Abstract
The key gluconeogenic enzyme fructose-1,6-bisphosphatase (FBPase) is synthesized when cells of the yeast Saccharomyces cerevisiae are grown on a non-fermentable carbon source. After shifting the cells to glucose-containing medium, in a process called catabolite degradation, FBPase is selectively and rapidly broken down. We have isolated gid mutants, which are defective in this glucose-induced degradation process. When complementing the defect in catabolite degradation of FBPase in gid3-1 mutant cells with a yeast genomic library, we identified the GID3 gene and found it to be identical to UBC8 encoding the ubiquitin-conjugating enzyme Ubc8p. The in vivo function of Ubc8p (Gid3p) has remained a mystery so far. Here we demonstrate the involvement of Ubc8p in the glucose-induced ubiquitylation of FBPase as a prerequisite for catabolite degradation of the enzyme via the proteasome. Like FBPase, Ubc8p is found in the cytoplasmic fraction of the cell. We demonstrate cytoplasmic degradation of FBPase.
Keywords: catabolite degradation/fructose-1,6-bisphosphatase/GID/Ubc8p/ubiquitin
Introduction
Fructose-1,6-bisphosphatase (FBPase), a key gluconeogenetic enzyme, is synthesized when cells are grown on non-fermentable carbon sources like ethanol. When yeast cells are subsequently shifted to a glucose-containing medium, FBPase is rapidly inactivated and degraded (Gancedo, 1971; Holzer, 1976). This mechanism prevents a futile cycling of ATP hydrolysis, which would be induced by a simultaneous proceeding of the glycolytic reaction (formation of fructose-1,6-bisphosphate) and the gluconeogenic reaction (formation of fructose-6-phosphate). Inactivation of FBPase is triggered by rapid phosphorylation, which is followed by degradation of the enzyme (Funayama et al., 1980; Müller and Holzer, 1981; Mazon et al., 1982). The site of this so-called catabolite degradation of FBPase is the topic of numerous studies, but is still a matter of debate (Schork et al., 1994b). On one hand, the selective engulfment of FBPase in vesicular compartments and its import into the vacuole, followed by degradation through the vacuolar proteinases, has been reported (Chiang and Schekman, 1991; Chiang et al., 1996; Huang and Chiang, 1997; Chiang and Chiang, 1998). We have shown that after addition of glucose to cells, polyubiquitylation of FBPase occurs as a prerequisite for the degradation process (Schork et al., 1995). Furthermore, we demonstrated the stabilization of FBPase in mutants defective in the activity of the cytosolic proteasome, indicating this protease complex to be the machinery that degrades FBPase (Schork et al., 1994a,b, 1995). As the enzymes involved in ubiquitin–proteasome-dependent degradation are all localized in the cytosol, hydrolysis of FBPase is supposed to be a cytosolic event under the conditions tested (Schork et al., 1995). We also found the N-terminal proline to be essential for selective degradation of FBPase (Haemmerle et al., 1998). To gain deeper insight into the mechanism of catabolite-induced FBPase degradation, we isolated mutants with a defect in glucose-induced degradation of the enzyme (gid mutants) (Haemmerle et al., 1998). Here we report the isolation of the GID3 gene, which we identified as identical to UBC8. So far, no in vivo function could be attributed to Ubc8p, which is a ubiquitin-conjugating enzyme (Ubc) (Qin et al., 1991; Kaiser et al., 1994). We found a nearly complete stabilization of FBPase in ubc8Δ cells and a lack of ubiquitin conjugation onto the enzyme. This identifies Ubc8p as a central component of the ubiquitylation machinery necessary to tag FBPase for proteasomal degradation. Localization studies identify Ubc8p as a cytosolic enzyme. Under the conditions tested, FBPase is not engulfed in vesicles during the inactivation process, underlining a cytoplasmic localization of the catabolite degradation process.
Results
Isolation and chromosomal deletion of theGID3/UBC8 gene
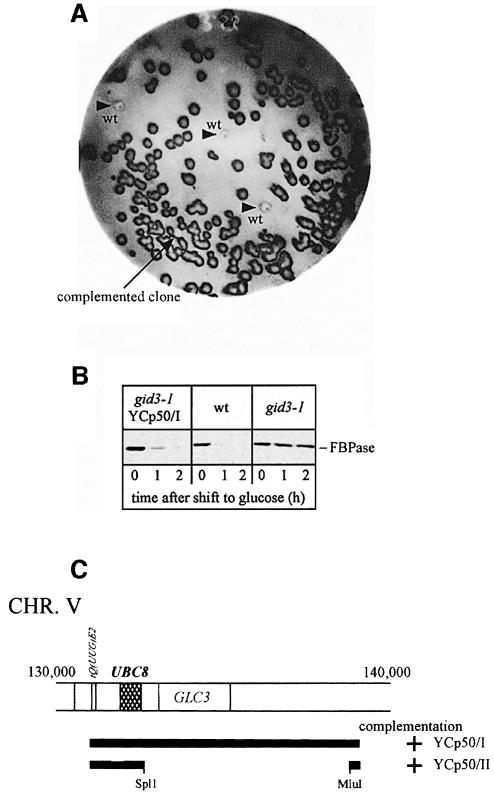
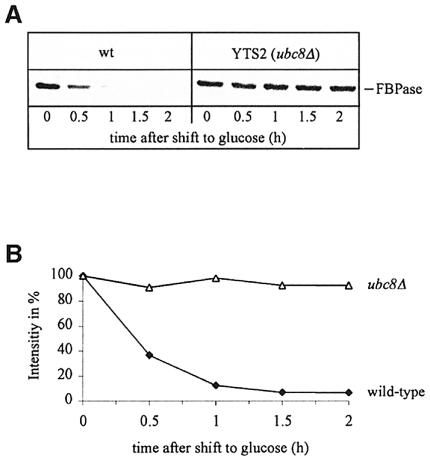
In a previous study, gid3-1 mutant cells had been isolated on the basis of a defect in glucose-induced degradation of a fusion protein consisting of the N-terminal part (291 amino acids) of FBPase and β-galactosidase (Haemmerle et al., 1998). To identify mutant cells, the alteration of β-galactosidase activity after addition of glucose had been measured using a microtitre plate reader (Haemmerle et al., 1998). This method could not be used to identify complementing plasmids after transformation of gid3-1 mutant cells with a genomic library (not shown). We therefore established a screening procedure using antibodies directed against FBPase. After transformation of gid3-1 mutant cells with a YCp50-based yeast genomic library (Rose et al., 1987), transformants were plated onto medium containing glucose. The colonies were replica plated onto selective plates containing 2% ethanol and grown for 2 days. Then cells were transferred onto nitrocellulose sheets and placed on filters soaked with yeast extract/peptone/dextrose (YPD) medium for 2 h. Thereafter, colonies were lysed and the concentration of FBPase was determined using specific antibodies. Non-complemented colonies of gid3-1 mutant cells appear dark due to their high level of FBPase, while wild-type cells and colonies harbouring a complementing plasmid are significantly brighter due to FBPase degradation (Figure 1A). As a control, wild-type colonies were placed on each filter (Figure 1A, arrowheads). One positive clone was found among the 20 000 colonies screened. After rescuing the respective plasmid and retransformation in gid3-1 mutant cells the wild-type-like glucose-induced degradation of FBPase was confirmed by immunoblotting (Figure 1B). Sequencing localized the complementing genomic fragment on chromosome V (Figure 1C). Subcloning (Figure 1C) suggested that GID3 might be identical to UBC8. We constructed a chromosomal deletion of UBC8 by integrating a LoxP-KanR-LoxP cassette (Wach et al., 1994; Güldener et al., 1996), generated by PCR, into this locus of wild-type cells. Correct gene replacement was confirmed by Southern hybridization (not shown). Immunoblot analysis of gid3-1 ubc8Δ diploid cells confirmed the identity of GID3 with UBC8 (not shown). As expected, ubc8Δ cells exhibit a strong defect in glucose-induced degradation of FBPase. Pulse–chase measurements unravelled a significant stabilization of FBPase (half-life >10 h) in ubc8Δ cells compared with wild-type cells (half-life ∼20 min) (Figure 2A and B).
Fig. 1. (A) Colony screen for the detection of gid3-1-complementing genomic DNA fragments. Cells were transferred on nitrocellulose sheets as described in Materials and methods and probed with antibodies against FBPase. Further details are given in the text. gid mutant cells appear significantly darker than wild-type cells (arrowheads) due to their higher FBPase level. (B) Complementation of gid3-1 cells was verified in immunoblots following FBPase immunogenic material after retransformation of the plasmid, isolated from the clone shown in (A). The complementing plasmid re-introduces the FBPase degradation phenotype. (C) Subcloning identifies UBC8 as the complementing ORF.
Fig. 2. Pulse–chase analysis of glucose-induced degradation of FBPase in wild-type and ubc8Δ cells (A). Quantification unravels a half-life of FBPase of ∼20 min in wild-type cells and >10 h in ubc8Δ cells (B).
UBC8 is required for glucose-induced ubiquitylation of FBPase in vivo
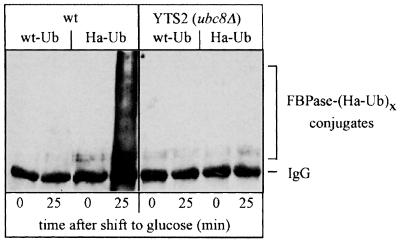
UBC8 had initially been cloned by chance and was identified as a Ubc (E2) capable of modifying histones in vitro (Qin et al., 1991; Kaiser et al., 1994). So far, no specific phenotypes could be attributed to UBC8 deleted cells and no in vivo substrate had been identified. The lack of phenotypes of UBC8 deleted cells has led to speculation that the biological function of the expressed enzyme may overlap with other Ubcs or that it is required for a more specialized purpose. We checked the fidelity of the glucose-induced ubiquitin conjugation of FBPase by overexpressing a haemagglutinin (HA)-tagged ubiquitin species (Ellison and Hochstrasser, 1991; Schork et al., 1995). As shown previously (Schork et al., 1995), in wild-type cells a large amount of FBPase–ubiquitin conjugates are detectable 25 min after induction of degradation (Figure 3). Most interestingly, the formation of these conjugates is significantly reduced in ubc8Δ cells, suggesting that Ubc8p is required for the glucose-induced ubiquitylation of FBPase in vivo.
Fig. 3. Glucose-induced ubiquitylation of FBPase is significantly affected in ubc8Δ cells. Crude extracts from wild-type and ubc8Δ cells overexpressing wt-Ub or HA-Ub were immunoprecipitated with anti-FBPase antibodies. After SDS–PAGE, proteins were transferred to a nitrocellulose membrane and ubiquitin conjugates were detected with anti-HA antibodies.
Ubc8p is not involved in the N-end rule pathway
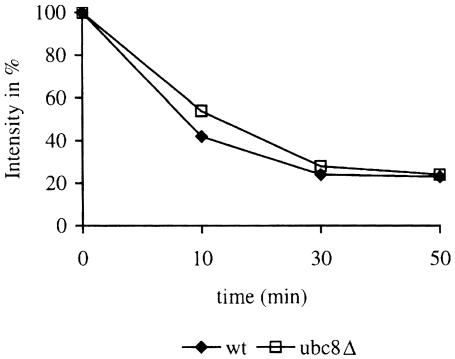
Measurement of steady-state levels of plasmid-expressed β-galactosidase fusion proteins (Bachmair et al., 1986) in wild-type and gid3-1 mutant cells had indicated a 21-fold increase in the steady-state level of Arg-β-gal and a 2-fold increase in the level of Ub-Pro-β-gal in mutant cells (Haemmerle et al., 1998). We repeated these experiments with mutant cells carrying the UBC8 deletion and performed pulse–chase analysis of β-galactosidase to follow its degradation. As can be seen, there is no significant difference in degradation of Arg-β-gal in wild-type and ubc8Δ mutant cells (Figure 4). Also, degradation of Ub-Pro-β-gal was not altered in ubc8Δ mutant cells (not shown). This strongly supports the notion that Ubc8p is not involved in the N-end rule pathway.
Fig. 4. Degradation of the N-end rule substrate Arg-β-gal is not affected in ubc8Δ cells. Degradation of Arg-β-gal was followed in a pulse–chase experiment and the remaining antigenic β-galactosidase material was quantified. Antigenic material at time zero was set to 100%.
Localization of Ubc8p
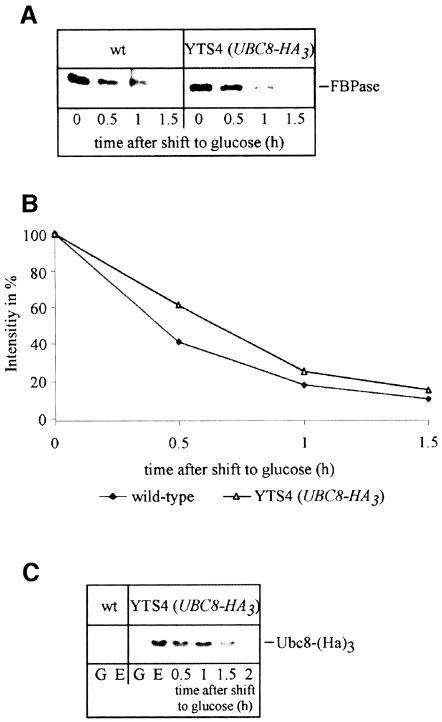
To learn more about the intracellular localization and function of Ubc8p, we generated a chromosomally integrated, C-terminally HA-tagged version of Ubc8p under the control of its native promoter. A PCR fragment containing a triple HA-tag and carrying the Schizosaccharomyces pombe HIS5 gene as a selectable marker for the his3 mutant allele was introduced into the chromosome of Saccharomyces cerevisiae (see Materials and methods). Correct integration at the UBC8 locus was confirmed by Southern analysis (not shown). Biological activity of the HA-tagged version of Ubc8p was confirmed by pulse–chase analysis of FBPase breakdown induced by glucose, which remained at wild-type rate (Figure 5A and B). The steady-state level of Ubc8-(HA)3 itself was significantly increased in cells grown on ethanol. After shifting the cells to glucose-containing medium the steady-state level decreased with time (Figure 5C).
Fig. 5. HA-tagged Ubc8p is functional. (A) Pulse–chase analysis demonstrates the biological activity of a chromosomally integrated Ubc8-(HA)3-tagged fusion protein. (B) Quantification of FBPase immunogenic material in Ubc8p and Ubc8-(HA)3 protein expressing strains. (C) Steady-state levels of Ubc8-(HA)3 in ethanol-containing medium and after glucose addition. Crude extracts of cells grown in glucose- (G) or ethanol (E)-containing media were probed with HA antibodies. Ethanol-grown cells were shifted to glucose medium and aliquots were analysed at the indicated times.
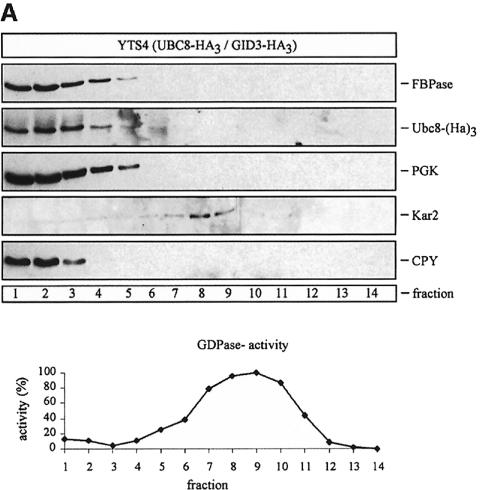
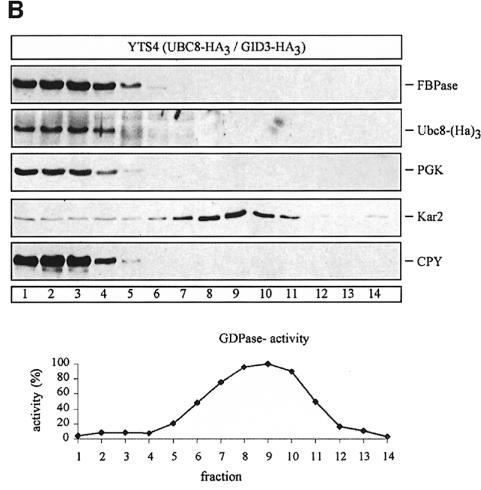
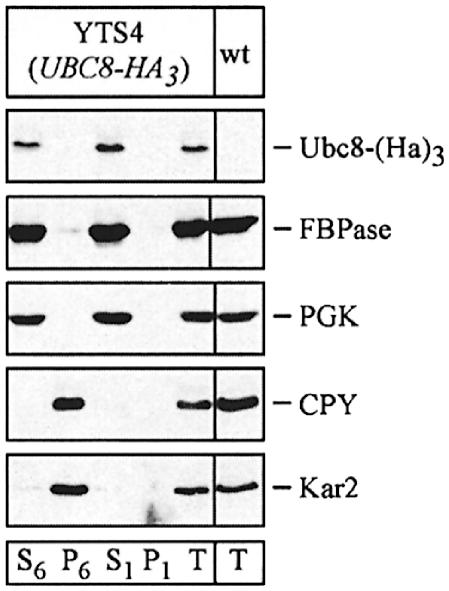
Since there is an ongoing debate about the site of catabolite inactivation and the involvement of vesicular intermediates in the catabolite-induced degradation of FBPase (Schork et al., 1994b), we used sucrose density gradient centrifugation to localize the Ubc8-(HA)3 protein. We analysed cells immediately before shifting them to glucose medium and cells treated for 30 min with glucose to induce catabolite degradation of FBPase (Figure 6A and B). Under both conditions the Ubc8-(HA)3 protein and FBPase were detectable in fractions 1–5, co-sedimenting with the cytosolic marker 3-phospho-glycerokinase (PGK) and the vacuolar proteinase carboxypeptidase yscY (CPY) (Figure 6A and B). Since we were further interested in whether the Ubc8-(HA)3 protein is localized in the cytosol or the vacuole, we additionally fractionated hypotonically lysed spheroplasts taken 30 min after the shift to glucose medium (Figure 7), using a previously described procedure (Lang et al., 1998). We started with a centrifugation step at 6000 g, which yielded a P6 pellet fraction enriched for the endoplasmic reticulum (ER) and the vacuole, as shown by enrichment of Kar2p (BiP), an ER lumenal chaperone, and CPY, respectively (Figure 7). In the P6 pellet fraction almost no FBPase and Ubc8-(HA)3 protein were detectable. Then the 6000 g supernatant fraction (S6) was centrifuged at 100 000 g, yielding a P1 pellet fraction and an S1 supernatant fraction. FBPase and Ubc8-(HA)3 were detectable predominantly in the S1 fraction. Almost no FBPase and Ubc8-(HA)3 were found in the P1 pellet fraction, suggesting the localization of both proteins in the soluble fraction.
Fig. 6. Sucrose density gradient fractionation of cell extracts from cells expressing a chromosomally integrated Ubc8-(HA)3 fusion protein. Cells were grown for 18 h on ethanol medium and then transferred to glucose medium. Aliquots were analysed before (A) and 30 min after (B) the transfer. Immunoreactive material of proteins (PGK, cytosol; CPY, vacuole; Kar2, ER and GDPase activity, Golgi) were measured.
Fig. 7. Thirty minutes after addition of glucose, FBPase localizes in the soluble fraction together with cytosolic PGK, different from CPY (vacuole) and Kar2 (ER). Thirty minutes after transfer of yeast cells expressing a chromosomally integrated HA-tagged Ubc8p to glucose medium, the cells were spheroplasted and, after hypotonic lysis, fractionated by centrifugation as indicated in the text.
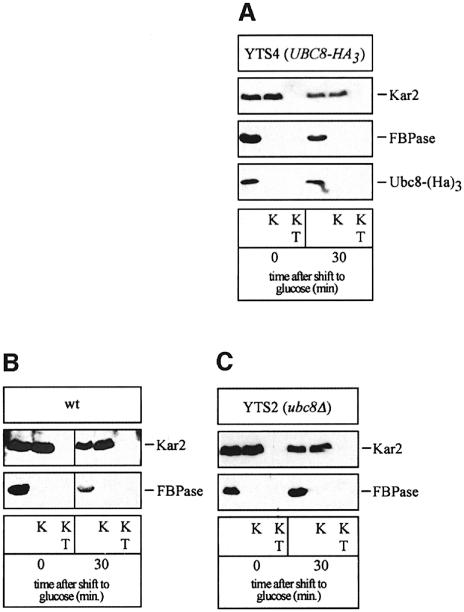
To check whether some Ubc8-(HA)3 or FBPase became membrane protected during glucose-induced degradation, we further performed a proteinase protection experiment using hypotonically lysed spheroplasted cells (Figure 8). As a control we checked the lumenal ER protein Kar2p. For both Ubc8-(HA)3 and FBPase no significant membrane protection was detected (Figure 8A). To check for the accumulation of FBPase-containing vesicular intermediates in UBC8 deleted cells, we also measured the accessibility of FBPase to proteinase K in these cells in a proteinase protection experiment with wild type as a control (Figure 8B and C). As shown in Figure 8A and C, no membrane protection of FBPase or Ubc8-(HA)3, respectively, was detectable. This indicates that under the derepression conditions tested, FBPase is not engulfed in a vesicular compartment. FBPase and all components of the degradation machinery, including Ubc8p, are in the soluble compartment of the cell, the cytosol.
Fig. 8. FBPase is not vesicle engulfed under inactivation conditions. At the times indicated after shift to glucose media, spheroplasted and lysed cells were incubated with proteinase K (K) and proteinase K with Triton X-100 (KT). Western blots were analysed with antibodies directed against Kar2p (ER), Ubc8-(HA)3 and FBPase, respectively. Protein protection experiment of Ubc8-(Ha)3 (A) and FBPase in wild-type (B) and ubc8Δ (C) cells.
Discussion
So far, 13 Ubcs have been identified in the yeast genome (Hochstrasser, 1996; Scheffner et al., 1998). Specific in vivo functions have been uncovered only for part of these Ubcs. UBC8 was initially cloned by chance and identified as encoding a Ubc due to its sequence homology (Qin et al., 1991). It has been found to conjugate ubiquitin to histones in vitro (Kaiser et al., 1994). No specific phenotypes have been detected for UBC8 deleted cells so far. This led to speculation that functionally overlapping enzymes may exist in yeast or that Ubc8p may be required for a more specialized in vivo function (Kaiser et al., 1994). When searching for mutants that are defective in catabolite degradation of FBPase, we found a mutation, gid3-1 (Haemmerle et al., 1998), which nearly completely blocked degradation of the enzyme. Cloning of the respective wild-type gene uncovered GID3 as being identical to UBC8. This now allows us to describe the first in vivo function of Ubc8p. Our findings demonstrate that Ubc8p is essential for conjugating ubiquitin onto FBPase after shifting gluconeogenic cells to medium containing glucose, thus initiating degradation of the enzyme by the proteasome. FBPase degradation is almost completely abolished in ubc8Δ cells. With a half-life of >10 h, compared with 20 min in wild-type cells, FBPase is stabilized >30-fold. In a previous study, we described partial stabilization of FBPase in ubc1 (2-fold) and ubc4/ubc5 double mutant cells (4-fold) (Schork et al., 1995). This partial stabilization suggests that FBPase is also modified to a lesser degree by this family of Ubcs known to be involved in the degradation of abnormal and short-lived proteins (Hershko and Ciechanover, 1998; Scheffner et al., 1998), and that Ubc8p is indeed the major enzyme polyubiquitylating FBPase.
It has been shown previously that different Ubcs can act on a certain substrate without being additive in their effect (Biederer et al., 1996; Hiller et al., 1996). This may be due to considerably different Km or vmax values of different Ubcs for a particular substrate. Also, complex formation of a Ubc with other Ubcs or other proteins may be necessary for its activity (Chen et al., 1993; Biederer et al., 1997). The non-additivity of their effects also holds true for ubiquitin conjugation of FBPase by Ubc1p, Ubc4p, Ubc5p (Schork et al., 1995) and Ubc8p (this paper). The mechanism behind this phenomenon awaits further elucidation. In contrast to our previous study, where we measured activities of steady-state levels of plasmid-encoded β-galactosidase fusions (Haemmerle et al., 1998), pulse–chase experiments following the degradation of β-galactosidase protein show unequivocally that Ubc8p does not function in the N-end rule pathway. Even though in other instances comparison of steady-state levels and degradation rates of N-end rule substrates had given reliable results (Richter-Ruoff et al., 1992), this does not seem to hold true for all cases. Depending on the strain background, overexpression of the plasmid-encoded β-galactosidase constructs might, for instance, cause changes in cellular metabolism, finally leading to uneven downregulation of β-galactosidase or to uneven plasmid loss in wild-type and mutant cells.
The important function of Ubc8p during the catabolite-induced degradation of FBPase is further illustrated by the enhanced steady-state level of the protein in cells grown on ethanol (Figure 5C). After shifting the cells to glucose-containing medium the steady-state level of Ubc8p decreases with time, which is in accordance with the action of the protein during catabolite degradation of FBPase.
The site of catabolite degradation of FBPase has been under debate for some time (Chiang and Schekman, 1991; Schork et al., 1994a,b, 1995; Shieh and Chiang, 1998). Degradation by the ubiquitin–proteasome pathway (Schork et al., 1994a, 1995; Haemmerle et al., 1998) and vesicle-mediated uptake into the vacuole followed by degradation by the vacuolar proteinases (Huang and Chiang, 1997; Chiang and Chiang, 1998) have been reported. Under the conditions we tested (derepression of FBPase on ethanol as non-fermentable carbon source for between 5 and 18 h), the enzyme reaches highest activities and catabolite degradation appears to be completely cytosolic. Only cytoplasmic components, the known Ubcs and the proteasome consist of the degradation machinery (Schork et al., 1994a, 1995; Haemmerle et al., 1998). We thus also searched for the location of Ubc8p, which we discovered here as the most important Ubc to deliver FBPase to proteasomal degradation. For this purpose we used a biologically active HA-tagged version of Ubc8p. We especially checked for the involvement of vesicular intermediates using cell fractionations and proteinase protection experiments (Figure 8). Our experiments clearly suggest a non-membranous, cytosolic environment for Ubc8p. As expected, FBPase was found in the same non-membranous, cytosolic fraction as Ubc8p. Also, in ubc8 null mutant cells, no hints for the accumulation of FBPase-containing vesicular intermediates were found (Figure 8C). This again indicates ubiquitin conjugation of FBPase and subsequent proteasomal breakdown in the cytosol under the conditions tested.
It will be of great interest to follow the glucose-induced signals, which finally render FBPase accessible to the Ubcs. The N-terminal proline of the enzyme has previously turned out to be necessary for the ubiquitylation and degradation process (Haemmerle et al., 1998). It can therefore be assumed that the N-terminus of FBPase plays a crucial role in the signalling process.
Materials and methods
Yeast strains and media
Yeast strains used in this study were W303-1B (MATα ade2 leu2-3 112 his3 trp1 ura3), WAY.5-4A (MATa his3-Δ1 ura3-52) and WAY.5-4A/C3 (MATa his3-Δ1 ura3-52 gid3-1). Cells were grown in synthetic medium (CM; 0.67% Difco yeast nitrogen base without amino acids) containing 2% glucose or, for derepression of FBPase, 2% ethanol as the sole carbon source, and required supplements. Radiolabelling for pulse–chase experiments was carried out in pulse medium (0.17% Difco yeast nitrogen base without amino acids and ammonium sulfate, 0.5% proline, 100 µM ammonium sulfate, 2% ethanol and required supplements).
Plasmids
YEp96 is a 2-µm-based S.cerevisiae–Escherichia coli shuttle vector carrying a synthetic version of yeast ubiquitin (Ub) under the control of the copper-inducible CUP1 promoter (Hochstrasser et al., 1991). YEp112 encodes yeast ubiquitin carrying an epitope from the HA of influenza virus attached to the N-terminus of ubiquitin (HA-Ub) (Ellison and Hochstrasser, 1991).
Identification of UBC8/GID3
The gid3-1 mutant strain was transformed with a yeast genomic library based on the CEN URA3 shuttle vector YCp50 (Rose et al., 1987), spread on CM plates without uracil (2% glucose) and grown for 3 days at 30°C. The colonies were then replica plated onto selective CM plates without uracil containing 2% ethanol for 2 days, replica plated onto nitrocellulose membranes and transferred for 2 h at 30°C onto filters soaked with 3 ml of YPD medium. Afterwards the cells were lysed with 3 ml of lysis buffer (0.1% SDS, 0.2 M NaOH, 35 mM dithiothreitol) for 30 min and washed off the membranes. Nitrocellulose sheets containing cellular lysates were subsequently incubated with 10 ml of 2% milk in TBS-T (20 mM Tris–HCl pH 7.6, 137 mM NaCl, 0.1% Tween-20) for 1 h, and anti-FBPase antibody (1:5000) in TBS-T for another hour. After several washes with TBS-T, the membranes were incubated with the secondary goat anti-rabbit antibody (Medac, Hamburg, Germany) (1:5000) coupled to peroxidase, which was diluted in 10 ml of TBS-T buffer. Membranes were washed three times with TBS-T and the peroxidase activity was detected using the ECL system (Amersham, Braunschweig, Germany).
One positive clone was found among 20 000 colonies screened. After plasmid rescue of the respective plasmid YCp50/I an ∼6 kb DNA fragment was released from the genomic insert by digestion with MluI–BsiWI, and subsequent religation yielded plasmid YCp50/II. Molecular biological procedures were performed according to Ausubel et al. (1987).
Generation of strains YTS2 and YTS4
Strain YTS2 was obtained by chromosomal deletion of the UBC8 gene in W303-1B cells. Using oligonucleotides Δgid3 KAN1 (5′-TAATGAAACAGTAGCTCTAAAAGAAGGATCGAGACAGATGTTCAGCTGAAGCTTCGTACGC-3′), Δgid3 KAN2 (5′-ACATACATATATATAT ATATATATATATGTGTGTGCTCGAAAAGCATAGGCCACTAGTGGATCG-3′) and plasmid pUG6, a DNA fragment for the chromosomal replacement of UBC8 with a LoxP-KANR-LoxP cassette was created by PCR. Strain YTS2 was generated by transforming the single LoxP-KANR-LoxP cassette into the wild-type strain and selecting for colonies on YPD plates containing kanamycin.
YTS4 was generated by chromosomal integration of a 1.5 kb fragment, consisting of a triple HA-tag and an S.pombe HIS5 marker (Cottarel, 1995) upstream of the UBC8 coding region. The 1.5 kb fragment was synthesized by PCR using oligonucleotides GID3-HA (TGTCAGTGATGACGATGACTACGACGAAGTCGCAAATCAAGGAGCAGGGGCGGGTGC), GID3-HIS5 (5′-ATACATATATATATATATATATAT ATGTGTGTGCTCGAAAGAGGTCGACGGTATCGATAAG-3′) and plasmid p3xHA HIS5 (S.Munro, Cambridge, UK) as template. After transformation of the PCR product into wild-type cells, positive transformants were selected on plates lacking histidine. Correct gene replacement was confirmed by Southern blotting (not shown).
Pulse–chase analysis, immunoprecipitation and Western blotting
Pulse–chase experiments were performed as described by Schork et al. (1995), using specific antibodies against FBPase. Protein bands were quantitated with a PhosphorImager (Molecular Dynamics). Immuno detection of FBPase or FBPase–HA-ubiquitin conjugates was carried out according to Schork et al. (1995), using FBPase or HA antibodies [Boehringer Mannheim (Roche), Germany; clone 12CA5].
Degradation of the N-end rule substrates Arg-β-gal and Ub-Pro-β-gal was followed by pulse–chase analysis in wild-type and ubc8Δ strains. The experiments were carried out according to Richter-Ruoff et al. (1992). Cells were grown to an OD600 of 3, labelled with radioactive [35S]methionine for 10 min and shifted onto chase medium. Samples were taken at the time points indicated. Crude extracts were treated with specific anti-β-galactosidase antibodies for the immunoprecipitation of Arg-β-gal and Ub-Pro-β-gal. Proteins were then separated by SDS–PAGE and protein bands were quantitated with a PhosphorImager (Molecular Dynamics).
Cell fractionation
Subcellular fractionations were carried out as described by Lang et al. (1998) with the following modifications. Cells expressing either the wild-type or HA-tagged Ubc8p were cultured in YPD medium containing 2% glucose to an A600 of 5–6. After harvesting, cells were resuspended in YP medium containing 2% ethanol to an A600 of 0.2 and incubated for 16 h to derepress FBPase. Thirty minutes after addition of glucose (final concentration 2%), cells (A600 = 60) were harvested, converted to spheroplasts and lysed by an osmotic procedure. Unlysed cells were removed by an additional 300 g centrifugation step. The cytosol-enriched S6 fraction was obtained by a second centrifugation step at 6000 g for 20 min. A further centrifugation of the S6 fraction at 100 000 g for 3 h yielded an S1 supernatant and a P1 pellet fraction. The fractions were precipitated with 10% trichloroacetic acid (TCA), washed twice in cold acetone, and then solubilized in SDS sample buffer. Proteins were separated by SDS–PAGE (10%) and transferred to nitrocellulose membranes. Fractions were analysed by immunoblotting with a monoclonal antibody against the HA-tag (Boehringer Mannheim, Germany), anti-FBPase antibody (1:5000), anti-PGK (1:5000) (Molecular Probes, Leiden, The Netherlands), anti-CPY (1:10 000) (Molecular Probes, Leiden, The Netherlands) and anti-Kar2 antibody (1:5000) (R.Schekman, Berkeley, CA), followed by goat anti-rabbit (Medac, Hamburg, Germany) or goat anti-mouse (Boehringer Mannheim, Germany) antibody coupled to peroxidase (1:5000). Peroxidase activity was detected using the ECL system (Amersham, Braunschweig, Germany).
Sucrose density gradient fractionation
Subcellular fractionation was performed as described by Antebi and Fink (1992) and Egner et al. (1995). Cells of the yeast strain YTS4 expressing the HA-tagged Ubc8p were grown in YPD medium (2% glucose) to an A600 of 5–6, shifted to YP medium containing 2% ethanol to an A600 of 0.2 and incubated for 16 h to derepress FBPase. After addition of glucose (2%), cells (A600 = 1000) were harvested at the times indicated (0 and 30 min), converted to spheroplasts and then homogenized in the presence of proteinase inhibitors using a Dounce homogenizer. Afterwards cell lysates were centrifuged at 2000 g for 10 min at 4°C to remove cell debris. The supernatants were then loaded onto the top of 10 ml of an 18–54% sucrose gradient and centrifuged to equilibrium at 100 000 g for 3 h at 4°C. Fractions (0.6 ml) were collected from the top. Part of each fraction was precipitated with 10% TCA, washed twice in cold acetone, solubilized in SDS sample buffer and analysed as described above. Guanosine diphosphatase (GDPase) activity was measured as described by Abeijon et al. (1989).
Proteinase K treatment
Cells of wild-type, YTS2 (ubc8Δ) and YTS4 (UBC8-HA3) strains were grown, derepressed for FBPase and lysed before and after shift to glucose for 30 min, as described above. Total cell extracts (A600 = 60) were treated with proteinase K (1 mg/ml) in the absence or presence of Triton X-100 (2%) for 30 min on ice. The reactions were stopped by the addition of 10% TCA and the samples were processed as described above.
Acknowledgments
Acknowledgements
The authors thank M.Hochstrasser, J.Hegemann, S.Munro and R.Schekman for providing plasmids and antibodies. The work was supported by a grant of the Deutsche Forschungsgemeinschaft (Bonn, Germany) and the Fonds der Chemischen Industrie (Frankfurt, Germany).
References
- Abeijon C., Orlean,P., Robbins,P.W. and Hirschberg,C.B. (1989) Topography of glycosylation in yeast: characterization of GDPmannose transport and lumenal guanosine diphosphatase activities in Golgi-like vesicles. Proc. Natl Acad. Sci. USA, 86, 6935–6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antebi A. and Fink,G.R. (1992) The yeast Ca2+-ATPase homologue, PMR1, is required for normal Golgi function and localizes in a novel Golgi-like distribution. Mol. Biol. Cell, 3, 633–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F.M., Brent,R., Kingston,R.E. and Moore,D.D. (1987) Current Protocols in Molecular Biology. Greene Publishing Associates, New York, NY. [Google Scholar]
- Bachmair A., Finley,D. and Varshavsky,A. (1986) In vivo half-life of a protein is a function of its amino-terminal residue. Science, 234, 179–186. [DOI] [PubMed] [Google Scholar]
- Biederer T., Volkwein,C. and Sommer,T. (1996) Degradation of subunits of the Sec61p complex, an integral component of the ER membrane, by the ubiquitin–proteasome pathway. EMBO J., 15, 2069–2076. [PMC free article] [PubMed] [Google Scholar]
- Biederer T., Volkwein,C. and Sommer,T. (1997) Role of Cue1p in ubiquitination and degradation at the ER surface. Science, 278, 1806–1809. [DOI] [PubMed] [Google Scholar]
- Chen P., Johnson,P., Sommer,T., Jentsch,S. and Hochstrasser,M. (1993) Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MATα2 repressor. Cell, 74, 357–369. [DOI] [PubMed] [Google Scholar]
- Chiang H.L. and Schekman,R. (1991) Regulated import and degradation of a cytosolic protein in the yeast vacuole. Nature, 350, 313–318. [DOI] [PubMed] [Google Scholar]
- Chiang H.L., Schekman,R. and Hamamoto,S. (1996) Selective uptake of cytosolic, peroxisomal and plasma membrane proteins into the yeast lysosome for degradation. J. Biol. Chem., 271, 9934–9941. [DOI] [PubMed] [Google Scholar]
- Chiang M.C. and Chiang,H.L. (1998) Vid24p, a novel protein localized to the fructose-1,6-bisphosphatase-containing vesicles, regulates targeting of fructose-1,6-bisphosphatase from the vesicles to the vacuole for degradation. J. Cell Biol., 140, 1347–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottarel G. (1995) The Saccharomyces cerevisiae HIS3 and LYS2 genes complement the Schizosaccharomyces pombe his5-303 and lys1-131 mutations, respectively: new selectable markers and new multi-purpose multicopy shuttle vectors, pSP3 and pSP4. Curr. Genet., 28, 380–383. [DOI] [PubMed] [Google Scholar]
- Egner R., Mahe,Y., Pandjaitan,R. and Kuchler,K. (1995) Endocytosis and vacuolar degradation of the plasma membrane-localized Pdr5 ATP-binding cassette multidrug transporter in Saccharomyces cerevisiae. Mol. Cell. Biol., 15, 5879–5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison M.J. and Hochstrasser,M. (1991) Epitope-tagged ubiquitin. A new probe for analyzing ubiquitin function. J. Biol. Chem., 266, 21150–21157. [PubMed] [Google Scholar]
- Funayama S., Gancedo,J.M. and Gancedo,C. (1980) Turnover of yeast fructose-bisphosphatase in different metabolic conditions. Eur. J. Biochem., 109, 61–66. [DOI] [PubMed] [Google Scholar]
- Gancedo C. (1971) Inactivation of fructose-1,6-diphosphatase by glucose in yeast. J. Bacteriol., 107, 401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güldener U., Heck,S., Fielder,T., Beinhauer,J. and Hegemann,J.H. (1996) A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res., 24, 2519–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haemmerle M., Bauer,J., Rose,M., Szallies,A., Thumm,M., Dusterhus,S., Mecke,D., Entian,K.D. and Wolf,D.H. (1998) Proteins of newly isolated mutants and the amino-terminal proline are essential for ubiquitin–proteasome-catalyzed catabolite degradation of fructose-1,6-bisphosphatase of Saccharomyces cerevisiae. J. Biol. Chem., 273, 25000–25005. [DOI] [PubMed] [Google Scholar]
- Hershko A. and Ciechanover,A. (1998) The ubiquitin system. Annu. Rev. Biochem., 67, 425–479. [DOI] [PubMed] [Google Scholar]
- Hiller M.M., Finger,A., Schweiger,M. and Wolf,D.H. (1996) ER degradation of a misfolded luminal protein by the cytosolic ubiquitin–proteasome pathway. Science, 273, 1725–1728. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. (1996) Ubiquitin-dependent protein degradation. Annu. Rev. Genet., 30, 405–439. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M., Ellison,M.J., Chau,V. and Varshavsky,A. (1991) The short-lived MATα2 transcriptional regulator is ubiquitinated in vivo. Proc. Natl Acad. Sci. USA, 88, 4606–4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzer H. (1976) Catabolite inactivation in yeast. Trends Biochem. Sci., 1, 178–181. [Google Scholar]
- Huang P.H. and Chiang,H.L. (1997) Identification of novel vesicles in the cytosol to vacuole protein degradation pathway. J. Cell Biol., 136, 803–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser P., Seufert,W., Hofferer,L., Kofler,B., Sachsenmaier,C., Herzog,H., Jentsch,S., Schweiger,M. and Schneider,R. (1994) A human ubiquitin-conjugating enzyme homologous to yeast UBC8. J. Biol. Chem., 269, 8797–8802. [PubMed] [Google Scholar]
- Lang T., Schaeffeler,E., Bernreuther,D., Bredschneider,M., Wolf,D.H. and Thumm,M. (1998) Aut2p and Aut7p, two novel microtubule-associated proteins are essential for delivery of autophagic vesicles to the vacuole. EMBO J., 17, 3597–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazon M.J., Gancedo,J.M. and Gancedo,C. (1982) Inactivation of yeast fructose-1,6-bisphosphatase. In vivo phosphorylation of the enzyme. J. Biol. Chem., 257, 1128–1130. [PubMed] [Google Scholar]
- Müller D. and Holzer,H. (1981) Regulation of fructose-1,6-bisphosphatase in yeast by phosphorylation/dephosphorylation. Biochem. Biophys. Res. Commun., 103, 926–933. [DOI] [PubMed] [Google Scholar]
- Qin S., Nakajima,B., Nomura,M. and Arfin,S.M. (1991) Cloning and characterization of a Saccharomyces cerevisiae gene encoding a new member of the ubiquitin-conjugating protein family. J. Biol. Chem., 266, 15549–15554. [PubMed] [Google Scholar]
- Richter-Ruoff B., Heinemeyer,W. and Wolf,D.H. (1992) The proteasome/multicatalytic-multifunctional proteinase. In vivo function in the ubiquitin-dependent N-end rule pathway of protein degradation in eukaryotes. FEBS Lett., 302, 192–196. [DOI] [PubMed] [Google Scholar]
- Rose M.D., Novick,P., Thomas,J.H., Botstein,D. and Fink,G.R. (1987) A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene, 60, 237–243. [DOI] [PubMed] [Google Scholar]
- Scheffner M., Smith,S. and Jentsch,S. (1998) The ubiquitin-conjugation system. In Peters,J.M., Harris,J.R. and Finley,D. (eds), Ubiquitin and the Biology of the Cell. Plenum Press, New York, NY, pp. 65–98. [Google Scholar]
- Schork S., Bee,G., Thumm,M. and Wolf,D.H. (1994a) Catabolite inactivation of fructose-1,6-bisphosphatase in yeast is mediated by the proteasome. FEBS Lett., 349, 270–274. [DOI] [PubMed] [Google Scholar]
- Schork S., Bee,G., Thumm,M. and Wolf,D.H. (1994b) Site of catabolite inactivation. Nature, 369, 283–284. [DOI] [PubMed] [Google Scholar]
- Schork S.M., Thumm,M. and Wolf,D.H. (1995) Catabolite inactivation of fructose-1,6-bisphosphatase of Saccharomyces cerevisiae. Degradation occurs via the ubiquitin pathway. J. Biol. Chem., 270, 26446–26450. [DOI] [PubMed] [Google Scholar]
- Shieh H.L. and Chiang,H.L. (1998) In vitro reconstitution of glucose-induced targeting of fructose-1,6-bisphosphatase into the vacuole in semi-intact yeast cells. J. Biol. Chem., 273, 3381–3387. [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat,A., Pohlmann,R. and Philippsen,P. (1994) New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast, 10, 1793–1808. [DOI] [PubMed] [Google Scholar]