Abstract
The AAA-ATPase, p97/Cdc48p, has been implicated in many different pathways ranging from membrane fusion to ubiquitin-dependent protein degradation. Binding of the p47 complex directs p97 to act in the post-mitotic fusion of Golgi membranes. We now describe another binding complex comprising mammalian Ufd1 and Npl4. Yeast Ufd1p is required for ubiquitin-dependent protein degradation whereas yeast Npl4p has been implicated in nuclear transport. In rat liver cytosol, Ufd1 and Npl4 form a binary complex, which exists either alone or bound to p97. Ufd1/Npl4 competes with p47 for binding to p97 and so inhibits Golgi membrane fusion. This suggests that it is involved in another cellular function catalysed by p97, the most likely being ubiquitin-dependent events during mitosis. The fact that the binding of p47 and Ufd1/Npl4 is mutually exclusive suggests that these protein complexes act as adapters, directing a basic p97 activity into different cellular pathways.
Keywords: AAA-ATPase/CDC48/DiGeorge/ubiquitin/UFD1L
Introduction
Proteins of the AAA family (ATPases associated with different cellular activities) are, as their name implies, involved in a large number of cellular processes, including membrane fusion, organelle biogenesis, protein degradation and cell cycle regulation (reviewed in Patel and Latterich, 1998). They are characterized by a common motif that is defined by a conserved sequence of 230–250 amino acids. It includes the Walker type A and B cassettes, which are important for ATP binding and hydrolysis, and other regions of similarity unique to AAA proteins.
One extensively studied AAA-ATPase is mammalian p97 (first termed VCP, for valosin-containing protein; Koller and Brownstein, 1987) and its highly conserved homologues identified in Saccharomyces cerevisiae (Cdc48p) (Moir et al., 1982) (Fröhlich et al., 1991), Xenopus laevis (Peters et al., 1990), Thermoplasma acidophilum (VAT) (Pamnani et al., 1997) and many other organisms (see Patel and Latterich, 1998). Several lines of evidence implicate p97/Cdc48p in a variety of cellular processes. Its gene was first identified in yeast as a cell division cycle (CDC) mutant, which causes an arrest in mitosis with large budded cells and elongated nuclei spanning the mother–daughter junctions (Moir et al., 1982). At the biochemical level it has been proposed to function in homotypic membrane fusion events, including fusion of the endoplasmic reticulum in yeast (Latterich et al., 1995) and the reassembly of Golgi cisternae in mammals from fragments generated by mitotic cytosol (Rabouille et al., 1995a) or specific drugs (Acharya et al., 1995). Another line of evidence connects p97/Cdc48p to ubiquitin-dependent protein degradation. In yeast, it has been shown that Cdc48p is necessary for the degradation of a ubiquitin fusion reporter protein (Ghislain et al., 1996), which was first used to identify a ubiquitin-dependent degradation pathway (UFD) involving several other genes, termed UFD1 to UFD5 (Johnson et al., 1995). In mammalian cells, it has been reported that p97 is involved in the degradation of IκBα and copurifies with the proteasome. This has led to the speculation that p97 provides a link between the ubiquitylation step and the final degradation of proteins by the proteasome (Dai et al., 1998).
The remarkable functional diversity of p97/Cdc48p is most likely due to the deployment of one basic activity in a broad range of cellular processes. This basic activity may be protein unfolding or disassembly of protein complexes (Patel and Latterich, 1998). p97/Cdc48p is a type II AAA-ATPase with two AAA domains, D1 and D2, which bind ATP, and an N-terminal (N) domain. It forms a hexameric, barrel-shaped structure (Peters et al., 1992) somewhat reminiscent of the bacterial GroEL chaparonin (Xu et al., 1997). Recently, it was shown that the archaeal homologue of the eukaryotic p97/Cdc48p, VAT, is not only able to fold but also to unfold a model substrate in vitro in an ATP-dependent manner (Golbik et al., 1999). Further more, the N-domain is sufficient to refold a permissive substrate independent of any ATPase activity. It is interesting that this domain forms a groove that could serve to bind substrate peptides (Coles et al., 1999). Unfolding activity has also been shown for NSF (NEM-sensitive factor), another AAA-ATPase involved in membrane fusion with a similar structure to p97 (Whiteheart et al., 1994; Hanson et al., 1997). It unravels highly stable SNARE complexes (Söllner et al., 1993), a mechanism that has also been proposed for p97 (Rabouille et al., 1998).
If p97 provides an unfoldase or disassembly activity, the question arises as to how substrate specificity and recruitment of that activity to such diverse pathways as membrane fusion and protein degradation is achieved. In the fusion of mitotic Golgi fragments, p47 links p97 to its substrate, the t-SNARE, syntaxin 5, which it is then thought to unfold (Rabouille et al., 1998). p47 can thus be regarded as a membrane fusion-specific adapter for p97. Alternative adapters could then explain the role of p97 in other unfolding events. In yeast, two other physically interacting proteins of Cdc48p have been identified that are both involved in the UFD pathway (Johnson et al., 1995) and are therefore potential factors to direct Cdc48p to act in ubiquitin-dependent degradation. The first is the E4 polyubiquitylation factor, Ufd2p (Koegl et al., 1999), and the second, Ufd3p, a protein of unknown biochemical function (Ghislain et al., 1996). However, it is not clear whether these complexes exist in mammals and whether they have a structure similar to that of the p97/p47 complex. Furthermore, it is not clear that p97 binds to these proteins in a mutually exclusive fashion as demanded by the adapter hypothesis proposed above.
Here we present the identification and characterization of a complex comprising Ufd1 and Npl4. We show that the Ufd1/Npl4 complex binds to p97 in a mutually exclusive way with p47. We propose a model in which p47 and Ufd1/Npl4 represent alternative adapter modules that direct p97 to different pathways and help explain its functional diversity. With the identification of the Ufd1/Npl4 complex, we have also revealed a link between ubiquitylation and nuclear transport processes.
Results
Isolation of p97 binding proteins from rat liver cytosol
Cytosolic p97 migrates on gel filtration columns with an apparent Mr of 600–800 kDa, ∼150 kDa larger than the purified protein (Kondo et al., 1997). This difference suggested that p97 was bound to other cytosolic proteins and we were able to identify the homotrimeric p47 as one of the binding partners (Kondo et al., 1997). To identify others we isolated the high molecular weight fraction from cytosol, containing p97 complexes, and treated this fraction with high salt to release the bound proteins. After adding immobilized recombinant p97 and then lowering the salt, we reasoned that the released proteins would now bind to the immobilized p97, from which they could then be eluted by further salt treatment.
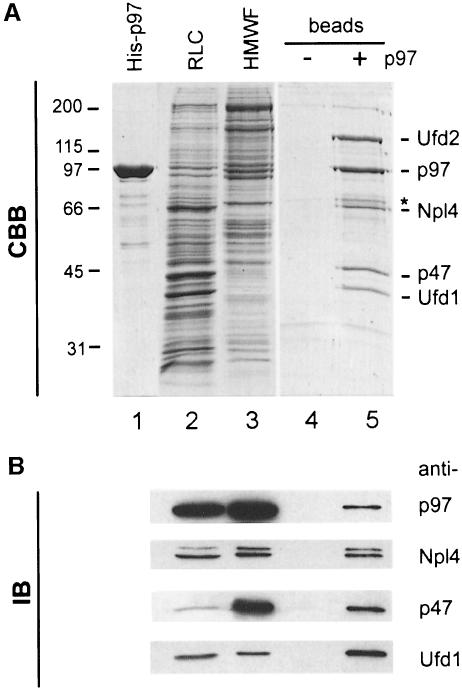
Immobilized p97 was prepared using recombinant His-tagged p97 (Figure 1A, lane 1). Recombinant p97 was structurally and functionally the same as endogenous p97. It was hexameric by gel filtration and negative staining (data not shown) and it catalysed the regrowth of Golgi cisternae from mitotic Golgi fragments (see below). It was immobilized after biotinylation to streptavidin beads. Rat liver cytosol (RLC, Figure 1A, lane 2) was subjected to gel filtration on a Superose 6 column and the high molecular weight fraction (HMWF, Figure 1A, lane 3) containing endogenous p97 was collected. This fraction was treated with 750 mM KCl to release the proteins bound to endogenous p97 and immobilized recombinant p97 was then added. The binding proteins were then transferred to the immobilized p97 by lowering the concentration of KCl to 200 mM. The p97 beads were then washed, bound proteins were eluted with 1 M KCl and TCA-precipitated (Figure 1A, lane 5). The same procedure was carried out using streptavidin beads alone as a control (Figure 1A, lane 4). Five major proteins were consistently eluted from p97 beads but not from control beads. These had apparent molecular weights of 130, 97, 67, 47 and 42 kDa. In addition, a weaker band at Mr of 71 kDa could be observed (asterisk in Figure 1A, lane 5). These bands were excised and subjected to tryptic digestion followed by mass spectrometry.
Fig. 1. Affinity purification of cytosolic binding proteins using His-tagged p97. (A) Coomassie Blue-stained gel (CBB) of the purification steps. Recombinant His-tagged p97 (1 µg in lane 1) was biotinylated and immobilized on streptavidin beads. Rat liver cytosol (RLC, 10 µg in lane 2) was fractionated by gel filtration. The high molecular weight fraction (HMWF, 10 µg in lane 3) containing the p97 peak was salt-treated to separate proteins bound to endogenous p97 and then incubated with immobilized His-p97 or streptavidin beads alone. After lowering the salt concentration to permit binding to immobilized p97, the bound proteins were eluted with salt, TCA-precipitated and fractionated using 10% SDS–PAGE (lanes 4 and 5). The bands indicated were excised and analysed by mass spectrometry. Those identified are indicated: p97, p47, Ufd1 (42 kDa), Ufd2 (130 kDa) and a 67 kDa protein with high homology to yeast Npl4p. See text for details. (B) Identification of the p97 binding proteins by immunoblotting (IB). Ten micrograms of RLC, 5 µg HMWF and 5% of the eluates were subjected to SDS–PAGE as decribed in (A). Proteins were transferred to nitrocellulose and incubated with antibodies against p97, p47 and with antibodies generated against Ufd1 and Npl4 (see Figure 3). The anti-Npl4 antibody recognized a doublet of 67 and 71 kDa [asterisk in (A)].
As expected, the 47 and 97 kDa proteins turned out to be p47 and p97, respectively, the latter being either endogenous p97 or recombinant p97 washed off the column. Sequencing of tryptic peptides from the 130 kDa band yielded a peptide (LAGGQTSQPTTPLTSPQ) that matched Ufd2 (DDBJ/EMBL/GenBank accession No. AAD02233), a human homologue of yeast Ufd2p that is involved in the UFD ubiquitylation pathway (see Introduction). Yeast Ufd2p has been shown to interact with Cdc48p (Koegl et al., 1999). We therefore conclude that Ufd2 interacts with p97 in mammals as well. The tryptic peptide pattern of the 42 kDa protein matched Ufd1, the mouse homologue of yeast Ufd1p, again involved in the UFD pathway. Its mouse and human homologues have been cloned in connection with the DiGeorge syndrome, a congenital developmental disorder (Pizzuti et al., 1997).
The peptide pattern of the 67 kDa protein did not match with any known protein in the database. The amino acid sequence of one of its peptides, however, matched two EST sequences from mouse (see Materials and methods) with high homology to yeast Npl4p, a protein known to be required for nuclear membrane integrity and nuclear transport (DeHoratius and Silver, 1996). Since neither of the two proteins, Ufd1 and Npl4, or their homologues had previously been shown to interact with p97 or Cdc48p, we decided to study this interaction in detail.
Rat Npl4 is highly conserved, ubiquitously expressed and contains a RanBP2 zinc finger
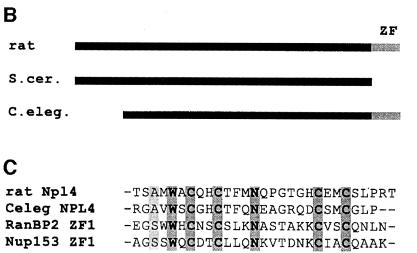
In order to clone the 67 kDa protein, primers were designed to amplify a DNA segment from a rat liver cDNA library representing the EST sequences identified using peptide 1. The amplification product was then used to screen a rat liver λgt11 cDNA library. Four clones were isolated, one of which was 4.5 kb long and encoded the entire open reading frame of rat Npl4. The sequence of the entire coding region and the deduced amino acid sequence are shown in Figure 2A. The peptides matching the data from mass spectrometric analysis in sequence or mass are shaded or underlined, respectively. The amino acid sequence is highly homologous to Npl4p from S.cerevisisae (33% similarity, 37% identity) but in addition contains a single zinc finger at the C-terminus (boxed in Figure 2A) not found in yeast Npl4p. However, the Caenorhabditis elegans gene F59E12.5 encoding a homologue of NPL4 contains the same zinc finger (Figure 2B). This motif has been classified as a RanBP2 zinc finger in the Prosite data bank (PS50199). It has homology to zinc fingers of RanBP2 and Nup153 (Figure 2C), nuclear pore proteins that can bind RanGDP via this motif (Nakielny et al., 1999; Yaseen and Blobel, 1999). Northern blot analysis of multiple tissue mRNA showed that rat Npl4 is ubiquitously expressed with a single mRNA of 4.5 kb in all tissues, though with varying expression levels (data not shown).
Fig. 2. The rat Npl4 sequence. (A) Nucleotide and predicted amino acid sequence of rat Npl4. Peptide 1 (boxed and shaded) was identified by mass spectrometric analysis of the 67 kDa band and matched two mouse EST clones with high homology to yeast Npl4. The EST sequence was used to screen a rat liver λgt11 cDNA library to isolate the full length rat Npl4 clone shown here. Peptides matching in sequence (shaded) or mass (underlined) with data obtained from the 67 kDa band are indicated. A zinc finger motif is boxed (see text). (B) Rat Npl4 (rat) shares high homology with yeast (S.cer) Npl4p and C.elegans (C. eleg.) gene F59E12.5 throughout the entire sequence (33 and 37% identities, 38 and 29% similarities, respectively), but only rat and the C.elegans sequence contain a zinc finger (ZF). (C) Alignment of homologous zinc fingers of rat Npl4 and C.elegans F59E12.5 with those of RanBP2 and Nup153.
Expression of recombinant Npl4 and Ufd1 and generation of antibodies
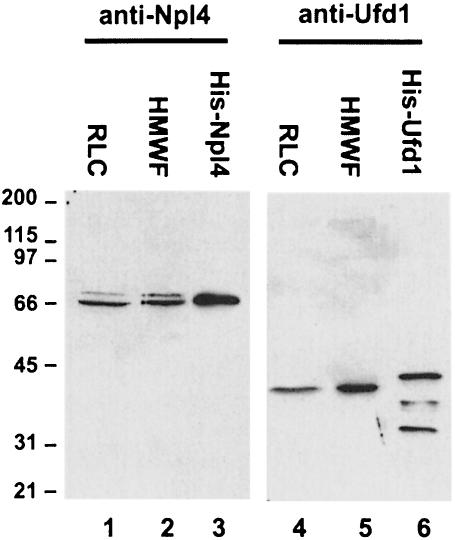
Mammalian Ufd1 and Npl4 were expressed in bacteria as GST- and His-tagged fusion proteins, which were then used for production and affinity purification of polyclonal antibodies. Anti-Npl4 antibodies recognized a major band in RLC and HMWF at 67 kDa (Figure 3, lanes 1 and 2) as well as a minor band at 71 kDa. The same doublet was observed in tissue culture cell lysates prepared with an instantly denaturing lysis buffer (data not shown) making it unlikely that proteolysis is the cause. Immunoisolation of both proteins with the anti-Npl4 antibody followed by mass spectrometric analysis revealed overlapping tryptic peptide patterns. It is therefore likely that the 71 kDa protein represents an alternative form of Npl4 in cytosol.
Fig. 3. Expression of recombinant Npl4 and Ufd1 and generation of antibodies. Rat Npl4 and mouse Ufd1 were expressed both as GST fusion proteins and as His-tagged proteins. Antibodies were raised in rabbits against the proteins fused to GST and affinity purified using His-tagged antigens. RLC (10 µg, lanes 1 and 4), HMWF (10 µg, lanes 2 and 5), His-Npl4 (20 ng, lane 3) and His-Ufd1 (20 ng, lane 6) were fractionated using SDS–PAGE and blots probed using anti-Npl4 (lanes 1–3) and anti-Ufd1 (lanes 4–6) antibodies. Anti-Npl4 recognizes a doublet at 67 and 71 kDa in RLC and HMWF. Anti-Ufd1 recognizes a single band at 42 kDa. Both recombinant proteins are slightly bigger than the endogenous proteins because of the His-tag and the His-Ufd1 fraction contains proteolytic degradation products.
The antibodies against Ufd1 recognized a single band at 42 kDa in RLC and HMWF (Figure 3, lanes 4 and 5) that is bigger than the calculated 34.5 kDa deduced from the amino acid sequence. Both recombinant proteins were slightly bigger than the endogenous proteins because of the His tag (lanes 3 and 6). Both antibodies were used to confirm the identity of Npl4 and Ufd1 eluted from immobilized p97 in the original affinity purification (Figure 1B).
p97, Npl4 and Ufd1 form a ternary complex
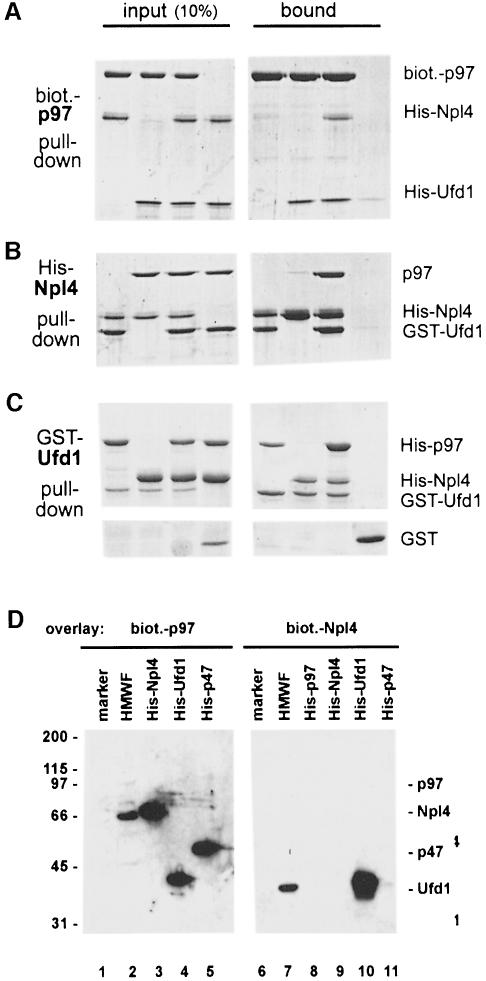
The affinity isolation of mammalian Npl4 and Ufd1 using p97 beads did not reveal whether these proteins interacted directly or indirectly with p97. To address this question, binding experiments with purified components were carried out in which one of the proteins was pulled down via a specific tag and the bound proteins analysed (Figure 4A–C). Biotinylated p97 immobilized on streptavidin beads pulled down Ufd1 (Figure 4A), but only very little or no Npl4. Npl4 bound to p97 only when Ufd1 was present. Similarly, His-tagged Npl4 on nickel beads did not bind p97 (Figure 4B). However, His-Npl4 bound GST–Ufd1, and in the presence of GST–Ufd1, p97 bound as well. Consequently, when GST–Ufd1 was incubated with either p97 or Npl4 and precipitated using glutathione beads, it bound both proteins independently (Figure 4C). Interestingly, when both p97 and Npl4 were present, p97 could be pulled down more efficiently indicating that Npl4 not only binds Ufd1, but also increases its affinity for p97.
Fig. 4. Complexes of p97, Npl4 and Ufd1. (A–C) Pull down experiments with purified proteins followed by SDS–PAGE and Coomassie Blue staining. The panel on the left shows 10% of input, the right panel the material pulled down. (A) Biotinylated His-p97 was incubated with His-Npl4, His-Ufd1 or His-Npl4/His-Ufd1 and pulled down with streptavidin beads. His-Npl4/His-Ufd1 alone served as the control. (B) His-Npl4 was incubated with GST–Ufd1, rat liver p97 or GST–Ufd1 and rat liver p97 together and pulled down with Ni-NTA beads. GST–Ufd1 and p97 together served as a control. (C) GST–Ufd1 was incubated with His-p97, His-Npl4, or His-p97 and His-Npl4 together and pulled down with glutathione beads. GST alone with His-p97 and His-Npl4 served as the control. Note that more p97 was pulled down in the presence of His-Npl4. (D) Far-Western blots of p97 binding proteins probed with biotinylated p97 or biotinylated Npl4 in the overlay. The cytosolic HMWF (5 µg, lanes 2 and 7) and recombinant proteins as indicated (50 ng in lanes 3–5, 20 ng in lanes 8–11) were fractionated by SDS–PAGE, transferred onto nitrocellulose and overlaid with biotinylated p97 (lanes 1–5) or biotinylated Npl4 (lanes 6–11). Bound probes were visualized using streptavidin-HRP followed by chemiluminescence. p97 recognized recombinant Npl4, Ufd1 and p47, but only Npl4 from cytosol. Npl4 in the overlay bound endogenous Ufd1 in HMWF and the recombinant protein, but not p97, Npl4 or p47. Marker proteins served as a negative control (lanes 1 and 6, myosin, β-galactosidase, phosphorylase b, serum albumin, ovalbumin, carbonic anhydrase, 100 ng each).
Binding was also examined by far-Western blotting with biotinylated p97 and Npl4 in the overlay probing HMWF and purified recombinant proteins transferred onto a filter (Figure 4D). Consistent with the results from pull-down experiments, Npl4 in the overlay interacted with cytosolic and recombinant Ufd1 (lanes 7 and 10), but could not directly interact with p97 (lane 8), with p47 (lane 11) nor with itself (lane 9). When p97 was used as a probe in the overlay, it interacted as expected with recombinant Ufd1 and p47 (lanes 4 and 5). However, reaction with the endogenous, cytosolic forms of both proteins was very weak and only visible after long exposure (lane 2). In contrast, and surprisingly, p97 interacted very strongly with recombinant and cytosolic Npl4 (lanes 2 and 3). Npl4 therefore contains a strong binding site for p97, which is accessible in the denatured Npl4 on the filter, and it suggests that Npl4 can bind directly to p97 also in solution under certain conditions.
Cytosol has two independent complexes containing Npl4 and Ufd1
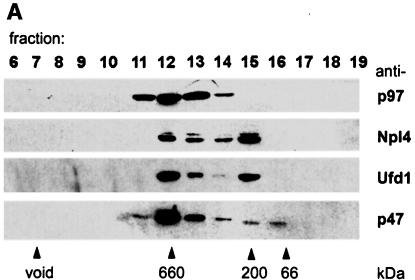
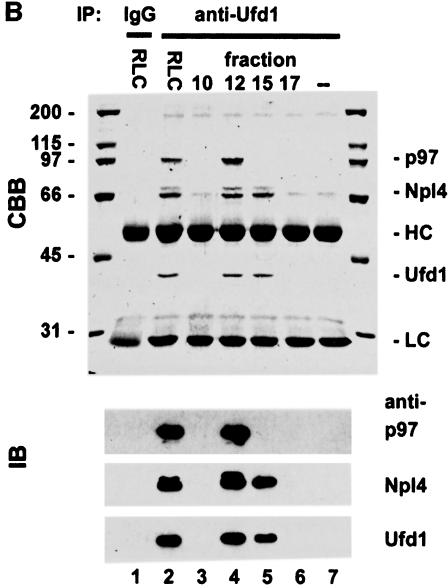
Most if not all of the p47 in cytosol is bound to p97 (Kondo et al., 1997). To find out if this was the case for Npl4 and Ufd1, fractions from gel filtration of RLC were analysed by immunoblotting with antibodies against p97 and its binding proteins. As expected, Npl4, Ufd1 and p47 all co-fractionated with p97 at an apparent Mr of 600–800 kDa (Figure 5A). However, the distribution of Npl4 and Ufd1 was biphasic with a second peak at ∼200 kDa, indicating a second complex lacking p97. Neither Npl4 nor Ufd1 could be found in later fractions where monomeric forms of the two proteins would be expected. To study these two complexes, Ufd1 was immunoprecipitated from the two peak fractions under low stringency conditions and analysed by Coomassie Blue-stained SDS–PAGE gels and immunoblotting (Figure 5B). When Ufd1 was precipitated from the p97 peak fraction (fraction 12), a complex containing Ufd1, Npl4 and p97 could be isolated (Figure 5B, lane 4). When precipitated from the 200 kDa peak (fraction 15), only Npl4 co-precipitated with Ufd1 (lane 5), indicating an independent complex containing Npl4 and Ufd1. When precipitated from total cytosol, all three proteins were present, though the relative amounts reflected the fact that both complexes were present (lane 2). As observed earlier, Npl4 appeared as a doublet in both complexes.
Fig. 5. Cytosol contains two independent complexes: a binary Ufd1/Npl4 and a ternary p97/Ufd1/Npl4 complex. (A) Rat liver cytosol (RLC) was fractionated by gel filtration on a Superose 6 column. Aliquots were separated by SDS–PAGE and p97, and its binding proteins analysed by immunoblotting. Almost all the p47 comigrated with the p97 peak at 600–800 kDa. In contrast, Npl4 and Ufd1 ran together exhibiting a biphasic distribution, the first peak co-migrating with p97, the second at ∼200 kDa. (B) Immunoprecipitation of Ufd1 complexes from RLC gel filtration fractions. Total RLC and selected gel filtration fractions identified in (A) were subjected to low stringency immunoprecipitation with a monoclonal anti-Ufd1 antibody. As controls, immunoprecipitation was performed from total RLC using non-immune mouse IgG (lane 1) and from buffer using anti-Ufd1 (lane 7). Precipitates were analysed by SDS–PAGE followed by Coomassie Blue-staining (CBB) or immunoblotting (IB) using specific rabbit antibodies. Npl4 and p97 were both co-immunoprecipitated with Ufd1 from total RLC (lane 2). When precipitated from the high molecular weight peak (lane 4), Ufd1 pulled down both Npl4 and p97, whereas precipitation from the 200 kDa peak (lane 5), bound only Npl4. Note that Ufd1 was not precipitated from fraction 17 (lane 6) where monomeric Ufd1 would be expected. HC, IgG heavy chain; LC, IgG light chain.
Ufd1/Npl4 and p47 compete for p97 binding and form alternative complexes in cytosol
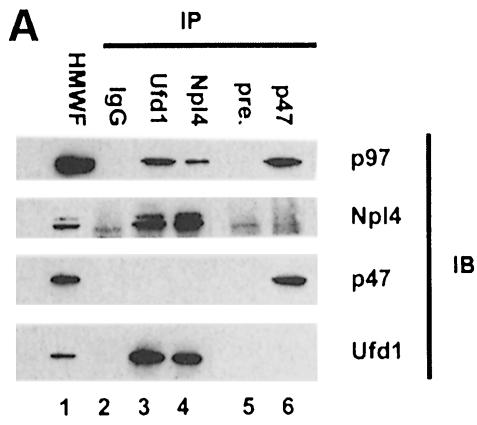
Since both p47 and Ufd1/Npl4 form stable complexes with p97 in cytosol, we wanted to find out whether they could bind at the same time. Two types of experiments were performed. One involved immunoprecipitation of all three binding proteins from cytosol using specific antibodies and analysis of co-precipitating components by immunoblotting (Figure 6A). Consistent with the previous results, anti-Ufd1 and anti-Npl4 antibodies pulled down complexes containing Ufd1, Npl4 and p97, but not p47 (lanes 3 and 4). When an anti-p47 serum was used, only p47 and p97, but not Ufd1 or Npl4, could be detected in the precipitates (lane 6). Cytosolic p97 therefore comprises at least two alternative complexes: p97/p47 and p97/Ufd1/Npl4.
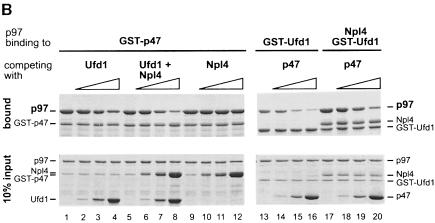
Fig. 6. Ufd1/Npl4 and p47 compete for p97 binding and form alternative complexes in cytosol. (A) Immunoprecipitation of p97 complexes from RLC and analysis of their components. Total RLC was subjected to low stringency immunoprecipitation (IP) using purified IgG, purified antibodies against Ufd1 or Npl4, preimmune-serum (pre-) or anti-serum against p47. Precipitates were fractionated using SDS–PAGE, then analysed by immunoblotting (IB) with antibodies against p97, Npl4, Ufd1 and p47. HMWF (5 µg) was loaded as a reference (lane 1). Note that anti-Ufd1 precipitated Npl4 and p97 but not p47, whereas anti-p47 precipitated p97, but not Npl4 or Ufd1. Rabbit antibodies were used for IP, mouse antibodies for immunodetection, apart from anti-Npl4, which explains the high background. (B) Competition experiments using purified proteins. GST–p47 (lanes 1–12) or GST–Ufd1 with or without Npl4 (lanes 13–20) were incubated with p97 in the absence or presence of increasing amounts of the alternative binding partners (molar excess: 0, 1, 5 and 25 times) as indicated. GST fusion proteins were pulled down with glutathione beads and the bound complexes were analysed by SDS–PAGE followed by Coomassie Blue staining. The lower panel shows 10% of the input, the upper panel shows bound protein. Note the amount of p97 pulled down in each lane.
The second type of experiment was a competition experiment. GST–p47 was incubated with p97 in the presence of increasing amounts of Ufd1, Ufd1/Npl4 or Npl4 alone. In parallel, GST–Ufd1 was incubated with p97 in the absence or presence of Npl4 and increasing amounts of p47 as the competitor. The GST fusion proteins were pulled down and precipitates analysed using Coomassie Blue-stained SDS–PAGE gels (Figure 6B). Ufd1 alone competes with p47 for binding to p97 (lanes 1–4), but in the presence of Npl4 it does so more efficiently (lanes 5–8). Npl4 alone does not influence the binding of p47 to p97 (lanes 9–12). Conversely, when GST–Ufd1 was used, p47 could inhibit binding of Ufd1 to p97 very efficiently (lanes 13–16). The efficiency was lowered when Npl4 was present (lanes 17–20). Ufd1/Npl4 had the same effect on p97/p47 formation as p47 had on p97/Ufd1/Npl4 formation. We conclude that p47 and the Ufd1/Npl4 complex compete for p97 binding with comparable affinities. Ufd1 is sufficient for binding to p97 but the binding is enhanced when it is in a complex with Npl4.
Ufd1/Npl4 does not function in p97-mediated mitotic Golgi membrane fusion
The need for p47 in the p97-mediated cisternal regrowth from mitotic Golgi fragments (MGFs) is well established (Kondo et al., 1997). Since Ufd1/Npl4 forms a similar complex with p97 as p47, we wanted to know whether it was able to promote Golgi membrane fusion as well. To circumvent the need to purify p97 from rat liver cytosol, we used the bacterially expressed p97 in the Golgi reassembly assay. Comparative experiments showed that the recombinant protein was functional and behaved in a very similar manner to the endogenous protein (data not shown).
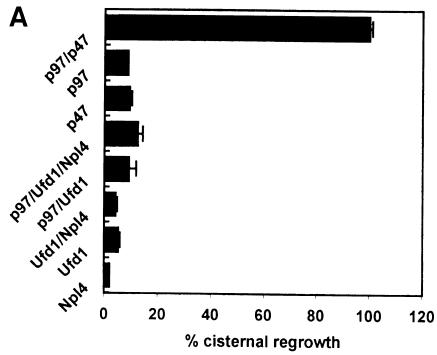
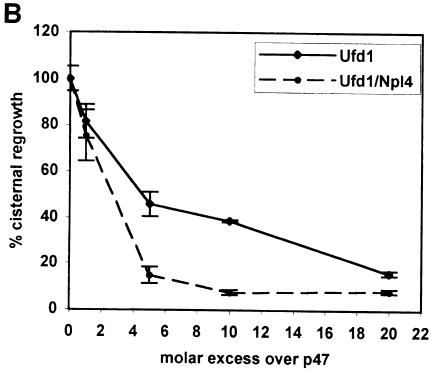
MGFs were generated from rat liver Golgi by incubation with mitotic HeLa cytosol. They were sedimented through a 0.5 M sucrose cushion to remove endogenous factors and incubated at 37°C for 60 min with different combinations of recombinant p97 and its binding proteins. Reactions were processed for electron microscopy (EM) and the amount of cisternal regrowth determined (Figure 7A). Only a combination of p97 and p47 could promote reassembly. Neither p97 with Ufd1 nor Ufd1/Npl4 had any significant effect. Neither did Ufd1/Npl4 nor the individual proteins alone. Under the same conditions, cisternal regrowth in the presence of p97/p47 was inhibited when increasing amounts of Ufd1 or Ufd1/Npl4 were added (Figure 7B). This observation is best explained as a competition between the two protein complexes for p97, with only p97/p47 being able to promote fusion. This effect was stronger with Ufd1/Npl4 than with Ufd1 alone, confirming its higher affinity for p97. Therefore, of the two complexes, only p47 can direct p97 to mediate membrane fusion.
Fig. 7. Ufd1/Npl4 does not function as a p97 co-factor in Golgi membrane fusion. (A) MGFs isolated through a 0.5 M sucrose cushion were incubated with p97/p47, p97, p47, p97/Ufd1/Npl4, p97/Ufd1, Npl4 or Ufd1 for 60 min at 37°C. Incubations were processed for EM, and the percentage of membrane in cisternae determined. Results are presented as the percentage cisternal regrowth, where 0% represents starting MGFs and 100% represents the effect of p97/p47. Values represent means ± SEM (n = 3). (B) MGFs were incubated as in (A) with p97/p47 in the presence of increasing amounts of Ufd1 alone or Ufd1/Npl4 (1, 5, 10 and 20 molar excess over p47).
Subcellular localization in tissue culture cells
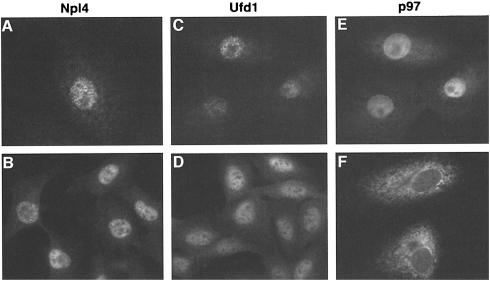
The subcellular distribution of Npl4, Ufd1 and p97 was determined in exponentially growing tissue culture cells by immunofluorescence microscopy using several cell lines and fixation conditions (Figure 8). Npl4 had the same staining pattern in both BSC-1 and NRK cells (Figure 8A and B), with most of the staining restricted to the nucleus. This is the same pattern that was observed for Npl4p in S.cerevisiae (DeHoratius and Silver, 1996). Ufd1 had a very similar staining pattern in the same cell lines using both monoclonal (Figure 8C and D) and polyclonal (data not shown) antibodies.
Fig. 8. Immunofluorescence microscopy showing the localization of Npl4, Ufd1 and p97. Monkey BS-C-1 cells (A, C, E and F) and Normal rat kidney cells (B and D) were stained with affinity-purified antibodies against Npl4 and p97, and a monoclonal antibody against Ufd1 as indicated. Cells were either fixed with paraformaldehyde/methanol (A, C, D and E) or methanol (B and F). C and E is a double staining of Ufd1 and p97 in the same cells. Note that the staining pattern for p97 depends on the fixation method (see text).
The p97 staining pattern depended on the fixation conditions, as has been reported before (Peters et al., 1990). Using paraformaldehyde (Figure 8E), the staining pattern was similar to that for Ufd1 and Npl4 though there was more staining in the cytoplasm. Using methanol fixation (Figure 8F), the staining of the cytoplasm was enhanced relative to the nucleus and had the characteristic reticular structure of the ER including the nuclear envelope. These data suggest that Ufd1 and Npl4 are mostly restricted to the nucleus whereas p97 is, in addition, found in the cytoplasm.
Discussion
To understand the mechanism by which p97 can function in different cellular pathways, we set out to isolate binding proteins in addition to its co-factor in membrane fusion, p47. We wanted to see whether alternative binding complexes exist that are responsible for directing p97 to other processes. We exploited the salt sensitivity of p97 binding to other proteins to set up a purification system that resulted in the isolation of three proteins, Npl4, Ufd1 and Ufd2. Npl4 was a previously unknown protein in mammals (see below).
The binding of Npl4 and Ufd1 to p97 was studied in detail. Binding experiments with recombinant proteins revealed that Npl4 and Ufd1 form a binary complex on their own that can bind p97 to form a ternary complex. In solution, Ufd1 can bind Npl4 as well as p97 independently, whereas Npl4 and p97 do not bind in the absence of Ufd1. Ufd1 therefore forms a bridge in the p97/Ufd1/Npl4 complex. However, two lines of evidence indicate that this interaction is more complex. First, Npl4 enhances the affinity of Ufd1 for p97 in pull-down experiments. Secondly, Npl4 contains a binding site to p97, which was only revealed in overlay assays when denatured Npl4 on the filter was probed with p97. One possibility is that Ufd1 brings p97 and Npl4 together, thereby exposing a binding site on Npl4 for p97.
The binary Ufd1/Npl4 and the ternary p97/Ufd1/Npl4 exist as independent complexes in cytosol. This was shown by gel filtration followed by immunoblotting and immunoprecipitation of Ufd1 complexes. Unlike p47, Npl4 and Ufd1 had a biphasic distribution. The complex immunoprecipitated from the 600–800 kDa peak contained p97/Ufd1/Npl4, the complex at ∼200 kDA contained only Ufd1/Npl4. It will be interesting to find out why and how the binding of Ufd1/Npl4 to p97 is regulated, especially since the cellular pool of p47 appears to be almost completely bound to p97. Preliminary experiments have been carried out to measure the stoichiometry of binding of Ufd1 to Npl4 alone and in combination with p97. Immunoprecipitated complexes were fractionated by SDS–PAGE and stained with Coomassie Blue. Densitometric analysis yielded a ratio of p97:Ufd1:Npl4 of 6:3:3. Further biophysical measurements will be needed to confirm this stoichiometry but it is worth noting that this ratio is the same as that for p97/p47 (6:3; Kondo et al., 1997), suggesting a similar structural organization.
Ufd1/Npl4 and p47 form mutually exclusive complexes with cytosolic p97. Antibodies against Ufd1 pulled down Npl4 and p97 from cytosol but not p47. Antibodies against p47 pulled down p97 but not Ufd1 or Npl4. Experiments with recombinant proteins showed that Ufd1 could displace p47 from p97, especially in the presence of Npl4. p47 could displace both Ufd1 and Npl4 from p97. These data also showed that p47 and Ufd1/Npl4 bound p97 with comparable affinities. It will be interesting to extend these experiments to other potential adapter complexes to see whether they also bind in a mutually exclusive manner. Our preliminary experiments with the mammalian Ufd2p homologue, which also binds p97, show that it is not found in complexes of p97 with either Ufd1/Npl4 or p47.
In contrast to p97/p47, the p97/Ufd1/Npl4 complex does not function in post-mitotic Golgi fusion. Neither p97/Ufd1/Npl4 nor p97/Ufd1 promoted cisternal regrowth from mitotic Golgi fragments. When Ufd1/Npl4 or Ufd1 were titrated into the assay, fusion catalysed by p97/p47 was inhibited. This observation excludes a function for p97/Ufd1/Npl4 in the reassembly pathway, at least in vitro, and is best explained by competition between the two adapters. We therefore suggest that Ufd1/Npl4 represents an alternative adapter to p47 that directs p97 to another cellular pathway in the same way that p47 directs it to act in Golgi membrane fusion.
The pathway to which Ufd1/Npl4 directs p97 is still unknown, but a large body of work on yeast and mammalian Ufd1p and yeast Npl4p raises some obvious possibilities. In yeast, Ufd1p along with four other proteins (Ufd2p–Ufd5p) was identified in a genetic screen for a pathway that recognizes ubiquitin as a degradation signal (Johnson et al., 1995). A non-physiological fusion protein comprising an N-terminal ubiquitin fused to β-galactosidase was used as the reporter for degradation. The ubiquitin fusion degradation (UFD) proteins were shown to be required at specific steps in the pathway of ubiquitylation and degradation of this reporter protein. The same screen also showed that the p97 homologue, Cdc48p, was required (Ghislain et al., 1996). Two proteins in the pathway, Ufd2p and Ufd3p, have already been shown to interact physically with Cdc48p. Ufd2p is required for efficient polyubiquitylation (Koegl et al., 1999) though it is not clear why it binds to Cdc48p, since Cdc48p is not required for polyubiquitylation either in vitro (Koegl et al., 1999) or in vivo (Ghislain et al., 1996). The function of Ufd3p is not fully understood, but it somehow influences the level of free ubiquitin. It will be interesting to compare these Cdc48p/Ufd complexes with those found in mammals. So far, we have evidence that a mammalian homologue of Ufd2p binds to p97, but not a Ufd3p homologue.
Though the exact biochemical function of Ufd1p and Cdc48p in the UFD pathway is unclear, they both act late in the pathway following polyubiquitylation. Intriguingly, both proteins are essential for viability and conditional mutants result in formation of interconnected daughter cells late in mitosis (Moir et al., 1982; Johnson et al., 1995). Given these observations and the fact that they interact with each other in mammals, it seems likely that they both act together in a ubiquitin-dependent process that occurs during mitosis. The fact that Npl4 is also essential in yeast suggests that it might be involved in the same process. To our knowledge, however, Npl4 has not been tested for its involvement in the UFD pathway. Ufd2p and Ufd3p are the other two known binding proteins for Cdc48p. However, since they are not essential (Johnson et al., 1995), they cannot account for the essential role of Cdc48p in mitosis.
In humans, the gene coding for Ufd1, UFD1L, has been linked to the DiGeorge syndrome (Pizzuti et al., 1997; Yamagishi et al., 1999), a congenital developmental disorder with cardiac and craniofacial defects. Though controversial, the product of UFD1L has been proposed to regulate the amount of an unknown crucial substrate by protein degradation in neural crest cells of the third and fourth pharyngeal arches. A defect in that pathway would then cause malformations in this area (reviewed by Baldini, 1999; Schinke and Izumo, 1999). A homozygous deletion of UFD1L in mice is lethal (Lindsay et al., 1999). If Ufd1 is linked to the disease, it will be interesting to find out whether p97 and Npl4 are involved as well.
Intriguingly, the third protein of the novel complex, Npl4, has never been linked to either p97/Cdc48p or to ubiquitin-dependent degradation. Yeast Npl4p is essential for nuclear transport, both import of proteins and export of mRNA (DeHoratius and Silver, 1996). Its structural and functional features led to the conclusion that it is a component of the nuclear pore complex (NPC). One possibility, therefore, is that Npl4 is involved in the import of p97/Ufd1 into the nucleus. This would be consistent with the presence of all three proteins in the nucleus as shown by immunofluorescence microscopy. In yeast the nuclear import of Cdc48p is dependent on its phosphorylation and exposure of a nuclear localization signal (Madeo et al., 1998). Perhaps Npl4 is part of this mechanism.
Since Npl4 also exists as a complex with Ufd1 in the absence of p97, this further suggests that the Ufd1/Npl4 complex might be involved in the other nuclear transport processes.
Ufd1p has been reported to bind Uba2p in yeast (del Olmo et al., 1997) and Uba2p has been shown to activate the ubiquitin-like protein Smt3p (Johnson et al., 1997). The attachment of the mammalian homologue of Smt3p, SUMO-1, to RanGAP directs the latter to the nuclear pore complex (Mahajan et al., 1997; Matunis et al., 1998). It is therefore possible that Ufd1 not only functions in ubiquitin-dependent degradation but also in other pathways involving ubiquitin-like proteins including the regulation of nucleo-cytoplasmic transport through Smt3p/SUMO-1. In this case, the interaction of Ufd1 and Npl4 might help explain the role of Npl4 in nuclear transport processes. Further work will be needed to explore these possibilities.
We also noted an interesting difference between yeast and mammalian Npl4. Mammalian Npl4, as well as its homologue in C.elegans, contains a single copy of a zinc finger motif at the very C-terminus that is not found in the yeast protein. This motif, termed the RanBP zinc finger, is also found in other NPC proteins including RanBP2 and Nup153. It has been shown to be the binding site of the small GTPase Ran in its GDP-bound form (Yaseen and Blobel, 1999; Nakielny et al., 1999). Whether or not Npl4 can bind Ran and whether it links it to an activity of p97 remains to be determined.
The identification and characterization of the new p97 binding complex Ufd1/Npl4 supports a model in which p97 is directed to different cellular pathways through alternative adapters. Our present working hypothesis is that p97/Ufd1/Npl4 is involved in ubiquitin-dependent processes and that its activity is required and essential in mitosis. Further work will concentrate on the identification of its targets in mitosis and how this process is linked to the function of p97/p47 in mitotic Golgi reassembly.
Materials and methods
Purification of binding proteins
Rat liver cytosol (RLC, 400 mg) (Hui et al., 1998) was fractionated by gel filtration in RB buffer [25 mM Tris–HCl pH 7.4, 150 mM KCl, 2 mM MgCl2, 1 mM dithiothreitol (DTT), 5% (v/v) glycerol] using a Superose 6 HR 10/30 column (Pharmacia Biotech). A HMWF containing the p97 peak as determined by Western blotting was collected. Recombinant p97 (1 mg, see below) was biotinylated using EZ-Link (Pierce) according to the manufacturer’s instructions, bound to streptavidin beads (UltraLink, Pierce) and washed with 1 M KCl in RBT (RB with 1% Triton X-100). HMWF (10 mg) was adjusted to 750 mM KCl and immobilized p97 was added. The mixture was diluted to 200 mM KCl with RBT (without KCl) and incubated with agitation for 2 h at 4°C. The same procedure was performed using beads alone as the control. The beads were then washed with RBT and eluted with 1 M KCl in RBT. Eluted proteins were trichloroacetic acid (TCA)-precipitated, washed with ice-cold acetone and solubilized in reducing SDS–PAGE sample buffer. The samples were separated on a 10% SDS–PAGE gel, Coommassie Blue-stained and protein bands of interest were excised.
Mass spectrometric analysis of proteins
The stained protein bands were excised from the gel and digested with trypsin essentially according to Shevchenko et al. (1996). Following overnight digestion 0.5–1 µl aliquots were sampled directly from the digest supernatant for peptide-mass fingerprint (PMF) analysis using a TofSpec 2E laser desorption time-of-flight MS (Micromass, UK). Experimental peptide masses were screened against the OWL non-redundant composite protein database (∼350 000 entries) using the MASCOT search program (Bleasby and Wootton, 1990) (Perkins et al., 1999). The remaining digested peptides (>90% of total digest) were eluted with successive washes (30 µl each) of 50% (v/v) aq. acetonitrile/5% (v/v) trifluoroacetic acid (three times) followed by 3× 30 µl washes of acetonitrile. Pooled washes were dried in vacuo and derivatized with N-succinimidyl-2-morpholine acetate (SMA) in order to enhance b-ion abundance and facilitate sequence analysis by tandem MS (Sherman et al., 1995; Hoss et al., 1999). Derivatized peptides were sequenced de novo by low-energy collision-activated dissociation (CAD) using an LCQ Ion-Trap MS (ThermoQuest, CA) fitted with a nanoelectrospray source (Hunt et al., 1986; Wilm and Mann, 1996). CAD was performed using 1 mTorr helium with collisional offset voltages between 22 and 38% of full voltage (5 V).
Cloning of the rat Npl4 cDNA
The sequence of peptide 1 (HVDNIMFENHTVADR) obtained from the analysis of the 67 kDa band matched two overlapping mouse EST sequences (DDBJ/EMBL/GenBank accession Nos AA015556 and AA020051). A segment of 350 bp, termed NPL4A, was amplified by nested PCR from a mouse liver, first strand cDNA library (Origene, MD, USA) using primers NPL4-1 (5′-GATCGATCAATACCTCAGC-3′), NPL4-2 (5′-CTGGTTTCCGGTCTTTCTCC-3′), NPL4-3 (5′-CGAGAC CCACAGCTATGCCGC-3′) and NPL4-4 (5′-AGAAGTCAAGGAAGC GGTCAC-3′). This segment (representing bp 453–802 in Figure 2A) was used as a probe to screen a rat liver λgt11 cDNA library (Clontech, CA, USA). Screening 0.75 million plaques resulted in the isolation of nine positive clones, four of which were sequenced and shown to match the probe. One clone was 4.5 kb in length and encoded the entire open reading frame of rat Npl4 (DDBJ/EMBL/Genbank accession No. AF234600).
Expression and purification of p97, p47, Npl4 and Ufd1
The open reading frame (ORF) of mouse p97 was PCR amplified and cloned into the bacterial expression vector pQE9 (Qiagen). Recombinant His-tagged p97 was expressed in E.coli after induction with IPTG. Cells were lysed by sonication in LyB (500 mM KCl, 100 mM Tris pH 7.4, 5 mM MgCl2, 1 mM ATP, 5% glycerol, 2 mM β-mercaptoethanol, 20 mM imidazole) and bound to Ni-NTA agarose (Qiagen). The resin was washed with 25 ml WB (150 mM KCl, 50 mM HEPES pH 7.4, 5 mM MgCl2, 1 mM ATP, 5% glycerol, 2 mM β-mercaptoethanol, 20 mM imidazole) and with 5 ml 1 M KCl in WB. Protein was eluted with 350 mM imidazole in WB, loaded on a sucrose gradient (5–30% in WB) and centrifuged in a VTi65.1 rotor (Beckman) for 75 min at 65 000 r.p.m. Peak fractions were collected, exchanged into storage buffer (150 mM KCl, 20 mM HEPES pH 7.4, 1 mM MgCl2, 5% glycerol, 1mM DTT) on a PD10 column (Pharmacia) and snap frozen. p97 from rat liver cytosol was purified as described previously (Kondo et al., 1999). All buffers contained a protease inhibitor cocktail (completeTM, EDTA-free, Boehringer Mannheim).
The ORF of Ufd1 (Pizzuti et al., 1997) was PCR amplified from a mouse liver, first strand cDNA library (Origene, MD, USA) and cloned into pBluescript II KS+ (Stratagene) for sequence verification. The ORFs of Ufd1 and Npl4 were cloned into the pGEX-4T (Pharmacia) and pQE9 (Qiagen) expression vectors using the BamHI sites in both cases in order to generate GST- and His-tagged versions of both proteins. The ORF of p47 was cloned into pGEX-4T using the BamHI site. Recombinant GST- and His-tagged proteins were expressed in E.coli and purified as for His-p97 without the sucrose gradient step. GST-tagged proteins were bound to glutathione Sepharose (Pharmacia) and eluted with 25 mM glutathione in WB. His-p47 was prepared as described previously (Kondo et al., 1997).
Antibodies
Polyclonal antisera against Ufd1, Npl4 and p47 were raised in rabbits by injecting the GST fusion proteins according to Harlow and Lane (1988). Specific antibodies against Ufd1 and Npl4 were purified using the His-tagged antigens immobilized on AffiGel (Bio-Rad). The monoclonal antibodies 1E6 (immunofluorescence) and 5E2 (immunoprecipitations) were generated against GST–Ufd1. The polyclonal anti-p97 antibody N5 has been described previously (Müller et al., 1999). A monoclonal antibody against p47 was kindly provided by Hisao Kondo. A monoclonal antibody against p97 was kindly provided by J.M.Peters.
Pull-down experiments with purified proteins
Binding experiments were performed with purified proteins carrying different tags. Tagged proteins and their interacting partners were precipitated by tag-specific beads. The combinations of proteins and resins were as follows: (i) biotinylated His-p97 with His-Npl4 and His-Ufd1 using streptavidin–Sepharose (Pierce); (ii) His-Npl4 with rat liver p97 and GST–Ufd1 using magnetic Ni-NTA agarose (Qiagen); (iii) GST–Ufd1 with His-p97 and His-Npl4 using glutathione–Sepharose (Pharmacia). Binding was performed with 1–3 µg of each protein in 200 µl of BB (25 mM Tris–HCl pH 8.0, 200 mM KCl, 2 mM MgCl2, 1 mM ATP, 1 mM DTT, 5% glycerol, 1% Triton X-100, 1 mg/ml soybean trypsin inhibitor) for 1 h at 4°C. Ten microlitres of beads were added and incubated for 30 min. Beads were washed with BB, bound proteins were eluted with sample buffer and analysed by SDS–PAGE followed by Coomassie Blue staining.
Competition experiments were carried out in the same way. Two micrograms of GST–p47 or GST–Ufd1 was incubated with 4 µg His-p97 in the presence of 0, 1, 5 or 25 molar excess of competitors and pulled down with glutathione–Sepharose. The combinations were as follows: GST–p47 competing with His-Ufd1, His-Ufd1/His-Npl4 or His-Npl4; GST–Ufd1 with His-p47; GST–Ufd1/His-Npl4 with His-p47.
SDS–PAGE and Western blotting
Protein samples were solubilized in reducing SDS–PAGE sample buffer, boiled for 3 min, and analysed using SDS–polyacrylamide gels followed by staining with Coomassie Brilliant Blue R-250. Western blotting on nitrocellulose was performed using a semi-dry blotter. Blocking and antibody incubations were performed in PBS plus 5% (w/v) low-fat milk powder. All secondary antibodies were HRP conjugates (Tago, Buckingham, UK) detected using ECL (Amersham Life Science, UK).
Overlay experiments
Protein samples were separated by SDS–PAGE and transferred onto nitrocellulose by semi-dry blotting. Membranes were blocked in OB [5% (w/v) low-fat milk powder, 0.5% Triton X-100, 5 mM MgCl2 in PBS] overnight, changing the buffer several times. They were then incubated with biotinylated p97 or biotinylated Npl4 at 10 µg/ml in OB for 2 h and washed. The bound probe was detected by incubation with streptavidin-conjugated HRP (Vector, CA, USA) diluted 1:2000 for 1 h and, after washing, visualized with ECL Plus (Amersham Life Science, UK). All steps were carried out in OB at 4°C.
Immunoprecipitations
Fractions of interest were adjusted to 500 µl with IPB (25 mM Tris–HCl pH 7.4, 200 mM KCl, 2 mM MgCl2, 1 mM DTT, 5% glycerol, 1% Triton X-100). Purified antibody (1 µg), 5 µl of antiserum or 100 µl of hybridoma cell supernatant was added and incubated for 1 h at 4°C. Rabbit IgG was bound to protein A, mouse IgG to protein G–Sepharose for 30 min. The Sepharose was washed with IPB, proteins were eluted with sample buffer and analysed by SDS–PAGE.
Golgi reassembly assay
The Golgi reassembly assay was performed as described previously (Rabouille et al., 1995b) with modifications (Shorter and Warren, 1999). Briefly, MGFs were prepared from purified rat liver Golgi stacks and mitotic HeLa cytosol, and centrifuged through a 0.5 M sucrose cushion to remove endogenous p97 and its binding proteins. MGFs were incubated for 60 min at 37°C with preformed complexes of p97 (70 ng/µl), p47 (35 ng/µl), Ufd1 (35 ng/µl) and Npl4 (49 ng/µl) in combinations indicated in the figure legends. Samples were fixed and processed for EM. Cisternal regrowth was quantitated using standard stereological procedures (Shorter and Warren, 1999).
Immunolocalization
NRK cells (normal rat kidney) were grown in DMEM/10% fetal calf serum (Gibco-BRL, Gaithersburg, MD, USA) and BS-C-1 cells (normal African green monkey kidney) in MEM/10% fetal calf serum. The cells were treated by two different methods: (i) fixed for 15 min in 4% paraformaldehyde in PBS, incubated for 15 min in 50 mM NH4Cl in PBS and permeabilized for 8 min in –20°C cold methanol; or (ii) fixed and permeabilized in ice-cold methanol. The cells were incubated with the following antibodies: affinity-purified rabbit antibodies against p97 (N5), affinity-purified rabbit antibodies against Npl4 or a monoclonal antibody against Ufd1 (1E6), followed by the secondary antibodies Alexa Fluor 488 goat anti-rabbit, Alexa Fluor 594 goat anti-rabbit or Alexa Fluor 594 goat anti-mouse (Molecular Probes, Eugene, OR, USA), respectively. Fluorescence analysis was performed using a Zeiss Axiovert 100M inverted microscope (Carl Zeiss, Oberkochen, Germany) and images were captured on an Orca 100 CCD camera (Hamamatsu, Hamamatsu City, Japan) using the software package Openlab 2.1 (Improvision, Coventry, UK).
Acknowledgments
Acknowledgements
We thank Brian Burke and Al Price for critical reading. This work was supported by postdoctoral fellowships to H.H.M. initially from the Deutsche Forschungsgemeinschaft and latterly from the Human Frontiers Science Program. J.S. was supported by a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft.
References
- Acharya U., Jacobs,R., Peters,J.-U., Watson,N., Farquhar,M.G. and Malhotra,V. (1995) The formation of Golgi stacks from vesiculated Golgi membranes requires two distinct fusion events. Cell, 82, 895–904. [DOI] [PubMed] [Google Scholar]
- Baldini A. (1999) Is the genetic basis of DiGeorge syndrome in HAND? Nature Genet., 21, 246–247. [DOI] [PubMed] [Google Scholar]
- Bleasby A.J. and Wootton,J.C. (1990) Construction of validated, non-redundant composite protein sequence databases. Protein Eng., 3, 153–159. [DOI] [PubMed] [Google Scholar]
- Coles M., Diercks,T., Liermann,J., Groger,A., Rockel,B., Baumeister,W., Koretke,K.K., Lupas,A., Peters,J. and Kessler,H. (1999) The solution structure of VAT-N reveals a ‘missing link’ in the evolution of complex enzymes from a simple βαββ element. Curr. Biol., 9, 1158–1168. [DOI] [PubMed] [Google Scholar]
- Dai R., Chen,E., Longo,D., Gorbea,C. and Li,C. (1998) Involvement of valosin-containing protein, an ATPase co-purified with IκBα and 26 S proteasome, in ubiquitin-proteasome-mediated degradation of IκBα. J. Biol. Chem., 273, 3562–3573. [DOI] [PubMed] [Google Scholar]
- DeHoratius C. and Silver,P.A. (1996) Nuclear transport defects and nuclear envelope alterations are associated with mutation of the Saccharomyces cerevisiae NPL4 gene. Mol. Biol. Cell, 7, 1835–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Olmo M., Mizrahi,N., Gross,S. and Moore,C.L. (1997) The Uba2 and Ufd1 proteins of Saccharomyces cerevisiae interact with poly(A) polymerase and affect the polyadenylation activity of cell extracts. Mol. Gen. Genet., 255, 209–218. [DOI] [PubMed] [Google Scholar]
- Fröhlich K.-U., Fries,H.-W., Rudiger,M., Erdmann,R., Botstein,D. and Mecke,D. (1991) Yeast cell cycle protein cdc48p shows full-length homology to the mammalian protein VCP and is a member of a protein family involved in secretion, peroxisome formation and gene expression. J. Cell Biol., 114, 443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislain M., Dohmen,R., Levy,F. and Varshavsky,A. (1996) Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin-mediated proteolysis in Saccharomyces cereviae. EMBO J., 15, 4884–4899. [PMC free article] [PubMed] [Google Scholar]
- Golbik R., Lupas,A., Koretke,K., Baumeister,W. and Peters,J. (1999) The Janus face of the archaeal Cdc48/p97 homologue VAT: protein folding versus unfolding. Biol. Chem., 380, 1049–1062. [DOI] [PubMed] [Google Scholar]
- Hanson P.I., Roth,R., Morisaki,H., Jahn,R. and Heuser,J.E. (1997) Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell, 90, 523–535. [DOI] [PubMed] [Google Scholar]
- Harlow E. and Lane,D. (1988) Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor Laboratory, NY. [Google Scholar]
- Hoss M., Robins,P., Naven,T.J.P., Pappin,D.J.C., Sgouros,J. and Lindahl,T. (1999) A human DNA editing enzyme homologous to the Escherichia coli DNaQ/MutD protein. EMBO J., 18, 3868–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui N., Nakamura,N., Slusarewicz,P. and Warren,G. (1998) Purification of rat liver Golgi stacks. In Celis,J. (ed.), Cell Biology. A Laboratory Handbook. Academic Press, San Diego, CA, Vol. 2, pp. 46–55. [Google Scholar]
- Hunt D.F., Yates,J.R., Shabanowitz,J., Winston,S. and Hauer,C.R. (1986) Protein sequencing by tandem mass spectrometry. Proc. Natl Acad. Sci. USA, 84, 6233–6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E.S., Ma,P.C., Ota,I.M. and Varshavsky,A. (1995) A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem., 270, 17442–17456. [DOI] [PubMed] [Google Scholar]
- Johnson E.S., Schwienhorst,I., Dohmen,R.J. and Blobel,G. (1997) The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J., 16, 5509–5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koegl M., Hoppe,T., Schlenker,S., Ulrich,H., Mayer,T. and Jentsch,S. (1999) A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell, 96, 635–644. [DOI] [PubMed] [Google Scholar]
- Koller K. and Brownstein,M. (1987) Use of a cDNA clone to identify a supposed precursor containing valosin. Nature, 325, 542–545. [DOI] [PubMed] [Google Scholar]
- Kondo H., Rabouille,C., Newman,R., Levine,T.P., Pappin,D., Freemont,P. and Warren,G. (1997) p47 is a cofactor for p97-mediated membrane fusion. Nature, 388, 75–78. [DOI] [PubMed] [Google Scholar]
- Latterich M., Fröhlich,K.-U. and Schekman,R. (1995) Membrane fusion and the cell cycle: cdc48p participates in the fusion of ER membranes. Cell, 82, 885–893. [DOI] [PubMed] [Google Scholar]
- Lindsay E., Botta,A., Jurecic,V., Carattini-Rivera,S., Cheah,Y., Rosenblatt,H., Bradley,A. and Baldini,A. (1999) Congenital heart disease in mice deficient for the DiGeorge syndrome region. Nature, 401, 379–383. [DOI] [PubMed] [Google Scholar]
- Madeo F., Schlauer,J., Zischka,H., Mecke,D. and Fröhlich,K. (1998) Tyrosine phosphorylation regulates cell cycle-dependent nuclear localization of Cdc48p. Mol. Biol. Cell, 9, 131–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan R., Delphin,C., Guan,T., Gerace,L. and Melchior,F. (1997) A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell, 88, 97–107. [DOI] [PubMed] [Google Scholar]
- Matunis M.J., Wu,J. and Blobel,G. (1998) SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J. Cell Biol., 140, 499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir D., Stewart,S., Osmond,B. and Botstein,D. (1982) Cold-sensitive cell-division-cycle mutants of yeast: isolation, properties and pseudoreversion studies. Genetics, 100, 547–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J.M.M., Meyer,H.H., Ruhrberg,C., Stamp,G.W., Warren,G. and Shima,D.T. (1999) The mouse p97 (CDC48) gene: genomic structure, definition of transcriptional regulatory sequences, gene expression and characterization of a pseudogene. J. Biol. Chem., 274, 10154–10162. [DOI] [PubMed] [Google Scholar]
- Nakielny S., Shaikh,S., Burke,B. and Dreyfuss,G. (1999) Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J., 18, 1982–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamnani V., Tamura,T., Lupas,A., Peters,J., Cejka,Z., Ashraf,W. and Baumeister,W. (1997) Cloning, sequencing and expression of VAT, a CDC48/p97 ATPase homologue from the archaeon Thermoplasma acidophilum. FEBS Lett., 404, 263–268. [DOI] [PubMed] [Google Scholar]
- Patel S. and Latterich,M. (1998) The AAA team: related ATPases with diverse functions. Trends Cell Biol., 8, 65–71. [PubMed] [Google Scholar]
- Perkins D.N., Pappin,D.J., Creasey,D.M. and Cottrell,J.S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis, 20, 3551–3567. [DOI] [PubMed] [Google Scholar]
- Peters J.-M., Walsh,M.J. and Franke,W.W. (1990) An abundant and ubiquitous homo-oligomeric ring-shaped ATPase particle related to the putative vesicle fusion proteins, Sec18p and NSF. EMBO J., 9, 1757–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J.-M., Harris,J.R., Lustig,A., Müller,S., Engel,A., Volker,S. and Franke,W.W. (1992) Ubiquitous soluble Mg2+-ATPase complex: a structural study. J. Mol. Biol., 223, 557–571. [DOI] [PubMed] [Google Scholar]
- Pizzuti A., Novelli,G., Ratti,A., Amati,F., Mari,A., Calabrese,G., Nicolis,S., Silvani,V., Marino,B., Scarlato,G., Ottolenghi,S. and Dallapiccola,B. (1997) UFD1L, a developmentally expressed ubiquitination gene, is deleted in CATCH 22 syndrome. Hum. Mol. Genet., 6, 259–265. [DOI] [PubMed] [Google Scholar]
- Rabouille C., Levine,T.P., Peters,J.M. and Warren,G. (1995a) An NSF-like ATPase, p97 and NSF mediate cisternal regrowth from mitotic Golgi fragments. Cell, 82, 905–914. [DOI] [PubMed] [Google Scholar]
- Rabouille C., Misteli,T., Watson,R. and Warren,G. (1995b) Reassembly of Golgi stacks from mitotic Golgi Fragments in a cell-free system. J. Cell Biol., 129, 605–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouille C., Kondo,H., Newman,R., Hui,N., Freemont,P. and Warren,G. (1998) Syntaxin 5 is a common component of the NSF- and p97-mediated reassembly pathways of the Golgi cisternae from mitotic Golgi fragments in vitro. Cell, 92, 603–610. [DOI] [PubMed] [Google Scholar]
- Schinke M. and Izumo,S. (1999) Getting to the heart of DiGeorge syndrome. Nature Med., 5, 1120–1121. [DOI] [PubMed] [Google Scholar]
- Sherman N.E., Yates,N.A., Shabanowitz,J., Hunt,D.F., Jeffery,W.A., Bartlet-Jones,M. and Pappin,D.J.C. (1995) A novel N-terminal derivative designed to simplify peptide fragmentation. 43rd ASMS Conference on Mass Spectrometry and Allied Topics, Atlanta, GA, pp. 626–627. [Google Scholar]
- Shevchenko A., Wilm,M., Vorm,O. and Mann,M. (1996) Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem., 68, 850–858. [DOI] [PubMed] [Google Scholar]
- Shorter J. and Warren,G. (1999) A role for the vesicle tethering protein, p115, in the post-mitotic stacking of reassembling Golgi cisternae in a cell-free system. J. Cell Biol., 146, 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söllner T., Bennett,M.K., Whiteheart,S.W., Scheller,R.H. and Rothman,J.E. (1993) A protein assembly–disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation and fusion. Cell, 75, 409–418. [DOI] [PubMed] [Google Scholar]
- Whiteheart S.W., Rossnagel,K., Buhrow,S.A., Brunner,M., Jaenicke,R. and Rothman,J.E. (1994) N-ethylmaleimide-sensitive fusion protein—a trimeric ATPase whose hydrolysis of ATP is required for membrane-fusion. J. Cell Biol., 126, 945–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm M. and Mann,M. (1996) Analytical properties of the Nanoelectrospray ion source. Anal. Chem., 68, 1–8. [DOI] [PubMed] [Google Scholar]
- Xu Z., Horwich,A.L. and Sigler,P.B. (1997) The crystal structure of the asymmetric GroEL–GroES–(ADP)7 chaperonin complex. Nature, 388, 741–750. [DOI] [PubMed] [Google Scholar]
- Yamagishi H., Garg,V., Matsuoka,R., Thomas,T. and Srivastava,D. (1999) A molecular pathway revealing a genetic basis for human cardiac and craniofacial defects. Science, 283, 1158–1161. [DOI] [PubMed] [Google Scholar]
- Yaseen N.R. and Blobel,G. (1999) Two distinct classes of Ran-binding sites on the nucleoporin Nup-358. Proc. Natl Acad. Sci. USA, 96, 5516–5521. [DOI] [PMC free article] [PubMed] [Google Scholar]