Abstract
Apolipoprotein E (apoE), first described in 1973, is a truly fascinating protein. While studies initially focused on its role in cholesterol and lipid metabolism, one apoE isoform (apoE4) is a major risk factor for development of late onset Alzheimer's disease. Yet the difference between apoE3, the common form, and apoE4 is a single amino acid of the 299 in this 34 kDa protein. Structure determination of the two domain full length apoE3 protein was only accomplished in 2011 and supports the notion that mutations in the N-terminal domain can be propagated through the structure to the C-terminal domain. Understanding the structural differences between apoE3 and apoE4 is critical for finding ways to modulate the deleterious effect of apoE4.
Keywords: Alzheimer's disease, apoE isoforms, structural changes, salt bridges, domain interactions
Introduction
Human plasma contains a variety of proteins associated with lipoprotein particles. In the 1970s, Shore and Shore investigated the heterogeneity of proteins associated with very low density lipoproteins (VLDLs) and showed that there were mixtures of proteins associated with these particles.1 One such protein family was classified as the apolipoprotein E (apoE). Using isoelectric focusing, Utermann et al. showed that the apoE proteins consisted primarily of three isoforms2 which were later termed apoE2, apoE3, and apoE4 by Zannis et al.3
In 1981, Weisgraber et al.4 demonstrated that the three major apoE isoforms, of molecular weight 34 kDa, differed only by simple arginine to cysteine changes: apoE4 does not contain cysteine while the other isoforms did. In 1982, Rall et al.5 found the location of the cysteine in apoE3 (Cys112) and those in apoE2 (Cys112,158). In 1982, Weisgraber et al. showed that the receptor binding function was isoform-specific although apoE3 and apoE4 appeared to be essentially identical.6 The complete sequence of apoE2 (299 amino acids) was determined by Rall et al. in 19825 and that there were two structural domains in apoE3 was suggested by the Weisgraber group in 1988.7 Structure determination of the full-length protein, either by crystallography or NMR, was not possible due to aggregation of the protein to high molecular weight forms as investigated by Perugini et al.8 Even at low nanomolar concentrations, oligomers, dimers, and tetramers form.9 By isolation of fragments of apoE, however, it was established that aggregation occurs through the C-terminal domain since the N-terminal domain remains monomeric even at high concentrations10 while the isolated C-terminal domain aggregates.
The first determination of the structure of the N-terminal domain of apoE3 reported in 1991 as an elongated four-helical bundle.11 Other structures of the N-terminal domain were subsequently determined by NMR for apoE312 and by X-ray crystallography for both apoE313 and apoE4.14 Perhaps the most unusual aspect of this structure is the regions between helices were rather flexible loops rather than tight turns. Not unexpectedly, polar groups in helix 2 (residues 55–79) and regions of helix 4 (residues 140–158) were all solvent exposed. Since it was not possible to determine the structure of the full length protein there were efforts to determine the structure of the C-terminal domain using structural prediction programs. For example, Chou–Fasman predictions15 suggested that the C-terminal domain was highly helical, a prediction supported by circular dichroism data.10
While lipoproteins were considered to be important for cholesterol metabolism, it is now clearly recognized that they have multiple functions related to lipid metabolism.16 The role of apoE in lipid and cholesterol metabolism was recognized early, but it was not until 1993, in a classic paper, that Corder et al. found apoE4 to be the major risk factor in late-onset Alzheimer's disease.17 This observation made the determination of the full length structure, and structural differences between apoE3 and apoE4, critically important.
The Proposed Structure of the Full Length Protein. The Arg61 Mutant
Based on mutational data, specifically an Arg61Thr mutation in apoE4, Weisgraber and coworkers18–20 suggested that Arg61 in helix 2 of the N-terminal domain interacted with the C-terminal domain of apoE4. Hatters et al. discussed the interaction between the N- and C-terminal domains for apoE3 and apoE4 suggesting the importance of a salt bridge between Arg61 and Glu255 in apoE4 that was proposed not to exist in apoE3.21
Dong and Weisgraber showed that mutation of Glu255 alters the preference of apoE4 from VLDL to high density lipoprotein (HDL) similar to what would be expected for apoE3.19 They concluded that the preferential association of apoE4 for VLDL is an intrinsic property of apoE4, likely related to domain interaction, and that domain interaction is probably mediated by a salt bridge between arginine 61 and glutamic acid 255. Their proposed structures of full length apoE3 and apoE4 are shown in Figure 1.
Figure 1.
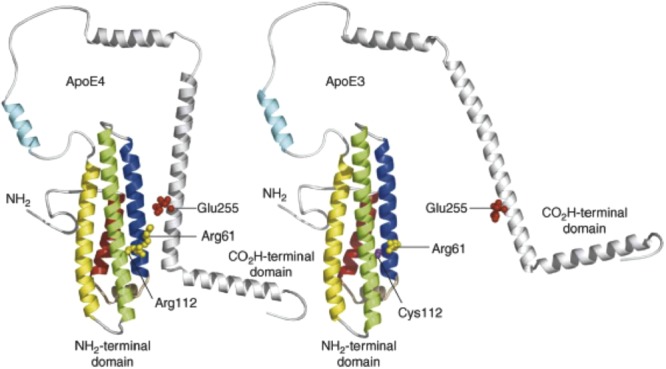
Proposed structures of full length apoE3 and apoE4. Reproduced with permission from Ref.21 from Elsevier Trends Journals. The authors proposed that a salt bridge in the apoE4 structure, Glu255 of the C-terminal domain to Arg61 in helix 2 (blue), occurs in apoE4 but not in apoE3. Helices 1–4 are colored red, blue, green, and yellow, respectively.
The Full Length apoE Structure
In 2004, Fan et al. found that changing 4–5 residues in the isolated C-terminal domain (residues 200–299) yielded material that did not aggregate and showed a well defined helical structure22 as predicted by Chou and Fasman.15 This mutant replaced five bulky hydrophobic residues in the region of residues 253–289 with either smaller hydrophobic or polar/charged residues (F257A, W264R, V269A, L279Q, and V287E). Zhang et al. using the same mutations23 then found that apoE3 existed as a monomer at high enough concentrations for NMR studies. They also concluded that this monomeric form retained many of the biological functions as did wild-type apoE3. This observation opened the door for the determination of the structure of the full length apoE3.
In 2011, Chen et al., published the full length structure of the mutated apoE3.24 This structure, determined by NMR, is shown in Figure 2. One of the surprising results was that Arg61, which had been suggested to interact with Glu255, did not, in fact, interact with that residue. Rather, Glu255 forms a salt bridge with Lys95. This result is critical to the overall structure because the C-terminal domain does not interact with helix 2 but rather with both helices 3 and 4. Surprisingly, the polar groups of helix 4 which were solvent exposed in the isolated N-terminal domain are completely buried by regions of the C-terminal domain in the full length protein. One could, and should, legitimately question whether mutations made to determine the NMR structure affect one or more of these interactions. The chances, however, that the mutant monomeric structure differs markedly from the wild-type structure are minimal since the five mutated residues are probably solvent exposed.
Figure 2.
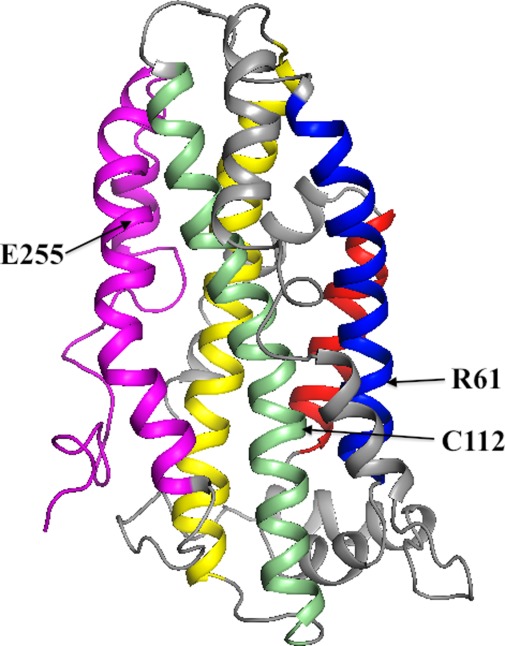
The average NMR structure of apoE3 as determined by Chen et al.24 The helices in the N-terminal domain are shown in the same colors and similar orientation as in Figure 1. The C-terminal domain is colored magenta. To determine this structure, the authors made five mutations in the C-terminal domain to prevent apoE3 aggregation as discussed in the text.
But if arginine 61 is located distant from glutamate 255, how does one explain the results of the mutational data described above? The explanation appears to be similar to that proposed by Frieden and Garai in their discussion of structural differences between apoE3 and apoE4.25 These authors suggested, based on hydrogen-deuterium exchange results26 that the charge difference between cysteine and arginine at position 112 (the difference between apoE3 and apoE4) was propagated through the highly charged helix 4 resulting in structural changes of specific residues in the C-terminal domain. Figure 2, based on the apoE3 structure determined by Chen et al., shows that the side chains of the solvent exposed Arg61and Cys112 are within 8 Å separated by mostly hydrophobic residues. Thus, mutating the Arg61 to a non-charged or oppositely charged amino acid may result in exactly the same change in structural differences noted for apoE3 relative to apoE4. That is, the change in behavior as a consequence of the mutation of Arg61 is not a direct effect on the interaction between the N- and C-terminal domains, but an indirect, allosteric, effect. Figure 3 shows one possible path for how the mutation of Arg61 in helix 2 can be propagated to the region 271–287 of the C-terminal domain. In this particular example, there are no contacts between helix 3 and residues 271–287. There are, however, numerous interactions between helices 3 and 4 and the C-terminal domain of apoE324 suggesting that there will be many structural changes in the C-terminal domain in apoE4 compared to apoE3 as a consequence of the cysteine to arginine change at position 112.
Figure 3.
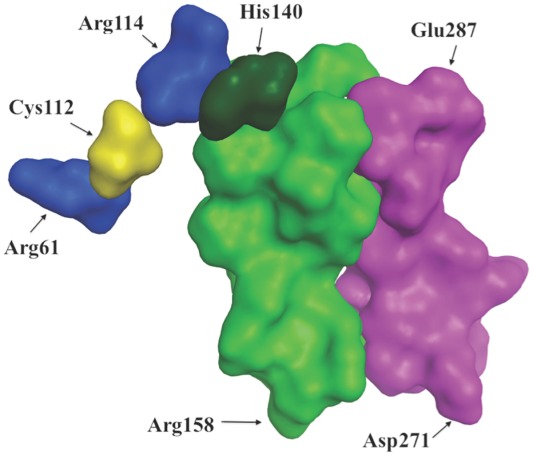
A proposal for those portions of the protein involved in the propagation of structural changes in apoE3 for the Arg61Thr mutant. The proposal details how the Arg61Thr change in the N-terminal domain may be propagated to a region in the C-terminal domain. The proposal is based on the structure of the full length apoE3 as determined by Chen et al.24 Shown are Arg61, Cys112 (yellow), Arg114, His140, a region of helix 4 (residues 140–158) and a region of the C-terminal domain (residues 271–287). Helix 2 (not shown) does not contact this region of the C-terminal domain.
Changing the Behavior of apoE4
ApoE binds to multiple ligands. In order to understand these interactions it is essential to use the full length structure determined by Chen et al.24 As noted above, Frieden and Garai suggested that the arginine/cysteine change at position 112 was propagated through the structure to regions of the C- and N-terminal domains distant from residue 112. They discussed the possibility of targeting these latter regions to alter the behavior of apoE4 relative to apoE3. Another possibility, based on the above discussion, exists: to target the region near arginines 61 and 112 in apoE4. The region around position 112 might be considered as an obvious target for compounds to change the behavior of apoE4. It is, but not for the obvious reason. The rationale for this approach is based on the same argument used by Frieden and Garai to explain how distant regions of the protein were affected by the arginine to cysteine change at position 112 via the highly charged, but buried, helix 4 as shown in Figure 3. Targeting this region would be easier than targeting regions of the C-terminal domain because there is a known structure of the N-terminal domain of apoE4 while no full length structure of apoE4 currently exists. While there may be structural differences in the C-terminal domain between apoE3 and apoE4, as suggested by the hydrogen/deuterium exchange data,26 the expectation is that the domain interactions are similar in both apoE4 and apoE3. Structural differences in the C-terminal domain between apoE3 and apoE4 may influence uptake of Aβ via cell surface receptors such as LRP1.27
Since the arginine residue of apoE4 is positively charged compared to the cysteine residue of apoE3, compounds that might be useful in changing the behavior of apoE4 might be those that specifically alter the positive charge of arginine at position 112. Thus, such compounds need to bind specifically to the region around 112 and carry a negative charge.
What Behavior Should be Changed?
In vitro: It was recognized as early as 1993 that apoE interacts with Aβ.28,29 The mechanism of binding, the form of Aβ that binds, and the regions of apoE that bind remain to be definitively clarified. In vitro experiments using recombinant apoE show that apoE interacts with Aβ oligomers30 but these binding experiments have been carried out under widely varying conditions by different investigators and the mechanism of the interaction is unclear. Many investigators have found isoform dependent differences in Aβ aggregation and this could be a valid test for determining whether the behavior of apoE4 resembles that of apoE3.
In vivo: Considerable evidence has accumulated suggesting apoE affects Aβ clearance31–33 from cells and that apoE isoforms differentially affect Aβ clearance with clearance by apoE4 being slower than that by apoE3 or apoE2.32,34 For example, LaDu and coworkers find that Aβ accumulation is apoE isoform dependent in a transgenic mouse model.35,36 The ultimate test for whether a potential therapeutic agent alters the effect of apoE4 may be measuring the clearance of Aβ from cells. Chen et al.37 have found several small molecule effectors (called correctors) that abolish detrimental effects of apoE4 in cultured neurons. Alternatively, immunotherapy targeting apoE4 specifically may be an effective therapeutic approach. Kim et al. have shown that intraperitoneal administration of a specific monoclonal apoE antibody HJ6.3 dramatically decreased the amyloid plaque load in mouse brain possibly by promoting clearance of Aβ associated with the plaques.38 Mahley et al. suggest other possible differences between apoE isoforms in that apoE4 is less effective at redistributing lipids among cells in the central nervous system (CNS)39,40 or in mitochondrial function.41
Comments on apoE2
The difference between apoE3 and apoE2 is yet again a single amino change (Arg158Cys). While not discussed here, similar arguments can be made for this change as for the change at position 112. For example, polar residues distant in the sequence but especially close to the buried residue at position 158 include Lys92, Glu96, and Arg260, the former two in helix 2 and the latter in the C-terminal domain.
Conclusion
The recent determination of the full length structure of apoE3 has not yet revealed why apoE4, which differs by a single amino acid from apoE3, is the major risk factor for late onset Alzheimer's disease. The structure does show, however, that apoE is a surprisingly complex protein in which the consequence of the single amino changes between the apoE isoforms can be propagated throughout the structure. Small molecular weight compounds that bind to specific regions of the protein may exert effects on the structure that are distant from the binding site of the compound. Such compounds could alter the behavior of the apoE4, the major risk factor for late onset Alzheimer's disease.
Acknowledgments
Authors thank Dr. Scott Wildman for Figure 3.
References
- 1.Shore VG, Shore B. Heterogeneity of human plasma very low density lipoproteins. Separation of species differing in protein components. Biochemistry. 1973;12:502–507. doi: 10.1021/bi00727a022. [DOI] [PubMed] [Google Scholar]
- 2.Utermann G, Hees M, Steinmetz A. Polymorphism of apolipoprotein E and occurrence of dysbetalipoproteinaemia in man. Nature. 1977;269:604–607. doi: 10.1038/269604a0. [DOI] [PubMed] [Google Scholar]
- 3.Zannis, Breslow JL, Utermann G, Mahley RW, Weisgraber KH, Havel RJ, Goldstein JL, Brown MS, Schonfeld G, Hazzard WR, Blum C. Proposed nomenclature of apoE isoproteins, apoE genotypes, and phenotypes. J Lipid Res. 1982;23:911–914. [PubMed] [Google Scholar]
- 4.Weisgraber KH, Rall SC, Jr, Mahley RW. Human E apoprotein heterogeneity. Cysteine-arginine interchanges in the amino acid sequence of the apo-E isoforms. J Biol Chem. 1981;256:9077–9083. [PubMed] [Google Scholar]
- 5.Rall SC, Jr, Weisgraber KH, Mahley RW. Human apolipoprotein E. The complete amino acid sequence. J Biol Chem. 1982;257:4171–4178. [PubMed] [Google Scholar]
- 6.Weisgraber KH, Innerarity TL, Mahley RW. Abnormal lipoprotein receptor-binding activity of the human E apoprotein due to cysteine-arginine interchange at a single site. J Biol Chem. 1982;257:2518–2521. [PubMed] [Google Scholar]
- 7.Wetterau JR, Aggerbeck LP, Rall SC, Jr, Weisgraber KH. Human apolipoprotein E3 in aqueous solution. I. Evidence for two structural domains. J Biol Chem. 1988;263:6240–6248. [PubMed] [Google Scholar]
- 8.Perugini MA, Schuck P, Howlett GJ. Self-association of human apolipoprotein E3 and E4 in the presence and absence of phospholipid. J Biol Chem. 2000;275:36758–36765. doi: 10.1074/jbc.M005565200. [DOI] [PubMed] [Google Scholar]
- 9.Garai K, Frieden C. The association–dissociation behavior of the ApoE proteins: kinetic and equilibrium studies. Biochemistry. 2010;49:9533–9541. doi: 10.1021/bi101407m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aggerbeck LP, Wetterau JR, Weisgraber KH, Wu CS, Lindgren FT. Human apolipoprotein E3 in aqueous solution. II. Properties of the amino- and carboxyl-terminal domains. J Biol Chem. 1988;263:6249–6258. [PubMed] [Google Scholar]
- 11.Wilson C, Wardell MR, Weisgraber KH, Mahley RW, Agard DA. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science. 1991;252:1817–1822. doi: 10.1126/science.2063194. [DOI] [PubMed] [Google Scholar]
- 12.Sivashanmugam A, Wang J. A unified scheme for initiation and conformational adaptation of human apolipoprotein E N-terminal domain upon lipoprotein-binding and for receptor-binding activity. J Biol Chem. 2009;284:14657–14666. doi: 10.1074/jbc.M901012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong LM, Parkin S, Trakhanov SD, Rupp B, Simmons T, Arnold KS, Newhouse YM, Innerarity TL, Weisgraber KH. Novel mechanism for defective receptor binding of apolipoprotein E2 in type III hyperlipoproteinemia. Nat Struct Biol. 1996;3:718–722. doi: 10.1038/nsb0896-718. [DOI] [PubMed] [Google Scholar]
- 14.Dong J, Peters-Libeu CA, Weisgraber KH, Segelke BW, Rupp B, Capila I, Hernaiz MJ, LeBrun LA, Linhardt RJ. Interaction of the N-terminal domain of apolipoprotein E4 with heparin. Biochemistry. 2001;40:2826–2834. doi: 10.1021/bi002417n. [DOI] [PubMed] [Google Scholar]
- 15.Chou PY, Fasman GD. Prediction of protein conformation. Biochemistry. 1974;13:222–245. doi: 10.1021/bi00699a002. [DOI] [PubMed] [Google Scholar]
- 16.Mahley RW. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- 17.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 18.Dong LM, Wilson C, Wardell MR, Simmons T, Mahley RW, Weisgraber KH, Agard DA. Human apolipoprotein E. Role of arginine 61 in mediating the lipoprotein preferences of the E3 and E4 isoforms. J Biol Chem. 1994;269:22358–22365. [PubMed] [Google Scholar]
- 19.Dong LM, Weisgraber KH. Human apolipoprotein E4 domain interaction. Arginine 61 and glutamic acid 255 interact to direct the preference for very low density lipoproteins. J Biol Chem. 1996;271:19053–19057. doi: 10.1074/jbc.271.32.19053. [DOI] [PubMed] [Google Scholar]
- 20.Hatters DM, Peters-Libeu CA, Weisgraber KH. Engineering conformational destabilization into mouse apolipoprotein E. A model for a unique property of human apolipoprotein E4. J Biol Chem. 2005;280:26477–26482. doi: 10.1074/jbc.M503910200. [DOI] [PubMed] [Google Scholar]
- 21.Hatters DM, Peters-Libeu CA, Weisgraber KH. Apolipoprotein E structure: insights into function. Trends Biochem Sci. 2006;31:445–454. doi: 10.1016/j.tibs.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 22.Fan D, Li Q, Korando L, Jerome WG, Wang J. A monomeric human apolipoprotein E carboxyl-terminal domain. Biochemistry. 2004;43:5055–5064. doi: 10.1021/bi035958w. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Vasudevan S, Sojitrawala R, Zhao W, Cui C, Xu C, Fan D, Newhouse Y, Balestra R, Jerome WG, Weisgraber K, Li Q, Wang J. A monomeric, biologically active, full-length human apolipoprotein E. Biochemistry. 2007;46:10722–10732. doi: 10.1021/bi700672v. [DOI] [PubMed] [Google Scholar]
- 24.Chen J, Li Q, Wang J. Topology of human apolipoprotein E3 uniquely regulates its diverse biological functions. Proc Natl Acad Sci USA. 2011;108:14813–14818. doi: 10.1073/pnas.1106420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frieden C, Garai K. Structural differences between apoE3 and apoE4 may be useful in developing therapeutic agents for Alzheimer's disease. Proc Natl Acad Sci USA. 2012;109:8913–8918. doi: 10.1073/pnas.1207022109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang RYC, Garai K, Frieden C, Gross ML. Hydrogen/deuterium exchange and electron-transfer dissociation mass spectrometry determine the interface and dynamics of apolipoprotein E oligomerization. Biochemistry. 2011;50:9273–9282. doi: 10.1021/bi2010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verghese PB, Castellano JM, Garai K, Wang Y, Jiang H, Shah A, Bu G, Frieden C, Holtzman DM. ApoE influences amyloid-beta (Abeta) clearance despite minimal apoE/Abeta association in physiological conditions. Proc Natl Acad Sci USA. 2013;110:E1807–E1816. doi: 10.1073/pnas.1220484110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci USA. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wisniewski T, Golabek A, Matsubara E, Ghiso J, Frangione B. Apolipoprotein E: binding to soluble Alzheimer's beta-amyloid. Biochem Biophys Res Commun. 1993;192:359–365. doi: 10.1006/bbrc.1993.1423. [DOI] [PubMed] [Google Scholar]
- 30.Cerf E, Gustot A, Goormaghtigh E, Ruysschaert J-M, Raussens V. High ability of apolipoprotein E4 to stabilize amyloid-β peptide oligomers, the pathological entities responsible for Alzheimer's disease. FASEB J. 2011;25:1585–1595. doi: 10.1096/fj.10-175976. [DOI] [PubMed] [Google Scholar]
- 31.Basak JM, Kim J, Pyatkivskyy Y, Wildsmith KR, Jiang H, Parsadanian M, Patterson BW, Bateman RJ, Holtzman DM. Measurement of apolipoprotein E and amyloid beta clearance rates in the mouse brain using bolus stable isotope labeling. Mol Neurodegen. 2012;7:14. doi: 10.1186/1750-1326-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castellano JM, Kim J, Stewart FR, Jiang H, DeMattos RB, Patterson BW, Fagan AM, Morris JC, Mawuenyega KG, Cruchaga C, Goate AM, Bales KR, Paul SM, Bateman RJ, Holtzman DM. Human apoE isoforms differentially regulate brain amyloid-beta peptide clearance. Sci Transl Med. 2011;3:89ra57. doi: 10.1126/scitranslmed.3002156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wildsmith KR, Holley M, Savage JC, Skerrett R, Landreth GE. Evidence for impaired amyloid beta clearance in Alzheimer's disease. Alzheimers Res Ther. 2013;5:33. doi: 10.1186/alzrt187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holtzman DM, Herz J, Bu G. Apolipoprotein e and apolipoprotein e receptors: normal biology and roles in Alzheimer disease. Cold Spring Harb Perspect Med. 2012;2:a006312. doi: 10.1101/cshperspect.a006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Youmans KL, Tai LM, Nwabuisi-Heath E, Jungbauer L, Kanekiyo T, Gan M, Kim J, Eimer WA, Estus S, Rebeck GW, Weeber EJ, Bu G, Yu C, LaDu MJ. APOE4-specific changes in Aβ accumulation in a new transgenic mouse model of Alzheimer disease. J Biol Chem. 2012;287:41774–41786. doi: 10.1074/jbc.M112.407957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tai LM, Bilousova T, Jungbauer L, Roeske SK, Youmans KL, Yu C, Poon WW, Cornwell LB, Miller CA, Vinters HV, Van Eldik LJ, Fardo DW, Estus S, Bu G, Gylys KH, Ladu MJ. Levels of soluble apolipoprotein E/amyloid-beta (Abeta) complex are reduced and oligomeric abeta increased with APOE4 and Alzheimer disease in a transgenic mouse model and human samples. J Biol Chem. 2013;288:5914–5926. doi: 10.1074/jbc.M112.442103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen HK, Liu Z, Meyer-Franke A, Brodbeck J, Miranda RD, McGuire JG, Pleiss MA, Ji ZS, Balestra ME, Walker DW, Xu Q, Jeong DE, Budamagunta MS, Voss JC, Freedman SB, Weisgraber KH, Huang Y, Mahley RW. Small molecule structure correctors abolish detrimental effects of apolipoprotein E4 in cultured neurons. J Biol Chem. 2012;287:5253–5266. doi: 10.1074/jbc.M111.276162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim J, Eltorai AE, Jiang H, Liao F, Verghese PB, Stewart FR, Basak JM, Holtzman DM. Anti-apoE immunotherapy inhibits amyloid accumulation in a transgenic mouse model of Abeta amyloidosis. J Exp Med. 2012;209:2149–2156. doi: 10.1084/jem.20121274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahley RW, Weisgraber KH, Huang Y. Apolipoprotein E4: a causative factor and therapeutic target in neuropathology, including Alzheimer's disease. Proc Natl Acad Sci USA. 2006;103:5644–5651. doi: 10.1073/pnas.0600549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahley RW, Huang Y. Alzheimer disease: multiple causes, multiple effects of apolipoprotein E4, and multiple therapeutic approaches. Ann Neurol. 2009;65:623–625. doi: 10.1002/ana.21736. [DOI] [PubMed] [Google Scholar]
- 41.Chang S, ran Ma T, Miranda RD, Balestra ME, Mahley RW, Huang Y. Lipid- and receptor-binding regions of apolipoprotein E4 fragments act in concert to cause mitochondrial dysfunction and neurotoxicity. Proc Natl Acad Sci USA. 2005;102:18694–18699. doi: 10.1073/pnas.0508254102. [DOI] [PMC free article] [PubMed] [Google Scholar]


