Abstract
Recombination induced by double-strand breaks (DSBs) in yeast leads to a higher proportion of expansions to contractions than does replication-associated tract length changes. Expansions are apparently dependent on the property of the repeat array to form hairpins, since DSB repair of a CAA87 repeat induces only contractions of the repeat sequence. DSB-repair efficiency is reduced by 40% when DNA synthesis must traverse a CAG98 array, as compared with a CAA87 array. These data indicate that repair- associated DNA synthesis is inhibited by secondary structures formed by CAG98 and that these structures promote repeat expansions during DSB repair. Overexpression of Mre11p or Rad50p suppresses the inhibition of DSB repair by CAG98 and significantly increases the average size of expansions found at the recipient locus. Both effects are dependent on the integrity of the Mre11p–Rad50p–Xrs2p complex. The Mre11 complex thus appears to be directly involved in removing CAG or CTG hairpins that arise frequently during DNA synthesis accompanying gene conversion of these trinucleotide repeats.
Keywords: double-strand break repair/MRE11/recombination/trinucleotide repeats/yeast
Introduction
A growing number of human diseases involving trinucleotide repeat expansions have been discovered (Ashley and Warren, 1995; Reddy and Housman, 1997; Richards and Sutherland, 1997). This class of microsatellites has the property of undergoing rapid and sometimes massive expansion of their repeat number, during germline transmission or early embryogenesis. One striking feature of all expansion-associated microsatellites is their propensity to form secondary structures. CAG/CTG trinucleotide repeats have been shown to form stable hairpins in vitro (Gacy et al., 1995; Mitas et al., 1995b; Yu and Mitas, 1995), as do CCG/CGG trinucleotide repeats (Gacy et al., 1995; Mitas et al., 1995a; Yu et al., 1997). GAA repeats involved in Friedreich’s ataxia (Campuzano et al., 1996) were shown to form triple helices in vitro (Gacy et al., 1998; Sakamoto et al., 1999). The ability to form stable secondary structures is thought to play a role during the expansion process, perhaps by slowing down or stalling replication (Samadashwily et al., 1997). Replication pausing at trinucleotide repeats has been shown in vitro for CCG (Usdin and Woodford, 1995) and GAA (Gacy et al., 1998), and for CTG and CCG in vivo (Samadashwily et al., 1997).
The mechanism by which trinucleotide secondary structures mediate expansions is not well understood. Recent data suggest that several mechanisms may be needed to give rise to the full spectrum of large expansions observed in human disease (reviewed by Wells, 1996; McMurray, 1999; Richard et al., 1999a). In model systems, trinucleotide repeats can undergo tract length changes during replication, as shown in both Escherichia coli (Kang et al., 1995) and Saccharomyces cerevisiae (Maurer et al., 1996; Freudenreich et al., 1997; Miret et al., 1998; Schweitzer and Livingston, 1998). However, in wild-type strains, starting with CAG templates up to 100 repeats, the vast majority of tract length changes are deletions (Maurer et al., 1996; Freudenreich et al., 1997; Miret et al., 1998; Schweitzer and Livingston, 1998).
In contrast, in mutant strains carrying mutations in proteins involved in DNA replication or repair, expansions are frequently seen. In S.cerevisiae, the absence of the Rad27/FEN-1 nuclease that processes Okazaki fragments during DNA replication causes increases in the size of CAG repeats (Freudenreich et al., 1998; Schweitzer and Livingston, 1998; Spiro et al., 1999). Rad27/FEN-1 is also implicated in long-patch excision repair (Gary et al., 1999).
In E.coli, CAG repeats were dramatically expanded in a strain carrying a mutation in the SbcC gene (Sarkar et al., 1998). The SbcC–SbcD protein complex exhibits 3′–5′ double-strand DNA exonucleolytic and single-strand DNA endonucleolytic activities in vitro (Connelly and Leach, 1996). In addition, the SbcC–SbcD complex cleaves DNA hairpins in vitro (Connelly et al., 1998). The SbcC–SbcD nuclease is highly conserved through evolution, from bacteria to humans. Yeast and mammalian homologues of SbcC and SbcD, the Rad50 and Mre11 proteins, respectively, exhibit in vitro biochemical activities similar to their bacterial counterparts (Paull and Gellert, 1998; Usui et al., 1998). However, RAD50 and MRE11 are involved in many different recombinational processes (reviewed by Haber, 1998), and their possible roles in trinucleotide expansions in eukaryotic cells remain to be elucidated.
Although CAG expansions certainly occur with a low frequency during replication, it is important to characterize microsatellite length changes that arise during recombination, especially given the association between repeat expansions in humans with transmission of the germline. Recently, Jakupciak and Wells (1999) have shown that recombination occurred between two DNA molecules carrying CAG/CTG repeats in E.coli. These recombination events led to contractions and expansions of the repeats, were RecA- and RecBC-dependent, and were accompanied by cross-overs. In addition, Freudenreich et al. (1998) showed that long CAG/CTG repeats were fragile sites in yeast during vegetative growth, and Jankowski et al. (2000) showed that meiotic double-strand breaks (DSBs) are formed within CAG/CTG repeats in yeast. In order to understand by which mechanism(s) triplet repeats may be expanded in eukaryotic cells, it is thus relevant to study trinucleotide repeat instability during DSB repair.
We have developed a system to assess the instability of repeated sequences arising during gene conversion in S.cerevisiae. A DSB is created on a chromosome, such that the ends of the DSB are homologous to donor sequences located on a plasmid. The donor also contains a set of repeated sequences that DNA polymerases associated with repair synthesis must traverse (Figure 1A). Using this system, when the donor contained eight 375-bp repeats, Pâques et al. (1998) found that almost 50% of the gene conversions exhibited expansions or contractions (from one to 13 copies) and virtually all the tract length changes were found in the recipient locus. These results were interpreted in terms of a synthesis-dependent strand annealing (SDSA) mechanism of gene conversion in which the two newly synthesized DNA strands are displaced from the intact template and are both found in the recipient locus (reviewed by Pâques and Haber, 1999). A similar set of experiments were performed by Richard et al. (1999b), studying the fate of short CAG repeats (<40 repeats). In that study, ∼20% of DSB-induced gene conversions led to contraction of the repeat, again virtually all in the recipient locus.
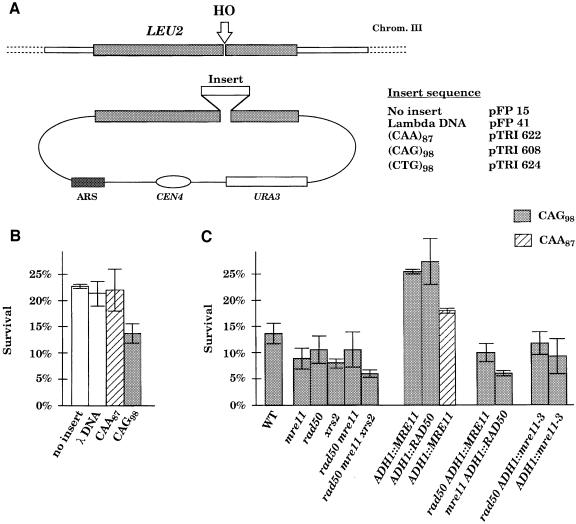
Fig. 1. Survival of yeast cells after DSB induction. (A) Experimental system. The HO recognition site was inserted on chromosome III, inside the LEU2 gene. Cells were transformed with a centromeric plasmid containing the LEU2 gene. Different inserts were cloned inside the plasmid-borne LEU2 gene. After HO induction, the DSB was repaired by gene conversion using the plasmid-borne LEU2 template. Analysis of recombinants after DSB repair was carried out with or without the plasmid template, in order to distinguish between plasmid-borne and chromosome-borne repeats. For each repeat tract studied, sequence of the Watson (top) strand is shown. (B and C) Frequencies of survival after DSB repair. Yeast cells were plated in parallel on glucose and galactose plates. After 4–5 days at 30°C, colonies were scored on both plates. The frequency of survival was calculated as the ratio of the number of colonies growing on galactose plates over the number of colonies growing on glucose plates. The average frequencies and the standard errors were calculated from a set of 2–4 independent experiments. (B) Survival of a wild-type strain containing different templates after HO induction. (C) Survival of different mutant strains containing either a CAG98 or CAA87.
In the present work, we examined HO-induced gene conversions of longer CAG triplet repeats, and tested the effects of the Mre11p–Rad50p–Xrs2p protein complex on trinucleotide tract length changes, during gene conversion-associated DNA synthesis. We show that gene conversion involving the copying of a CAG98 array produces both expansions and contractions, whereas only deletions were found during replication. Moreover, CAG sequences, but not CAA sequences, impede the efficiency of DNA synthesis during gene conversion. This sequence-specific effect can be suppressed by overexpression of the Mre11 and Rad50 proteins. This suggests that the Mre11p–Rad50p–Xrs2p complex is responsible for the removal of hairpin structures that arise during repair-associated DNA synthesis. We conclude that DNA synthesis during recombinational repair of DSBs can be an important source of expansions of CAG repeats.
Results
An HO endonuclease recognition site was inserted into the LEU2 gene of chromosome III, in strain YFP17. Homology to the HO cleavage site at MAT, HML and HMR was deleted (Pâques et al., 1998). Expression of the HO endonuclease gene was controlled by a galactose-inducible promoter (Sandell and Zakian, 1993). A DSB is created within the LEU2 gene of chromosome III, such that sequences near the ends of the DSB are homologous to donor sequences located on a plasmid (Figure 1A). Repair of the DSB requires the presence of the homologous sequences on the plasmid. Thus, when HO was expressed in the absence of the donor plasmid, 99.7% of cells died, as they were unable to repair the DSB. However, when a plasmid containing an intact LEU2 gene was introduced to provide homology to the HO-cleaved LEU2, ∼22% of cells were able to repair the DSB using the plasmid sequence as a template (Pâques et al., 1998, and Figure 1B).
We then supplied different templates in which the cells survive by a gene conversion event in which the repair DNA polymerases must traverse an ∼300 bp insertion in the template (Figure 1A). When the template contained 300 bp of phage λ DNA, 21.3% of the cells survived (Figure 1B); similarly a donor containing CAA87 gave 22.0% survival, very similar to results when there was no insertion in the template. However, when the donor carried CAG98, survival dropped to 13.8%. We conclude that CAG repeats, but not CAA repeats, reduce efficient gene conversion. This is consistent with the idea that CAG repeats, but not CAA repeats, are capable of forming hairpin structures that impede new DNA synthesis during repair (Samadashwily et al., 1997; McMurray, 1999). We have performed several experiments to confirm that CAA/GTT repeats form no hairpins. In contrast to CAG, CTG, CGG and GAC, DNA-melting analysis shows that repeating GTT has no melting transition consistent with the absence of structure (data not shown). Additionally, GTT25 sequences on polyacrylamide gels migrate at a position that is consistent only with an unstructured strand and not with a hairpin. The lack of structure has also been confirmed functionally. Spiro et al. (1999) showed that repeating GTT has a 1000-fold lower expansion rate relative to hairpin-forming sequences such as CTG or GAC. The low mutation rate of GTT was identical in frequency to random sequences with no secondary structure. In addition, Moore et al., (1999) showed that heteroduplex DNA containing CAG or CTG repeats is not efficiently repaired in vivo, whereas CAA loops are repaired. These results strongly suggest that CAA repeats do not form the same structures in vivo as CAG or CTG repeats.
If secondary structures in CAG98 cause the reduction in survival, then proteins involved in cleaving or unwinding these secondary structures might increase the efficiency of successfully traversing these sequences and thus increase survival. We, therefore, evaluated cell survival under conditions when MRE11 was overexpressed. MRE11, fused to the ADH1 promoter, was carried on a centromere-containing plasmid in an mre11Δ strain. MRE11 overexpression suppressed the CAG-dependent recombination defect, so that survival occurred at the same level as for the other 300 bp inserts (Figure 1C). MRE11 overexpression did not affect repair of the CAA template. This suppression of CAG-specific inhibition of repair was dependent on having an intact Mre11–Rad50–Xrs2 complex, because a rad50Δ mre11Δ double mutant strain carrying the overexpression MRE11 plasmid had the same low survival as rad50Δ or rad50Δ mre11Δ (Figure 1C). Similarly, overexpressing RAD50 suppressed the defect in gene conversion of CAG repeats, but this suppression disappeared in a mre11Δ rad50Δ strain (Figure 1C). Thus overexpression of two members of the Mre11–Rad50–Xrs2 complex suppressed the CAG-specific reduction in successful recombination, presumably by removing a secondary structure in DNA (presumably a hairpin) that would impair DNA synthesis during DSB repair.
There is also an effect of deleting the genes encoding the Mre11–Rad50–Xrs2 complex. Deletions of mre11Δ, rad50Δ or xrs2Δ, as well as a triple mutant (rad50Δ mre11Δ xrs2Δ) reduced the efficiency of gene conversion with the CAG98 template, relative to wild type (Figure 1C). However, this effect is not CAG-specific, as we found a similar reduction in strains carrying LEU2 or LEU2::λ300 templates (data not shown).
If MRE11 is required to remove hairpins, then expression of a defective mre11 mutant should prevent the suppression of the recombination defect in cells using the CAG template. The Mre11-3 protein carries a leucine substitution at a histidine residue conserved in all phosphoesterases of the SbcD/Mre11 family (Bressan et al., 1998). The mre11-3 (H125L, D126V) mutant is similar to mre11-H125N, which has been shown to be defective in in vitro nuclease assays (Moreau et al., 1999). Curiously, both mre11-3 and mre11-H125N mutants have proven to be nearly wild-type in many mitotic phenotypes that are affected by an mre11Δ mutation, including spontaneous mitotic recombination, radiation sensitivity, mating-type switching, 5′ to 3′ resection of a DSB, non-homologous end-joining and telomere length (Bressan et al., 1998; Moreau et al., 1999; S.E.Lee, D.Bressan, J.H.J.Petrini and J.E.Haber, manuscript submitted). The mre11-3 mutant does not disrupt the physical interaction between Rad50p and Mre11-3p (Bressan et al., 1998). In the assay described here, we found that overexpression of Mre11-3p in an mre11Δ strain did not relieve the CAG-dependent inhibition of gene conversion, giving the same results as for the mre11Δ strain (Figure 1C). Hence this mutation, which shows no significant defect in a variety of other mitotic phenotypes, is defective in suppressing CAG-mediated inhibition of DSB repair. This implies that the nuclease activity of Mre11p seen in vitro may be important in hairpin removal, or possibly hairpin unwinding (see below). We also note that Bressan et al. (1998) showed that Mre11-3p was expressed at the same level as wild-type Mre11p, so that the absence of an effect in this system cannot be explained by the instability of this mutant protein.
Recombination produces expansions of CAG98
If secondary structure inhibits gene conversion, it may also affect the fidelity of copying CAG repeats. Changes in the number of repeated sequences may arise by replication slippage and/or by the pairing of newly synthesized DNA strands, both of which can be influenced by secondary structures that impede repair-associated DNA synthesis and perhaps promote dissociation of DNA polymerases (Pâques et al., 1998). We evaluated changes in repeat length accompanying gene conversion by determining the size of the trinucleotide repeats in both the recipient array at the LEU2 chromosomal locus and in the plasmid template (see Materials and methods). The sensitivity of our measurements was calculated to be ± 8 triplets. Hence, we were not able to detect small changes in size of <8 triplets in the following experiments. Without HO induction, only 8% of the templates carried on the plasmid showed tract length changes of the repeat, and all of them were contractions. Thus replication in the absence of DSB repair exhibited the same frequency of rearrangements, with deletions greatly outnumbering expansions, that have been seen in previous studies in yeast analyzing tract length changes of CAG repeats of comparable sizes (Maurer et al., 1996; Schweitzer and Livingston, 1997). Moreover, the frequency of tract length changes in the template sequences remained constant, again yielding mostly contractions, after HO-induced recombination (Table I).
Table I. Spontaneous and induced trinucleotide repeats rearrangements, in wild-type and mutant strains.
| Strain | Genotype | Template | Template contractionsa | Gene conversions of the recipient | ||||
|---|---|---|---|---|---|---|---|---|
| |
|
|
Before HOinduction |
After HOinduction |
Without tractlength change |
With tractexpansion |
With tractcontraction |
Total |
| GFY510 | WT | CAG98 | 8% | 12% | 49 (57%) | 11 (13%) | 26 (30%) | 86 (100%) |
| GFY549 | WT | CTG98 | 72% | 6 (27%) | 2 (9%) | 14 (64%) | 22 (100%) | |
| GFY529 | WT | CAA87 | 3% | 43 (90%) | 0 | 5 (10%) | 48 (100%) | |
| GFY508 | rad50 | CAG98 | 11% | 43 (67%) | 9 (14%) | 12 (19%) | 64 (100%) | |
| GFY513 | mre11 | CAG98 | 6% | 35 (65%) | 10 (18%) | 9 (17%) | 54 (100%) | |
| GFY515 | rad50 mre11 | CAG98 | 2% | 27 (53%) | 10 (20%) | 14 (27%) | 51 (100%) | |
| GFY527 | mre11 ADH1::MRE11 | CAG98 | 29% | 18%b | 46 (66%) | 5 (7%) | 19 (27%) | 70 (100%) |
| GFY528 | rad50 mre11 ADH1::MRE11 | CAG98 | 12% | 34 (67%) | 9 (17%) | 8 (16%) | 51 (100%) | |
WT: wild type.
aTemplate rearrangements were scored either during vegetative growth of independent cultures in the absence of HO induction and/or as the number of rearranged templates following HO induction and recovery of the survivors. All template rearrangements were contractions.
bFor strain GFY527, 15% of the templates recovered after HO induction showed expansions. None of the templates analyzed before HO induction showed expansions.
In contrast, induction of DSB repair by HO endonuclease resulted in a significant number of both contractions and expansions. All the tract length changes were found in the recipient locus, as predicted by SDSA models of gene conversion (Pâques et al., 1998; Pâques and Haber, 1999). Examples are shown in Figure 2. Only 57% of the cells completing gene conversion in the wild-type strain faithfully copied 98 CAG triplets from the template into the chromosome. The remaining 43% contained either contractions of the CAG repeat (30%) or expansions (13%; Table I). When the same analysis was performed on the strain containing CAA87 as template, only 10% showed tract length changes after DSB repair. All tract length changes were contractions, all in the recipient locus (Table I). This argues that the CAG trinucleotide expansions we observe are apparently dependent on their ability to form hairpins during DSB repair.
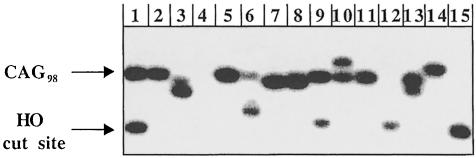
Fig. 2. Changes in CAG repeats during gene conversion. Following HO induction of gene conversion between a chromosomal leu2 locus and a URA3-containing plasmid containing leu2::(CAG98), individual colonies were isolated. Cells lacking the URA3-marked plasmid were selected on 5-FOA and DNA was analyzed by Southern blots. The examples shown are from wild-type cells. Lane 1: uninduced control cells; lane 4: non-homologous recombination event giving rise to partial deletion of the LEU2 locus; lanes 2, 5, 11: gene conversion that faithfully copies CAG98 into the chromosomal locus; lanes 3, 6, 7, 8, 12, 15: gene conversion that yields a deletion in the recipient; lane 14: gene conversion that leads to an expansion of CAG repeats in the recipient; lanes 9, 10, 13: gene conversion in which the recipient contained heteroduplex DNA, producing a colony with two different-sized CAG repeats of equal intensity.
Since overexpression of MRE11 increased survival of strains carrying the CAG-containing template, we wished to know if higher levels of MRE11 would also affect the proportions or sizes of tract length changes. The proportions of tract length changes after DSB repair when MRE11 was overexpressed with the CAG98-containing donor (GFY527) were 27% contractions and 7% expansions, which is statistically indistinguishable by a contingency χ2 test from the 30% deletions and 13% expansions that we observed with the wild-type strain GFY510. In the rad50 derivative overexpressing MRE11 (GFY528), there were 16% deletions and 17% expansions (Table I). These numbers are not significantly different from an mre11 or a rad50 strain alone (Table I). However, the proportions of expansions and contractions among the wild-type strains (GFY510 and GFY527) are statistically significantly different from those strains that are mutant for rad50 and/or mre11 (GFY508, GFY513, GFY515 and GFY528), by a contingency χ2 test (p <0.025). The absence of mre11 or rad50 appears to nearly double the proportion of expansions, from the average wild-type value of 26% of tract length changes to 47%. We conclude that overexpression of MRE11 did not change the ratio of contractions over expansions recovered after DSB repair, but the absence of either Mre11p or Rad50p does increase the proportion of expansions.
We also examined the stability of the template when MRE11 was overexpressed, in the absence of HO induction. Out of 17 independent clones, five were found to carry a contraction of the CAG98 repeat, two of them exhibiting a double band, suggesting that contraction occurred during the vegetative growth of the clone. This result indicates that MRE11 overexpression does increase the instability of the CAG98 array during growth, presumably during repair-associated DNA synthesis, but does not induce spontaneous expansions of the template.
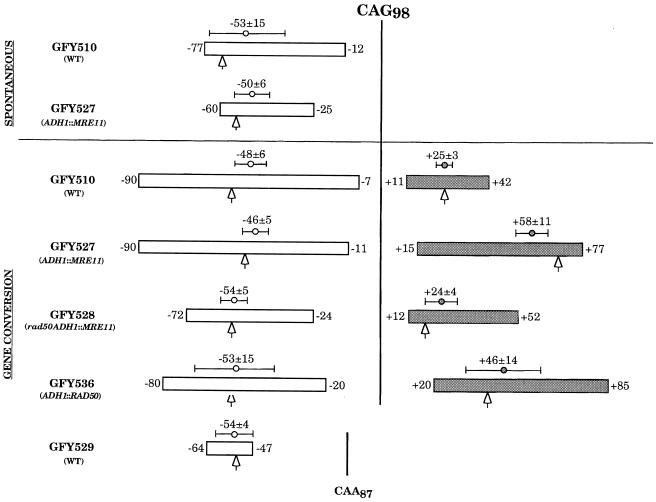
Although MRE11 overexpression does not change the proportion of expansions after DSB repair, compared with wild-type, it did affect the size of expansions. Compared with an average expansion size of 25 ± 3 triplets in the wild-type strain, overexpressing MRE11 increased this average to 58 ± 11 triplets (Figure 3). All expansions except one were significantly larger (p <0.05 using a Student’s t-test) than in the wild-type strain. This important increase was suppressed in a rad50 strain, in which expansion sizes were comparable to wild-type (Figure 3). The sizes of contractions after gene conversion were not significantly different in the five strains studied (Figure 3). The interpretation of this result will be discussed later.
Fig. 3. Trinucleotide repeat size changes after gene conversion using CAG98 or CAA87 as templates. For each strain, the white rectangle indicates the range of contractions detected after DSB repair at the recipient locus on the chromosome. Sizes of the shortest and largest contractions are indicated, respectively, right and left of the rectangle, along with the average contraction size and the standard error. Similarly, the gray rectangle indicates the range of expansions detected after DSB repair. Sizes of the shortest and largest expansions are indicated, left and right of the rectangle, respectively, along with the average expansion size and the standard error. The vertical black lines indicate the original size of the CAG98 or of the CAA87 repeats. The arrows represent the median value of each of the distributions. For CAG98 repeats, the changes in the array arising spontaneously on the donor plasmid sequence are also shown.
As a control, we also overexpressed MRE11 in a strain containing CAA87 as the template. Out of 36 survivors analyzed, all of them were faithfully repaired, and no expansion was detected. A short contraction of the repeat was found on one template. We concluded that the effect of MRE11 overexpression on expansions was specific to CAG repeats, thus probably dependent on their ability to form secondary structures that we presume to be hairpins.
Orientation-independence of recombination-induced CAG/CTG tract length changes
Several authors reported that the frequency of CAG/CTG tract length changes during replication in yeast is orientation-dependent, relative to the direction of the replication fork (Maurer et al., 1996; Freudenreich et al., 1997; Miret et al., 1998). In our experimental system, strand invasion and repair-associated DNA synthesis of the CAG98 template can theoretically occur from both ends (Figure 4). However, if for some reason, invasion occurs preferentially from one end, the orientation of the repeat could affect tract length changes during gene conversion. To address this question, we flipped the CAG98 insert in the plasmid template. The resulting CTG98 template was transformed into yeast (GFY549, Table I). There was a much higher proportion of tract length changes in the template (72%)—again, all contractions—than with the template in the CAG98 orientation (12%). These results confirm previous studies showing that the stability of a CAG/CTG repeat carried on a centromeric plasmid in yeast is orientation-dependent (Maurer et al., 1996; Freudenreich et al., 1997; Miret et al., 1998).
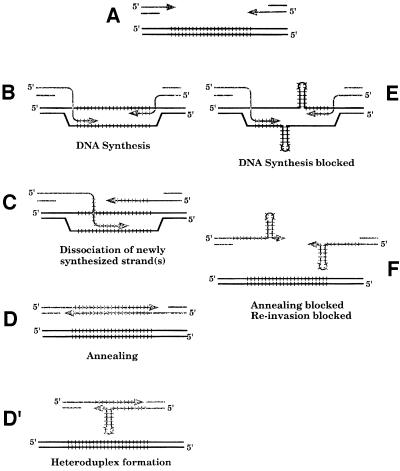
Fig. 4. Possible roles of Mre11p during gene conversion of a CAG trinucleotide repeat. After HO induction and processing of the 3′ ends of the DSB (A), strand invasion may occur and DNA synthesis proceeds through the repeats (B). If secondary structures are formed on the template, DNA synthesis will be stopped (E). Unwinding of one or both strands may occur (C), allowing annealing between the two newly synthesized strands (D), or re-invasion of the template. Formation of a hairpin on one of the newly synthesized strands would give rise to a heteroduplex DNA molecule that is resistant to mismatch repair (D′). Formation of a hairpin on each of the newly synthesized strands may block re-invasion and annealing, leading to cell death (F). Mre11p may help to bypass hairpins and allow DNA synthesis to proceed further. Alternatively, Mre11p may help to unwind or process the hairpin(s) formed on the newly synthesized strand(s), favoring re-invasion.
When HO was induced and the CTG98 sequences were used as a template for gene conversion, there was also a statistically significantly higher frequency of tract length changes at the recipient locus than with CAG98 (p <0.01) (Table I). For this analysis we only considered the 22 cases where the template size was not apparently changed prior to induction of HO. Despite the increase in the frequency of tract length changes, the proportion of events that were expansions was not significantly different using CTG98 than with CAG98. Thus, we observed recombination-dependent expansions and contractions with both orientations of the template. The higher proportion of instability in the CTG98 orientation suggests that the DSB repair process is not entirely symmetrical, because if recombination proceeded the same way from each end, the orientation should not have had any effect. It is possible that strand invasion is preferentially directed from one end, as envisioned in several different versions of SDSA models (reviewed by Pâques and Haber, 1999). Preferential invasion from one end could lead to differences in stability when repair DNA synthesis must synthesize CAG versus CTG triplets. Preferential strand invasion could be caused by differences in the lengths of the non-homologous tails of the HO cleavage site, which must be removed in order to initiate new DNA synthesis during repair, because of sequence-specific differences in the flanking regions of homology that influence strand invasion, or because HO endonuclease remains bound to one side of the break after cleavage, a property found for another endonuclease of the same family (Perrin et al., 1993).
In support of the hypothesis that there is some asymmetry in which end is used to initiate DNA repair synthesis, we observed a significantly higher survival with CTG98 (22.7% ± 1.1%). This could be explained by preferential strand invasion from one side of the break, leading to preferential synthesis of CAGs or CTGs by the repair-associated machinery. Previously, we have shown that HO-induced gene conversion can occur independently of S phase (Holmes and Haber, 1999), and therefore the difference in the outcomes of CAG and CTG orientations cannot be attributed to the position of these sequences relative to a nearby origin of DNA replication.
Heteroduplex DNA formation involving CAG or CTG sequences during gene conversion
Gene conversion of CAG repeats frequently involves formation of heteroduplex DNA. Approximately 20% of the colonies analyzed after DSB repair of the CAG98 template in the wild-type strain contained two bands of equal intensity in the recipient locus, corresponding to CAG repeats of different sizes (Figure 2). When CAA87 was used as a template, only one mixed colony was detected out of 47 analyzed. Thus mixed colonies are most likely to result from stable formation of CAG or CTG hairpins in heteroduplex DNA that is not repaired (Figure 4), rather than the result of two separate repair events in a G2-phase cell. This conclusion is supported by recent evidence that heteroduplex DNA containing CAGs or CTGs is not efficiently repaired, whereas CAA loops are repaired (Moore et al., 1999).
The number of mixed colonies was also computed in different mutant strains (Table II). When we added together the number of mixed colonies detected in mre11, rad50 and mre11 rad50 double-mutant strains, we found that it was significantly lower than in the wild-type strain using a contingency χ2 test (p <0.05). In the strain overexpressing MRE11, there is no significant difference as compared with wild-type. We suggest that Mre11p cannot process CAG or CTG hairpins when they are part of a DNA heteroduplex molecule even though it affects earlier steps in recombination (Figure 4).
Table II. Mixed colonies detected after DSB repair.
| Strain | |||||||
|---|---|---|---|---|---|---|---|
| GFY510 | GFY529 | GFY508 | GFY513 | GFY515 | GFY527 | GFY528 | |
| Genotype | WT | WT | rad50 | mre11 | rad50 | mre11 ADH1::MRE11 | rad50 mre11 ADH1::MRE11 |
| Template | CAG98 | CAA87 | CAG98 | CAG98 | CAG98 | CAG98 | CAG98 |
| Single band | 63 | 46 | 60 | 40 | 42 | 52 | 43 |
| Double band | 15 | 1 | 4 | 7 | 5 | 9 | 4 |
| Total colonies | 78 | 47 | 64 | 47 | 47 | 61 | 47 |
WT: wild type.
Discussion
We present evidence that repair-associated DNA synthesis across CAG repeats can lead to their expansion. We conclude that it is the propensity of the CAG trinucleotide repeat sequence to form secondary structures that impedes repair, increases their likelihood of rearrangement during recombination, and causes expansions during gene conversion. Our data further suggest that the Mre11–Rad50–Xrs2 complex plays a role in removing a block to the completion of recombination, presumably by removing inhibitory secondary structures.
Trinucleotide repeat expansions occur during gene conversion
CAG expansions occur with a high frequency (1/3 of all tract length changes) during gene conversion of a CAG98. This contrasts with several previous studies in yeast, during vegetative growth, where CAG triplets of comparable sizes yielded mainly deletions (Maurer et al., 1996; Freudenreich et al., 1997; Miret et al., 1997; Schweitzer and Livingston, 1997). In our experimental system, only contractions of CAG repeats were found during normal replication, even when MRE11 was overexpressed (Figure 3). We conclude that DNA synthesis occurring during HO endonuclease-induced DSB repair is more prone to give rise to CAG repeat expansions than S-phase DNA synthesis. Our conclusions are supported by the contemporaneous finding that gene conversions of CAG repeats that occur during yeast meiosis, where DSBs are initiated by the Spo11 protein, also yield both expansions and contractions (Jankowski et al., 2000).
CAG repeats impede the completion of gene conversion
Studies in bacteria have shown that CTG repeats cause a weak pause in normal replication in bacteria, but it is not sufficient to prevent eventual completion of replication (Samadashwily et al., 1997). In our assay, where DNA repair polymerases must traverse ∼300 bp of trinucleotide repeats, the presence of CAG98, but not CAA87, prevents the completion of DSB repair in 40% of the cases. This is most likely to reflect an effect on the ability to complete repair-associated DNA synthesis, although the secondary structures formed by CAG repeats might also affect later steps in recombination (see below). The result fits well with the conclusions of our previous studies where we showed that processivity of the repair-associated DNA synthesis machinery was considerably less efficient than during normal replication (Richard et al., 1999b). We suggest that during repair-associated DNA synthesis, polymerases encounter hairpins that form in single-stranded regions ahead of the polymerase. Repair-associated DNA synthesis may be blocked or the polymerase (perhaps still associated with a partially synthesized DNA strand) may dissociate from the template.
CAG repeats are more prone to expansions than CAA repeats
The dissociation of repair polymerases at hairpins may also explain how both expansions and deletions are formed (Figure 4). The dissociated polymerase may re-invade the template at a different point, either adding additional units or deleting sequences (Figure 4). The fact that repair-associated DNA synthesis across CAA repeats only produces deletions, as does normal S-phase replication across CAG repeats, may suggest that deletions can also form by a hairpin-independent process of replication slippage that preferentially yields contractions.
In a previous study using a CAG39 template, we found that tract length changes accompanying gene conversion were about half as frequent as found here with CAG98 and, importantly, virtually all tract length changes were contractions (Richard et al., 1999b). This suggests there is a threshold number of CAG repeats, >39 and <98, which is required to create expansions, just as has been described for CAG expansions in humans (Ashley and Warren, 1995).
The MRE11–RAD50–XRS2 complex affects recombination-induced expansions of CAG repeats
Another important conclusion is that Mre11p and Rad50p play a sequence- or structure-specific role in trinucleotide repeat expansions during gene conversion-associated DNA synthesis. Suppression of the inhibition of gene conversion by CAG98 depends on both Rad50p and Mre11p, and is also likely to include Xrs2p (Haber, 1998). This complex may exonucleolytically remove secondary structures such as hairpins during replication, or may unwind such structures, allowing DNA synthesis to proceed. In Figure 4 two different steps where this might occur are illustrated. In Figure 4E hairpin unwinding would allow polymerase to proceed; in Figure 4F hairpin unwinding or cleavage would free up the 3′ end for efficient re-invasion of the template. These two hypotheses are not mutually exclusive. Re-invasion of the dissociated DNA strand at different sites within the repeated array can produce both deletions and expansions. Because overexpression of either MRE11 or RAD50 has a similar effect, it seems that the increase in abundance of either protein shifts the equilibrium toward a more stable Mre11p–Rad50p complex. Overexpression of MRE11 only increases the abundance of the protein by a factor of two (S.E.Lee, D.Bressan, J.H.J.Petrini and J.E.Haber, manuscript submitted).
In addition to suppressing the CAG-specific defects in recombination, overexpression of Mre11p or Rad50p does not increase the proportion of expansions, but increases the average size of the CAG expansions. The mean size increase of an expansion increased from 25 repeats to 58 repeats, and a few events produced a near doubling in size. In vitro experiments have shown that the Mre11p complex has a single-strand endonuclease activity, a high affinity for hairpins and is able to hydrolyze the single-stranded loop of a DNA hairpin (Paull and Gellert, 1998; Usui et al., 1998). The intact human Mre11–Rad50–NBS1 complex is also able to unwind a short (17 bp) DNA duplex, but not longer duplexes (34 bp) (Paull and Gellert, 1999). It seems to us very likely that this unwinding activity could also be found for short hairpins. We suggest that hairpins are unwound rather than cleaved, as cleavage of the template might prevent recombination from being completed, and removal of hairpins in displaced strands should favor making deletions. Hairpins may also be cleaved, at least during replication, as suggested by results of Freudenreich et al. (1998), who showed that spontaneous DSBs arising within long CAG repeats were reduced in a rad50 mutant. We propose that the Mre11 complex is able to process short hairpins, but not longer ones. Long hairpins would thus escape processing by the complex, giving rise to larger expansions. Alternatively, larger expansions could be the result of successive rounds of invasion–dissociation–reinvasion (Figure 4), favored by hairpin cleavage by the Mre11 complex. Overexpression of MRE11 or RAD50 would not increase the proportion of expansions, but their size.
It is significant that half of the expansions that were recovered when MRE11 or RAD50 was overexpressed were found as mixed colonies, as would be expected if the product of gene conversion contained a large heteroduplex structure. Such heteroduplexes are insensitive to repair (Moore et al., 1999) and hence would be found as mixed progeny of a single event. Apparently the Mre11 complex does not remove these structures, which on average have 23 trinucleotide repeats, by cleavage or unwinding. This is consistent with the idea that overexpression of Mre11p or Rad50p would fail to remove longer duplexes.
The effect of deleting MRE11 or RAD50 also affects recombination of a CAG-containing template. First, there was a reduction in successful repair, but this effect was not specific to CAG-containing templates and probably reflects a more general requirement for these proteins in homologous recombination. Secondly, there is an increase in the proportion of expansions among all tract length changes without these gene products. This can be explained if some hairpins that would have been removed, remain and lead to expansions. However, the average size of the expansions was not significantly affected by deleting these genes.
We have also made the important finding that the mre11-3 mutation behaves like mre11Δ in terms of suppressing the CAG-specific reduction in recombination. In all previous assays of mitotic recombination, DNA end-joining, telomere maintenance or checkpoint response to DNA damage, this mutation appeared nearly wild-type, despite its lack of in vitro nuclease activity (Bressan et al., 1998; Moreau et al., 1999; S.E.Lee, D.Bressan, J.H.J.Petrini and J.E.Haber, manuscript submitted). The fact that Mre11-3p is unable to improve the survival of cells using the CAG template for DSB repair suggests that this activity is more directly related to the endonuclease and DNA unwinding activities that have been reported for the Mre11 complex in vitro, whereas many of the other attributes may simply depend on an intact Mre11–Rad50–Xrs2 (or NBS1) complex that could interact with other proteins.
The model system we have established will allow us to investigate other aspects of recombination-based expansions of triplet repeats. For example, we wish to know the behavior of donor templates containing GAA repeats that are reported to form different secondary structures (Gacy et al., 1998; Sakamoto et al., 1999). Another important question concerns the threshold at which CAG repeats begin to show expansions as well as contractions and which proteins may be involved in that step. We have shown that copying a template containing CAG39 only produced contractions (Richard et al., 1999b), whereas one with CAG98 yielded 1/3 expansions. Very recently Cohen et al. (1999) have examined CAG stability during yeast meiosis, using CAG52–61 templates. Instability was increased ∼4-fold over mitotic rates, but almost only deletions were found. With larger CAG arrays expansions could also be found (Jankowski et al., 2000). We note that it is possible to use our system, with an HO endonuclease under control of a meiosis-specific promoter (Malkova et al., 1996), to compare the same event during mitosis and meiosis.
Materials and methods
Strains
All strains used in this study are isogenic to YFP17 strain (Pâques et al., 1998), whose genotype is MATaΔ::hisG hmlΔ::ADE1 hmrΔ::ADE1 ade1 lys5 ura3-52 trp1 ade3::GAL10::HO leu2::HOcs. RAD50 was disrupted with pJH372 (RAD50::hisG::URA3::hisG). MRE11 was disrupted with pJH1432 (MRE11::hisG::URA3::hisG). XRS2 was replaced by LEU2 with pEI40 (Ivanov et al., 1994).
Plasmids
pFP15 was constructed by inserting the LEU2 gene into the Ted plasmid (CEN4, URA3). pFP36 was constructed by cloning pBluescript polylinker (Stratagene) into the KpnI site of pFP15 LEU2 gene (Pâques et al., 1998). pFP41 was constructed by inserting a 300 bp of lambda DNA into pFP36 polylinker, inside LEU2. Plasmid pTRI608 was constructed by cloning a HindIII–XbaI DNA fragment from pRW3216 (Kang et al., 1995) containing a CAG98 repeat into the BamHI–XbaI sites of pFP36. pTRI624 was constructed by cloning the same CAG98 fragment into the XhoI–XbaI sites of pFP36, in the opposite orientation. pTRI622 was constructed by cloning a NdeI–XbaI fragment from pcDNA3-CAA87 into the XhoI–XbaI sites of pFP36. Overexpression of MRE11, mre11-3 and RAD50 was performed using, respectively, pScMRE11, pScmre11-3 and pTRI102. MRE11, mre11-3 and RAD50 were placed under the control of the ADH1 promoter on centromeric plasmids carrying the TRP1 selection marker (Bressan et al., 1998). MRE11 and mre11-3 were overexpressed in mre11 and mre11 rad50 strains. RAD50 was overexpressed in rad50 and mre11 rad50 strains.
HO inductions
Cultures were grown overnight in uracil dropout dextrose (or in uracil–tryptophan double dropout dextrose when MRE11 or mre11-3 were overexpressed), to a concentration of 107–108 cells/ml. Cells were washed in water and resuspended in YEP-lactate for 5 h, to a final concentration of 106–107 cells/ml, before plating in parallel on uracil dropout galactose and dextrose plates (or uracil–tryptophan double drop out). After growth, efficiency of repair (survival) was scored as the ratio of colonies growing on galactose plates as compared with colonies growing on glucose plates. Survivors were replica plated on 5-fluoroorotic acid (5-FOA)-containing plates to select for loss of the URA3-containing plasmid (Boeke et al., 1984). Each induction was performed at least twice from independent colonies.
Molecular analysis of the survivors
Genomic DNA was extracted from survivors, digested by SspI and StyI restriction enzymes, loaded on a 20 cm gel along with DNA extracted from an uninduced control strain and run overnight at 1.6 V/cm for 14 h. A 1 kb LEU2 DNA fragment was randomly radio-labeled (Hodgson and Fisk, 1987) and used as a probe to detect LEU2 bands containing trinucleotide repeats. When variations in size were detected, as compared with the uninduced control, size measurements were performed to estimate the size of contractions or expansions, according to molecular weight markers of known sizes. On average, 1 mm corresponded to a size increase or decrease of 7.4 ± 0.3 triplets (22 ± 1 nucleotide). The error on measurement was estimated to be 0.5 mm, or 3.7 triplets (11 nucleotides). All the values given in Figure 3 are thus subject to a variation of ± 3.7 triplets.
Acknowledgments
Acknowledgements
We wish to thank R.D.Wells, J.H.J.Petrini and F.Pâques for the generous gift of plasmids and yeast strains. We also thank all the members of the Haber Laboratory for enthusiastic and stimulating discussions. This work was supported by NIH grant GM 20056.
References
- Ashley C.T. Jr and Warren,S.T. (1995) Trinucleotide repeat expansion and human disease. Annu. Rev. Genet., 29, 703–728. [DOI] [PubMed] [Google Scholar]
- Boeke J.D., Lacroute,F. and Fink,G.R. (1984) A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet., 197, 345–346. [DOI] [PubMed] [Google Scholar]
- Bressan D.A., Olivares,H.A., Nelms,B.E. and Petrini,J.H.J. (1998) Alteration of N-terminal phosphoesterase signature motifs inactivates Saccharomyces cerevisiae Mre11. Genetics, 150, 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campuzano V. et al. (1996) Friedreich’s Ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science, 271, 1423–1427. [DOI] [PubMed] [Google Scholar]
- Cohen H., Sears,D.D., Zenvirth,D., Hieter,P. and Simchen,G. (1999) Increased instability of human CTG repeat tracts on yeast artificial chromosomes during gametogenesis. Mol. Cell. Biol., 19, 4153–4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly J.C. and Leach,D.R.F. (1996) The SbcC and SbcD genes of Esherichia coli encode a nuclease involved in palindrome inviability and genetic recombination. Genes Cells, 1, 285–291. [DOI] [PubMed] [Google Scholar]
- Connelly J.C., Kirkham,L.A. and Leach,D.R.F. (1998) The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc. Natl Acad. Sci. USA, 95, 7969–7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freudenreich C.H., Stavenhagen,J.B. and Zakian,V.A. (1997) Stability of a CTG/CAG trinucleotide repeat in yeast is dependent on its orientation in the genome. Mol. Cell. Biol., 17, 2090–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freudenreich C.H., Kantrow,S.M. and Zakian,V.A. (1998) Expansion and length-dependent fragility of CTG repeats in yeast. Science, 279, 853–856. [DOI] [PubMed] [Google Scholar]
- Gacy A.M., Goellner,G., Juranic,N., Macura,S. and McMurray,C.T. (1995) Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell, 81, 533–540. [DOI] [PubMed] [Google Scholar]
- Gacy A.M. et al. (1998) GAA instability in Friedreich’s ataxia shares a common, DNA-directed and intra-allelic mechanism with other trinucleotide diseases. Mol. Cell, 1, 583–593. [DOI] [PubMed] [Google Scholar]
- Gary R., Kim,K., Cornelius,H.L., Park,M.S. and Matsumoto,Y. (1999) Proliferating cell nuclear antigen facilitates excision in long-patch base excision repair. J. Biol. Chem., 274, 4354–4363. [DOI] [PubMed] [Google Scholar]
- Haber J.E. (1998) The many interfaces of Mre11. Cell, 95, 583–586. [DOI] [PubMed] [Google Scholar]
- Hodgson C.P. and Fisk,R.Z. (1987) Hybridization probe size control: optimized ‘oligolabelling’. Nucleic Acids Res., 15, 6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes A. and Haber,J.E. (1999) Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell, 96, 415–424. [DOI] [PubMed] [Google Scholar]
- Ivanov E.L., Sugawara,N., White,C.I., Fabre,F. and Haber,J.E. (1994) Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol. Cell. Biol., 14, 3414–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakupciak J.P. and Wells,R.D. (1999) Genetic instabilities in (CTG.CAG) repeats occur by recombination. J. Biol. Chem., 274, 23468–23479. [DOI] [PubMed] [Google Scholar]
- Jankowski C., Nasar,F. and Nag,D.K. (2000) Meiotic instability of CAG repeat tracts occurs by double-strand break repair in yeast. Proc. Natl Acad. Sci. USA, 97, 2134–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S., Jaworski,A., Ohshima,K. and Wells,R.D. (1995) Expansion and deletion of CTG repeats from human disease genes are determined by the direction of replication in E.coli. Nature Genet., 10, 213–217. [DOI] [PubMed] [Google Scholar]
- Malkova A., Ross,L., Dawson,D., Hoekstra,M.F. and Haber,J.E. (1996) Meiotic recombination initiated by a double-strand break in rad50 delta yeast cells otherwise unable to initiate meiotic recombination. Genetics, 143, 741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer D.J., O’Callaghan,B.L. and Livingston,D.M. (1996) Orientation dependance of trinucleotide CAG repeat instability in Saccharomyces cerevisiae. Mol. Cell. Biol., 16, 6617–6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray C.T. (1999) DNA secondary structure: a common and causative factor for expansion in human disease. Proc. Natl Acad. Sci. USA, 96, 1823–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miret J.J., Pessoa-Brandao,L. and Lahue,R.S. (1997) Instability of CAG and CTG trinucleotide repeats in Saccharomyces cerevisiae. Mol. Cell. Biol., 17, 3382–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miret J.J., Pessoa-Brandao,L. and Lahue,R.S. (1998) Orientation-dependent and sequence-specific expansions of CTG/CAG trinucleotide repeats in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 95, 12438–12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitas M. (1997) Trinucleotide repeats associated with human diseases. Nucleic Acids Res., 25, 2245–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitas M., Yu,A., Dill,J. and Haworth,I.S. (1995a) The trinucleotide repeat sequence d(CGG)15 forms a heat-stable hairpin containing Gsyn.Ganti base pairs. Biochemistry, 34, 12803–12811. [DOI] [PubMed] [Google Scholar]
- Mitas M., Yu,A., Dill,J., Kamp,T.J., Chambers,E.J. and Haworth,I.S. (1995b) Hairpin properties of single-stranded DNA containing a GC-rich triplet repeat: (CTG)15. Nucleic Acids Res., 23, 1050–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore H., Greenwell,P.W., Liu,C.-P., Arnheim,N. and Petes,T.D. (1999) Triplet repeats form secondary structures that escape DNA repair in yeast. Proc. Natl Acad. Sci. USA, 96, 1504–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau S., Ferguson,J.R. and Symington,L.S. (1999) The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell. Biol., 19, 556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pâques F. and Haber,J.E. (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev., 63, 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pâques F., Leung,W.-Y. and Haber,J.E. (1998) Expansions and contractions in a tandem repeat induced by double-strand break repair. Mol. Cell. Biol., 18, 2045–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull T.T. and Gellert,M. (1998) The 3′ to 5′ exonuclease activity of Mre11 facilitates repair of DNA double-strand breaks. Mol. Cell, 1, 969–979. [DOI] [PubMed] [Google Scholar]
- Paull T.T. and Gellert,M. (1999) Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev., 13, 1276–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin A., Buckle,M. and Dujon,B. (1993) Asymmetrical recognition and activity of the I-Sce I endonuclease on its site and on intron–exon junction. EMBO J., 12, 2939–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronis A. (1996) Genomic imprinting in unstable DNA diseases. BioEssays, 18, 587–590. [DOI] [PubMed] [Google Scholar]
- Reddy P.S. and Housman,D.E. (1997) The complex pathology of trinucleotide repeats. Curr. Opin. Cell Biol., 9, 364–372. [DOI] [PubMed] [Google Scholar]
- Richard G.-F., Hennequin,C., Thierry,A. and Dujon,B. (1999a) Trinucleotide repeats and other microsatellites in yeasts. Res. Microbiol., 150, 589–602. [DOI] [PubMed] [Google Scholar]
- Richard G.-F., Dujon,B. and Haber,J. (1999b) High frequency of rearrangements of short CAG/CTG trinucleotide repeats in yeast induced by double-strand break repair. Mol. Gen. Genet., 261, 871–882. [DOI] [PubMed] [Google Scholar]
- Richards R.I. and Sutherland,G.R. (1997) Dynamic mutation: possible mechanisms and significance in human disease. Trends Biochem. Sci., 22, 432–436. [DOI] [PubMed] [Google Scholar]
- Sakamoto N., Chastain,P.D., Parniewski,P., Ohshima,K., Pandolfo,M., Griffith,J.D. and Wells,R.D. (1999) Sticky DNA: self-association properties of long GAA.TTC repeats in R.R.Y triplex structures from Friedreich’s ataxia. Mol. Cell, 3, 465–475. [DOI] [PubMed] [Google Scholar]
- Samadashwily G., Raca,G. and Mirkin,S.M. (1997) Trinucleotide repeats affect DNA replication in vivo. Nature Genet., 17, 298–304. [DOI] [PubMed] [Google Scholar]
- Sandell L.L. and Zakian,V.A. (1993) Loss of a yeast telomere: arrest, recovery and chromosome loss. Cell, 75, 729–739. [DOI] [PubMed] [Google Scholar]
- Sarkar P.S., Chang,H.-C., Boudi,F.B. and Reddy,S. (1998) CTG repeats show bimodal amplification in E.coli. Cell, 95, 531–540. [DOI] [PubMed] [Google Scholar]
- Schweitzer J.K. and Livingston,D.M. (1997) Destabilization of CAG trinucleotide repeat tracts by mismatch repair mutations in yeast. Hum. Mol. Genet., 6, 349–355. [DOI] [PubMed] [Google Scholar]
- Schweitzer J.K. and Livingston,D.M. (1998) Expansions of CAG repeat tracts are frequent in a yeast mutant defective in Okazaki fragment maturation. Hum. Mol. Genet., 7, 69–74. [DOI] [PubMed] [Google Scholar]
- Spiro C., Pelletier,R., Lahue,R.S., Gupta,G., Chen,X., Rolfsmeier,M.L., Dixon,M.J. and McMurray,C.T. (1999) Inhibition of flap processing by DNA secondary structure at trinucleotide repeats. Mol. Cell, 4, 1079–1085. [DOI] [PubMed] [Google Scholar]
- Usdin K. and Woodford,K.J. (1995) CGG repeats associated with DNA instability and chromosome fragility form structures that block DNA synthesis in vitro. Nucleic Acids Res., 23, 4202–4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui T., Ohta,T., Oshiumi,H., Tomizawa,J.-I., Ogawa,H. and Ogawa,T. (1998) Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell, 95, 705–716. [DOI] [PubMed] [Google Scholar]
- Wells R.D. (1996) Molecular basis of genetic instability of triplet repeats. J. Biol. Chem., 271, 2875–2878. [DOI] [PubMed] [Google Scholar]
- Yu A. and Mitas,M. (1995) The purine-rich trinucleotide repeat sequences d(CAG)15 and d(GAC)15 form hairpins. Nucleic Acids Res., 23, 4055–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A., Barron,M.D., Romero,R.M., Christy,M., Gold,B., Dai,J., Gray,D.M., Haworth,I.S. and Mitas,M. (1997) At physiological pH, d(CCG)15 forms a hairpin containing protonated cytosines and a distorted helix. Biochemistry, 36, 3687–3699. [DOI] [PubMed] [Google Scholar]