Abstract
We describe a simple and robust method to construct complex three-dimensional (3D) structures using short synthetic DNA strands that we call “DNA bricks”. In one-step annealing reactions, bricks with hundreds of distinct sequences self-assemble into prescribed 3D shapes. Each 32-nucleotide brick is a modular component; it binds to four local neighbors and can be removed or added independently. Each 8-base-pair interaction between bricks defines a voxel with dimensions 2.5 nanometers by 2.5 nanometers by 2.7 nanometers, and a master brick collection defines a “molecular canvas” with dimensions of 10 by 10 by 10 voxels. By selecting subsets of bricks from this canvas, we constructed a panel of 102 distinct shapes exhibiting sophisticated surface features as well as intricate interior cavities and tunnels.
Self-assembly of nucleic acids (DNA and RNA) provides a powerful approach for constructing sophisticated synthetic molecular structures and devices (1–31). Structures have been designed by encoding sequence complementarity in DNA strands in a manner such that, by pairing up complementary segments, the strands self-organize into a prescribed target structure under appropriate physical conditions (1). From this basic principle, researchers have created diverse synthetic nucleic acid structures (27–30) such as lattices (4, 6, 8–10, 25), ribbons (15), tubes (6, 15, 25, 26), finite two-dimensional (2D) and three-dimensional (3D) objects with defined shapes (2, 9–11, 13, 16–19, 22, 23, 26), and macroscopic crystals (20). In addition to static structures, various dynamic systems have been constructed (31), including switches (5), walkers (7, 14, 21), circuits (12, 14, 24), and triggered assembly systems (14). Additionally, as DNA and RNA can be interfaced with other functional molecules in a technologically relevant fashion, synthetic nucleic acid structures promise diverse applications; researchers are using nucleic acid structures and devices to direct spatial arrangement of functional molecules (6, 25, 32–34), to facilitate protein structure determination (35), to develop bioimaging probes (33, 34), to study single-molecule biophysics (36), and to modulate biosynthetic and cell-signaling pathways (25, 37).
An effective method for assembling megadalton nanoscale 2D (11) and 3D shapes (16–19, 23) is DNA origami (29), in which a long “scaffold” strand (often a viral genomic DNA) is folded to a predesigned shape via interactions with hundreds of short “staple” strands. However, each distinct shape typically requires a new scaffold routing design, and the synthesis of a different set of staple strands. In contrast, construction from standardized small components (e.g. DNA tiles) that each can be included, excluded, or replaced without altering the rest of the structure – i.e. modular assembly – offers a simpler approach to constructing shapes. In addition, if all components are short strands that can be chemically synthesized, the resulting structures would have greater chemical diversity than DNA origami, which typically contains half biological material (the scaffold) in mass and half synthetic material (the staples). A variety of structures have been assembled using DNA (3, 4, 6, 8, 10, 13, 15, 20) and RNA (9, 22, 25) tiles, including periodic (4, 6, 25) and algorithmic (8) 2D lattices, extended ribbons (15) and tubes (6, 15, 25), 3D crystals (20), polyhedra (13, 22), and finite two-dimensional shapes (9, 10). However, modular self-assembly of finite-size, discrete DNA structures has generally lacked the complexity that DNA origami can offer.
Only recently have researchers demonstrated finite complex 2D shapes (26) self-assembled from hundreds of distinct single-stranded tiles (SST) (15). Unlike a traditional multi-stranded tile (3, 4, 6, 8–10, 13, 20, 25), which is a well-folded, compact structure displaying several sticky ends, an SST is a floppy single-strand DNA composed entirely of concatenated sticky ends. In one-pot reactions, hundreds of SST self-assemble into desired target structures mediated by inter-tile binding interactions; no scaffold strand is required. The simplicity and modularity of this approach allowed the authors to build more than 100 distinct shapes by selecting subsets of tiles from a common 2D molecular “canvas”. This latest success has challenged previous thinking that modular components, such as DNA tiles, are not suitable for assembling complex, uniquely addressable shapes (38). This presumption was largely based on a supposed, technically challenging requirement for perfect strand stoichiometry (i.e. the relative ratio of the strands). Deviations from equality were expected to result in predominating partial structure formation (38). The surprising success of SST assembly may have bypassed this challenge via putative slow and sparse nucleation followed by fast growth (26), such that a large number of particles complete their formation well before depletion of the component strand pool.
Here we generalize the concept of single-stranded “tiles” to “bricks” and thus extend our modular-assembly method from 2D to 3D. A canonical DNA brick is a 32-nucleotide single-strand with four 8-base binding domains (sticky ends). In simple one-step annealing reactions, prescribed target 3D structures self-assemble robustly from hundreds of unpurified brick strands that are mixed together with no tight control of stoichiometry. The modularity of our method enabled the construction of 102 distinct structures by simply selecting subsets of bricks from a common 3D cuboid “molecular-canvas” consisting of 1000 voxels (39, fig. S1); each voxel fits 8 base-pairs and measures approximately 2.5 nanometers by 2.5 nanometers by 2.7 nanometers. These structures include solid shapes with sophisticated geometries and surface patterns, and hollow shapes with intricate tunnels and enclosed cavities. Additionally, we have constructed structures with alternative packing geometries or using non-canonical brick motifs, demonstrating the method’s versatility. The work here thus establishes DNA bricks as a simple, robust, modular, and versatile framework for constructing complex 3D nano-structures using only short synthetic DNA strands. More generally, it demonstrates how complex 3D molecular structures can be assembled from small, modular components mediated strictly by local binding interactions.
Design of DNA-brick structures and a 3D “molecular canvas”
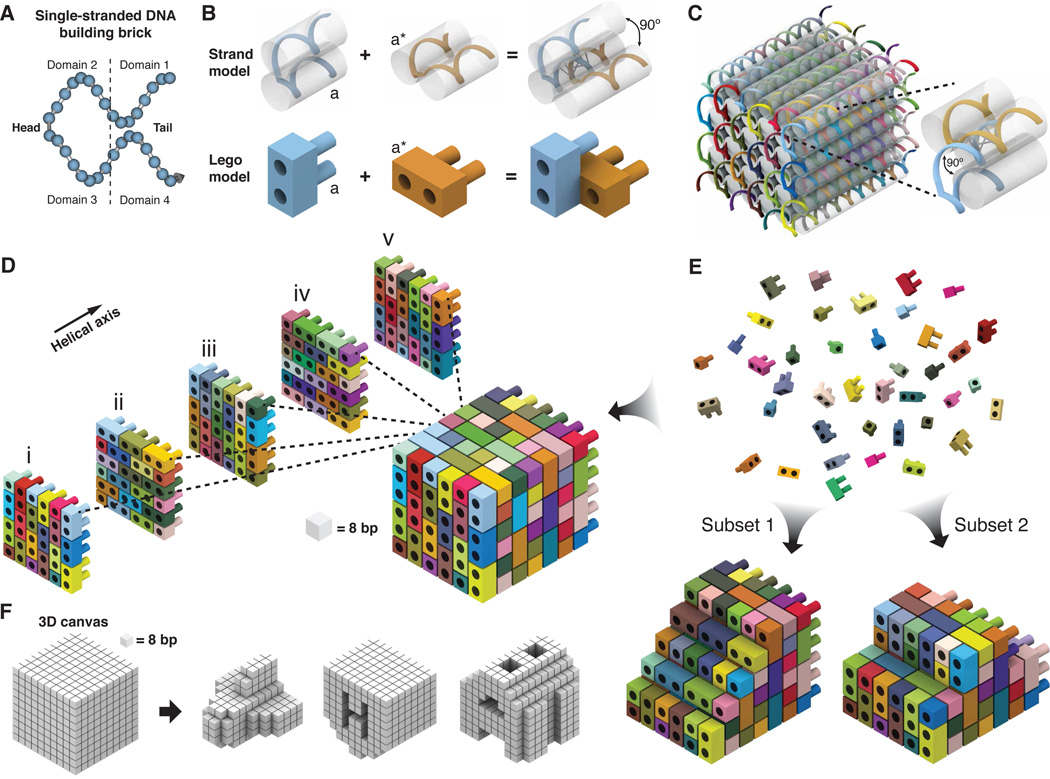
In our design, a DNA brick is a 32-nucleotide strand that we conceptualize as four consecutive 8-nucleotide domains (Fig. 1A). Each DNA brick bears a distinct nucleotide sequence. All DNA bricks adopt an identical shape when incorporated into the target structure: two 16-nucleotide antiparallel helices joined by a single phosphate linkage. The two domains adjacent to the linkage are designated as “head” domains and the other two are designated as “tail” domains. A DNA brick with a tail domain bearing sequence “a” can interact productively with a neighboring brick with a complementary “a*” head domain in a stereospecific fashion. Each pairing between bricks defines three parallel helices packed to produce a 90° dihedral angle (Fig. 1B, top); this angle derives from the approximate 3/4 right-handed helical twist of 8 base-pairs of DNA.
Fig. 1.
Design of DNA brick structures. (A) A 32-nucleotide four-domain single-stranded DNA brick. Each domain is 8 nucleotides in length. The connected domains 2 and 3 are “head” domains; domains 1 and 4 are “tail” domains. (B) Each two-brick assembly forms a 90° dihedral angle via hybridization of two complementary 8-nucleotide-domains “a” and “a∗”. (C) A molecular model that shows the helical structure of a 6H × 6H × 48B cuboid 3D DNA structure. Each strand has a unique sequence, as indicated by a distinct color. The inset shows a pair of bricks. (D) A Lego-like model of the 6H × 6H × 48B cuboid. Each brick has a unique sequence. The color use is consistent with (B). Half-bricks are present on the boundary of each layer. (E) The 6H × 6H × 48B cuboid is self-assembled from DNA bricks. The bricks are not interchangeable during self-assembly because of the distinct sequence of each brick. Using the 6H × 6H × 48B as a 3D “molecular canvas”, a smaller shape can be designed using a subset of the bricks. (F) 3D shapes designed from a 10 × 10 × 10 voxel “3D canvas”; each voxel fits 8 base-pairs (2.5 nm × 2.5 nm × 2.7 nm).
We introduce a Lego-like model to depict the design in a simple manner (Fig. 1B, bottom). The model intentionally overlooks the detailed helical structure and strand polarity but preserves the aspect ratios and some of the orientational constraints on interactions between DNA bricks: the two protruding round plugs, pointing in the same direction as the helical axes, represent the two tail domains; the two connected cubes with recessed round holes represent the two head domains. A brick must adopt one of two classes of orientation, horizontal or vertical (Fig. 1B). The two bricks connect to form a 90° angle via hybridization, represented as the insertion of a plug into a hole. An insertion is only allowed between a plug and a hole that carry complementary sequences with matching polarity (which is not graphically depicted in the current model for expositional simplicity). In fig. S2, we present a more detailed Lego-like model that explicitly tracks the polarity of the DNA bricks and their stereospecific interaction pattern.
Structural periodicities of the design are illustrated in a 6H(helix) × 6H(helix) × 48B(bp) cuboid structure (Figs. 1C, D). Bricks can be grouped into 8-base-pair layers that contain their head domains. Bricks follow a 90° counterclockwise rotation along successive 8-base-pair layers resulting in a repeating unit with consistent brick orientation and arrangement every four layers. For example, the first and fifth 8-base-pair layers in Fig. 1D share the same arrangement of bricks. Within an 8-base-pair layer, all bricks share the same orientation and form a staggered arrangement to tile the layer. On the boundary of each layer, some DNA bricks are bisected to half-bricks, representing a single helix with two domains. The cuboid is self-assembled from DNA bricks in a one-step reaction. Each brick carries a unique sequence that directs it to fit only to its predesigned position. Because of its modular architecture, a predesigned DNA brick structure can be used for construction of smaller custom shapes assembled from subsets of DNA bricks (Fig. 1E). See figs. S3 and S4 for detailed strand diagrams for the DNA brick structures.
3D “molecular canvas”
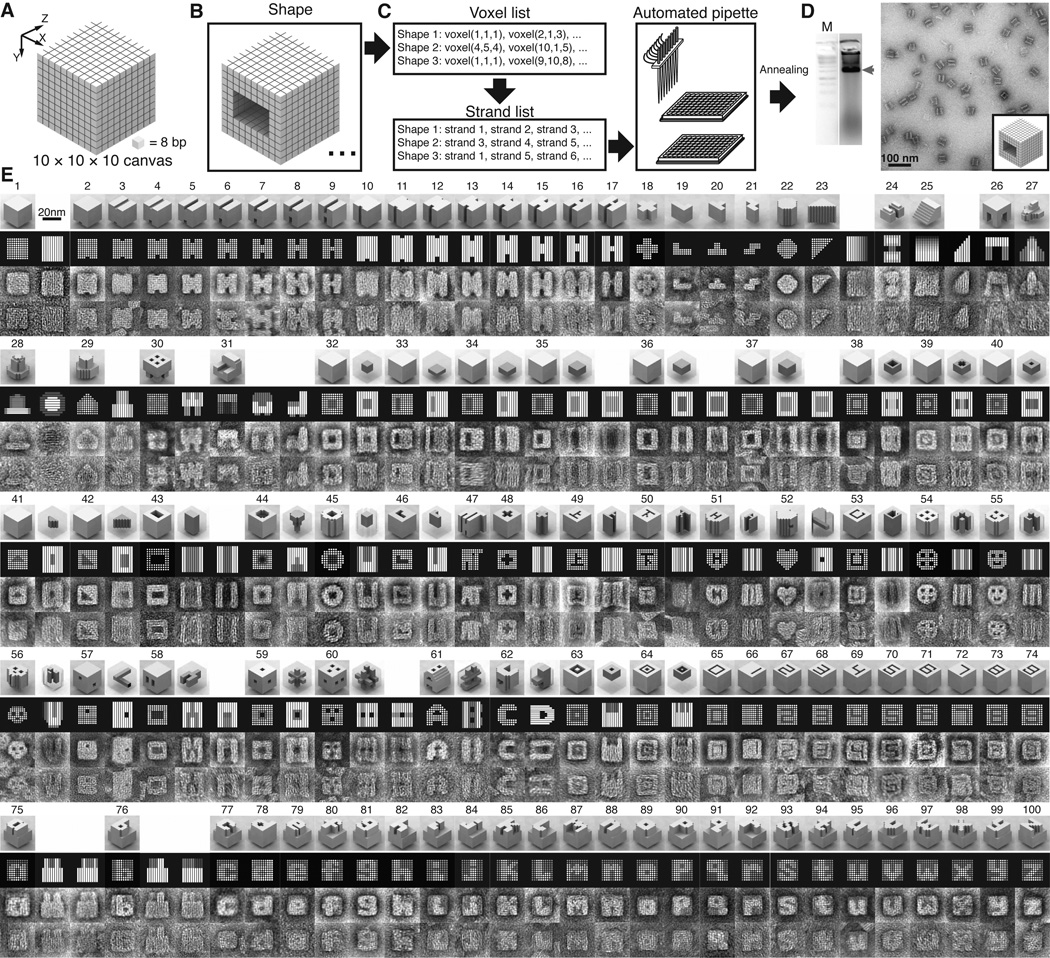
The Lego-like model can be further abstracted to a 3D model that contains only positional information of each 8-base-pair duplex. A 10H × 10H × 80B cuboid is conceptualized as a 3D “molecular canvas” that contains 10 × 10 × 10 voxels. Each voxel fits an 8-base-pair duplex and measures 2.5 nm × 2.5 nm × 2.7 nm (Fig. 1F). Based on the 3D canvas, a computer program first generates a full set of DNA bricks, including full-bricks and half-bricks that can be used to build a prescribed custom shape. Using 3D modeling software, a designer then needs only to define the target shapes by removing unwanted voxels from the 3D canvas – a process resembling 3D sculpting. Subsequently, the computer program analyzes the shape and automatically selects the correct subset of bricks for self-assembly of the shape.
Self-assembly of DNA-brick cuboid structures
Using the above design strategy, we constructed a wide range of DNA brick structures. The detailed Methods and Materials can be found in Supplementary Material I, and the DNA sequences can be found in Supplementary Tables S1–S20 in Supplementary Materials II (39). We first constructed 3D cuboid structures of a variety of sizes and aspect ratios (Fig. 2).
Fig. 2.
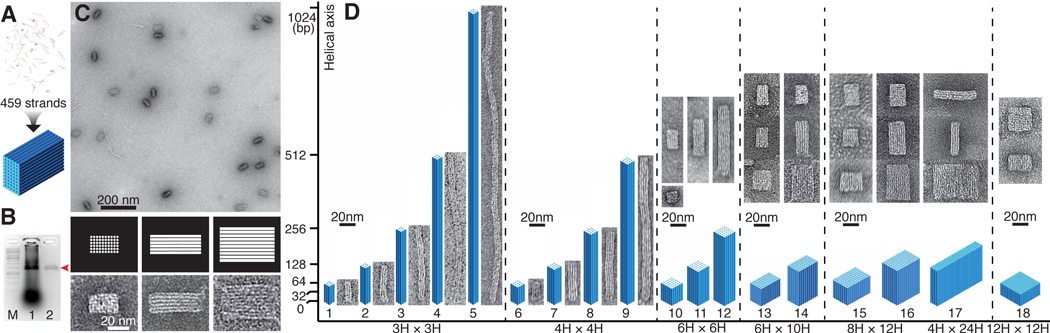
Cuboid structures self-assembled from DNA bricks. (A) DNA bricks self-assembled into a 6H × 10H ×128B cuboid in a one-step thermal annealing process. (B) Agarose gel electrophoresis showing 50% purification recovery efficiency of the 6H × 10H × 128B cuboid. Lane M contains the 1 kb ladder. Lanes 1 and 2 contain unpurified and purified 6H × 10H × 128B cuboid structures, respectively. The red arrow points to the cuboid product band. (C) TEM images of gel purified 6H × 10H × 128B cuboid. Zoomed-in images (bottom) and corresponding computer-generated graphics (middle) show three different projection views. (D) Designs and TEM images of 18 cuboids of a variety of dimensions. Horizontal axis is labeled with the cross-section dimensions of the cuboids; vertical axis is labeled with the lengths of the constituent helices. The lengths are 48B (shape 18), 64B (shapes 1, 6, 10, 13, 15), 120B (shapes 16,17), 128B (shapes 2, 7, 11, 14), 256B (shapes 3, 8, 12), 512B (shapes 4, 9), and 1,024B (shape 5). Each 3D cylinder model is drawn proportionally to the relative dimensions of the cuboid; corresponding TEM images are shown on the right or above.
Random sequence design
The sequences of DNA bricks were designed by random assignments of base pairs (A-T, C-G) to 3D structures. We first tested two versions of a 6H × 6H × 64B cuboid, with either random sequences or specially designed sequences (designed by smoothing binding energy, minimizing undesired secondary structure, and reducing sequence symmetry), and observed comparable self-assembly yields (fig. S5). We also tested three sets of random sequences using a 4H × 12H × 120B cuboid and again observed similar assembly yields (figs. S6, S7, see fig. S8 for more discussion on domain similarity of random sequence design). Thus, random sequences were applied to all subsequent designs.
Protector bricks
Including unpaired single-strands at the ends of DNA duplexes has proven to be effective for mitigating unwanted aggregation that results from blunt-end stacking (11). An 8-nucleotide single-stranded domain protruded out from every 5' or 3' end of all DNA duplexes in our 3D structure designs (Fig. 1C). The sequences of these 8-nucleotide domains were replaced with eight continuous thymidines to further prevent undesired non-specific binding interactions between exposed single-stranded domains. DNA bricks with modified head or tail poly-T domains are named “head protectors” or “tail protectors,” respectively.
Boundary bricks
A 16-nucleotide half-brick could be merged with a preceding 32-nucleotide full-brick along the direction of its helix, to form a 48-nucleotide strand (figs. S9–S11). We observed a 1.4-fold improvement in assembly yield for a 6H × 6H × 64B cuboid when this 48-nucleotide boundary-strand design was implemented, possibly reflecting accelerated nucleation of target structure formation. Hence, this merge strategy was applied to all our 3D structures.
Assembly and characterization of 6H × 10H × 128B cuboid
For a detailed characterization study, we constructed a 6H × 10H × 128B cuboid (Fig. 2A). It consists of 459 strands (7,680 base-pairs, with a comparable molecular weight to an M13-based DNA origami, see figs. S12, S13 for design details). Unpurified DNA strands were mixed together at nominally equal ratios without careful adjustment of stoichiometry (39). To determine the optimal assembly conditions, we tested two annealing ramps (24-hour annealing and 72-hour annealing), two strand concentrations (100 and 200 nM per strand), and eight MgCl2 concentrations (10, 20, 30, 40, 50, 60, 70, 80 mM). Equal amounts of each sample (2 pmol per strand) were then subjected to EtBr-stained 2% agarose gel electrophoresis (fig. S14). The best gel yield (about 4% as calculated by Yield = Measured mass of product/Mass of all strands) was achieved at the following conditions: 200 nM per strand, 72-hour annealing, 40 mM MgCl2 (fig. S15). It is worth noting that the above gel yield reflects only an approximate estimate for the incorporation ratio of the monomer strands (also see discussion in (26)).
For comparison, 4% to 14% gel yield was reported for 3D DNA origami with similar size and aspect ratios (e.g. the 10H × 6H × 98B and other origami cuboids in (40)). It is worth noting that the origami gel yield was estimated as Yield = (Scaffold strands incorporated into product/Total scaffold strands); the loss of excessive staple strands (normally 5- to 10-fold more than the scaffold strand) was not taken into account. For DNA bricks, the optimal 40 mM MgCl2 was higher than the optimal MgCl2 concentration for 3D origami folding, which typically is below 30 mM (18). Column-purified DNA bricks product (~50% recovery efficiency, Fig. 2B) migrated as a single band on agarose gel and appeared under transmission electron microscopy (TEM) with expected morphology (Fig. 2C) and measured dimensions of 0.34 nm (± 0.01 nm SD) per base pair and 2.5 nm (± 0.2 nm SD) per helix width. For the gel-purified product, “the percentage of intact structures” was estimated at 55% by counting the ratio of intact particles over all the particles in TEM images (fig. S16). This percentage of intact structures is comparable to the previously reported percentages of 3D square-lattice DNA origami (27% for a 6H × 12H× 80B cuboid, 59% for an 8H × 8H × 96B cuboid) (41).
Special designs can be applied to increase the assembly yield of the 6H × 12H× 80B cuboid. “Head protectors” and “tail protectors” appeared especially unstable because half of their 8-nucleotide domains are unpaired. By merging “head protectors” of the 6H × 10H × 128B cuboid with their neighboring strands (figs. S17, S18), a modified version 6H × 10H × 128B-M cuboid was obtained and showed 190% improvement in gel assembly yield and 17% improvement in the percentage of intact structures under TEM over the standard 6H × 10H × 128B cuboid (fig. S19). Thus, 3D structures can be further stabilized using special design rules such as this merging strategy. However, this modification requires deletions of crossovers between helices, which may potentially create global or local deformations, and was not used for constructions in the remainder of the paper.
Structures of different sizes
Eighteen distinct cuboid structures that contain 9, 16, 36, 60, 96, and 144 helices were designed, annealed using the optimal conditions previously identified for the 6H × 10H × 128B cuboid self-assembly, and characterized by gel and TEM (Fig. 2D, fig. S20). Additional TEM images are shown in figs. S21–S27. Measured dimensions of intact particles for each structure agree with the designs (fig. S28). Gel yields varied from less than 1% to about 80% (figs. S20C, S28). For structures with the same number of helices, smaller cuboids exhibited higher assembly yields. The highest yield (80%) was observed for the smallest object, the 3H × 3H × 64B cuboid; the lowest yields (< 1%) were observed for the 8H × 12H × 120B, 4H × 24H × 120B, and 12H × 12H × 48B cuboids. The biggest DNA objects constructed in this paper are an 8H × 12H × 120B cuboid (formed by 728 strands) and a 4H × 24H × 120B cuboid (formed by 710 strands), which are identical in molecular weight (24,576-nucleotide, 8-megadalton, 60% more massive than an M13-based DNA origami). Increasing the concentration for the brick strands helped to increase the yield for a small cuboid, 4H × 4H × 128B (fig. S29). In some cases, higher molecular weight bands can be detected above the product band; these bands are likely multimers caused by non-specific interactions between assembled products. For example, for the 6H × 10H × 64B structure, TEM revealed an upper band contained dimers of the cuboids (fig. S30). Cuboids with 32-base-pair (32B) helices were also tested but failed to assemble (fig. S20). This is likely due to the fact that these cuboids contained only one crossover between each pair of neighboring helices, and hence were less stable.
Complex shapes made from a 10 × 10 × 10 voxel 3D canvas
Using the 10 × 10 × 10 voxel 3D canvas (Fig. 1F, Fig. 3A, fig. S31), we next constructed 102 distinct shapes (Fig. 3), demonstrating the modularity of the DNA brick strategy.
Fig. 3.
Shapes made from a 3D “molecular canvas”. (A) A 10 × 10 × 10 voxel 3D canvas. Z-axis is the helical axis. Each voxel (8 base-pairs) measures 2.5 nm × 2.5 nm × 2.7 nm. (B) Shapes are designed by editing voxels using 3D modeling software. (C) A computer program recognizes the voxel composition of each shape and generates a list of strands to form this shape. The list then is used to direct an automated liquid-handling robot to mix the strands. (D) After annealing, the shapes are characterized by agarose gel electrophoresis and TEM imaging. Lane M contains the 1 kb ladder. The product band is indicated by the red arrow. (E) Computer-generated models and TEM images of shapes. The top row for each shape depicts a 3D model, followed by a computer-generated projection view, an image averaged from 6 different particles visualized using TEM, and a representative raw TEM image. More raw images are shown in figs. S38–54. In a number of cases, multiple projections are presented. Some shapes with cavities or tunnels are depicted with additional transparent 3D views that highlight the deleted voxels (colored dark gray). For example, the top-right model of shape 32 shows the enclosed cuboid cavity.
DNA bricks and derivatives
Any brick in the 3D canvas can become either a boundary half brick (i.e. exposed at the edges of a layer and bisected, Fig. 1D), a protector brick (i.e. located in the first or last layer along Z-axis), or even both at same time, in a custom shape design. Thus, modified versions of each brick were generated with all combinations of domain-deletion (i.e. bisect to a half-brick), polythymidine-sequence-substitution (i.e. change to protector bricks), and boundary-brick-merger (i.e. change to 48-nucleotide boundary bricks) to accommodate the possibilities (fig. S32, note that two types of strands with low occurring frequency and four types of strands with only one binding domain were excluded in our implementation). Overall, a master collection of 4,455 strands (with a total of 138,240 nucleotides) were generated by a computer program to guarantee that a designed shape could be assembled with head/tail protector bricks and 48-nucleotide boundary bricks. Custom shapes were assembled via selecting subsets of the master collection without synthesizing new strands.
Automated design process
By rendering the 3D canvas using 3D modeling software, we can edit voxels and visualize a shape using a graphical user interface (Fig. 3B). Then, the voxel information of multiple shapes is interpreted by a custom program to generate a list of strands involved in the formation of each shape. This list is subsequently processed to direct an automated liquid-handling robot to select DNA strands from source plates and pipette them to the wells of a product plate, mixing strands for many shapes in a high throughput manner (Fig. 3C). The strands will be subsequently annealed in separate test tubes to produce the desired structures (Fig. 3D). The complete design workflow is shown in figs. S33, S34. To utilize existing computational tools previously developed by other researchers, we can also convert shapes to caDNAno files (40). Each shape’s conformation then can be simulated using CanDo (42), a software tool for computing 3D structures of DNA origami (fig. S35).
Using the 3D canvas and following the automated design process, we successfully constructed 102 distinct shapes (gels in fig. S36, S37; TEM images of shapes 1–100 in Fig. 3E; raw TEM images for all the shapes in fig. S38–S54).
Shapes 1–17
The basic design constraints were studied using a group of shapes containing two 4H × 10H × 80B blocks connected by a middle “connecting block” (shapes 2–17). The connecting blocks were two-voxel wide along X-axis and systematically designed to possess decreasing numbers of voxels along Y-axis (shapes 2–9) or Z-axis (shapes 10–17). Eliminating voxels along the X-axis should have the same effect as eliminating voxels along the Y-axis because of the shape symmetry. Agarose gel electrophoresis revealed that, in both systems, as the connector became overly thin, the gel yields for the intact structures decreased and partial structures (putative unconnected 4H × 10H × 80B blocks) became more prominent (e.g. in lanes for shapes 8, 9, 15–17 in fig. S36). However, reducing the number of voxels along the Z-axis appeared to decrease the yield more significantly than along the Y-axis. Shape 9, which contained only a 2-voxel connection along the Y-axis, gave 6% gel yield. In contrast, the yield for shape 17 (2-voxel along the Z-axis) dropped to 1%. Overall, these observations suggest safe design criteria of at least two continuous voxels along the X-axis or Y-axis (2 helices) and three Z-axis voxels (24 base-pairs) for stable features. However, as demonstrated in following experiments, smaller features (e.g. two Z-axis voxels, shapes 33–37; one X-axis or Y-axis voxel, shapes 64–74) can still stably exist in certain shapes where these features are presumably reinforced by other voxels in close proximity.
Solid shapes 18–31
A number of solid shapes were designed including Z-direction extrusions of simple geometric shapes (shapes 18–23) and more intricate objects (shapes 24–31; also see shape 102 in fig. S54). Gel yields and TEM images of these objects provided more knowledge of the design space of our methodology. For example, shapes 26 and 27, which both contained 3-helix thick appendages anchored only on one edge, were occasionally found without these protrusions or with them but containing defects (data not shown). Thus, such thin features, though obeying our design criteria, appeared to be less stable than the better-supported or thicker features.
Closed-cavity shapes 32–42
Previously, a few examples of 3D DNA origami with closed cavities were demonstrated, including a box (16), a tetrahedron (17), a sphere and an ellipsoid (23). Here we created a series of “empty boxes” with different sizes of cuboid cavities (shapes 32–37) as well as more intricate cavity shapes (e.g. square ring, cross, and triangle; shapes 38–42).
Open-cavity shapes 43–62
We constructed shapes with a single open-cavity (i.e. tunnel) of varying width, depth, and geometry (shapes 43–53) and multiple-parallel cavities (shapes 54–56). Shapes with non-crossing perpendicular tunnels (shape 57), turning and branching tunnels (shape 58), and crossing tunnels (shapes 59, 60; also see shape 101 in fig. S54) were also demonstrated. Furthermore, we constructed tunnel-containing cuboids with modified outer surfaces to create varying external views from different angles, as demonstrated by shapes 60–62.
Features-on-solid-base shapes 63–100
Sophisticated features were designed on a solid base, including a full set of 10 Arabic numerals (shapes 65–74) and 26 lowercase letters for the English alphabet (shapes 75–100). Two concentric ring structures (shapes 63, 64) and the numerals (shapes 65–74) contained features as thin as one voxel (2.5 nm), suggesting that the design criteria (e.g. thin structures tend to fail) are contingent on the surrounding environment of a particular feature. These shapes also highlight the capacity of creating extruded features that would otherwise be unattainable via 2D assembly (26).
For most shapes, assembly yields were between a few percent and 30% (figs. S36, S37; by comparison, yields of five 3D DNA origami structures were reported as 7% to 44% (18)). Only five shapes had assembly yields higher than 30%; three shapes had assembly yields lower than 1%.
In spite of our success in making a variety of intricate 3D shapes, some shapes exhibited undesired properties. For example, shapes 60–62 only showed less than 1% of intact particles in TEM images; some fine features of a shape (e.g. the two wings of shape 27) could be damaged or even completely missing if the shape was extracted from an agarose gel band. We also observed four failed designs that did not produce clear product bands on agarose gels (fig. S55A). Two features-on-solid-base designs showed strong bands on agarose gels (fig. S55B), and were of the expected size in TEM images. However, their features were not clearly resolved under TEM, suggesting that the shapes may have formed but the features were too subtle to be visualized.
Generality of DNA brick self-assembly
To explore the generality of the DNA brick assembly framework, we constructed structures with brick motifs other than the 32-base canonical brick motif. These structures include those with alternative lattice geometries that have been previously demonstrated by DNA origami (11, 18, 43).
Single-layer (2D) structures
Conceptually, a single-layer structure can be constructed by “extraction” of a layer from a 3D brick structure (Fig. 4A, also see fig. S56 for comparison with a 2D single-stranded tile rectangle design (26)). A 30H × 1H × 126B rectangle was intentionally modified to be 10.5 base pairs per turn instead of 10.67 base pairs per turn (for 3D design), in order to get a relatively flat structure (fig. S57). Gel yield was estimated to be 18% (fig. S58), comparable to 2D single-stranded tile structures (26). TEM (Fig. 4B) and AFM (Fig. 4C) revealed expected rectangle structures. Based on AFM images, the dimensions were measured as 0.31 nm (± 0.01 nm SD) per base-pair and 2.6 nm (± 0.3 nm SD) per helix width.
Fig. 4.
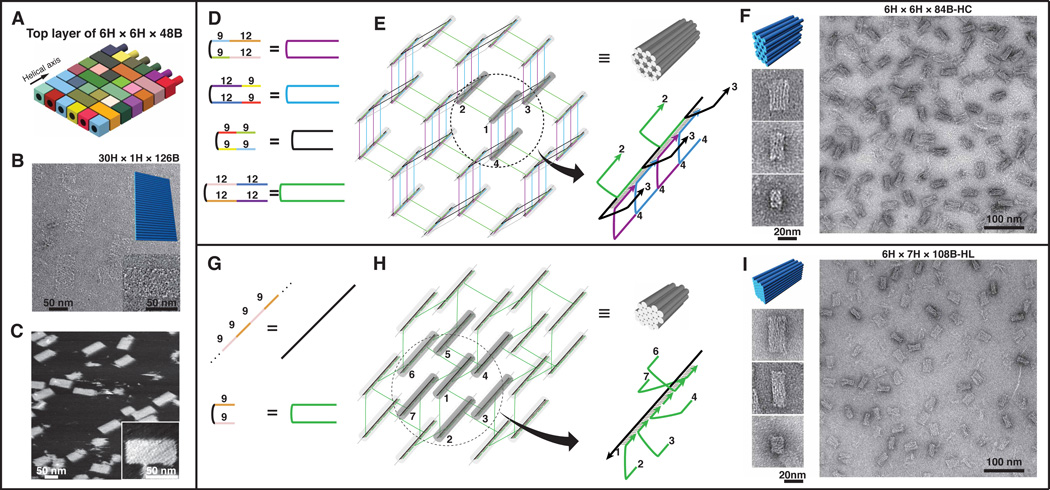
Generality of DNA brick self-assembly. (A–C) depict the design and construction of a single-layer brick structure. (A) DNA bricks of the top layer of the 6H × 6H × 48B cuboid in Fig. 1D, with the crossovers to the layer below removed. (B) TEM images of a 30H × 1H × 126B rectangle. Top-right inset shows the model of the design. Bottom-right inset contains a zoomed-in image of the structure. (C) AFM images of the 30H × 1H × 126B rectangle. Inset contains a zoomed-in image of the structure. (D–I) depict the designs and constructions of 3D honeycomb-lattice (D–F) and hexagonal-lattice (G–I) brick structures. (D, G) The strands used for honeycomb-lattice (D) and hexagonal-lattice (G) self-assembly. Numbers in the left panels indicate the number of nucleotides in a domain. (E, H) Strand diagrams of an 84-base-pair honeycomb-lattice structure (E) and a 54-base-pair hexagonal-lattice structure (H). The right-bottom image depicts an enlarged image of the circled helix bundle. Strand colors match those described in the right side of (D) or (G). Numbers indicate DNA helices. Note helix 1 and helix 5 are not directly connected in (H). The 9-base half-bricks were merged with their neighboring bricks to form longer strands. (F, I) TEM images of a 6H × 6H × 84B-HC hexagonal-lattice structure (F) and a 6H × 7H × 108B-HL 3D hexagonal-lattice structure (I). 3D model and zoomed-in images of different projection views are shown on the left.
3D honeycomb-lattice structures
We then created 10.8-base-pair/turn (33.3° twist per base-pair) honeycomb-lattice (HC) and hexagonal-lattice (HL) DNA structures. Four types of four-domain DNA strands were designed for honeycomb-lattice structures (Figs. 4D, E). A 6H × 6H × 84B-HC structure was successfully constructed and characterized (Fig. 4F, fig. S59). Particles in TEM images were measured to be 13 nm (± 0.9 nm SD) × 22 nm (± 1.0 nm SD) × 29 nm (± 1.2 nm SD). Assembly yield was estimated to be 30% (fig. S60).
3D hexagonal-lattice DNA structures
Two types of strands are used to build a hexagonal-lattice structure: a linear strand with multiple 9-nucleotide domains and an 18-nucleotide strand with two 9-nucleotide domains that are connected by a crossover (Figs. 4G, H). A 6H × 7H × 108B-HL structure was constructed and characterized (Fig. 4I, fig. S61). Particles in TEM images were measured to be 13 nm (± 0.8 nm SD) × 18 nm (± 1.1 nm SD) × 35 nm (± 2.2 nm SD). Assembly yield was estimated to be 26% (fig. S62).
Other brick motifs
We also constructed a 6H × 10H × 64B cuboid that arranges brick strands in an alternating fashion between layers (figs. S63, S64), and two 6H × 6H × 64B cuboids that implement two other brick motif designs (figs. S65, S66). One design is based on “chopping” the scaffold of a DNA origami to short strands (fig. S65A). The other adopts standardized motifs that are each 32-nucleotide long and has two crossovers (fig. S65B). These designs further demonstrate the versatility of DNA brick self-assembly.
Discussion
DNA bricks provide a simple, modular, and robust framework for assembling complex structures from short strands. Simplicity. A canonical brick is a standardized 32-nucleotide single strand composed of four 8-nucleotide binding domains; bricks interact via simple local binding rules. Modularity. With no scaffold present, an assembly of bricks has a modular architecture; each brick can be added or removed independently. Robustness. The assembly process is robust to variations in sequence composition (random sequences are used), strand synthesis (unpurified strands suffice) and stoichiometry (no tight control is required). Together, the simple and standardized motif, modular architecture, and robust performance permit straightforward automation of the design and construction process. A software tool takes as input a 3D shape specification and directs a liquid-handling robot to select and mix pre-synthesized brick strands to form the shape. Using a 1000-voxel canvas, 102 diverse shapes were rapidly prototyped. These shapes demonstrate a new level of geometrical sophistication, as exemplified by the intricate tunnel and cavity features.
The DNA brick framework is not restricted to the canonical 32-nucleotide motif and can be generalized to include various other motifs (Fig. 4), enabling the construction of 3D structure with diverse lattice packing geometries. In addition, previously demonstrated single-stranded tiles (15, 26) can be viewed as a special case of bricks where each pair of neighboring bricks form a 180° angle. For comparison, in hexagonal-, square-, and honeycomb-lattice structures, neighboring bricks form 60°, 90°, and 120° angles, respectively. These different angles are achieved by changing the domain lengths of bricks. Furthermore, neighboring bricks may be merged into a longer strand, which may facilitate nucleation or strengthen structurally weak positions. The DNA brick (and single-stranded tile) method differs from previous multi-stranded tiles in that each brick monomer is a floppy single-strand and only folds into a brick-like shape when incorporated into the assembly. It also differs from DNA origami by not using a scaffold strand. However, DNA origami can also be related to the brick framework, where half of the bricks are concatenated into a long scaffold (fig. S65A). The successes of constructions that use only short strands (as in bricks) and those that includes a long scaffold (as in origami) together suggest a full spectrum of motif possibilities with strands of diverse lengths: longer strands may provide better structural support, and shorter ones finer modularity and features; the eclectic use of both may lead to the most rapid progression towards greater complexity.
The DNA brick structures constructed here are still far below the size limit allowed by sequence uniqueness. Making the conservative assumption (by neglecting the contribution of cooperativity) that every domain must display a different sequence, a structure using canonical 32-nucleotide, 4-domain bricks could potentially reach a size of 8-nucleotide × 48 (524,288 nucleotides). In our experiments, the assembly process appeared to tolerate (sparse) identical domains (fig. S8), further expanding the potential obtainable size. Further exponential increase in size could potentially be achieved by using bricks with longer domains, or by encoding algorithmic growth patterns (8) in the assembly. However, in practice, low yields were already observed for larger designs (up to 24,576 nucleotides attempted thus far). Solving this challenge may require improvements in structure and sequence design, enzymatic synthesis for higher-quality strands, optimized thermal or isothermal (44) annealing conditions, and a detailed understanding and perhaps explicit engineering of the kinetic assembly pathways (8, 14, 44) of DNA brick structures.
The DNA brick structure, with its modular architecture, sophisticated geometry control, and synthetic nature, will further expand the range of applications and challenges that nucleic acid nanotechnology has already started to address, e.g. to arrange technologically relevant guest molecules into functional devices (6, 25, 32–34), to serve as programmable molecular probes and instruments for biological studies (33, 34, 36), to render spatial control for biosynthesis of useful products (25), to function as smart drug delivery particles (37), and to enable high-throughput nanofabrication of complex inorganic materials for electronics or photonics applications (6, 32). The modularity of the brick structure may facilitate rapid prototyping of diverse functional nano-devices. Its sophisticated and refined geometrical control may enable applications that require high precision arrangements of guest molecules. As the brick structure is composed entirely of short synthetic strands (i.e. no biologically-derived scaffold), it is conceivable to make bricks using synthetic informational polymers other than the natural form of DNA. Such polymers may include L-DNA, DNA with chemically modified backbones or artificial bases, chemically synthesized or in vitro (or even in vivo) transcribed RNA. This material diversity may potentially produce nanostructures with not only prescribed shapes but also designer (bio-)chemical properties (e.g. nuclease resistance, reduced immunogenicity) that would be useful for diverse applications requiring the structure to function robustly in complex environments, e.g. in living cells or organisms.
Supplementary Material
Acknowledgments
The authors thank M. Dai for technical assistance; E. Winfree and B. Wei for discussions; D. Pastuszak for assistance in draft preparation. This work is supported by an ONR Young Investigator Program Award N000141110914, an ONR Grant N000141010827, an ARO Grant W911NF1210238, an NSF CAREER Award CCF1054898, an NIH Director’s New Innovator Award 1DP2OD007292, and a Wyss Institute Faculty Startup Fund to P.Y., and by a Wyss Institute Faculty Grant, ONR Grants N000014091118 and N000141010241, and an NIH Director’s New Innovator Award 1DP2OD004641 to W.M.S.. L.L.O. is supported by an NSF graduate research fellowship.
Footnotes
Y.K. conceived the project, designed and performed the experiments, analyzed the data, and wrote the paper; L.L.O. designed and performed the experiments, analyzed the data, and wrote the paper; W.M.S. conceived the project, discussed the results, and wrote the paper; P.Y. conceived, designed, and supervised the study, interpreted the data, and wrote the paper. The DNA sequences for the nanostructures can be found in Supplementary Materials II. A provisional patent has been filed based on this work.
Supplementary Materials I
Materials and Methods
Supplementary Text
Figs. S1 to S66
Supplementary Materials II
Tables S1 to S20
References and Notes
- 1.Seeman NC. Nucleic acid junctions and lattices. J. Theor. Biol. 1982;99:237. doi: 10.1016/0022-5193(82)90002-9. [DOI] [PubMed] [Google Scholar]
- 2.Chen J, Seeman NC. Synthesis from DNA of a molecule with the connectivity of a cube. Nature. 1991;350:631. doi: 10.1038/350631a0. [DOI] [PubMed] [Google Scholar]
- 3.Fu TJ, Seeman NC. DNA double-crossover molecules. Biochemistry. 1993;32:3211. doi: 10.1021/bi00064a003. [DOI] [PubMed] [Google Scholar]
- 4.Winfree E, Liu F, Wenzler LA, Seeman NC. Design and self-assembly of two-dimensional DNA crystals. Nature. 1998;394:539. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- 5.Yurke B, Turberfield AJ, Mills AP, Jr, Simmel FC, Neumann JL. A DNA-fuelled molecular machine made of DNA. Nature. 2000;406:605. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- 6.Yan H, Park SH, Finkelstein G, Reif JH, LaBean TH. DNA-templated self-assembly of protein arrays and highly conductive nanowires. Science. 2003;301:1882. doi: 10.1126/science.1089389. [DOI] [PubMed] [Google Scholar]
- 7.Sherman WB, Seeman NC. A precisely controlled DNA biped walking device. Nano Lett. 2004;4:1203. [Google Scholar]
- 8.Rothemund PWK, Papadakis N, Winfree E. Algorithmic self-assembly of DNA Sierpinski triangles. PLoS Biol. 2004;2:e424. doi: 10.1371/journal.pbio.0020424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chworos A, et al. Building programmable jigsaw puzzles with RNA. Science. 2004;306:2068. doi: 10.1126/science.1104686. [DOI] [PubMed] [Google Scholar]
- 10.Park SH, et al. Finite-size, fully-addressable DNA tile lattices formed by hierarchical assembly procedures. Angew. Chem. Int. Ed. 2006;45:735. doi: 10.1002/anie.200503797. [DOI] [PubMed] [Google Scholar]
- 11.Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 12.Seelig G, Soloveichik D, Zhang DY, Winfree E. Enzyme-free nucleic acid logic circuits. Science. 2006;314:1585. doi: 10.1126/science.1132493. [DOI] [PubMed] [Google Scholar]
- 13.He Y, et al. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature. 2008;452:198. doi: 10.1038/nature06597. [DOI] [PubMed] [Google Scholar]
- 14.Yin P, Choi HMT, Calvert CR, Pierce NA. Programming biomolecular self-assembly pathways. Nature. 2008;451:318. doi: 10.1038/nature06451. [DOI] [PubMed] [Google Scholar]
- 15.Yin P, et al. Programming DNA tube circumferences. Science. 2008;321:824. doi: 10.1126/science.1157312. [DOI] [PubMed] [Google Scholar]
- 16.Andersen ES, et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature. 2009;459:73. doi: 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]
- 17.Ke Y, et al. Scaffolded DNA origami of a DNA tetrahedron molecular container. Nano. Lett. 2009;9:2445. doi: 10.1021/nl901165f. [DOI] [PubMed] [Google Scholar]
- 18.Douglas SM, et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature. 2009;459:414. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dietz H, Douglas SM, Shih WM. Folding DNA into twisted and curved nanoscale shapes. Science. 2009;325:725. doi: 10.1126/science.1174251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng J, et al. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature. 2009;461:74. doi: 10.1038/nature08274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Omabegho T, Sha R, Seeman NC. A bipedal DNA Brownian motor with coordinated legs. Science. 2009;324:67. doi: 10.1126/science.1170336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Severcan I, et al. A polyhedron made of tRNAs. Nat. Chem. 2010;2:772. doi: 10.1038/nchem.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han D, et al. DNA origami with complex curvatures in three-dimensional space. Science. 2011;332:342. doi: 10.1126/science.1202998. [DOI] [PubMed] [Google Scholar]
- 24.Qian L, Winfree E. Scaling up digital circuit computation with DNA strand displacement cascades. Science. 2011;332:1196. doi: 10.1126/science.1200520. [DOI] [PubMed] [Google Scholar]
- 25.Delebecque CJ, Lindner AB, Silver PA, Aldaye FA. Organization of intracellular reactions with rationally designed RNA assemblies. Science. 2011;333:470. doi: 10.1126/science.1206938. [DOI] [PubMed] [Google Scholar]
- 26.Wei B, Dai M, Yin P. Complex shapes self-assembled from single-stranded DNA tiles. Nature. 2012;485:623. doi: 10.1038/nature11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin C, Liu Y, Rinker S, Yan H. DNA tile based self-assembly: building complex nanoarchitectures. Chem Phys Chem. 2006;7:1641. doi: 10.1002/cphc.200600260. [DOI] [PubMed] [Google Scholar]
- 28.Leontis NB, Lescoute A, Westhof E. The building blocks and motifs of RNA architecture. Curr. Opin. Struct. Biol. 2006;16:279. doi: 10.1016/j.sbi.2006.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shih WM, Lin C. Knitting complex weaves with DNA origami. Curr. Opin. Struct. Biol. 2010;20:276. doi: 10.1016/j.sbi.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seeman N. Nanomaterials based on DNA. Annu. Rev. Biochem. 2010;79:65. doi: 10.1146/annurev-biochem-060308-102244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang DY, Seelig G. Dynamic DNA Nanotechnology using Strand Displacement Reactions. Nat. Chem. 2011;3:103. doi: 10.1038/nchem.957. [DOI] [PubMed] [Google Scholar]
- 32.Kuzyk A, et al. DNA-based self-assembly of chiral plasmonic nanostructures with tailored optical response. Nature. 2012;483:311. doi: 10.1038/nature10889. [DOI] [PubMed] [Google Scholar]
- 33.Choi HMT, et al. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat. Biotechnol. 2010;28:1208. doi: 10.1038/nbt.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin C, et al. Sub-micrometre geometrically encoded fluorescent barcodes self-assembled from DNA. Nat. Chem. 2012;4:832. doi: 10.1038/nchem.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berardi MJ, Shih WM, Harrison SC, Chou JJ. Mitochondrial uncoupling protein 2 structure determined by NMR molecular fragment searching. Nature. 2011;476:109. doi: 10.1038/nature10257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Derr ND, et al. Tug of war in motor protein ensembles revealed with a programmable DNA origami scaffold. Science. 2012 doi: 10.1126/science.1226734. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Douglas SM, Bachelet I, Church GM. A logic-gated nanorobot for targeted transport of molecular payloads. Science. 2012;335:831. doi: 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- 38.Rothemund PWK, Andersen ES. Nanotechnology: The importance of being modular. Nature. 2012;485:584. doi: 10.1038/485584a. [DOI] [PubMed] [Google Scholar]
- 39.Materials and methods and supplementary figures and texts are available as Supplementary Materials I and DNA sequences in Supplementary Materials II on Science Online.
- 40.Douglas SM, et al. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 2009;37:5001. doi: 10.1093/nar/gkp436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ke Y, et al. Multilayer DNA origami packed on a square lattice. J. Am. Chem. Soc. 2009;131:15903. doi: 10.1021/ja906381y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castro CE, et al. A primer to scaffolded DNA origami. Nat. Methods. 2011;8:221. doi: 10.1038/nmeth.1570. [DOI] [PubMed] [Google Scholar]
- 43.Ke Y, Voigt NV, Gothelf KV, Shih WM. Multilayer DNA origami packed on hexagonal and hybrid lattices. J. Am. Chem. Soc. 2012;134:1770. doi: 10.1021/ja209719k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schulman R, Yurke B, Winfree E. Robust self-replication of combinatorial information via crystal growth and scission. Proc. Natl. Acad. Sci. USA. 2012;109:6405. doi: 10.1073/pnas.1117813109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.