RRF (Yun et al., 2000). The crystal structure was determined by multiple isomorphous replacement with anomalous scattering (MIRAS) and density modification including solvent flipping (Table I). Phase quality, which was not good enough for model building due to the non-isomorphism among crystals, was remarkably improved by addition of anomalous data to the phase calculation (Table I). The amino acid registration in the model was confirmed by the position of a mercury atom at Cys16, and the heptad repeat in the coiled-coil. The refined model at 2.3 Å resolution consists of residues 1–185 of E.coli RRF. No residues lie outside the allowed regions in the Ramachandran plot. The structure evaluation with the programs ERRAT (Colovos and Yeates, 1993) and PROCHECK (Laskowski et al., 1993) indicates that the refined model has good geometry.
Table I. Data collection, phasing and refinement statistics.
| Native1 | Native2 | Hg1a | Hg2b | Pt1c | |
|---|---|---|---|---|---|
| Data collection | |||||
| X-ray source | ALS | CuKα | CuKα | ALS | NSLS |
| Wavelength (Å) | 1.0074 | 1.5418 | 1.5418 | 1.0099 | 1.0713 |
| Resolution (Å) | 20.0–2.3 (2.38–2.30) | 30.0–2.9 (3.00–2.90) | 30.0–3.3 (3.42–3.30) | 30.0–2.3 (2.35–2.30) | 30.0–2.9 (3.00–2.90) |
| Rmerge (%)d | 4.8 (23.0) | 10.2 (46.9) | 17.1 (34.7) | 7.3 (23.4) | 9.0 (29.9) |
| Completeness (%) | 96.2 (97.7) | 95.5 (93.5) | 92.0 (98.1) | 98.1 (99.6) | 95.8 (93.0) |
| No. of unique reflections | 8691 (858) | 4367 (430) | 2907 (309) | 8846 (553) | 4332 (422) |
| Redundancy | 3.34 | 3.16 | 2.19 | 5.62 | 5.92 |
| Phasing (30.0–2.9 Å) | |||||
| Rderiv (%)e | 40.0 | 42.4 | 37.2 | ||
| No. of heavy atom sites | 6 | 1 | 7 | ||
| Anomalous refinement | No | Yes | Yes | ||
| Phasing power (acentric)f | 2.44 | 0.78 | 0.65 | ||
| Phasing power, anomalous | 2.72 | 1.30 | |||
| RCullis (acentric)g | 0.65 | 0.94 | 0.92 | ||
| RCullis, anomalous | 0.62 | 0.88 | |||
| Figure of merit, acentric | 0.596 | ||||
| Refinement | |||||
| Resolution (Å) | 20.0–2.3 | 30.0–2.9 | 30.0–2.9 | ||
| Rfactor/Rfree (%) | 22.8/29.7 | 22.9/32.2 | 23.3/32.9 | ||
| No. of reflections (2σ cut-off) | 8391 | 3939 | 4100 | ||
| No. of solvent atoms | 230 | 22 | 35 | ||
| No. of protein atoms | 1437 | 1437 | 1437 | ||
| No. of heteroatoms | 16 | 13 | 13 | ||
| Mean B-factor (Å2) | 53.9 | 39.3 | 48.6 | ||
| R.m.s.d. in bond length (Å) | 0.010 | 0.010 | 0.010 | ||
| R.m.s.d. in bond angle (°) | 1.63 | 1.63 | 1.66 |
Numbers in parentheses correspond to the highest resolution shell.
aHg1, HgCl2; bHg2, p-chloromercuribenzoate; cPt1, K2PtCl4.
dRmerge = Σ|I – <I>|/ΣI
eRderiv = Σ|FPH – FP|/ΣFPH
fPhasing power (acentric) = |FHcalc|/|FPH – |FP + FHcalc||
gRCullis (acentric) = Σ|FPH – |FP + FHcalc||/Σ|FPH – FP|
Overall structure of RRF
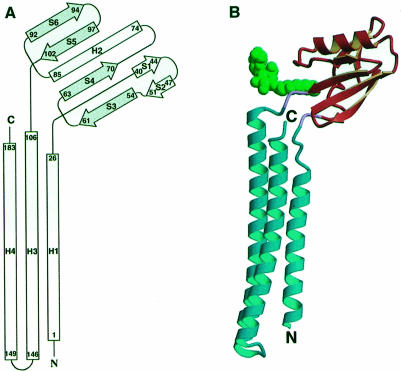
Escherichia coli RRF is an open L-shaped molecule composed of two domains (Figure 2A and B). The first domain (residues 1–30 and 104–185) contains three α-helices, and the second domain (residues 32–102) contains one α-helix and six β-strands. An N-terminal α-helix (H1, residues 1–26) in the first domain is followed by the second domain. Two antiparallel C-terminal α-helices (H3, residues 106–146; H4, residues 149–183) connected by a short U-turn are extended from the second domain and folded back to helix H1. Thus, the first domain (coil domain) consists of a three-stranded antiparallel coiled-coil ∼65 Å long, in which the seven-turn helix (H1) packs into the parallel 11-turn helix (H3) and the antiparallel 10-turn helix (H4). The two-turn overhang helices in the C-terminal end of H3 and the N-terminal end of H4 near the U-turn form a short two-stranded coiled-coil. The second domain (α/β domain) with approximate dimensions of 23 × 26 × 36 Å has an α/β topology containing a short β-hairpin (S1 and S2) and a single α-helix (H2) packed against one face of an antiparallel four-stranded β-sheet (S3–S6). Two domains are connected by two hinge residues (Arg31 and Pro103) with an approximate angle of 110° between the lines along helix H3 and strand S6 (Figure 2B).
Fig. 2. The crystal structure of E.coli RRF. (A) Topology diagram of E.coli RRF structure. β-strands are shown as arrows and α-helices as rectangles. The first and the last residues in each secondary structure are labeled. The four-stranded β-sheet is colored gray. (B) Ribbon diagram of RRF (Kraulis, 1991). The coil domain and α/β domain, colored sky-blue and orange, respectively, are connected by two hinge residues (pink). The arrows represent β-strands and the cylinders represent α-helices. The N- and C-termini are labeled. The detergent molecule is drawn in a green space-filling model.
Coiled-coil
The three-stranded coiled-coil in E.coli RRF shows the classical knobs-into-holes packing found in other coiled-coils, with a few discontinuities. In helix H4, one ‘skip’ residue (Ala167), which corresponds to an extra residue in the heptad pattern, causes a kink. As a result, helix H4 below the skip residue (residues 168–183) changes its direction close to helix H3 by an ∼5° rotation along an axis perpendicular to the plane containing the H1 and H4 helices (Figure 2B). However, the hydrophobic contacts in the core of the coiled coil are conserved. An unusual 3-4-4-3 periodicity (abc–defg–defg–abc) called a ‘shutter’ is found at one position in helix H1 and at two positions in helix H3 (Figure 1). These discontinuities decrease the extent of supercoiling in the coiled-coil and increase the number of residues per turn above the value found in other regular coiled-coils (Lupas, 1996). The breaks of the helix caused by the ‘skip’ and ‘shutter’ are also found in other three-stranded coiled-coils such as hemagglutinin (Bullough et al., 1994) and Ebola virus membrane fusion subunit GP2 (Weissenhorn et al., 1998).
α/β domain
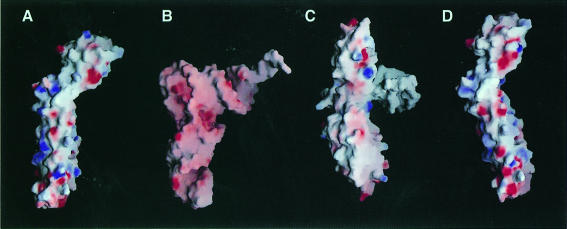
An exposed β-sheet on one side of the α/β domain found in RRF is reminiscent of several ribosomal and other RNA-binding proteins (Liljas and Garber, 1995). However, its topology is different from that in other RNA-binding proteins (Figure 2A) and it does not show the typical positive surface charge distribution necessary for binding to the phosphate backbone of RNA (Figure 3A and D). By using the DALI server software (Holm and Sander, 1993), it was found that the α/β domain of RRF has topology similar to that of the C-terminal domain of the arginine repressor (van Duyne et al., 1996) except for the substitution of a long loop and a β-hairpin in RRF for the helix in the repressor (Figure 4). RRF can be superimposed on the arginine repressor with a root-mean-square deviation (r.m.s.d.) of 1.7 Å for 39 of 48 Cα atoms (Figure 4).
Fig. 3. Structural comparisons of RRF with tRNA and EF-G. RRF superposed on (A) tRNA, (B) yeast tRNAPhe (Suddath et al., 1974) and (C) EF-G (Czworkowski et al., 1994), and (D) RRF superposed on EF-G are drawn by surface charge distribution with the program GRASP (Nicholls et al., 1991). The red and blue colors represent negatively and positively charged surfaces, respectively. For the superposition of RRF on tRNA (A), the coil domain of RRF is superimposed on the anticodon arm of tRNA and the α/β domain is oriented toward the acceptor arm in tRNA. The position of EF-G in (C) is determined by overlapping domains I and II of EF-G with the EF-Tu⋅GDP⋅tRNA complex (Nissen et al., 1995). For the superposition of RRF on EF-G (D), the coil and α/β domains of RRF are overlapped with domains IV and V of EF-G, respectively.
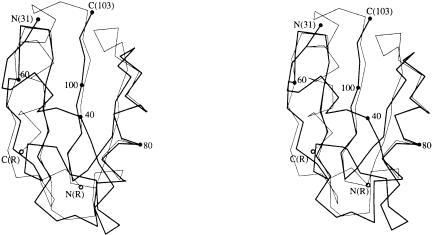
Fig. 4. Stereo Cα traces of the α/β domain in RRF superimposed onto those of the C-terminal domain in the arginine repressor (van Duyne et al., 1996). A total of 48 Cα atoms (residues 55–103) in RRF are used for overlapping the α/β domain of RRF (thick line) with the C-terminal domain of the arginine repressor (thin line). Cα positions of the first, last and every 20th residues in RRF are labeled.
Comparison with other A-site-binding molecules
To investigate the structural resemblance of RRF to other ribosomal A-site-binding molecules, the crystal structure of E.coli RRF has been compared with that of yeast tRNAPhe (Suddath et al., 1974) and EF-G (Czworkowski et al., 1994) (Figure 3). The orientations of EF-G and tRNA are determined by overlapping domains I and II of EF-G with EF-Tu in the tRNA⋅EF-Tu⋅GDPNP complex (Nissen et al., 1995). In this orientation, domains III, IV and V of EF-G correspond to the acceptor stem, the anticodon helix and the T stem of tRNA, respectively (Figure 3B and C). The L-shaped RRF structure can be superimposed on tRNA and EF-G by overlapping the α/β domain of RRF with the acceptor stem in tRNA and domain III in EF-G, and the coil domain with the anticodon helix in tRNA and domain IV in EF-G (Figure 3A). The overall shape of RRF and tRNA is not well matched because the α/β domain is connected to the coil domain by an obtuse angle. However, if the stretching of the L-shaped tRNA in the ribosomal A-site is considered (Stark et al., 1997a), their shapes may be more similar to each other than shown in Figure 3A and B. In this superposition, RRF lacks the domain corresponding to the T-stem of tRNA or the V domain in EF-G. Based on the similarities of domain shape and size, another superposition can be made by overlapping the coil and α/β domains of RRF with domains IV and V in EF-G, respectively (Figure 3D).
Hydrophobic cleft and domain movement
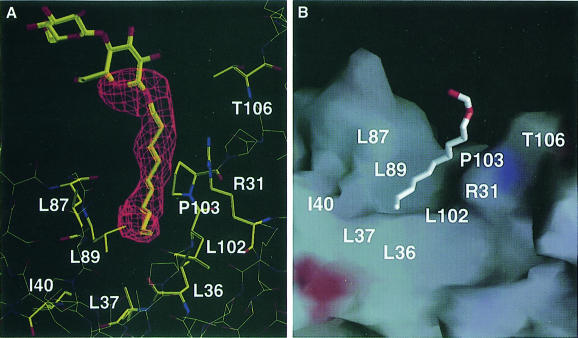
The sausage-shaped electron density (Figure 5A) found in the hinge between the coil domain and the α/β domain (Figure 2B) was assigned as the detergent, decyl-β-d-maltopyranoside. The alkyl chain of the detergent molecule is packed into the hydrophobic cleft formed by several residues from the coil domain (Thr106), the hinge (Arg31 and Pro103) and the α/β domain (Leu36, Leu37, Ile40, Leu87, Leu89 and Leu102) (Figures 1 and 5). Upon binding of the detergent to the cleft, 210 Å2 of the surface area are buried, which corresponds to ∼1.9% of the total surface area of RRF (11 291 Å2). The hydrophobic residues and the aliphatic chain of Arg31 involved in the interactions with detergent are well conserved among the species in the multiple sequence alignment (residues with asterisks in Figure 1). It can be inferred that the residues in the cleft could have specific interactions with proteins or other hydrophobic molecules in the absence of detergent in vivo. Alternatively, a domain movement might occur to minimize the exposure of hydrophobic residues in the absence of hydrophobic molecules. It is tempting to postulate a dramatic conformational movement including the change of secondary structure by exposing the hydrophobic surface, which is found in several structures such as lipases (Kim et al., 1997).
Fig. 5. The detergent molecule bound in a hydrophobic cleft. (A) A simulated-annealed omit electron density map calculated at 2.3 Å resolution with the detergent molecule omitted was drawn near the detergent-binding cleft. The detergent and surrounding residues were also drawn. (B) A stick model of the detergent located on a cleft of RRF drawn by surface charge distribution with the program GRASP (Nicholls et al., 1991). The white color represents the hydrophobic surface. The red and blue colors represent negatively and positively charged surfaces, respectively. The positions of the hydrophobic residues contacting the detergent are shown. The atoms outside the simulated-annealed omit map are excluded from the detergent model.
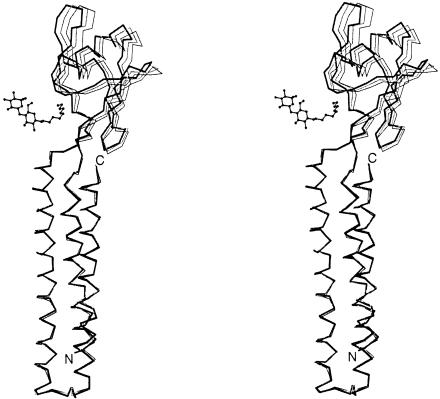
The non-isomorphism represented by high Rderiv values implies the occurrence of a structural change in RRF (Table I). Indeed, the expected domain movement is observed in RRF when three RRF structures are compared in Figure 6. While Cα atoms in the coil domains of Native1 and Native2 are superimposed with a 0.29 Å r.m.s.d., Cα atoms in each α/β domain deviate by 1.0 Å (Figure 6). When the two structures of Native2 and Pt1 are compared in the same way, the average deviation is 1.86 Å. Therefore, the two α/β domains of Native2 and Pt1 can be brought together by rotation of one domain ∼5° about an axis along the length of the detergent (Figure 6), suggesting that the coil domain and the α/β domain are connected by a hinge. This hinge motion could become larger in the absence of the detergent that links the two domains in our structure.
Fig. 6. Domain movement in E.coli RRF. Cα traces of Native1 (gray line), Native 2 (thin line) and Pt1 (thick line) structures are drawn by overlapping their coil domains (residues 1–30 and 104–185). The detergent molecule in Native1 is also shown as a ball-and-stick model. A total of 70 Cα atoms in the α/β domain show a deviation of 1.00 Å between Native1 and Native2, 0.89 Å between Native1 and Pt1, and 1.86 Å between Native2 and Pt1.
Discussion
The coil domain as a tRNA mimic
Several lines of biological evidence indicate that RRF binds to the A-site of the ribosome or a site overlapping the A-site where RF1 or tRNA binds. The first evidence is that RF1 is reported to be able to inhibit ribosomal recycling by associating with the ribosome and preventing RRF from entering the ribosomal A-site (Pavlov et al., 1997b; Grentzmann et al., 1998). The error reduction mechanism of RRF in the elongation step explained by the difference of binding affinity for the A-site among cognate aminoacyl-tRNA, RRF and non-cognate-aminoacyl tRNA (Janosi et al., 1996) also supports the A-site binding of RRF.
Translation factors that interact with the ribosomal A-site have been postulated to mimic the structure of A-site tRNA (Nakamura et al., 1996). Domains III–IV of EF-G were identified as the first tRNA mimic by the structural similarity of EF-G to the EF-Tu⋅GDP⋅tRNA complex (Nissen et al., 1995). RF1 and RF2 were also proposed to mimic tRNA based on their binding affinity for the A-site as well as their sequence homology with EF-G (Nakamura et al., 1996). This tRNA mimicry hypothesis was extended to IF1 and IF2 (Brock et al., 1998). In the same manner, it can be postulated that RRF also mimics tRNA since it can interact with the ribosomal A-site. However, when the RRF structure is compared with that of tRNA or EF-G, no remarkable structural similarity is observed except in overall shape (Figure 3). This structural discrepancy is also found in IF1 (Sette et al., 1997), which binds to the ribosomal A-site (Moazed et al., 1995) and is suggested to be another tRNA mimic (Brock et al., 1998). In addition, tRNA and EF-G are expected to bind to the ribosomal A-site with different binding modes since they show many different structural features in spite of overall structural similarity (Agrawal et al., 1996, 1998, 1999; Stark et al., 1997a; Cate et al., 1999; Figure 3B and C). These data suggest that specific interactions with the ribosomal A-site rather than structural resemblance to tRNA might be more essential for binding to the A-site, and those interactions are the conserved features among IF1, EF-G, RRF and tRNA. Taken together, the tRNA mimicry might be understood in functional as well as structural terms.
Among three mimicry domains of EF-G, only domain IV is known to share the binding site with the anticodon arm of A-site tRNA in the post-translocation state (Agrawal et al., 1996; Stark et al., 1997a), while the other domains of EF-G orient differently (Agrawal et al., 1998; Wilson and Noller, 1998). The critical role of domain IV in A-site binding has been proved by showing that a deletion mutant of EF-G lacking domain IV possessed a reduced affinity for the ribosome (Rodnina et al., 1997; Martemyanov and Gudkov, 1999). The fact that only the anticodon arm of tRNA bound to EF-Tu reaches into approximately the same position as that of A-site tRNA (Stark et al., 1997b) also supports the importance of the anticodon arm of tRNA and domain IV of EF-G for ribosomal A-site binding. Taking these results together, we propose that the A-site-binding proteins share the domain that mimics the anticodon arm of tRNA or domain IV of EF-G. With these assumptions, it can be suggested that the coil domain of RRF is the tRNA mimicry domain corresponding to the anticodon arm of tRNA. In RRF, a three-stranded coiled-coil forms a cylinder with approximate dimensions of 21 × 21 × 65 Å. The size and shape of the coil domain are comparable with those of the anticodon arm of tRNA and domain IV of EF-G, although the structure is not identical (Figure 3). The negative surface charge distribution present at the bottom of RRF, EF-G and tRNA in Figure 3 might be necessary for a specific interaction with the A-site in the 30S ribosomal subunit. As a matter of fact, only the tip of domain IV of EF-G showed substantial overlap with the binding site of the anticodon stem of the A-site tRNA (Agrawal et al., 1998). Therefore, the coil domain of RRF could be important for binding to the A-site and could be a candidate for molecular mimicry by having a structure or function similar to that of the anticodon arm of tRNA or domain IV of EF-G. The overall similarity between RRF and tRNA proposed in this study represents another example of RNA mimicry by a protein.
It was reported that mutations exhibiting a temperature-sensitive phenotype are found exclusively in the coil domain (Janosi et al., 1998; Figure 1). Since a coiled-coil is a thermally stable structure compared with other three-dimensional folds, it is reasonable to suggest that point mutations in the coil domain affect the thermostability of RRF. In temperature-sensitive mutants, the mutations of residues involved in the formation of the coil domain could not alter the overall fold of RRF at moderate temperature. In contrast, at high temperature, the coil domains in the mutants lose their shape and their specific interactions with the A-site, which terminates ribosome recycling. However, it should be noted that mutations of the residues interacting directly with the ribosome could not be screened in the experiment performed by Janosi et al. (1998), since those mutations must be lethal.
The role of the α/β domain
The α/β domain in RRF has an exposed β-sheet which is similar to the C-terminal domain of the arginine repressor (Figure 4). Since an open β-sheet found in the arginine repressor (van Duyne et al., 1996) and in ribosomal protein S8 (Nevskaya et al., 1998) participates in protein–protein interactions, it can be suggested that the α/β domain in RRF might also be involved in protein–protein interactions. Considering the proposed role of RRF in preventing EF-G from leaving the ribosome after GTP hydrolysis (Karimi et al., 1999), RRF might form a complex with EF-G via its α/β domain. Another possible partner of the α/β domain is a ribosomal protein in the 50S subunit. If the α/β domain of RRF can be superimposed on domain V of EF-G, as proposed in Figure 3D, it could be positioned near L7/L12 where EF-G makes contacts through domain V (Agrawal et al., 1998; Ban et al., 1999). In this orientation, RRF can link the 30S ribosomal subunit to the 50S ribosomal subunit through its α/β domain. One possible explanation for the absence of mutations conferring a temperature-sensitive phenotype on the α/β domain (Janosi et al., 1998; Figure 1) could be that the proposed interactions with the α/β domain are essential for cell viability. However, further experiments such as direct cross-linking between RRF and its binding partners or cryoelectron microscopy are necessary in order to elucidate more precisely the role of the α/β domain in RRF. This study reveals that the coil domain and the α/β domain of RRF are connected by an interdomain hinge. Considering putative roles for each domain in RRF and the domain movements around the hinge, it can be suggested that RRF might play a role as a flexible linker that connects their binding counterparts via its two domains.
Hydrophobic cleft as a drug target
RRF is regarded as a good target for antibacterial drug development (Kaji et al., 1998), because it is essential for bacterial growth (Janosi et al., 1994). Inhibitors of RRF targeted to the contact region between RRF and its counterparts can be candidates for designing a novel compound with antibacterial activity. A small molecule bound to the hydrophobic cleft may also inhibit RRF activity by preventing the domain movement that could be important for the role of RRF. Domain movements found in tRNA (Stark et al., 1997a) and EF-G (Agrawal et al., 1998, 1999) have been suggested to be necessary for their binding to the ribosomal A-site. In this respect, an analysis of the interaction between decyl-β-d-maltopyranoside and residues in the cleft of RRF could be useful for designing a novel compound as an RRF inhibitor.
The role of RRF in ribosome recycling
The most intrinsic question regarding RRF is the mechanism of decomposition of the termination complex and the role of RRF in this process. Basically, two different models that explain the mechanism of ribosome recycling have been proposed based on biochemical studies performed in different assay systems (Kaji et al., 1998; Karimi et al., 1999). The most fundamental differences between the proposed models are the role of EF-G and RRF in ribosome recycling. In the first model, EF-G moves RRF bound at the ribosomal A-site to the P-site, and ejects deacylated tRNA in a translocation-like mechanism (Janosi et al., 1996; Pavlov et al., 1997a). The role of RRF in this model is analogous to that of peptidyl-tRNA during the elongation step. In the second model, the role of EF-G in termination is considered to be different from that in elongation (Karimi et al., 1999). According to this model, RRF and EF-G in the presence of GTP form a stable complex and cause the dissociation of the 50S ribosomal subunit from the 30S⋅tRNA⋅mRNA complex after GTP is hydrolyzed (Karimi et al., 1999). In this model, the role of RRF is to prevent EF-G from leaving the ribosome after GTP hydrolysis (Karimi et al., 1999). Since the present crystal structure of RRF and previous biological data suggest the presence of the tRNA mimicry domain in RRF and its binding to the A-site, we propose that RRF binds to the ribosomal A-site after release of RF1 from the ribosome by RF3 in the recycling mechanism. When RRF is present in the A-site, further recycling steps will proceed by a mechanism that needs to be explored further. EF-G might push RRF to the P-site and release tRNA, as proposed for the translocation step (Nakamura et al., 1996; Pavlov et al., 1997a; Kaji et al., 1998). However, the role of RRF and EF-G in ribosome recycling must be reconsidered in order to explain the dissociation of the 70S ribosome into 30S and 50S subunits, since it was shown that tRNA is not ejected from the ribosome by EF-G and RRF (Karimi et al., 1999). Further structural and biochemical experiments based on current structural information are required to elucidate the precise mode of RRF binding to the ribosome and the details of the recycling mechanism.
Materials and methods
Crystallization and data collection
The E.coli RRF was cloned, purified and crystallized as described elsewhere (Yun et al., 2000). Crystals suitable for data collection were grown by the hanging drop vapor diffusion method at 14°C from a reservoir solution containing 0.1 M MES–NaOH pH 6.5, 10% polyethylene glycol (PEG) 350 MME and 12–14% PEG 400. Decyl-β-d-maltopyranoside was added to 1.8 mM concentration in the protein drop. Crystals were transferred to a freezing solution containing 20% PEG 400 in addition to the same chemicals in the reservoir solution and equilibrated for >48 h before flash-freezing and data collection. X-ray data from the native crystal (Native2) and one mercury derivative (Hg1) were collected on a MacScience 2030b image plate detector (Table I). Other diffraction data sets were collected with anomalous signals at the beam line X8-C of the National Synchrotron Light Source (NSLS), Brookhaven, and at the beam line 5.0.2 of the Advanced Light Source (ALS), Berkeley. The native and derivative data were processed and integrated by DENZO and scaled by SCALEPACK (Otwinowski and Minor, 1997). The space group of these crystals is P3121 and the unit cell dimensions are a = b = 48.06 Å, and c = 142.27 Å for Native1. There is one molecule in the asymmetric unit with a VM of 2.30 Å3/Da and a calculated solvent content of 53.4%.
Structure determination and refinement
Native2, two mercury and one platinum data sets were used for phase calculation by MIRAS (Table I). The first mercury position was identified in the difference Patterson map between the HgCl2 and native data. Further heavy atom sites in other derivatives were found in the difference Fourier maps calculated by using the phases from the first derivative with the program MLPHARE (Collaborative Computational Project Number 4, 1994). The heavy atom parameters were refined and the phases were calculated to 2.9 Å with the program SHARP (Fortelle and Bricogne, 1997). The initial MIR phases were improved by solvent flipping at the same resolution with the program SOLOMON (Collaborative Computational Project Number 4, 1994). After density modification, the electron density revealed two domains with a few backbone interruptions in the loop region. Amino acids were assigned for residues 1–185 using the program O (Jones et al., 1991). Several cycles of rigid-body refinement, positional refinement and simulated annealing were performed at 2.9 Å resolution with CNS (Brünger et al., 1998). The refinements were continued at 2.3 Å using the Native1 data. Although Native1 data were collected from a crystal soaked in 1 mM HgCl2 for 24 h, they have been used for the final refinement since they showed the lowest heavy atom peaks in the difference Fourier map and maximum diffraction limit (Table I). The orientation of Native1 was found by molecular replacement with EPMR (Kissinger et al., 1999), using the model of Native2. Successive refinement with temperature factors and addition of solvents and other heteroatoms resulted in an R-value of 22.8% and an Rfree of 29.7% with a bulk solvent correction and overall anisotropic thermal factor refinement. Rfree was calculated with 10% of the reflections. The current model includes residues 1–185, 230 water molecules, three mercury atoms with an occupancy of 0.2 and a detergent molecule identified as a decyl-β-d-maltopyranoside. Most of the atoms in the head group of the detergent were modeled with zero occupancy, since electron density was missing except for the C1 and O6 atoms (Figure 5A). A total of 91% of the non-glycine residues were in the most favorable region of the Ramachandran plot and 9% in the additionally allowed region. The crystal structures of Native2 and Pt1 were determined by the molecular replacement method with the program EPMR (Kissinger et al., 1999), using the refined model of Native1. The rigid body refinement followed by simulated annealing and temperature factor refinement was performed at 2.9 Å with the program CNS. Table I summarizes the phasing and refinement statistics. The final coordinates and structure factors have been deposited in the Protein Data Bank (accession No. 1EK8).
Acknowledgments
Acknowledgements
We thank S.-H.Kim and T.Earnest of the Berkeley Advanced Light Source, and L.Flaks of the National Synchrotron Light Source, Brookhaven, for data collection. We also thank J.-H.Moon, J.Y.Lee and J.-K.Yang for their help in data collection, M.J.Cho for providing research facilities, and E.Holbrook for reading the manuscript. This work was supported by an international cooperative research grant from the Korea Science and Engineering Foundation and a grant from the Molecular Medicine Research Group Program (98-J03-02-03-A-06) from the Ministry of Science and Technology (to K.K.K.) and a grant from the Korea Science and Engineering Foundation through the Center for Molecular Catalysis at Seoul National University (to S.W.S.).
References
- Agrawal R.K., Penczek,P., Grassucci,R.A., Li,Y., Leith,A., Nierhaus,K.H. and Frank,J. (1996) Direct visualization of A-, P-, and E-site transfer RNAs in the Escherichia coli ribosome. Science, 271, 1000–1002. [DOI] [PubMed] [Google Scholar]
- Agrawal R.K., Penczek,P., Grassucci,R.A. and Frank,J. (1998) Visualization of elongation factor G on the Escherichia coli 70S ribosome: the mechanism of translocation. Proc. Natl Acad. Sci. USA, 95, 6134–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal R.K., Heagle,A.B., Penczek,P., Grassucci,R.A. and Frank,J. (1999) EF-G dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nature Struct. Biol., 6, 643–647. [DOI] [PubMed] [Google Scholar]
- Ban N., Nissen,P., Hansen,J., Capel,M., Moore,P.B. and Steitz,T.A. (1999) Placement of protein and RNA structures into a 5 Å-resolution map of the 50S ribosomal subunit. Nature, 400, 841–847. [DOI] [PubMed] [Google Scholar]
- Brock S., Szkaradkiewicz,K. and Sprinzl,M. (1998) Initiation factors of protein biosynthesis in bacteria and their structural relationship to elongation and termination factors. Mol. Microbiol., 29, 409–417. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Bullough P.A., Hughson,F.M., Skehel,J.J. and Wiley,D.C. (1994) Structure of influenza haemagglutinin at the pH of membrane fusion. Nature, 371, 37–43. [DOI] [PubMed] [Google Scholar]
- Cate J.H., Yusupov,M.M., Yosupova,G.Z., Earnest,T.N. and Noller,H.F. (1999) X-ray crystal structure of 70S ribosome functional complexes. Science, 285, 2095–2104. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Colovos C. and Yeates,T.O. (1993) Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci., 2, 1511–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czworkowski J., Wang,J., Steitz,T.A. and Moore,P.B. (1994) The crystal structure of elongation factor G complexed with GDP, at 2.7 Å resolution. EMBO J., 13, 3661–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortelle E.D.L. and Bricogne,G. (1997) Maximum likelihood heavy atom refinement for multiple isomorphous replacement and multianomalous diffraction methods. Methods Enzymol., 276, 472–494. [DOI] [PubMed] [Google Scholar]
- Freistroffer D.V., Pavlov,M.Y., MacDougall,J., Buckingham,R.H. and Ehrenberg,M. (1997) Release factor RF3 in E.coli accelerates the dissociation of release factors RF1 and RF2 from the ribosome in a GTP-dependent manner. EMBO J., 16, 4126–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grentzmann G., Kelly,P.J., Laalami,S., Shuda,M., Firpo,M.A., Cenatiempo,Y. and Kaji,A. (1998) Release factor RF-3 GTPase activity acts in disassembly of the ribosome termination complex. RNA, 4, 973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 133, 123–138. [DOI] [PubMed] [Google Scholar]
- Janosi L., Shimizu,I. and Kaji,A. (1994) Ribosome recycling factor (ribosome releasing factor) is essential for bacterial growth. Proc. Natl Acad. Sci. USA, 91, 4249–4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janosi L., Ricker,R. and Kaji,A. (1996) Dual functions of ribosome recycling factor in protein biosynthesis: disassembling the termination complex and preventing translational errors. Biochimie, 78, 959–969. [DOI] [PubMed] [Google Scholar]
- Janosi L. et al. (1998) Evidence for in vivo ribosome recycling, the fourth step in protein biosynthesis. EMBO J., 17, 1141–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.-Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kaji A., Teyssier,E. and Hirokawa,G. (1998) Disassembly of the post-termination complex and reduction of translational error by ribosome recycling factor (RRF)—a possible new target for antibacterial agents. Biochem. Biophys. Res. Commun., 250, 1–4. [DOI] [PubMed] [Google Scholar]
- Kanai T., Takeshita,S., Atomi,H., Umemura,K., Ueda,M. and Tanaka,A. (1998) A regulatory factor, Fil1p, involved in derepression of the isocitrate lyase gene in Saccharomyces cerevisiae—a possible mitochondrial protein necessary for protein synthesis in mitochondria. Eur. J. Biochem., 256, 212–220. [DOI] [PubMed] [Google Scholar]
- Karimi R., Pavlov,M.Y., Buckingham,R.H. and Ehrenberg,M. (1999) Novel roles for classical factors at the interface between translation termination and initiation. Mol. Cell, 3, 601–609. [DOI] [PubMed] [Google Scholar]
- Kim K.K., Song,H.K., Shin,D.H., Hwang,K.Y. and Suh,S.W. (1997) The crystal structure of a triacylglycerol lipase from Pseudomonas cepacia reveals a highly open conformation in the absence of a bound inhibitor. Structure, 5, 173–185. [DOI] [PubMed] [Google Scholar]
- Kissinger C.R., Gehlhaar,D.K. and Fogel,D.B. (1999) Rapid automated molecular replacement by evolutionary search. Acta Crystallogr. D, 55, 484–491. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Laskowski R.A., Macarthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Liljas A. and Garber,M. (1995) Ribosomal proteins and elongation factor. Curr. Opin. Struct. Biol., 5, 721–727. [DOI] [PubMed] [Google Scholar]
- Lupas A. (1996) Coiled coils: new structures and new functions. Trends Biochem. Sci., 21, 375–382. [PubMed] [Google Scholar]
- Martemyanov K.A. and Gudkov,A.T. (1999) Domain IV of elongation factor G from Thermus thermophilus is strictly required for translocation. FEBS Lett., 452, 155–159. [DOI] [PubMed] [Google Scholar]
- Moazed D., Samaha,R.R., Gualerzi,C. and Noller,H.F. (1995) Specific protection of 16S rRNA by translational initiation factors. J. Mol. Biol., 248, 207–210. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Ito,K. and Isaksson,L.A. (1996) Emerging understanding of translation termination. Cell, 87, 147–150. [DOI] [PubMed] [Google Scholar]
- Nevskaya N., Tishchenko,S., Nikulin,A., al-Karadaghi,S., Liljas,A., Ehresmann,B., Ehresmann,C., Garber,M. and Nikonov,S. (1998) Crystal structure of ribosomal protein S8 from Thermus thermophilus reveals a high degree of structural conservation of a specific RNA binding site. J. Mol. Biol., 279, 233–244. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Nissen P., Kjeldgaard,M., Thirup,S., Polekhina,G., Reshetnikova,L., Clark,B.F. and Nyborg,J. (1995) Crystal structure of the ternary complex of Phe-tRNA, EF-Tu, and a GTP analog. Science, 270, 1464–1472. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Pavlov M.Y., Freistroffer,D.V., MacDougall,J., Buckingham,R.H. and Ehrenberg,M. (1997a) Fast recycling of Escherichia coli ribosomes requires both ribosome recycling factor (RRF) and release factor RF3. EMBO J., 16, 4134–4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov M.Y., Freistroffer,D.V., Heurgue-Hamard,V., Buckingham,R.H. and Ehrenberg,M. (1997b) Release factor RF3 abolishes competition between release factor RF1 and ribosome recycling factor (RRF) for a ribosome binding site. J. Mol. Biol., 273, 389–401. [DOI] [PubMed] [Google Scholar]
- Rodnina M.V., Savelsbergh,A., Katunin,V.I. and Wintermeyer,W. (1997) Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature, 385, 37–41. [DOI] [PubMed] [Google Scholar]
- Rolland N. et al. (1999) Plant ribosome recycling factor homologue is a chloroplastic protein and is bactericidal in Escherichia coli carrying temperature-sensitive ribosome recycling factor. Proc. Natl Acad. Sci. USA, 96, 5464–5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoji M., Berland,R. and Kaji,A. (1981) Reinitiation of translation from the triplet next to the amber termination codon in the absence of ribosome-releasing factor. Proc. Natl Acad. Sci. USA, 78, 5973–5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette M., van Tilborg,P., Spurio,R., Kaptein,R., Paci,M., Gualerzi,C.O. and Boelens,R. (1997) The structure of the translational initiation factor IF1 from E.coli contains an oligomer-binding motif. EMBO J., 16, 1436–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark H., Orlova,E.V., Rinke-Appel,J., Junke,N., Mueller,F., Rodnina,M., Wintermeyer,W., Brimacombe,R. and van Heel,M. (1997a) Arrangement of tRNAs in pre- and posttranslocational ribosomes revealed by electron cryomicroscopy. Cell, 88, 19–28. [DOI] [PubMed] [Google Scholar]
- Stark H., Rodnina,M., Rinke-Appel,J., Brimacombe,R., Wintermeyer,W. and van Heel,M. (1997b) Visualization of elongation factor Tu on the Escherichia coli ribosome. Nature, 389, 403–406. [DOI] [PubMed] [Google Scholar]
- Suddath F.L., Quigley,G.J., McPherson,A., Sneden,D., Kim,J.J., Kim,S.H. and Rich,A. (1974) Three-dimensional structure of yeast phenylalanine transfer RNA at 3.0 angstroms resolution. Nature, 248, 20–24. [DOI] [PubMed] [Google Scholar]
- van Duyne G.D., Ghosh,G., Maas,W.K. and Sigler,P.B. (1996) Structure of the oligomerization and l-arginine binding domain of the arginine repressor of Escherichia coli. J. Mol. Biol., 256, 377–391. [DOI] [PubMed] [Google Scholar]
- Weissenhorn W., Carfi,A., Lee,K.H., Skehel,J.J. and Wiley,D.C. (1998) Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol. Cell, 2, 605–616. [DOI] [PubMed] [Google Scholar]
- Wilson K.S. and Noller,H.F. (1998) Mapping the position of translational elongation factor EF-G in the ribosome by direct hydroxyl radical probing. Cell, 92, 131–139. [DOI] [PubMed] [Google Scholar]
- Yun J., Kim,W., Ha,S.C., Eom,S.-H., Suh,S.W. and Kim,K.K. (2000) Crystallization and preliminary crystallographic studies of ribosome recycling factor from Escherichia coli. Acta Crystallogr. D, 56, 84–85. [DOI] [PubMed] [Google Scholar]