Abstract
Signaling by transforming growth factor–β (TGF-β) is critical for various developmental processes and culminates in the activation of the transcription factors Smad2 and Smad3. MAN1, an integral protein of the inner nuclear membrane, inhibits TGF-β signalling by binding to Smad2 and Smad3. Depletion of the gene LEMD3 encoding MAN1 leads to developmental anomalies in mice, and heterozygous loss-of-function mutations in LEMD3 in humans cause sclerosing bone dysplasia. We modeled the three-dimensional structure of the MAN1-Smad2 complex from nuclear magnetic resonance and small angle x-ray scattering data. As predicted by this model, we found that MAN1 competed in vitro and in cells with the transcription factor FAST1 (forkhead activin signal transducer 1) for binding to Smad2. The model further predicted that MAN1 bound to activated Smad2-Smad4 or Smad3-Smad4 complexes, which was confirmed by in vitro experiments; however, in cells, MAN1 bound only to Smad2 and Smad3, and not to the Smad4-containing complexes. Overexpression of MAN1 led to dephosphorylation of Smad2 and Smad3, thus hindering their recognition by Smad4, and MAN1 bound directly in vitro to the phosphatase PPM1A, which catalyzes the dephosphorylation of Smad2/3. These results demonstrate a nuclear envelope-localized mechanism of inactivating TGF-β signaling in which MAN1 competes with transcription factors for binding to Smad2 and Smad3 and facilitates their dephosphorylation by PPM1A.
INTRODUCTION
The nuclear envelope functions as a signaling node in development and disease (1). Alterations in the structure and composition of the nuclear envelope occur during metazoan development and tissue differentiation and mutations in genes encoding proteins of the nuclear envelope cause various diseases that are mostly tissue-selective (2). Several inner nuclear membrane proteins regulate critical signaling pathways and can be viewed as intranuclear regulators of signaling pathways that receive and transduce signals from the extracellular matrix and cytoskeleton, participating in cellular responses to developmental cues or stresses (3, 4).
An example of a nuclear envelope protein that regulates a key cell signaling pathway is MAN1, an antigen that is recognized by auto-antibodies found in the human serum designated MAN of a patient with an ill-defined collagen vascular disease (5, 6). MAN1 is an integral protein of the inner nuclear membrane with two transmembrane segments oriented such that its N- and C-terminal regions are nucleoplasmic and its central region is located between the inner and outer nuclear membranes (5, 6). The first nucleoplasmic region contains a LEM (LAP2, emerin, MAN1) motif, which is shared by other integral proteins of the inner nuclear membrane (5, 7, 8). The second nucleoplasmic region directly binds to Smad2 and Smad3 (9–14). Receptors activated by cytokines of the transforming growth factor–β (TGF-β) superfamily interact with and catalyze the phosphorylation of receptor-regulated Smads (R-Smads), such as Smad1, Smad2, Smad3, Smad5 and Smad8 (15). Upon phosphorylation, two activated R-Smads form a complex with Smad4, which translocates to the nucleus where it interacts with various transcription factors, co-activators, or co-repressors to modulate gene expression (15). TGF-β signaling is inhibited by interaction of R-Smads with cytoplasmic or nuclear proteins including the inhibitor Smads (I-Smads) Smad6 and Smad7 (15–17). Similarly, interactions between MAN1 and Smad1 or Smad2 and Smad3 inhibit bone morphogenic protein (BMP) and TGF-β signaling, respectively (9–13).
Data from model organisms and human genetics studies support the function of MAN1 in inhibiting TGF-β signaling. MAN1-deficient mice die during embryogenesis because of abnormal vascularization of the yolk sac (18, 19) and they exhibit abnormal heart morphogenesis resulting from aberrant expression of left-side–specific genes encoding proteins responsible for left-right asymmetry (20), defects that are linked to enhanced TGF-β signaling. Similarly, tissue-specific defects resulting from deletion of the MAN1 homolog in Drosophila (21) and abnormalities caused by overexpression or knockdown of the Xenopus MAN1 homolog in embryos (9, 10) also appear to result from alterations in signaling by TGF-β family members. Heterozygous loss-of-function mutations in LEMD3, the gene encoding human MAN1, cause the sclerosing bone dysplasia osteopoikilosis, which is sometimes associated with connective tissue nevi in the skin (Buschke-Ollendorff syndrome), phenotypes that can be explained by enhanced TGF-β signaling in these tissues (11).
Although MAN1 directly interacts with the MH2 (MAD homology 2) domains of Smad2 and Smad3 (12, 14), the biophysical mechanism by which this inhibits TGF-β signaling is not clear. MAN1 could act as a “sink” for activated nuclear Smads, sequestering them at the inner nuclear membrane and competitively inhibiting their interactions with other nuclear factors involved in gene regulation. Another possibility is that MAN1 functions as a scaffold protein that recruits Smads and enzymes, catalyzing their dephosphorylation at the inner nuclear membrane. Although no supporting data exist, a function in recruiting an activated nuclear phosphoprotein and its deactivating phosphatase has been hypothesized for MAN1 (13) as well as for a paralog, NET25, which inhibit extracellular signal-regulated kinases (ERKs) (22). NET25 coimmunoprecipitates with the phosphatase PP1 in HeLa cells (23). Finally, it is possible that MAN1 competitively inhibits other proteins that bind to Smad2 and Smad3. To test these hypotheses, we used small-angle x-ray scattering (SAXS), isothermal calorimetry (ITC) and protein binding experiments to model the interaction of MAN1 with Smad2. We showed that the C-terminal region of MAN1 competed with the transcription factor forkhead activin signal transducer 1 (FAST1) for binding to Smad2, leading to dissociation of the FAST1-Smad2 complex. In addition, this region of MAN1 bound to the MH2 domain of Smad2/3 and phosphatase PPM1A, leading to enhanced dephosphorylation of the Smads.
RESULTS
Tyr366 and Trp368 of Smad2 are involved in binding to MAN1
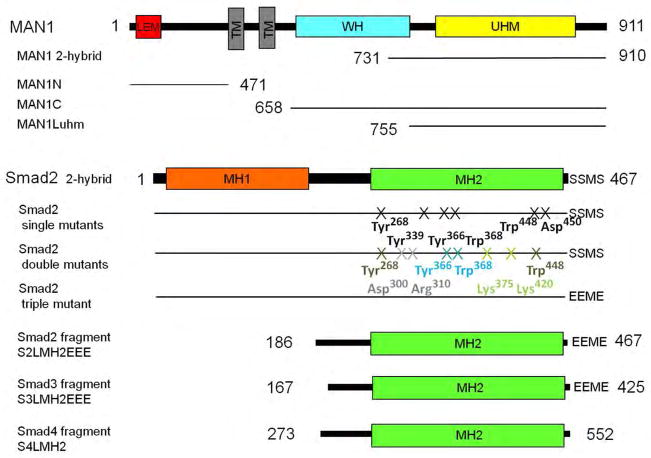
The C-terminal, nucleoplasmic region of MAN1 contains a winged helix (WH) domain, a linker region, a U2AF homology motif (UHM) and a short C-terminus (14). We previously showed that the linker region, UHM domain and C-terminus are involved in binding to Smad2 (14). Smad2 recognizes most of its protein-binding partners, including MAN1, through its MH2 domain (12, 14). To further define the Smad2 region involved in MAN1 binding, we generated cDNA constructs that encoded Smad2 with various amino acid substitutions, and we examined the binding of these proteins to MAN1.
We tested 10 Smad2 residues, which are involved in homotrimerization (Asp300, Arg310, Lys375, Lys420 and Asp450) (24), binding to the plasma-membrane Smad anchor for receptor activation (SARA) (Tyr268, Tyr339, Tyr366, Trp368 and Trp448) (25), binding to Smad4 after activation (Asp450) (24, 26) and binding to transcription factors in the nucleus (Tyr366 and Trp368) (26, 27). We introduced six single amino acid substitutions in Smad2 (Fig. 1): Y268A and Y339A (which abrogates binding to SARA), Y366A and W368A (which abrogates binding to SARA and transcription factors), W448A (which abrogates SARA-binding), and D450H (which abrogates binding to Smad4). Furthermore, we constructed 4 double mutants: D300H-R310H and K375A-K420A (which are unable to form homotrimers) (24), Y366A-W368A (which is unable to bind to the Xenopus transcription factors Mixer and FAST1 and corresponds to a patch on Smad2 involved in SARA recognition) (25–27) and Y268A-W448A (which corresponds to another patch on Smad2 that is involved in SARA recognition) (25). We also constructed a phosphomimetic triple mutant in which the C-terminal sequence SSMS, which is phosphorylated upon activation, was replaced by EEME (Fig. 1). Introduction of these three amino acid substitutions into a construct of the linker and MH2 domain of Smad2, which we called S2LMH2EEE (Fig. 1), stabilizes the trimeric form of Smad3, as observed previously by gel filtration analysis (28) and that of Smad2 in our purification conditions (14). Thus, the phosphomimetic form of the full-length Smad2 protein is predicted to behave as the trimeric Smad2 protein.
Fig. 1.
Schematic representation of the MAN1, Smad2, Smad3 and Smad4 constructs used in this study. MAN1 has an N-terminal LEM motif, two transmembrane (TM) segments, a WH domain, and a UHM domain. Smad2 protein contains an MH1 and MH2 domains.
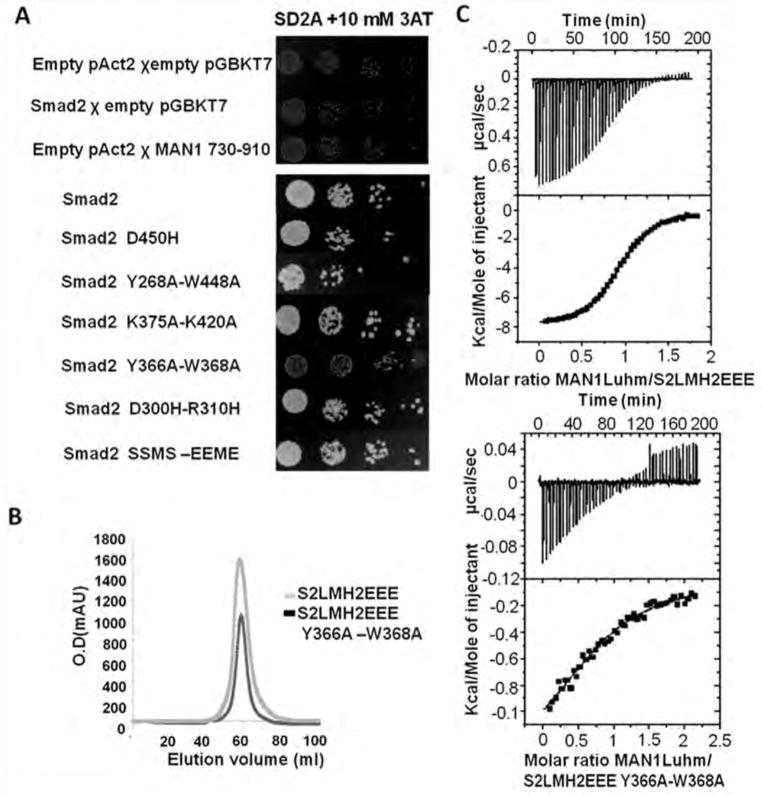
We performed yeast two-hybrid assays to assess the interaction between these Smad2 mutants and a fragment of MAN1 from amino acids 730 to 911, which binds to the MH2 domain (12). Similar to wild-type Smad2, the single mutant D450H, the double mutants D300H-R310H and K375A-K420A and the phosphomimetic triple mutant, which have different oligomerization behaviors, all bound to MAN1 (Fig. 2A). This was consistent with our previous finding that MAN1 recognizes both the monomeric and trimeric MH2 domain of Smad2 (14). Based on atomic resolution data on the Smad2 MH2 domain – SARA peptide complex (25), we then tested whether some of the three Smad2 patches involved in SARA recognition were also involved in MAN1 binding. Like the wild-type Smad2 MH2 domain, the five single mutants Y268A, Y339A, Y366A, W368A and W448A bound to MAN1 (fig. S1). The double mutant Y268A-W448A was also unaffected in its ability to bind MAN1; however, the double mutant Y366A-W368A did not interact with it (Fig. 2A). To verify that the Y366A and W368A mutations did not affect the 3D structure of Smad2, we checked by gel filtration the oligomeric state of the phosphomimetic triple mutant S2LMH2EEE with and without these mutations. Consistent with the solvent exposure of Tyr366 and Trp368 in the trimeric structure of the Smad2 MH2 domain (24), these mutations did not affect the global shape of the MH2 domain and its capacity to oligomerize (Fig. 2B). As measured by ITC, the affinity of the S2LMH2EEE fragment for the MAN1 fragment (amino acids 755 to 911; MAN1Luhm) was approximately 25 fold greater than that of the S2LMH2EEE fragment with the Y366A-W368A mutations (Fig. 2C and Table 1). These results are consistent with the involvement of the MH2 helix α2, which contains Tyr366, and the loop joining α2 and β8, which contains Trp368, in SMAD2 in binding to MAN1.
Fig 2.
Tyr366 and Trp368 in the Smad2 MH2 domain are involved in binding to MAN1. (A) Yeast two-hybrid assay results for binding of MAN1 fragments, which included fragments containing amino acid residues 730 to 910 and full-length Smad2 variants. Top panel, protein-coding or empty plasmids were used to transform yeast for the indicated control experiments. Bottom panel, results for interactions between the MAN1 fragments and the indicated wild-type and mutant Smad2 constructs. Physical interaction between a Smad2 variant and the Man1 fragment activated expression of HIS3, which enabled yeast cell growth. N=2 independent biological replicates. (B) Chromatography elution profiles of the two Smad2 fragments S2LMH2EEE and S2LMH2EEE W366A-Y368A. N=3 independent biological replicates. (C) Representative binding curves obtained by ITC for the MAN1 fragment containing amino acid residues 755 to 911 (MAN1Luhm) added to a phosphomimetic mutant of the MH2 domain of wild-type Smad2 (S2LMH2EEE, left), and its Y366A-W388A mutant (right). Fitting these curves yielded the Kd values in Table 1.
Table 1. Thermodynamic parameters deduced from analysis of ITC data.
Interactions between MAN1Luhm and different Smad fragment complexes were tested. The stoichiometry was always equal to 1. Standard errors were calculated from two independent ITC measurements. In the case of MAN1Luhm binding to a complex between Smad2 and a FAST1 peptide, only an apparent Kd value is provided because the ITC signal results from a competition. The error is evaluated from the deviation between the experimental ITC values and the calculated fitting curve.
| MAN1Luhm against: | Kd (μM) with their fitting errors | ΔH (kCal/Mol) | TΔS (kCal/Mol) |
|---|---|---|---|
| S2LMH2EEE | 1.27±0.04; 1.40±0.03 | −8.4; −8.4 | −0.5; −0.5 |
| S3LMH2EEE | 1.17±0.02; 1.35±0.04 | −8.4; −8.4 | −0.5; −0.5 |
| S2LMH2EEE/S4LMH2 | 1.25±0.04; 1.30±0.03 | −8.4; −8.4 | −0.5; −0.5 |
| S3LMH2EEE/S4LMH2 | 1.50±0.04; 1.37±0.03 | −8.7; −8.8 | −0.9; −0.9 |
| S2LMH2EEE Y366A-W368A | 37.9±6.3; 33.9±8.4 | −1.6; −1.4 | 4.3; 4.5 |
| S2LMH2EEE/SIM | 12.2±0.1; 5.92±0.09 |
The Smad2-MAN1 complex can be modeled from SAXS and mutagenesis data
The MAN1 fragment MAN1Luhm binds to the Smad2 fragment S2LMH2EEE through a surface involving the linker between its WH and UHM domain, the ULM-binding cavity of its UHM domain and its C-terminus (14). In particular, substituting alanines for Trp765 and Gln766 in the linker results in a large decrease in the affinity between MAN1LUHM and S2LMH2EEE as measured by fluorescence (14). Structurally, the MAN1 fragment consists of a globular UHM domain, a flexible N-terminus anchored onto the UHM domain and a disordered C-terminus (14). The Smad2 fragment is composed of a N-terminal segment, which is predicted to be unstructured, and the MH2 globular domain, which has been crystallized (24). We recorded SAXS data on the Smad2 fragment S2LMH2EEE alone and in complex with the MAN1 fragment MAN1Luhm. SAXS analysis performed on the free Smad2 fragment was consistent with the presence of a trimeric oligomer. The same analysis performed on the Smad2-MAN1 complex was consistent with the presence of a trimeric Smad2 oligomer bound to three MAN1 fragments. We calculated 6,000 models of the trimeric Smad2 fragment and 12,000 models of this same oligomer bound to three MAN1 fragments. In these models, the N-terminal part of the Smad2 fragment was in a random conformation, the MH2 trimer structure corresponded to that solved by X-ray crystallography (24) and the MAN1 fragment structure was that previously calculated from SAXS and NMR data (14). Furthermore, a distance constraint forced Trp765 and Gln766 in MAN1 to be close to Tyr366 and Trp368 in Smad2. Each model was analyzed by calculating a theoretical SAXS curve using CRYSOL (29) and comparing it to the experimental SAXS curve. For both complexes, we identified 20 models that were consistent with the mutagenesis and SAXS data (Fig. 3A and B).
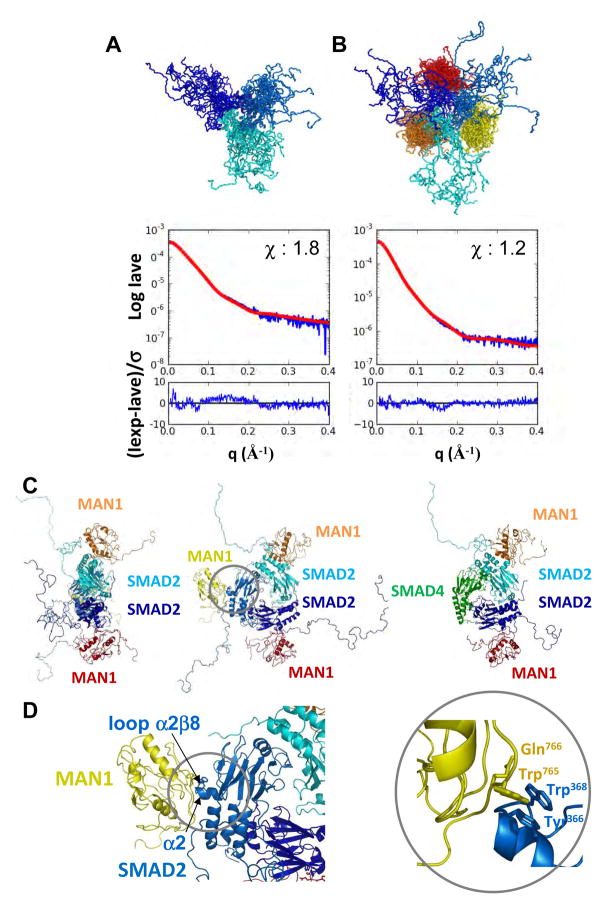
Fig. 3.
Architecture of the MAN1/Smad2 complex. (A to D) Models of the phosphomimetic Smad2 fragment S2LMH2EEE either free (A) or in complex (B) with the MAN1 fragment MAN1Luhm obtained from SAXS data. In each panel, 20 models of the trimeric Smad2 fragment are superimposed (each monomer is colored in a different shade of blue). In (B), a MAN1 fragment is bound to each Smad2 fragment (each MAN1 fragment is colored in yellow, orange and red, respectively). Curves show the corresponding fit between the calculated SAXS intensity averaged on the 20 models (Iave; red) and the experimental SAXS intensity (Iexp; blue). The chi value was 1.8 in (A) and 1.2 in (B), indicating that the deviations between the calculated and experimental intensities are close to the experimental error. The difference between the two intensities divided by the experimental error is also plotted as a function of the diffusion vector amplitude. In both panels, this difference is regularly distributed around 0 on the whole q interval, as expected for a random noise-like signal. (C) shows two orthogonal views of a typical MAN1-Smad2 complex, and an additional view in which one of the Smad2 MH2 monomers has been replaced by a Smad4 MH2 monomer, which suggest that Smad2 can simultaneously bind to MAN1 and Smad4. (D) shows the MAN1-Smad2 interface. Trp765 and Gln766 in MAN1 and Tyr366 and Trp368 in Smad2 are displayed as sticks. Because Tyr366 and Trp368 in Smad2 are crucial for binding to SIM-containing transcription factors, this interface suggests that Smad2 cannot simultaneously recognize MAN1 and these transcription factors.
In a typical 3D model of the MAN1-Smad2 complex consistent with the available heterogeneous experimental data, a MAN1 fragment is anchored onto the protruding helix α2 and loop α2β8 of each Smad2 fragment (Fig. 3C and D). This model predicted that there could be synergistic or competitive interactions with the MAN1-Smad2 complex. In particular, because the 3D structure of two Smad2 MH2 domains complexed to one Smad4 MH2 domain is similar to that of the trimeric Smad2 MH2 domain (PDB 1U7V (30)), we know that the presence of Smad4 in the trimer does not modify the Smad2 solvent-accessible surface. In this case, two MAN1 fragments can be docked onto the protruding α-helix and loop containing Tyr366 and Trp368 of the two Smad2 fragments within the Smad2-Smad4 heterotrimer (Fig 3C, right). This modeling suggests that Smad2 can simultaneously bind to both MAN1 and Smad4. Furthermore, transcription factors containing Smad interaction motifs (SIM) bind to Smad2 through Tyr366 and Trp368 (27, 31), which are buried at the interface in our model of the MAN1-Smad2 complex, suggesting that Smad2 cannot simultaneously bind to MAN1 and these transcription factors.
MAN1 and FAST1 compete for Smad2 binding
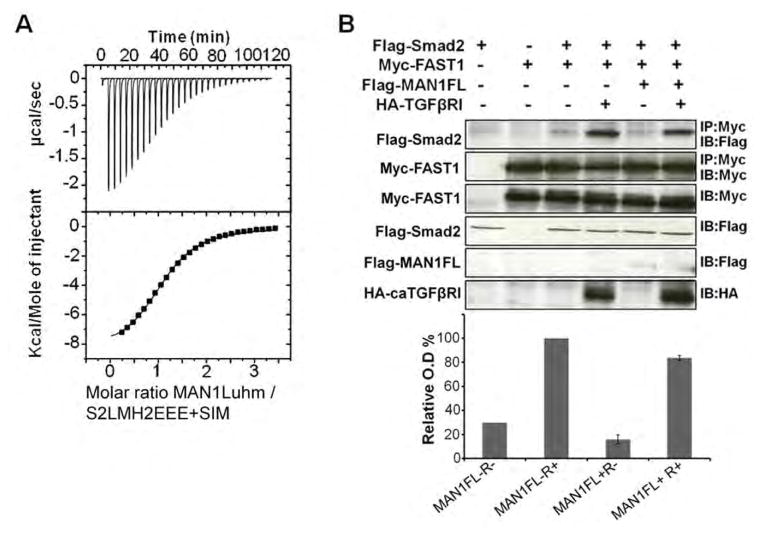
The Xenopus winged-helix forkhead factor FAST1, and other FAST-related proteins in zebrafish, mice, and humans interact with the Smad2 MH2 domain using a SIM sequence that recognizes Tyr366 and Trp368 in Smad2 (27, 31), resulting in transcriptional activation of target genes. Our yeast two-hybrid assay and ITC data demonstrated that similar to FAST-related proteins, MAN1 binds Smad2 through Tyr366 and Trp368. Our MAN1-Smad2 3D model also showed that a MAN1 molecule positioned on Tyr366 and Trp368 in Smad2 was consistent with the SAXS structural data (Fig. 3D). These results suggested competition between FAST1 and MAN1 for binding to Smad2. We tested the in vitro binding of the MAN1 fragment MAN1Luhm to the Smad2 phosphomimetic triple mutant S2LMH2EEE in the presence of a 25 residue peptide from human FAST1 (LDALFQGVPPNKSIYDVWVSHPRDL), which was chosen based on its homology to the Smad interacting motif of Xenopus FAST1 (34) and because the Xenopus peptide competes with full-length FAST1 for Smad2-Smad4 complex recognition (26). ITC analysis confirmed the binding of the FAST1 peptide to the Smad2 MH2 phosphomimetic domain (fig. S2) and indicated that the affinity of MAN1 for this Smad2 region was decreased in presence of the FAST1 peptide (Fig. 4A and Table 1). We then measured the effect of MAN1 overexpression on the formation of FAST1-Smad2 complexes in transfected 293T cells. Regardless of whether constitutively activated hemagglutinin (HA)-tagged TGF-β receptor was coexpressed, the amount of Flag-Smad2 bound to Myc-FAST1 was decreased when MAN1 was overexpressed (Fig. 4B). These results showed that MAN1 competes with FAST1 to bind Smad2 and that in cells MAN1 induces partial dissociation of Smad2 and FAST1.
Fig. 4.
Overexpression of MAN1 leads to partial Smad2-FAST1 complex dissociation. (A) Representative binding curve obtained by ITC when MAN1Luhm was added to S2LMH2EEE bound to the Smad Interacting Motif (SIM) peptide of FAST1. Fitting these curves yielded the Kd values in Table 1. (B) Representative immunoblots showing the amount of FAST1-bound Smad2 in 293T cells transfected with plasmids expressing Flag-Smad2, Myc-FAST1, Flag-MAN1 and HA-tagged constituvely active receptor caTGFβRI as indicated above the blot. Myc immunoprecipitates containing FAST1 (IP: Myc) were immunoblotted with anti-Flag antibodies to detect Smad2 (IB: Flag; top panel). Lysates were immunoblotted as indicated (bottom four panels). The histogram shows the amount of Smad2 bound to FAST1, normalized using the amount of immunoprecipitated Myc-FAST1, from 3 independent experiments, in cells overexpressing neither MAN1 nor caTGFβRI (MAN1FL−R−), only caTGFβRI (MAN1FL−R+), only MAN1 (MAN1FL+R−) and both MAN1 and caTGFβRI (MAN1FL+R+). The signal in the MAN1FL−R+ condition was set at 100%.
MAN1 does not interfere with the in vitro formation of Smad2/Smad4 and Smad3/Smad4 complexes
Smad4 binds to Smad2 and Smad3 and facilitates translocation of the heteromeric complexes into the nucleus (15). Smad2 and Smad3 exhibit highly similar sequences (84% identity and 88% similarity) and 3D architectures (fig. S4 and Fig. 3). X-ray structures of the heterotrimeric complexes between two phosphorylated Smad2 MH2 domains (S2MH2-P) and one Smad4 MH2 domain (S4MH2) or two phosphorylated Smad3 MH2 domains (S3MH2-P) and one S4MH2 indicate that these complexes are structurally organized in a similar manner to the homotrimeric forms of S2MH2-P and S3MH2-P (30). Our previous data (14) and yeast two hybrid experiments (Fig. 2A) indicated that the MAN1–Smad2 interaction is not regulated by the oligomerization state of Smad2, and our 3D modelling suggested that MAN1 binds to both Smad2 homotrimers and Smad2-Smad4 heterotrimers (Fig. 3C). To validate this model, we tested the in vitro binding of MAN1Luhm to activated Smad2 and Smad3 assembled into Smad4-containing heterotrimers.
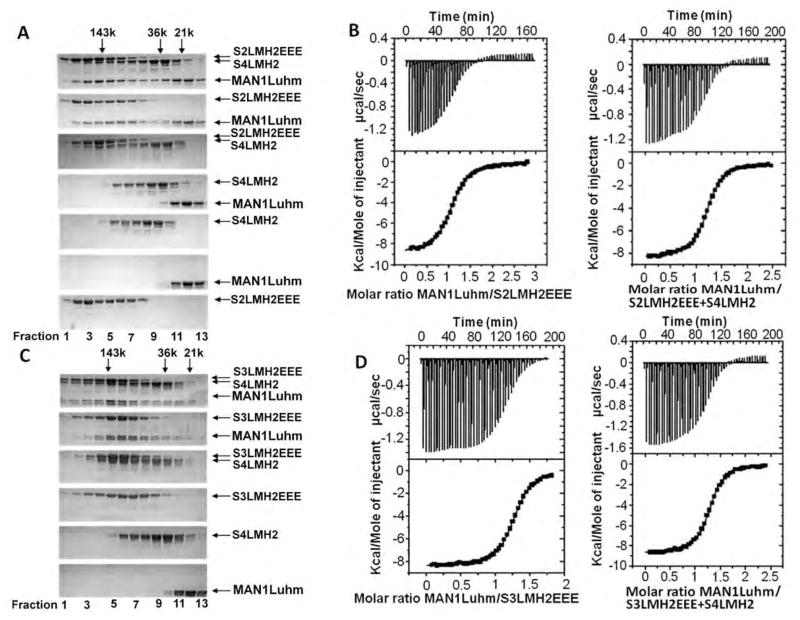
We mixed the phosphomimetic Smad2 fragment S2LMH2EEE and the fragment of Smad4 called S4LMH2, both of which contain the MH2 domain involved in heterotrimerization, in a 1:1 ratio and verified by gel filtration the formation of the S2LMH2EEE-S4LMH2 heterotrimer, the interaction between MAN1Luhm and S2LMH2EEE and the lack of interaction between MAN1Luhm and S4LMH2 (Fig. 5A). MAN1 bound to the Smad2-Smad4 heterotrimer, and ITC analysis indicated that MAN1Luhm had similar micromolar affinities for the Smad2-Smad4 heterotrimer and the Smad2 homotrimer (Fig. 5B and Table 1). Similarly, gel filtration showed that MAN1Luhm, the phosphomimetic Smad3 fragment (S3LMH2EEE) and S4LMH2 co-eluted (Fig. 5C). ITC analysis gave similar Kd values for the interaction between the MAN1 fragment and the Smad3-Smad4 heterotrimer and for the interaction with the Smad3 homotrimer (Fig. 5D and Table 1). Thus, similar to Smad2, MAN1 recognized both the Smad3 homotrimer and the Smad3-Smad4 heterotrimer with micromolar affinities.
Fig. 5.
The MAN1 fragment interacts with phosphomimetic Smad2 or Smad3 fragments complexed to a Smad4 fragment. (A) Size exclusion chromatography analysis of the interaction between MAN1Luhm (amino acids 755 to 911) and/or a phosphomimetic triple mutant of the Smad2 MH2 domain (S2LMH2EEE) and/or the Smad 4 MH2 domain (S4LMH2). N=2 independent biological replicates. (B) Representative binding curves obtained by ITC when MAN1Luhm was added to S2LMH2EEE,alone or in complex with S4LMH2. Fitting these curves yielded the Kd values in Table 1. (C) Size exclusion chromatography analysis of the interaction between MAN1Luhm and/or a phosphomimetic triple mutant of the Smad3 MH2 domain (S3LMH2EEE) and/or the MH2 of Smad4 (S4LMH2). Eluted fractions were separated on a denaturing gel and stained with Coomassie blue. N=2 independent biological replicates. (D) Representative binding curves obtained by ITC when MAN1Luhm at was added to S3LMH2EEE,alone or in complex with S4LMH2. Fitting these curves yielded the Kd values in Table 1.
MAN1 interferes with formation of active R-Smad-Smad4 heterotrimers and decreases phosphorylation of Smad2 and Smad3 in cells
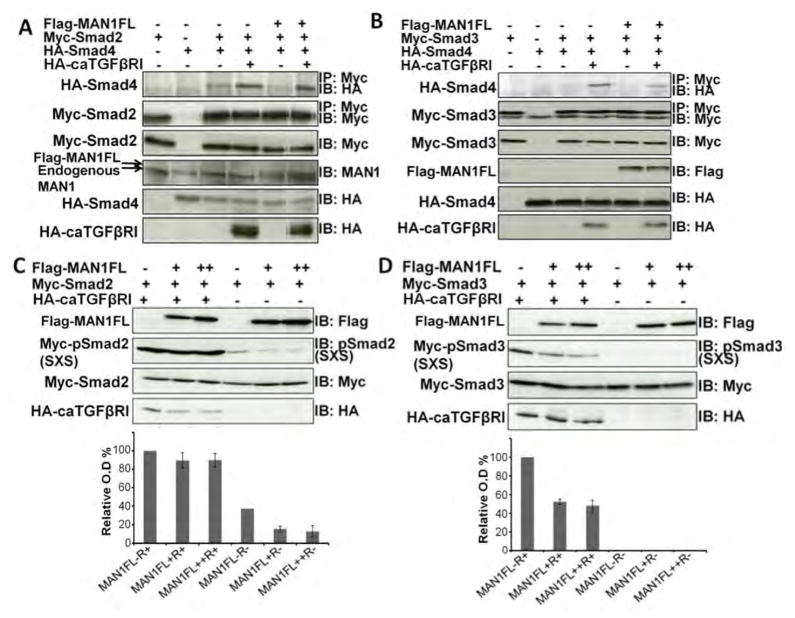
We next examined the interaction of MAN1 with the Smad2-Smad4 and Smad3-Smad4 heterotrimers in cells. Analysis of Myc-Smad2 immunoprecipitates indicated that overexpression of Flag-MAN1 induced partial dissociation of HA-Smad4 from Smad2 in 293T cells (Fig. 6A), which was observed mainly when the constitutively activated TGF-β receptor was overexpressed. Similarly, when Myc-Smad3 and HA-Smad4 were overexpressed, HA-Smad4 bound less efficiently to Myc-Smad3 after transfection with the Flag-MAN1 vector (Fig. 6B). Hence, MAN1 could induce partial Smad3-Smad4 dissociation in cells.
Fig. 6.
Overexpression of MAN1 leads to partial dissociation of Smad2/Smad4 and Smad3/Smad4 complexes and to Smad2/3 dephosphorylation. (A–B) Representative immunoblots showing the amount of Smad2/3 bound to Smad4 in 293T cells transfected as indicated. Cells overexpressed caTGFβRI (MAN1FL−R+), MAN1 (MAN1FL+R−), or both MAN1 and caTGFβRI (MAN1FL+R+), or did not express either MAN1 or caTGFβRI (MAN1FL−R−). Myc immunoprecipitates containing Myc−Smad2 or Myc−Smad3 (IP: Myc) were immunoblotted with anti-HA antibodies to detect HA-Smad4 (IB: HA; top panel). The amount of immunoprecipitated Myc-Smad2 or Myc-Smad3 (IP: Myc) was detected using anti-Myc antibodies. Lysates were immunoblotted as indicated. N=2 independent biological replicates; replicate blots are in fig. S5 (C–D) Representative immunoblots showing the amount of phosphorylated Myc-Smad2 or Myc-Smad3 in 293T cells transfected with plasmids expressing Myc-Smad2 or Myc-Smad3 in the presence of increasing concentration of FLAG-MAN1 (+ and ++) with or without the constitutively active receptor HA-caTGFβRI. Cell lysates were immunoblotted as indicated (bottom three panels). The histograms show the amount of phosphorylated Myc-Smad2 and Myc-Smad3, normalized using the amount of Myc-Smad2 or Myc-Smad3, from 3 independent experiments for each condition, in cells overexpressing neither MAN1 nor caTGFβRI (MAN1FL−R−), only caTGFβRI (MAN1FL−R+), only MAN1 (MAN1FL+R− and MAN1FL++R−) and both MAN1 and caTGFβRI (MAN1FL+R+ and MAN1FL++R+). The normalized signal from the MAN1FL−R+ condition was set at 100%.
Formation of Smad2-Smad4 and Smad3-Smad4 heterotrimers is regulated by phosphorylation of the C-terminal SSMS sequences of Smad2 and Smad3 (15). Dephosphorylation of these sequences leads to the dissociation of the heterotrimers. We therefore measured the amount of phosphorylated Smad2 and Smad3 with MAN1 overexpression in 293T cells. Cells were transfected with different concentrations of plasmid to give increasing Flag-MAN1 expression in the presence of constant amounts of expression of Myc-Smad2 and HA-tagged constitutively activated TGF-β receptor. Phosphorylation of Smad2 was decreased by overexpression of MAN1 when the constitutively activated TGF-β receptor was not expressed, an effect that was not systematically observed when the receptor was expressed, presumably because of the extensive phosphorylation of Smad2 (Fig. 6C). Phosphorylated Smad3 could not be detected when the receptor was not expressed, whereas phosphorylation of Smad3 was reduced by overexpression of MAN1 when the receptor was expressed (Fig. 6D). Thus, using conditions in which changes in the phosphorylation status of Smad2/3 could be experimentally detected, we demonstrated that MAN1 overexpression is associated with reduced phosphorylation of Smad2 and Smad3 in cells.
MAN1 binds to the Smad2/3 phosphatase PPM1A
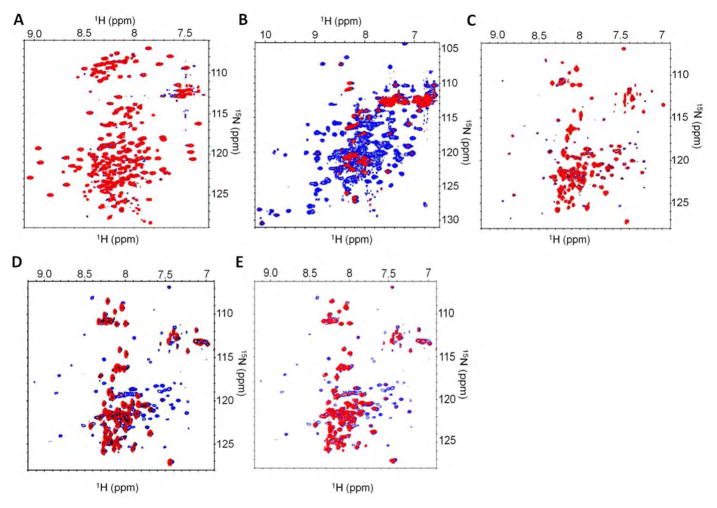
PPM1A catalyzes the dephosphorylation of Smad2 and Smad3, which promotes their nuclear export (35). MAN1 binds to Smad2 and Smad3 and its overexpression decreases their phosphorylation. This lead us to hypothesize that MAN1 could function as a scaffold that recruits PPM1A and Smad2/3 to the inner nuclear membrane to facilitate dephosphorylation. To test this hypothesis, we purified PPM1A and examined its interaction with MAN1 using NMR. Addition of PPM1A to a solution of 15N-labeled N-terminal nucleoplasmic region of MAN1 from amino acids 1 to 471 (MAN1N in Fig. 1) did not change the 1H-15N HSQC spectrum of MAN1N, showing that PPM1A did not bind to this region of MAN1 (Fig. 7A). In contrast, PPM1A added to a solution of 15N-labeled C-terminal nucleoplasmic region of MAN1 (amino acids 658 to 911; MAN1C) resulted in a progressive disappearance of the 1H-15N HSQC peaks corresponding to MAN1C (Fig. 7B). These results demonstrated that PPM1A bound specifically to the C-terminal region of MAN1. Addition of the C-terminal fragment, but not the N-terminal fragment, of MAN1 to 15N-labeled PPM1A caused a large set of weak peaks to disappear and globally decreased the intensity of all peaks in the 1H-15N TROSY-HSQC spectra (Fig. 7, C and D). Addition of the smaller Smad2/3 binding region of MAN1, MAN1Luhm, also evoked a large intensity decrease of the peaks corresponding to PPM1A (Fig. 7E). Thus, this region of MAN1 binds to both Smad2/3 and PPM1A.
Fig. 7.
PPM1A binds to the C-terminal domain of MAN1. 1H-15N 2D NMR spectra were recorded on 1:0 and 1:1 ratio samples of a 15N-labeled protein and its unlabelled partner. (A) Representative HSQC spectrum of 15N-labeled fragment of MAN1 from amino acid 1 to amino acid 471 (MAN1N) with (red) or without (blue) added PPM1A. N=2 independently generated replicates. (B) Representative HSQC spectrum of 15N-labeled fragment of MAN1 from amino acid 658 to amino acid 911 (MAN1C) with (red) or without (blue) added PPM1A. (C) Representative TROSY spectrum of 15N-labeled PPM1A with (red) or without (blue) addition of the MAN1 fragment from amino acid 1 to amino acid 471 (MAN1N). N=2 independently generated replicates. (D) Representative TROSY spectrum of 15N-labeled PPM1A with (red) or without (blue) addition of the MAN1 fragment from amino acid 658 to amino acid 911 (MAN1C). N=2 independently generated replicates. (E) Representative TROSY spectrum of 15N-labeled PPM1A with (red) or without (blue) addition of the MAN1 fragment from amino acid 755 to amino acid 911 (MAN1Luhm). N=2 independently generated replicates.
DISCUSSION
Our results provide evidence for a model of inactivation of TGF-β signaling at the nuclear envelope in which MAN1 has a dual function in repressing Smad-mediated transcriptional activation. First, MAN1 competes with transcription factors for Smad binding. Second, it binds to the Smad2/3 phosphatase PPM1A, thus providing a mechanism for enhancing dephosphorylation of Smad2/3 by recruiting both the activated, phosphorylated R-Smad and the enzyme catalyzing its dephosphorylation (inactivation) to the same location at the inner nuclear membrane. The model is consistent with previous observations that manipulating MAN1 expression in cultured cells as well as model organisms alters signaling by TGF-β (9, 13, 18, 21). In humans, it may be particular critical in bone and skin, because heterozygous loss-of-function mutation in the gene encoding MAN1 causes osteopoikilosis and Buschke-Ollendorff syndrome (11).
Our experimental data obtained in vitro and in cells show that MAN1 competes with the Smad interaction motif of FAST1 for binding to Smad2. Because members of the FAST and Mix families of transcription factors interact with the MH2 domains of Smad2 and Smad3 through a similar and conserved interaction motif (27, 34), MAN1 could compete with these transcription factors for binding. SARA, a Smad-binding protein anchored at the plasma membrane, also competes with these transcription factors for Smad2 and Smad3 binding using a proline-rich rigid coil sequence similar to the SIMs in the transcription factors (27) that interacts with a shallow hydrophobic pocket in the MH2 domain of Smad2 and Smad3 containing Tyr366 and Trp368 (25). We showed that MAN1 bound to Smad2 in a similar fashion through Tyr366 and Trp368, suggesting structural similarities between the recognition properties of the membrane proteins SARA and MAN1. Just as SARA competes with SIM-containing transcription factors for R-Smad binding at the plasma membrane, MAN1 competes with these transcription factors for R-Smad binding at the inner nuclear membrane. SARA recruits R-Smads for phosphorylation, which leads to their dissociation from SARA (36). We showed that MAN1 recruits R-Smads, possibly for dephosphorylation; however, the event triggering R-Smad dissociation from MAN1 remains unclear.
MAN1 overexpression led to decreased phosphorylation of Smad2 and Smad3 in cells, consistent with a previous report on the extent of Smad1 phosphorylation being lower after MAN1 overexpression (13). We have also shown that less Smad4 bound to either Smad2 or Smad3 after MAN1 overexpression, consistent with previous results showing that Smad3 bound to MAN1 does not bind to Smad4 (13). These results raise the possibility that in cells MAN1 facilitates dephosphorylation of the C-terminus of R-Smads and their release from Smad4.
PPM1A catalyzes the dephosphorylation of Smad2 and Smad3 (35). We showed that the C-terminal region of MAN1 interacted not only with Smad2/3, but also with PPM1A. The affinity of PPM1A for this region of MAN1 is between 10 μM and 500 μM because the PPM1A-MAN1 complex is not detected after gel filtration but is detected in the intermediate to slow exchange regime by NMR. We propose a model in which PPM1A interacts with the same region of MAN1 as Smad2/3, which would enable phosphorylated Smad2/3 to be preferentially targeted by PPM1A at the inner nuclear membrane. Because the affinity of PPM1A for MAN1 is weaker than that of Smad2/3, Smad2/3 would displace PPM1A from MAN1 if they compete for binding and are present at equal concentrations. However, additional interactions exist between Smad2/3 and PPM1A (35). After catalyzing the dephosphorylation of Smad2/3, PPM1A recruits RanBP3, which preferentially binds to dephosphorylated Smad2/3, to stimulate the nuclear export of Smad2/3 (37) (38). Release of unphosphorylated R-Smad from MAN1 could result from competition between MAN1 and RanBP3 for Smad2/3 binding. PPM1A also dephosphorylates RanBP3, which increases its nuclear exporter activity (37). It remains to be tested whether recruitment of RanBP3 to the inner nuclear membrane promotes its dephosphorylation by PPM1A.
In conclusion, we have described an inactivation mechanism for TGF-β signaling in which MAN1 competes with transcription factors for binding to Smad2 and Smad3 and also facilitates their association with their inactivating phosphatase. This mechanism is similar to that shown for inhibition of BAF activity by the transmembrane protein LEM4, which interacts with both the BAF-activating kinase and PP2A and is required for PP2A to dephosphorylate BAF during mitotic exit (23). The region of MAN1 responsible for the recruitment of both Smad2/3 and PPM1A is conserved through all coelomates (39), suggesting that this inactivation mechanism of TGF-β signaling at the nuclear envelope is evolutionarily conserved.
MATERIALS AND METHODS
Plasmids
Expression constructs for fragments of human MAN1 from amino acids 658 to 911 (MAN1C) and from amino acids 755 to 911 (MAN1Luhm) have been previously described (14). MAN1C was expressed as a 6His-ZZ-fusion protein, with a TEV site present between the ZZ and MAN1 region, and MAN1Luhm was expressed as a 6His-tagged protein. An expression construct for the fragment of MAN1 from amino acid 1 to amino acid 471 (MAN1N) was generated by synthesis of an optimized His-tagged MAN1 cDNA (Genscript) and insertion of this cDNA into pETM13-LIC vector (pETM13 expression vector modified in the laboratory for LIC cloning system). We also generated cDNA expression constructs in pETM10-LIC for fragments from amino acids 186 to 467 of human Smad2 (S2LMH2EEE) and from amino acids 167 to 425 of human Smad3 (S3LMH2EEE) containing the linker and the MH2 domains of the proteins. A nucleotide sequence changing SSMS to EEME was present in the primers used for cloning. Constructs coding for the Smad2 double mutants Y268A-W448A and Y366A-W368A were generated using the QuickChange Kit (Stratagen). We also generated a cDNA expression construct expressing the region human Smad4 from amino acids 273 to 552 (S4LMH2) in pETM10-LIC. An expression construct for human PPM1A was generated by synthesis of an optimized PPM1A cDNA (Genscript) and insertion into pETM13-LIC. Plasmid constructs expressing Myc-FAST1, Flag-Smad2, Myc-Smad2, Myc-Smad3, HA-Smad4, HA-caTGFβ RI for expression in 293T cells were provided by Céline Prunier (INSERM, Hôpital Saint-Antoine) (40). The construct expressing Flag-epitope tagged full-length MAN1 has been reported previously (5). For yeast two-hybrid assays, DNAs were generated by polymerase chain reactions and cloned into pGBKT7 and pACT2 (12, 14). Mutants were obtained using modified primers for PCR and after ligation in the pGBKT7 vector. All plasmid constructs were confirmed by DNA sequencing. Yeast two-hybrid assays were performed according to the instructions of the manufacturer (Clontech).
Yeast two-hybrid assays
Binding of the MAN1 fragment from amino acid 731 to amino acid 910 to Smad2 was measured. Five Smad2 single mutants, four Smad2 double mutants and a single Smad2 triple-mutant (SSMS to EEME) were tested. Cells containing the different plasmids were spotted four times by 10-fold serial dilutions onto SD2A plates containing 10 mM 3-aminotriazole and were incubated at 30°C for 3 days.
Protein expression and purification
Expression and purification of proteins used in previous studies were as described (14). The fragment of MAN1 from amino acids 1 to 471 (MAN1N), the Smad4 linker and MH2 domain construct (S4LMH2) and PPM1A were purified using a His trap column (GE Healthcare) and a gel filtration column (Superdex 200, Hi-Load, 120 mL, GE Healthcare). Protein concentrations were estimated from their absorbance at 280 nm, assuming that the ε at 280 nm was equal to the theoretical ε value.
Size-exclusion chromatography
Size-exclusion chromatography was performed using a Superdex 200 HR column on an Akta Explore 10 FPLC (Pharmacia Biotech). The column was equilibrated at 4°C with 100 mM Na2HPO4 (pH 8.0), 150 mM NaCl, and 1 mM tris(2-carboxyethyl)phosphine. Sample injection, elution and data analysis were performed using the UNICORN software (Pharmacia Biotech). To detect protein-protein interactions, samples were mixed at equal molar ratios and the mixtures were loaded on the column. The eluted fractions were then separated on a denaturing gel and stained with Coomassie blue. The column was calibrated with molecular weight standards blue dextran (2,000 kD), ferritin (440 kD), albumin (67 kD), chymotrypsin (25 kD), ribonuclease (13 kD) and vitamin B12 (1.36 kD). The flow rate was 0.8 ml.min−1 and the fraction volume was 0.6 ml.
ITC
Calorimetric titration experiments were performed as previously described (14). Purified FAST1 peptide was purchased from Genecust and dissolved in a buffer composed of 100 mM Na2HPO4 (pH 8.0), 150 mM NaCl and 1 mM tris(2-carboxyethyl)phosphine. All experiments were performed at 20°C.
To measure the binding of the MAN1LUHM fragment to S2LMH2EEE and its mutant Y366A-W368A, concentrations of the MAN1 fragment in the 280-μl syringe were 350 and 850 μM, respectively. Concentrations of the Smad2 fragments in the 2-ml sample cell were 35 and 80 μM, respectively. To test the impact of FAST1 on the MAN1/Smad2 interaction, a first experiment was carried out by injecting 280 μl of MAN1LUHM at 930 μM to 2 ml of S2LMH2EEE at 60 μM and the FAST1 peptide at 105 μM. In the second experiment, lower concentrations (MAN1Luhm 750 μM S2LMH2EEE 45μM peptide 90 μ were used. The resulting apparent Kd values depend on the protein and peptide concentrations. This is linked to the ten-times lower affinity of the FAST1 peptide for S2LMH2EEE, as compared to the affinity of MAN1Luhm for this same S2LMH2EEE fragment (fig. S2). When measuring the affinity of MAN1LUHM for heterotrimeric Smad2/Smad4 complexes, 280 μl of MAN1Luhm at 800 μ was added to 2 ml of S2LMH2EEE at 65 μalone or in complex with S4LMH2, at 130 μ. Similarly, in the case of Smad3, 280 μl of MAN1Luhm at 800 μM was added to 2 ml of S3LMH2EEE at 85 μ Malone or in complex with S4LMH2 at 170 μM.
SAXS
SAXS samples were prepared in 100 mM Na2HPO4 (pH 8.0), 150 mM NaCl, and 1 mM tris(2-carboxyethyl)phosphine buffer. Synchrotron radiation X-ray scattering data were collected at 17°C on the SWING line at Synchrotron SOLEIL (Gif-sur-Yvette, France). We recorded SAXS data on S2LMH2EEE and the MAN1Luhm-S2LMH2EEE complex at 50 μM and 30 μM concentrations, respectively (Table S1). Models for the trimeric Smad2 region from amino acids 186 to 467 were calculated using CNS (41), based on the X-ray structure of the MH2 domain of Smad2 (PDB ID 1KHX). We obtained 6,000 models in which the N-terminal region was in a random conformation. The theoretical SAXS curve was calculated for each of these models using CRYSOL (http://www.embl-hamburg.de/biosaxs/crysol.html). A chi value was obtained, that scored the fit between the experimental (Iexp) and calculated (Icalc) intensities and was calculated as the square root of 1/N*Σq((Iexp-Icalc)/σ)2, σ being the experimental error and N the number of points. The 20 final models were selected as those with the best chi values. The average chi value was 2.7 ± 0.1 for the 20 models. To correctly account for the conformational variability of the unfolded regions, we calculated an average SAXS intensity (Iave) curve from the 20 theoretical intensities (Icalc) curves. This average curve results from the presence of the 20 models within the population of Smad2. Comparison of Iave to Iexp yielded a chi value of 1.8. Models for the Smad2-MAN1 complexes were similarly calculated, and the structure of MAN1Luhm was consistent with previously described SAXS and NMR data (14). We used Pepsite (pepsite.russelllab.org) to predict the position of the poorly folded MAN1 fragment containing Trp765 and Gln766 onto the Smad2 MH2 domain. We observed that the MAN1 Trp765 and Gln766 residues were systematically positioned close to the Smad2 Tyr366 and Trp368 residues. Thus, to calculate models for the Smad2-MAN1 complex, we applied distance restraints between Smad2 Tyr366 and Trp368 and MAN1 Trp765 and Gln766. We obtained 12,000 models of the complex and calculated a theoretical SAXS curve for the 200 models with the lowest distance restraint energies using CRYSOL. The 20 final models were selected as those with the smallest chi values. The average chi value calculated for the 20 models was 2.0 ± 0.2. To correctly account for the conformational variability of the unfolded regions, we calculated an average SAXS intensity (Iave) curve from the 20 theoretical intensities (Icalc) curves. Comparison of Iave to Iexp yielded a chi value of 1.2. The chi value corresponding to the fit of the average theoretical intensity to the experimental intensity evolves with the number of models used to calculate the average intensity (fig. S3). Because of the large conformational variability of Smad2 N-terminal region, and perhaps also because of sampling biases, 20 models had to be taken into account to reproduce the experimental SAXS data obtained on Smad2 alone (fig. S3A). However, perhaps because of a reduced conformational variability of MAN1-bound Smad2, the chi value was already at its minimum using only 4 models for the Smad2-MAN1 complex (fig. S3B).
Immunoprecipitation and immunoblotting
293T cells were transfected with expression vectors by the Lipofectamine method and 48 h post-transfection were lysed at 4 °C in a 50 mM HEPES (pH 7.8), 500 mM NaCl, 5mM ethylenediaminetetraacetic acid, 3 mM dithiothreitol, 1% Nonidet P-40, 0.5 mM phenylmethanesulfonylfluoride buffer. In each experiment, a vector coding for β-galactosidase was transfected, β-galactosidase activity was measured using the Galacto-Star™ System kit (Applied BioSystems), and cell lysate aliquots containing identical β-galactosidase activities were analysed. For coimmunoprecipitation, these lysates were subjected to immunoprecipitation with the appropriate antibody followed by adsorption to Sepharose-coupled protein G over-night at 4°C. Immunoprecipitates were separated by SDS-PAGE and analyzed by immunoblotting. For determination of total protein amounts, aliquots of cell lysates were separated by SDS-PAGE and subjected to direct immunoblotting. Proteins were electrophoretically transferred to nitrocellulose membranes and probed with the indicated primary antibody. Bands were visualized by an enhanced chemiluminescence detection system according to the manufacturer’s instructions (ECL; Amersham Biosciences). Quantifications of western blots were carried out using the Image-J software. A representative blot is shown in the main figures. In Figs. 4B, 6C and 6D, histograms display the mean values and standard errors calculated from 3 independent biological replicates. Replicate blots corresponding to Figs. 6A and 6B are presented in Fig. S5.
NMR
15N-labeled samples of MAN1N, MAN1C, MAN1Luhm and PPM1A were obtained by cultivating E. coli BL21 (DE3) star (Invitrogen) in a minimal medium containing 0.5 g/l of 15NH4Cl as the sole nitrogen source. Purification was carried out as described for unlabelled proteins, and the last gel filtration step was carried out in 50 mM Tris (pH 6.7), 150 mM NaCl, 1 mM tris(2-carboxyethyl)phosphine, 1 mM ethylenediaminetetraacetic acid. Proteins were concentrated to 400 μM in 100 μl using AMICON ULTRA-15 3KDa (Fisher) and were diluted in either 100 μl of buffer or 100 μl of buffer containing the unlabelled partner concentrated at 400 μM. Finally, 20 μl of D2O was added to the samples so that the final sample volumes were 220 μl. 1H-15N HSQC and TROSY NMR spectra were recorded at 20°C on a 700 MHz Bruker spectrometer equipped with a TCI cryoProbe. They were analyzed using Topspin (Bruker) and Sparky (USCF) programs.
Supplementary Material
Fig. S1. Yeast two-hybrid assay results for the interactions between the Smad2 mutants and MAN1 730–910. In the top panel, empty vectors were used to perform control experiments as indicated. In the bottom panel, the interactions between MAN1 730–910 and wild-type and mutant Smad2 are shown. In both panels, cells containing the different plasmids were spotted four times using 10-fold serial dilutions onto SD2A plates containing 10 mM 3-aminotriazole and incubated at 30°C for 3 days, to test the expression of the first reporter gene HIS3 (dark background columns). Physical interaction between Smad2 and Man1 activated expression of HIS3 gene allowing yeast cell growth. No mutation hindered cell growth, thus demonstrating that all tested single mutants still bind to MAN1 730–910. N=2 independent biological replicates.
Fig. S2. Representative binding curve obtained by ITC for the FAST1 peptide and the phosphomimetic mutant of the MH2 domain of Smad2 (S2LMH2EEE). The concentrations used in this binding experiment were: S2LMH2EEE 60 μM in the cell, peptide 855 μM in the seringe. The resulting Kd yielded 11.4 ± 1.3 μM. In the replicate experiment, the concentrations were: S2LMH2EEE 48 μM in the cell, peptide 480 μM in the seringe. The resulting Kd yielded 15.2 ± 1.2 μM. The enthalpic ΔH values (0.9 and 1.3 kCal/Mol) and the entropic TΔS values (7.5 and 7.8 kCal/Mol) deduced from the 2 ITC experiments were always positive, showing that the interaction is totally entropy driven, unlike the interactions described in Table 1.
Fig. S3. Evolution of the chi value as a function of the number of models used in the calculation of the average SAXS intensity. This evolution is plotted for the trimeric Smad2 oligomer (A) and for the complex between trimeric Smad2 and three MAN1 fragments (B).
Fig. S4. Sequence alignment of human Smad2 and Smad3 proteins. The full-length sequences are 84% identical, and the Smad4 binding fragments (Smad2 207–467 and Smad3 167–425) are 92% identical.
Fig. S5. Replicate blots corresponding to Figs. 6A and 6B. These immunoblots show the amount of Smad2/3 bound to Smad4 in 293T cells transfected as indicated. Cells overexpressed caTGFβRI (MAN1FL−R+), MAN1 (MAN1FL+R−), or both MAN1 and caTGFβRI (MAN1FL+R+), or did not express either MAN1 or caTGFβRI (MAN1FL−R−). Myc immunoprecipitates containing Myc-Smad2 or Myc-Smad3 (IP: Myc) were immunoblotted with anti-HA antibodies to detect HA-Smad4 (IB: HA; top panel). The amount of immunoprecipitated Myc-Smad2 or Myc-Smad3 (IP: Myc) was detected using anti-Myc antibodies. Lysates were immunoblotted as indicated. In these replicate blots, Smad2/3 bound to Smad4 could not be detected when caTGFβRI was not overexpressed.
Table S1. SAXS data collection and scattering-derived parameters. Parameters specifically corresponding to experiments recorded on Smad2 and Smad2-MAN1 are referenced as a and b, respectively. * Partial specific volumes are calculated from the sequence based on (42).
Acknowledgments
We thank Céline Prunier for the mammalian expression vectors. We are also grateful to Marie-Bénédicte Barrault and Anne Peyroche for their help with the yeast two-hybrid assays. Funding: B.B., B.G., C.T.-L. and S.Z.-J. were supported by AFM grant 14849. H.J.W. was supported by grant R01AR048997 from the NIH/NIAMS. Author contributions: B.B. designed experiments, carried out protein purifications, chromatography, ITC, SAXS and NMR analyses and coimmunoprecitation experiments and wrote the manuscript. B.G. performed the modeling calculations from SAXS and mutagenesis data. C.T.-L. designed and generated plasmid constructs and carried out the double-hybrid experiments. C. Ö. and W. W. designed and generated plasmid constructs. J.P. supervised the SAXS experiments and their analysis. P. E. H. helped with the coimmunoprecipitation experiments. F.L. designed and supervised the coimmunoprecitation experiments. H.J.W. helped conceive the project, analyzed data and wrote the manuscript. S. Z.-J. conceived the project, analyzed data, supervised the entire project and wrote the manuscript. Competing interests: The authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Dauer WT, Worman HJ. The nuclear envelope as a signaling node in development and disease. Dev Cell. 2009;17:626–38. doi: 10.1016/j.devcel.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 2.Worman HJ, Bonne G. “Laminopathies”: a wide spectrum of human diseases. Exp Cell Res. 2007;313:2121–33. doi: 10.1016/j.yexcr.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andres V, Gonzalez JM. Role of A-type lamins in signaling, transcription, and chromatin organization. J Cell Biol. 2009;187:945–57. doi: 10.1083/jcb.200904124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barascu A, Le Chalony C, Pennarun G, Genet D, Imam N, Lopez B, Bertrand P. Oxidative stress induces an ATM-independent senescence pathway through p38 MAPK-mediated lamin B1 accumulation. Embo J. 2012;31:1080–94. doi: 10.1038/emboj.2011.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin F, Blake DL, Callebaut I, Skerjanc IS, Holmer L, McBurney MW, Paulin-Levasseur M, Worman HJ. MAN1, an inner nuclear membrane protein that shares the LEM domain with lamina-associated polypeptide 2 and emerin. J Biol Chem. 2000;275:4840–7. doi: 10.1074/jbc.275.7.4840. [DOI] [PubMed] [Google Scholar]
- 6.Paulin-Levasseur M, Blake DL, Julien M, Rouleau L. The MAN antigens are non-lamin constituents of the nuclear lamina in vertebrate cells. Chromosoma. 1996;104:367–79. doi: 10.1007/BF00337226. [DOI] [PubMed] [Google Scholar]
- 7.Laguri C, Gilquin B, Wolff N, Romi-Lebrun R, Courchay K, Callebaut I, Worman HJ, Zinn-Justin S. Structural characterization of the LEM motif common to three human inner nuclear membrane proteins. 2001;9:503–11. doi: 10.1016/s0969-2126(01)00611-6. [DOI] [PubMed] [Google Scholar]
- 8.Cai M, Huang Y, Ghirlando R, Wilson KL, Craigie R, Clore GM. Solution structure of the constant region of nuclear envelope protein LAP2 reveals two LEM-domain structures: one binds BAF and the other binds DNA. Embo J. 2001;20:4399–407. doi: 10.1093/emboj/20.16.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raju GP, Dimova N, Klein PS, Huang HC. SANE, a novel LEM domain protein, regulates bone morphogenetic protein signaling through interaction with Smad1. J Biol Chem. 2003;278:428–37. doi: 10.1074/jbc.M210505200. [DOI] [PubMed] [Google Scholar]
- 10.Osada S, Ohmori SY, Taira M. XMAN1, an inner nuclear membrane protein, antagonizes BMP signaling by interacting with Smad1 in Xenopus embryos. Development. 2003;130:1783–94. doi: 10.1242/dev.00401. [DOI] [PubMed] [Google Scholar]
- 11.Hellemans J, Preobrazhenska O, Willaert A, Debeer P, Verdonk PC, Costa T, Janssens K, Menten B, Van Roy N, Vermeulen SJ, Savarirayan R, Van Hul W, Vanhoenacker F, Huylebroeck D, De Paepe A, Naeyaert JM, Vandesompele J, Speleman F, Verschueren K, Coucke PJ, Mortier GR. Loss-of-function mutations in LEMD3 result in osteopoikilosis, Buschke-Ollendorff syndrome and melorheostosis. Nat Genet. 2004;36:1213–8. doi: 10.1038/ng1453. [DOI] [PubMed] [Google Scholar]
- 12.Lin F, Morrison JM, Wu W, Worman HJ. MAN1, an integral protein of the inner nuclear membrane, binds Smad2 and Smad3 and antagonizes transforming growth factor-beta signaling. Hum Mol Genet. 2005;14:437–45. doi: 10.1093/hmg/ddi040. [DOI] [PubMed] [Google Scholar]
- 13.Pan D, Estevez-Salmeron LD, Stroschein SL, Zhu X, He J, Zhou S, Luo K. The integral inner nuclear membrane protein MAN1 physically interacts with the R-Smad proteins to repress signaling by the transforming growth factor-{beta} superfamily of cytokines. J Biol Chem. 2005;280:15992–6001. doi: 10.1074/jbc.M411234200. [DOI] [PubMed] [Google Scholar]
- 14.Konde E, Bourgeois B, Tellier-Lebegue C, Wu W, Perez J, Caputo S, Attanda W, Gasparini S, Charbonnier JB, Gilquin B, Worman HJ, Zinn-Justin S. Structural analysis of the Smad2-MAN1 interaction that regulates transforming growth factor-beta signaling at the inner nuclear membrane. Biochemistry. 2010;49:8020–32. doi: 10.1021/bi101153w. [DOI] [PubMed] [Google Scholar]
- 15.Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 16.Nakao A, Afrakhte M, Moren A, Nakayama T, Christian JL, Heuchel R, Itoh S, Kawabata M, Heldin NE, Heldin CH, ten Dijke P. Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature. 1997;389:631–5. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- 17.Imamura T, Takase M, Nishihara A, Oeda E, Hanai J, Kawabata M, Miyazono K. Smad6 inhibits signalling by the TGF-beta superfamily. Nature. 1997;389:622–6. doi: 10.1038/39355. [DOI] [PubMed] [Google Scholar]
- 18.Ishimura A, Ng JK, Taira M, Young SG, Osada S. Man1, an inner nuclear membrane protein, regulates vascular remodeling by modulating transforming growth factor beta signaling. Development. 2006;133:3919–28. doi: 10.1242/dev.02538. [DOI] [PubMed] [Google Scholar]
- 19.Cohen TV, Kosti O, Stewart CL. The nuclear envelope protein MAN1 regulates TGFbeta signaling and vasculogenesis in the embryonic yolk sac. Development. 2007;134:1385–95. doi: 10.1242/dev.02816. [DOI] [PubMed] [Google Scholar]
- 20.Ishimura A, Chida S, Osada S. Man1, an inner nuclear membrane protein, regulates left-right axis formation by controlling nodal signaling in a node-independent manner. Dev Dyn. 2008;237:3565–76. doi: 10.1002/dvdy.21663. [DOI] [PubMed] [Google Scholar]
- 21.Pinto BS, Wilmington SR, Hornick EE, Wallrath LL, Geyer PK. Tissue-specific defects are caused by loss of the Drosophila MAN1 LEM domain protein. Genetics. 2008;180:133–45. doi: 10.1534/genetics.108.091371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huber MD, Guan T, Gerace L. Overlapping functions of nuclear envelope proteins NET25 (Lem2) and emerin in regulation of extracellular signal-regulated kinase signaling in myoblast differentiation. Mol Cell Biol. 2009;29:5718–28. doi: 10.1128/MCB.00270-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Asencio C, Davidson IF, Santarella-Mellwig R, Nga Ly-Hartig TB, Mall M, Wallenfang MR, Mattaj IW, Gorjanacz M. Coordination of Kinase and Phosphatase Activities by Lem4 Enables Nuclear Envelope Reassembly during Mitosis. Cell. 2012;150:122–135. doi: 10.1016/j.cell.2012.04.043. [DOI] [PubMed] [Google Scholar]
- 24.Wu JW, Hu M, Chai J, Seoane J, Huse M, Li C, Rigotti DJ, Kyin S, Muir TW, Fairman R, Massague J, Shi Y. Crystal structure of a phosphorylated Smad2. Recognition of phosphoserine by the MH2 domain and insights on Smad function in TGF-beta signaling. Mol Cell. 2001;8:1277–89. doi: 10.1016/s1097-2765(01)00421-x. [DOI] [PubMed] [Google Scholar]
- 25.Wu G, Chen YG, Ozdamar B, Gyuricza CA, Chong PA, Wrana JL, Massague J, Shi Y. Structural basis of Smad2 recognition by the Smad anchor for receptor activation. Science. 2000;287:92–7. doi: 10.1126/science.287.5450.92. [DOI] [PubMed] [Google Scholar]
- 26.Randall RA, Howell M, Page CS, Daly A, Bates PA, Hill CS. Recognition of phosphorylated-Smad2-containing complexes by a novel Smad interaction motif. Mol Cell Biol. 2004;24:1106–21. doi: 10.1128/MCB.24.3.1106-1121.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Randall RA, Germain S, Inman GJ, Bates PA, Hill CS. Different Smad2 partners bind a common hydrophobic pocket in Smad2 via a defined proline-rich motif. Embo J. 2002;21:145–56. doi: 10.1093/emboj/21.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chacko BM, Qin B, Correia JJ, Lam SS, de Caestecker MP, Lin K. The L3 loop and C-terminal phosphorylation jointly define Smad protein trimerization. Nat Struct Biol. 2001;8:248–53. doi: 10.1038/84995. [DOI] [PubMed] [Google Scholar]
- 29.Svergun DI, Barberato C, Koch MHJ. CRYSOL - a Program to Evaluate X-ray Solution Scattering of Biological Macromolecules from Atomic Coordinates. J Appl Cryst. 1995;28:768–773. [Google Scholar]
- 30.Chacko BM, Qin BY, Tiwari A, Shi G, Lam S, Hayward LJ, De Caestecker M, Lin K. Structural basis of heteromeric smad protein assembly in TGF-beta signaling. Mol Cell. 2004;15:813–23. doi: 10.1016/j.molcel.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 31.Chen YG, Hata A, Lo RS, Wotton D, Shi Y, Pavletich N, Massague J. Determinants of specificity in TGF-beta signal transduction. Genes Dev. 1998;12:2144–52. doi: 10.1101/gad.12.14.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen X, Rubock MJ, Whitman M. A transcriptional partner for MAD proteins in TGF-beta signalling. Nature. 1996;383:691–6. doi: 10.1038/383691a0. [DOI] [PubMed] [Google Scholar]
- 33.Attisano L, Silvestri C, Izzi L, Labbe E. The transcriptional role of Smads and FAST (FoxH1) in TGFbeta and activin signalling. Mol Cell Endocrinol. 2001;180:3–11. doi: 10.1016/s0303-7207(01)00524-x. [DOI] [PubMed] [Google Scholar]
- 34.Germain S, Howell M, Esslemont GM, Hill CS. Homeodomain and winged-helix transcription factors recruit activated Smads to distinct promoter elements via a common Smad interaction motif. Genes Dev. 2000;14:435–51. [PMC free article] [PubMed] [Google Scholar]
- 35.Lin X, Duan X, Liang YY, Su Y, Wrighton KH, Long J, Hu M, Davis CM, Wang J, Brunicardi FC, Shi Y, Chen YG, Meng A, Feng XH. PPM1A functions as a Smad phosphatase to terminate TGFbeta signaling. Cell. 2006;125:915–28. doi: 10.1016/j.cell.2006.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qin BY, Lam SS, Correia JJ, Lin K. Smad3 allostery links TGF-beta receptor kinase activation to transcriptional control. Genes Dev. 2002;16:1950–63. doi: 10.1101/gad.1002002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dai F, Shen T, Li Z, Lin X, Feng XH. PPM1A dephosphorylates RanBP3 to enable efficient nuclear export of Smad2 and Smad3. EMBO Rep. 2011;12:1175–81. doi: 10.1038/embor.2011.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dai F, Lin X, Chang C, Feng XH. Nuclear export of Smad2 and Smad3 by RanBP3 facilitates termination of TGF-beta signaling. Dev Cell. 2009;16:345–57. doi: 10.1016/j.devcel.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mans BJ, Anantharaman V, Aravind L, Koonin EV. Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle. 2004;3:1612–37. doi: 10.4161/cc.3.12.1316. [DOI] [PubMed] [Google Scholar]
- 40.Prunier C, Pessah M, Ferrand N, Seo SR, Howe P, Atfi A. The oncoprotein Ski acts as an antagonist of transforming growth factor-beta signaling by suppressing Smad2 phosphorylation. J Biol Chem. 2003;278:26249–57. doi: 10.1074/jbc.M304459200. [DOI] [PubMed] [Google Scholar]
- 41.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 42.Kharakoz DP. Partial volumes and compressibilities of extended polypeptide chains in aqueous solution: additivity scheme and implication of protein unfolding at normal and high pressure. Biochemistry. 1997;36:10276–85. doi: 10.1021/bi961781c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Yeast two-hybrid assay results for the interactions between the Smad2 mutants and MAN1 730–910. In the top panel, empty vectors were used to perform control experiments as indicated. In the bottom panel, the interactions between MAN1 730–910 and wild-type and mutant Smad2 are shown. In both panels, cells containing the different plasmids were spotted four times using 10-fold serial dilutions onto SD2A plates containing 10 mM 3-aminotriazole and incubated at 30°C for 3 days, to test the expression of the first reporter gene HIS3 (dark background columns). Physical interaction between Smad2 and Man1 activated expression of HIS3 gene allowing yeast cell growth. No mutation hindered cell growth, thus demonstrating that all tested single mutants still bind to MAN1 730–910. N=2 independent biological replicates.
Fig. S2. Representative binding curve obtained by ITC for the FAST1 peptide and the phosphomimetic mutant of the MH2 domain of Smad2 (S2LMH2EEE). The concentrations used in this binding experiment were: S2LMH2EEE 60 μM in the cell, peptide 855 μM in the seringe. The resulting Kd yielded 11.4 ± 1.3 μM. In the replicate experiment, the concentrations were: S2LMH2EEE 48 μM in the cell, peptide 480 μM in the seringe. The resulting Kd yielded 15.2 ± 1.2 μM. The enthalpic ΔH values (0.9 and 1.3 kCal/Mol) and the entropic TΔS values (7.5 and 7.8 kCal/Mol) deduced from the 2 ITC experiments were always positive, showing that the interaction is totally entropy driven, unlike the interactions described in Table 1.
Fig. S3. Evolution of the chi value as a function of the number of models used in the calculation of the average SAXS intensity. This evolution is plotted for the trimeric Smad2 oligomer (A) and for the complex between trimeric Smad2 and three MAN1 fragments (B).
Fig. S4. Sequence alignment of human Smad2 and Smad3 proteins. The full-length sequences are 84% identical, and the Smad4 binding fragments (Smad2 207–467 and Smad3 167–425) are 92% identical.
Fig. S5. Replicate blots corresponding to Figs. 6A and 6B. These immunoblots show the amount of Smad2/3 bound to Smad4 in 293T cells transfected as indicated. Cells overexpressed caTGFβRI (MAN1FL−R+), MAN1 (MAN1FL+R−), or both MAN1 and caTGFβRI (MAN1FL+R+), or did not express either MAN1 or caTGFβRI (MAN1FL−R−). Myc immunoprecipitates containing Myc-Smad2 or Myc-Smad3 (IP: Myc) were immunoblotted with anti-HA antibodies to detect HA-Smad4 (IB: HA; top panel). The amount of immunoprecipitated Myc-Smad2 or Myc-Smad3 (IP: Myc) was detected using anti-Myc antibodies. Lysates were immunoblotted as indicated. In these replicate blots, Smad2/3 bound to Smad4 could not be detected when caTGFβRI was not overexpressed.
Table S1. SAXS data collection and scattering-derived parameters. Parameters specifically corresponding to experiments recorded on Smad2 and Smad2-MAN1 are referenced as a and b, respectively. * Partial specific volumes are calculated from the sequence based on (42).