Abstract
Leucyl-, isoleucyl- and valyl-tRNA synthetases are closely related large monomeric class I synthetases. Each contains a homologous insertion domain of ∼200 residues, which is thought to permit them to hydrolyse (‘edit’) cognate tRNA that has been mischarged with a chemically similar but non-cognate amino acid. We describe the first crystal structure of a leucyl-tRNA synthetase, from the hyperthermophile Thermus thermophilus, at 2.0 Å resolution. The overall architecture is similar to that of isoleucyl-tRNA synthetase, except that the putative editing domain is inserted at a different position in the primary structure. This feature is unique to prokaryote-like leucyl-tRNA synthetases, as is the presence of a novel additional flexibly inserted domain. Comparison of native enzyme and complexes with leucine and a leucyl- adenylate analogue shows that binding of the adenosine moiety of leucyl-adenylate causes significant conformational changes in the active site required for amino acid activation and tight binding of the adenylate. These changes are propagated to more distant regions of the enzyme, leading to a significantly more ordered structure ready for the subsequent aminoacylation and/or editing steps.
Keywords: editing/leucyl-adenylate/leucyl-tRNA synthetase/X-ray crystallography/zinc binding domain
Introduction
Accurate protein synthesis requires that aminoacyl-tRNA synthetases discriminate against chemically similar, non-cognate amino acids by a factor of at least 104. This is difficult to achieve in one step, particularly for aliphatic hydrophobic amino acids where discrimination between, for example, valine and isoleucine, which differ by only one methyl group, can only be made on the basis of weak van der Waals interactions (Pauling, 1958). To overcome this problem some aminoacyl-tRNA synthetases have evolved a specific editing activity whereby they are capable of hydrolysing misactivated amino acids (‘pre-transfer editing’) and/or mischarged tRNAs (‘post-transfer editing’). This biologically essential activity has been extensively studied for several synthetases (for a review, see Jakubowski and Goldman, 1992). However, apart from the special case of homocysteine, whose activated form is edited in the synthetic active site of MetRS, LeuRS and IleRS to give homocysteine thiolactone (Jakubowski, 1995), the exact structural and enzymatic mechanism for editing in most cases still remains obscure.
A subfamily of class 1a aminoacyl-tRNA synthetases, leucyl-, isoleucyl- and valyl-tRNA synthetases (LeuRS, IleRS and ValRS, respectively), are particularly closely related and probably evolved from a common ancestor that did not discriminate between these three amino acids. The three enzymes are large monomers (∼100 kDa) and contain an unusually large insertion [often called CP1 (connective polypeptide 1); Starzyk et al., 1987] into the class 1 Rossmann-fold catalytic domain. The crystal structure of Thermus thermophilus IleRS first showed that the CP1 insert is a discrete domain of ∼200 residues, which contains a serine protease-like fold (Nureki et al., 1998). Based on the observation of a binding site for valine in this domain and on site-directed mutagenesis experiments, a putative hydrolytic editing active site was identified (Nureki et al., 1998). A recent crystal structure of Staphylococcus aureus IleRS (IleRSSA) complexed with an Escherichia coli tRNAile transcript and an inhibitor shows the first putative post-transfer editing complex. In this structure the 3′ end of the tRNA is directed towards the putative editing active site rather than the aminoacylation active site (Silvian et al., 1999).
LeuRS is the least well studied of this triad of editing enzymes. In vitro, it will edit hydroxy derivatives of leucine and homocysteine (Englisch et al., 1986). Unusually, a species-specific editing behaviour of LeuRS has been reported. The E.coli enzyme has a high initial substrate discrimination and deals with misactivated amino acids largely by post-transfer editing, whereas the yeast enzyme has a poorer initial discrimination but a very effective pre-transfer editing mechanism (Englisch et al., 1986). However, unlike the cases of ValRS and IleRS, which edit mischarged threonine and valine, respectively, it is not known which biologically significant non-cognate amino acids, other than homocysteine (Jakubowski, 1995), are edited by LeuRS in vivo. It is possible that non-protein amino acids such as norleucine or norvaline, which may be present in cells particularly in certain stress conditions, are edited (Jakubowski and Goldman, 1992).
A second interesting feature of LeuRS is that it is one of only three aminoacyl-tRNA synthetases that recognize tRNAs with long variable arms (class 2 tRNAs). However, unlike the other two such enzymes, SerRS (Cusack et al., 1996) and TyrRS, LeuRS surprisingly does not generally use the long variable arm of tRNAleu as an identity element (Asahara et al., 1993), with the recently identified exception of an archae LeuRS (Soma et al., 1999). Neither does LeuRS use the anti-codon triplet (Asahara et al., 1993). Another novel aspect of LeuRS is the involvement of certain mitochondrial LeuRS in splicing reactions. Saccharomyces cerevisiae mitochondrial leucyl-tRNA synthetase (NAM2 gene product) is an essential cofactor for splicing of pre-mRNAs that encode mitochondrial cytochrome b (cob) and a subunit of cytochrome oxidase (cox1) (Labouesse et al., 1985; Herbert et al., 1988; Dujardin and Herbert, 1997).
To give structural insight into the role of LeuRS in aminoacylation, editing and splicing, we have cloned and overexpressed the LeuS gene product (denoted LeuRSTT) from the hypothermophilic eubacterium T.thermophilus (M.Tukalo et al., unpublished results). We have found that this enzyme is capable of editing homocysteine, norleucine and norvaline, the latter activity being stimulated by addition of tRNAleu (M.Tukalo, S.Cusack and A.Yaremchuk, unpublished results). The crystal structure of LeuRSTT has been determined using seleno-methionated protein and the MAD method. Models have been refined of the ligand-free enzyme (2.0 Å resolution) and the enzyme with either leucine (2.3 Å resolution) or the sulfamoyl analogue of leucyl-adenylate (LeuAMS) bound (2.0 Å resolution). No structure of a class 1a synthetase with either ATP or aminoacyl-adenylate has been published previously.
Results and discussion
Overview of the structure
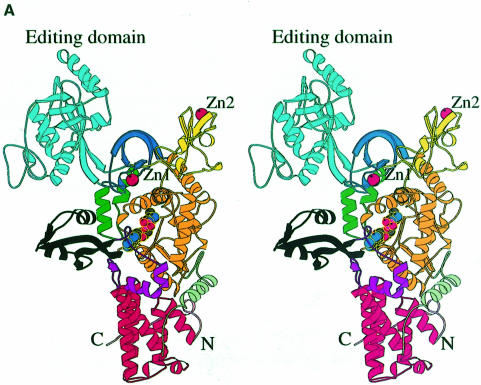
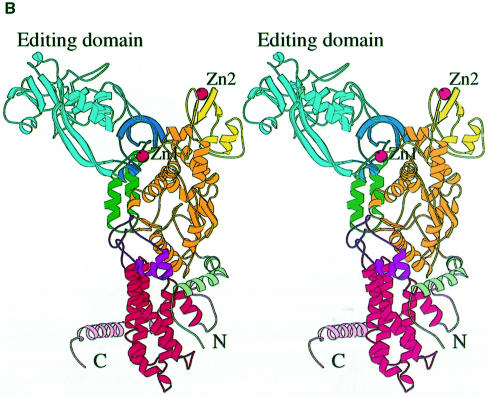
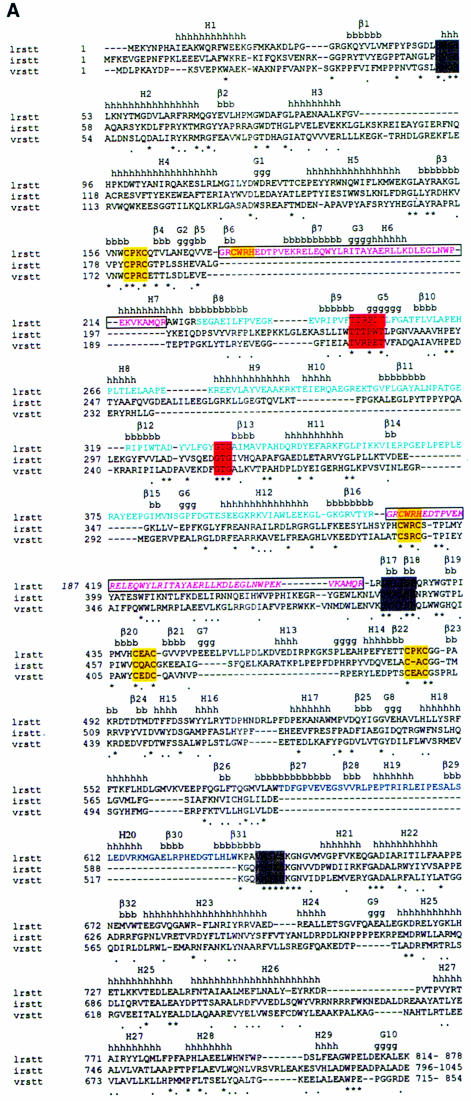
The overall structure of LeuRSTT has several architectural features in common with other class 1a synthetases (reviewed in Sugiura et al., 2000) and in particular with IleRSTT (Nureki et al., 1998), but there are also some notable differences (Figure 1). The bipartite Rossmann-fold catalytic domain (orange in Figure 1A) occupies the central part of the elongated molecule. On top of this are inserted two zinc binding modules denoted ZN-1 (dark blue, with zinc ligands Cys159X2Cys162X13Cys176X2His179, where X is any residue) and ZN-2 (yellow, with zinc ligands Cys439X2Cys442X41Cys484X2Cys487). After the second half of the Rossmann-fold there is a connecting module (purple) followed by, at the bottom, the tRNA anticodon binding domain (red) composed of a bundle of five α-helices characteristic of all class 1a synthetases IleRS (Nureki et al., 1998), ValRS, LeuRS, MetRS (Mechulam et al., 1999; Sugiura et al., 2000), CysRS and ArgRS (Cavarelli et al., 1998). The C-terminal residues 815–878 are invisible in the electron density. The equivalent domain was also disordered in the structure of apo-IleRSTT (Nureki et al., 1998), but was found ordered in the complex of IleRS with tRNAile where it makes contact with the tRNA anti-codon stem–loop (Silvian et al., 1999). The most complete model of LeuRSTT is that with LeuAMS bound, in which all residues 1–814 are well defined, with the exception of residues 293–300 in the putative editing domain, which are poorly ordered. A structure-based alignment of the primary sequences of T.thermophilus LeuRS, IleRS and ValRS, together with the secondary structure designations of LeuRS, are shown in Figure 2A.
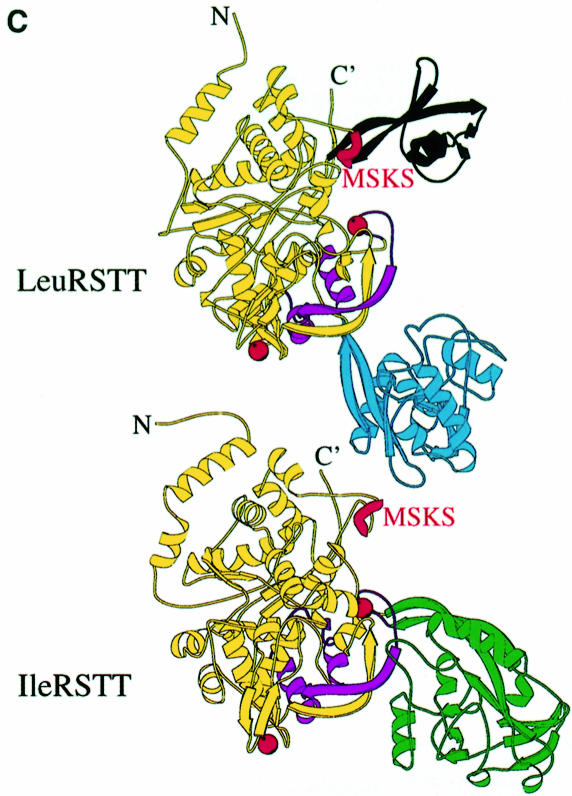
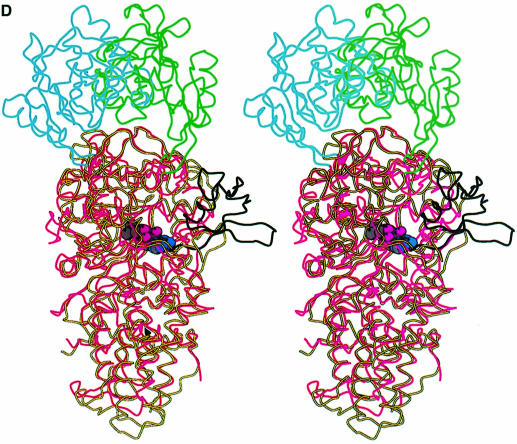
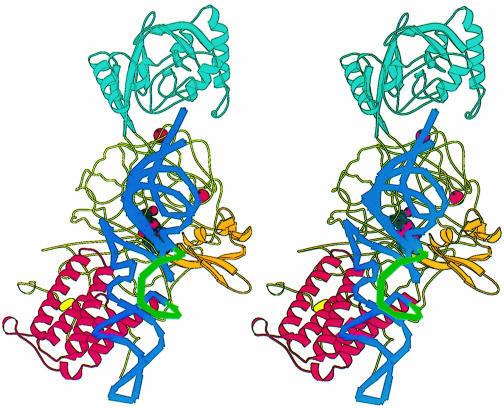
Fig. 1. (A) Stereo ribbon diagram of the structure of the complex between T.thermophilus leucyl-tRNA synthetase and a leucyl-adenylate analogue. The domains are coloured as follows: N-terminal extension (pale green), catalytic domain (Rossmann-fold, orange), ZN-1 domain (blue), helical hairpin insertion (green), editing domain (cyan), ZN-2 domain (yellow), leucyl-specific insertion domain (black), connecting module (purple), anti-codon binding domain (red) and C-terminal extension (pink). The zinc atoms are shown as red balls and the leucyl-adenylate analogue as a space-filling model in the centre. The N- and C-termini are marked. (B) Stereo ribbon diagram of the structure of the T.thermophilus isoleucyl-tRNA synthetase. Colouring as in (A). Note that in IleRS the ZN-1 domain is split by the inserted editing domain [see (C) and Figure 2]. (C) Comparison of point of insertion of extra domains in T.thermophilus leucyl- (top) and isoleucyl- (bottom) tRNA synthetases. For clarity, the enzymes are C-terminally truncated just before the anti-codon binding domain (C′). The purple polypeptide segment is homologous in each enzyme and contains the second half of the ZN-1 binding site [see also (A)]. In leucyl-tRNA synthetase, it precedes the editing domain (cyan), whereas in isoleucyl-tRNA synthetase it follows the editing domain (green). The disposition of the leucyl-specific insertion domain is shown in black. It is inserted just before the KMSKS loop (red segment in both enzymes). (D) Stereo view of the superposition of Cα traces of LeuRSTT and IleRSTT showing different points of insertion of the editing domain and the near 180° difference in rotational orientation. LeuRSTT is red with editing domain cyan, leucine-specific domain black and LeuAMS in the active site. IleRSTT is yellow with editing domain green. Considering the red and yellow traces, 437 Cα positions superpose within a cut-off of 3.8 Å with 115 identities and root-mean-square deviation (r.m.s.d.) of 1.99 Å. Superposing the two editing domains, the equivalent figures are 129 Cα positions with 37 identities and r.m.s.d. of 1.36 Å. In all, 69.5% of the 814 ordered residues in LeuRSTT can be superposed within 3.8 Å of equivalent residues in IleRSTT with 27% of these residues being identical.
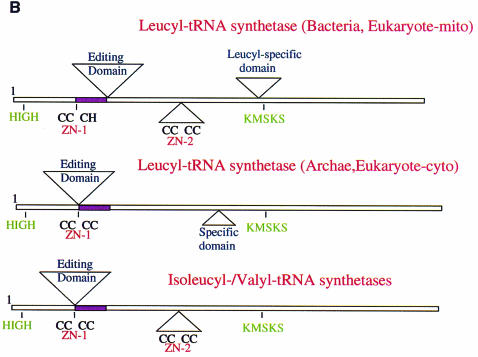
Fig. 2. (A) Primary sequence alignment of T.thermophilus leucyl-, isoleucyl- and valyl-tRNA synthetases. The alignment of LeuRSTT and IleRSTT is based on superposition of three-dimensional structures (see the legend to Figure 1D). An asterisk indicates amino acid identity and a stop indicates amino acid similarity. The secondary structure elements as defined by DSSP (Kabsch and Sander, 1983) are indicated for LeuRSTT (h, α-helix; g, 310 helix; β, β-strand). The editing domain of LeuRSTT is shown with cyan letters, the leucyl-specific domain with blue letters. The Class 1a conserved motifs, HIGH, DWLISR (Carter, 1993) and KMSKS, are boxed in grey. The conserved threonine-rich and GTG motifs of the editing domain are boxed in red. Zinc binding motifs are boxed in yellow. The boxed, pink region in LeuRS is shown twice: first in its actual position in the primary sequence (residues 174–211) and secondly (in italics) displaced to after the editing domain where it aligns with corresponding sequences in IleRS and ValRS. This peptide is also coloured purple in Figure 1C. Sequences are C-terminally truncated after the end of the visible region in the LeuRSTT structure. (B) Schematic diagram of the domain structure of leucyl-, isoleucyl- and valyl-tRNA synthetases, based on extensive sequence alignments. The positions of the two catalytically important class I motifs, HIGH and MSKS, are shown. The purple segment is homologous in each enzyme. Top: domain structure of T.thermophilus leucyl-tRNA synthetase, representative of eubacterial and mitochondrial leucyl-tRNA synthetases. Although the ZN-1 and ZN-2 domains are always conserved, the zinc ligands are not, so that there can be both zinc atoms (e.g. T.thermophilus, Helicobacter pylori), no zinc atoms (e.g. Borrelia burgdorferi, Mycoplasma), Zn-1 only (e.g. Haemophilus influenzae, E.coli) or Zn-2 only (human mitochondrial, Rickettsia prowazekii). A similar variability of zinc content is found in methionyl-tRNA synthetase (Mechulam, 1999). The specific insertion domain (black) ranges in size from 30 to 60 residues. Interestingly, the putative Caenorhabditis elegans mitochondrial leucyl-tRNA synthetase has a severely truncated and probably inactive editing domain. Middle: predicted domain structure of archae and eukaryotic cytoplasmic leucyl-tRNA synthetases. The editing domain splits the ZN-1 domain. Putative zinc ligands are present in the ZN-1 domain generally for archae enzymes, but not for eukaryotic cytoplasmic enzymes. Sequence alignments suggest that there is no ZN-2 domain, which normally precedes one of the leucine binding motifs (containing Phe501 and Ser504 in LeuRSTT), although there seems to be a short specific insertion after this leucine binding motif. Bottom: domain structure of T.thermophilus isoleucyl-tRNA synthetase (Nureki et al., 1998), representative of all isoleucyl- and valyl-tRNA synthetases.
A putative editing domain with a novel point of insertion
A particularly striking feature of the molecule is the large putative editing domain (residues 224–417; cyan in Figure 1A), which is inserted into the catalytic domain just after the ZN-1 module. The ZN-1 domain is also found in MetRS (Sugiura et al., 2000), but the editing domain is unique to IleRS, LeuRS and ValRS. However, as shown in Figure 1C and D, there is a significantly different point of insertion of the editing domain in LeuRSTT compared with IleRS, where it is inserted between the two halves of the ZN-1 binding site (Nureki et al., 1998). As a result, two structurally equivalent segments in IleRS and LeuRS are inverted in their respective primary structures (Figures 1C and 2). Detailed comparison of a large number of primary sequences of IleRS, ValRS and LeuRS shows that archae and eukaryotic cytoplasmic LeuRS and all ValRS and IleRS have the editing domain inserted between the two halves of the ZN-1 binding site, whereas bacterial and mitochondrial LeuRS have it inserted after the ZN-1 domain (Figures 1C and 2). This suggests that a module shuffling event has occurred in LeuRS only in the bacterial lineage, but before the mitochondrial endosymbiotic integration. The core of the editing domain including the putative active site has the same β-barrel fold as the equivalent domain of IleRS (Nureki et al., 1998), although the peripheral regions are quite different (see the alignment in Figure 2A). This corresponds with the fact that 65% of the residues in the editing domain of LeuRSTT have Cα atoms within 3.8 Å of those in IleRSTT after superposition, of which 28% are identical.
Biochemical and crystallographic work on IleRS (Nureki et al., 1998) has identified the hydrolytic active site to be a cleft in the editing domain formed by secondary structural elements β9–G5 and β13–H11 (Figure 2A). The first region contains a threonine-rich peptide highly conserved in LeuRS, IleRS and ValRS (Figure 2A). In LeuRSTT this peptide is T247TRPDT252 and in IleRSTT it is T228TTPWT233. Thr229 and nearby Asn237 in IleRSTT (Thr242 and Asn250, respectively, in E.coli IleRS) have been implicated in editing activity from mutagenesis experiments (Nureki et al., 1998, 1999; Nomanbhoy et al., 1999). The second part of the active site contains another highly conserved motif, G333TG in LeuRSTT and G314TG in IleRSTT (Figure 2A), and nearby His319 in IleRSTT has been postulated to be important in editing (Nureki et al., 1998). Asn237 and His319 in IleRSTT are structurally equivalent to Ala256 and Met338 in LeuRSTT, which are presumably not catalytically active. At this stage it is not clear which residues are particularly important in the putative active site of LeuRSTT. In all LeuRSTT structures examined there is strong, but uninterpretable, extra density in the putative editing active site adjacent to the highly conserved threonines Thr247, Thr248 and Tyr332. Attempts to clarify this density by soaking crystals with high concentrations of norvaline or homocysteine have so far been inconclusive.
The bulk of the editing domain has no contact with the rest of the enzyme, the only connection being via a flexible partially solvent-exposed β-ribbon. Flexibility in the orientation of the editing domain relative to the catalytic domain has been implicated in the editing mechanism. This is supported by the observation of a 47° rotation of the editing domain in the IleRS editing complex (Silvian et al., 1999) compared with the apo-enzyme (Nureki et al., 1998). If the functional positioning of the editing domain depends on tRNA interactions, this could plausibly explain why both pre- and post-transfer editing are generally tRNA dependent (Nomanbhoy et al., 1999). In the LeuRS structures reported here, the editing domain differs in rotational orientation by 173° from that found in the IleRSTT or IleRSSA editing complex after alignment of catalytic domains (Figure 1D). Given that the LeuRS structures are in the absence of tRNA it is possible that the editing domain position is affected by crystal packing, although in a second, distinct crystal form of LeuRSTT (Yaremchuk et al., 2000) a very similar orientation is found. However, it is clear from modelling the editing complex in LeuRSTT that the different point of insertion of the editing domain into the catalytic domain compared with IleRS requires this near 180° difference in orientation in order that the 3′ end of the tRNA can still enter the putative editing active site (Figures 1D and 3).
Fig. 3. Stereo diagram of a model of a putative LeuRSTT-tRNAleu editing complex obtained by superimposition of LeuRSTT on the IleRSSA-tRNAile editing complex (Silvian et al., 1999). The model tRNA used here is that of tRNAtyr with a long variable arm (S.Cusack, A.Yaremchuk and M.Tukalo, unpublished results). Its position has been manually adjusted to avoid steric clashes with the enzyme, notably with the ZN-1 domain, which is much closer to the active site in LeuRSTT than in the IleRSSA-tRNAile complex. The anticodon binding domain is in red, the editing domain in cyan. The model shows that the leucyl-specific domain (orange) could interact with the base of the acceptor stem and core of the tRNA. The long variable arm of the tRNA (green) points away from the enzyme and towards the viewer. This is consistent with it not being an important identity element, although it is possible that the disordered C-terminal domain of the enzyme makes some contact with this part of the tRNA. Given the flexibility of the various modules of the enzyme and of the tRNA itself, the aim of this figure is to give the general disposition of the various elements rather than to be an accurate model.
Leucyl-specific insertion domain
There is a second extra domain (residues 577–634, designated leucyl-specific domain) inserted into the catalytic domain of LeuRSTT (black in Figure 1), just before the catalytically important KMSKS motif (see below). This compact, well ordered module comprises five β-strands and two short α-helices, and is positioned at the entrance to the synthetic active site. It is connected to the catalytic domain via a β-ribbon, similarly to the editing domain, suggesting that it may be able to rotate flexibly. Sequence alignments show that this domain is unique to prokaryote-like LeuRS, but is not highly conserved in sequence or size. LeuRSTT has in fact one of the largest such modules, together with Aquifex aeolicus, whose gene for LeuRS is split into two pieces, the division being right in the middle of this putative domain (Deckert et al., 1998). At this stage we can only speculate on the function of this module. A model of tRNAleu docked to LeuRSTT, based on the IleRS–tRNAile editing complex structure (Silvian et al., 1999), suggests that it could interact with tRNAleu in the region of the base of the acceptor stem (Figure 3). The various architectural differences between prokaryote-like and eukaryote-/archae-like LeuRS (summarized in Figure 2) may explain the species differences observed in editing behaviour (Englisch et al., 1986; see above) and mode of recognition of tRNAleu (Soma et al., 1999).
Leucyl-adenylate binding site
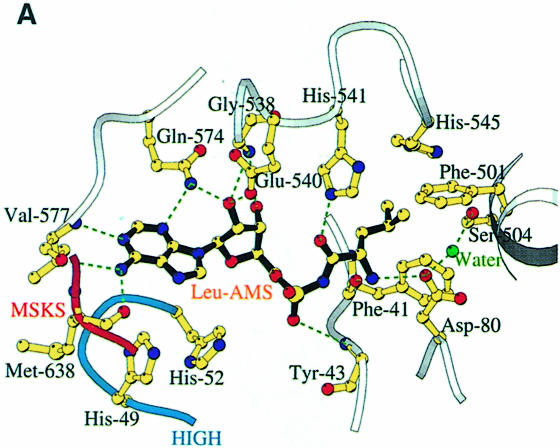
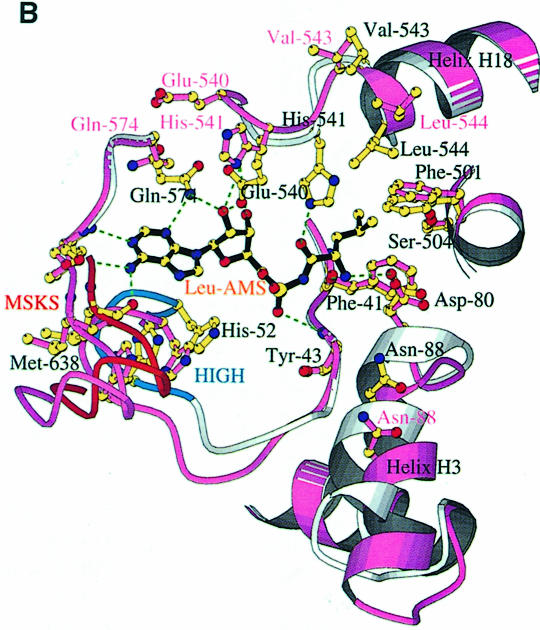
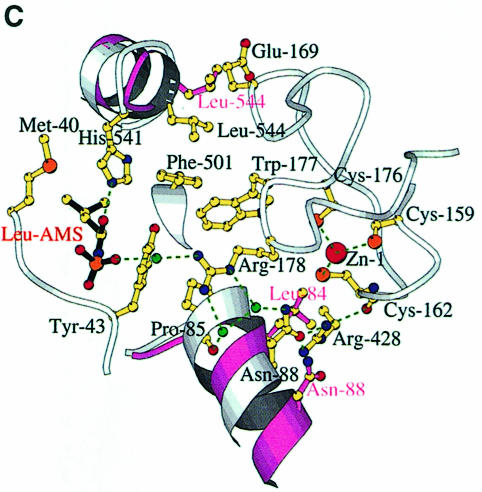
We have defined the leucine and adenosine binding site of LeuRSTT by determining the structure of the enzyme in complex with the non-hydrolysable, sulfamoyl analogue of leucyl-adenylate (LeuAMS). LeuAMS is bound tightly in the synthetic active site and the principal ligands are shown in Figure 4A. The largely hydrophobic pocket for the substrate leucine side chain is formed by Met40, Phe41, Tyr43, Phe501, Tyr507, His541 and His545, highly conserved residues in almost all LeuRS. The α-amino group of the amino acid makes hydrogen bonds to Asp80 (an interaction conserved in many class 1 synthetases) and the carbonyl oxygen of Phe41, whereas the carbonyl oxygen of the amino acid hydrogen bonds to NE1 of His541. A very well ordered water molecule, which is hydrogen bonded to the highly conserved Ser504 as well as to Tyr43, Asp80 and Thr507, is surprisingly close to the extremity of the leucine side chain (3.55 Å to CD2). This could explain the misactivation of γ-hydroxy-leucine by LeuRS (Englisch et al., 1986). The adenosine moiety of LeuAMS is specifically recognized by multiple hydrogen bonds to both the ribose and adenine base, the latter being stacked on Met576. Key conserved residues are Glu540, which makes hydrogen bonds to both ribose hydroxyl groups, and Gln574, whose NE1 hydrogen bonds to both the ribose O3′ and the adenine N3. The N1 and N2 positions of the adenine base hydrogen bond to main chain atoms of Val577 and Met638. Met638 is part of the class 1 characteristic KMSKS motif (V637MSKSK642 in LeuRSTT), which is immediately preceded by the leucine-specific insertion module described above (residues 577–634). The sulfate in LeuAMS (presumably at or very close to the α-phosphate position) hydrogen bonds to the main chain of Tyr43 and also via a water molecule to Arg178 (Figure 4C). The tip of Arg178 is only 5.5 Å from the closest sulfate oxygen. This arginine, which is part of the Zn-1 binding motif C176WRH, is highly conserved in almost all LeuRS, IleRS and ValRS (including those that do not have zinc ligands) and is positioned to play potentially an important role in the synthetic active site. This could help explain why mutation or chemical modification of zinc ligands in the CP1 of E.coli and T.thermophilus IleRS leads to unstable and inactive enzyme (Nureki et al., 1993; Landro et al., 1994) since the zinc is clearly required for the correct folding of the ZN-1 domain.
Fig. 4. (A) The LeuAMS binding site showing the major interacting residues. Hydrogen bonds are shown as dashed green lines and a tightly bound water as a green sphere. The catalytically essential class 1 motifs H49IGH and M638SKS are shown in cyan and red, respectively. The side chain of Tyr43 is omitted for clarity, but is visible in (C). (B) The conformational changes associated with LeuAMS binding. The view is the same as in (A). The pink ribbon diagram, pink side chains and pink labels correspond to the apo-structure (mercury derivative) and the grey ribbon and yellow side chains belong to the LeuAMS-bound structure. Upon binding of the adenosine moiety, the HIGH and MSKS loops towards the active centre, Gln574 and Glu540 move to bind the ribose tightly, and helices H18 and H3 refold to permit packing of the ZN-1 domain close to the active site [see the text and (C)]. A sulfate ion (not shown) is bound to His49 and His52 in the apo-structure, but not in the LeuAMS-bound structure. (C) Proximity of Arg178 to the active center in the LeuAMS complex. Colouring as in (B) with water molecules as green spheres and the Zn-1 atom as a red sphere. One of the zinc ligands (His179) and the adenosine moiety of the LeuAMS are omitted for clarity. The positions of Leu544 and Leu84 sterically prevent the packing of the ZN-1 domain close to the active site in the apo-structure.
Conformational changes due to leucyl-adenylate binding
The closed, highly ordered configuration of the active site just described reverts to a relaxed open state in the absence of substrates. The difference in order is reflected in the average protein B-factors, which are 30 Å2 for the LeuAMS-bound structure (20 Å2 for the LeuAMS itself) and 40–45 Å2 for the other two structures (Table I). The major conformational differences between the two states seem to be induced by the binding of the adenosine moiety of the adenylate (or presumably of ATP) rather than the leucine since the active site structures of apo- and leucine-bound enzyme are very similar. As shown in Figure 4B, adenosine binding induces a concerted movement of the two catalytically essential class 1-specific loops H49MGH and M638SKS towards the leucine binding site, which itself is hardly perturbed. These movements are presumably also induced by ATP binding and would appear to be necessary for catalysing activation of leucine. A similar movement has been reported upon comparison of the ligand-free and tryptophanyl-adenylate complexed forms of TrpRS (Ilyin et al., 2000). The movement of the KMSKS loop brings with it the entire leucine-specific domain in a rigid body fashion. The two halves of the active site are pulled together by the re-orientation and strong interaction of Glu540 and Gln574 with the ribose on one side and the main chain interactions with adenine base on the other, as described above (Figure 4B). One of the most remarkable aspects of this rearrangement is the refolding of the polypeptide G538GVE540H541AVLH545. In the absence of the constraints imposed by the binding of Gly538 and Glu540 to the ribose, this peptide adopts a conformation in which the amino acid alignment is shifted by up to one residue. For example, in the apo-structure, His541 and Ala542 occupy roughly the positions of Glu540 and His541 in the LeuAMS-bound structure (Figure 4B). Although in the apo- and leucine-bound structures the side chains of Glu540 and His541 are disordered, the clear positions of the other side chains in the peptide make this interpretation unambiguous. The shift in the position of His541 means that it can no longer hydrogen bond to the carbonyl oxygen of the substrate leucine in the absence of adenosine, thus presumably reducing the enzyme’s affinity for the free amino acid. This refolding, while hardly influencing the position of His545 (part of the leucine side chain hydrophobic pocket), leads to a significant movement of Val543 and Leu544, which form the first turn of the α-helix H18 in the apo-structure, but are unwound slightly in the LeuAMS-bound structure (Figure 4B). Another major change occurs in the orientation of the α-helix H3 and the following turn (residues 83–93) with notable shifts in position of Leu84, Pro85 and Asn88 (Figure 4B and C). The changes in helices H18 and H3 have important longer-range repercussions. In the apo- and leucine-bound structures, the positions of Leu544 (helix H18) and Leu84 (helix H3) sterically prevent the packing of the ZN-1 on the catalytic domain (Figure 4C) with a result that the electron density for the entire ZN-1 domain (residues 155–188) is very poor, if not untraceable (B-factors >100 Å2). In the LeuAMS structure, these steric hindrances are removed and the ZN-1 domain is well packed against the catalytic domain and highly ordered (<B> ∼35 Å2). For instance, the position previously occupied by Leu84 in the apo-structure becomes occupied by Asn88, which together with Pro85 and Arg428 and a number of water molecules forms a hydrogen bonding network stabilizing the Zn-1 ligand Cys162, the position of the ZN-1 domain in general and the side chain of Arg178 in particular (Figure 4C). In the apo-structure the side chain of Arg428 is disordered and the region around helix H13 (residues 459–470 in the ZN-2 domain), which packs against helix H3, is also slightly altered and less well ordered. As suggested above, one of the net results of rigidifying the ZN-1 domain close to the active centre may be to position Arg178 correctly for an important functional role. However, it remains to be seen from other structures, notably with ATP and cognate tRNA, the exact functional implications of these more extended conformational changes. In this respect we note that in the IleRSSA editing complex (which also has the large antibiotic mupirocin bound in the synthetic active site) the ZN-1 domain is swung a considerable distance away from the active site (Silvian et al., 1999).
Table I. Refinement statistics.
| LeuRSTT + leucine | LeuRSTT + Hg | LeuRSTT + LeuAMS | |
|---|---|---|---|
| Resolution (Å) | 20.0–2.3 | 30.0–2.0 | 20.0–2.0 |
| Work reflections | 53 149 | 81 920 | 83 980 |
| Test reflections | 2845 (4.6%) | 4276 (4.6%) | 4402 (4.6%) |
| Rfree | 0.260 | 0.255 | 0.229 |
| Rwork | 0.232 | 0.234 | 0.209 |
| No. of protein atoms | 6514 | 6335 | 6597 atoms |
| No. of substrate atoms | 9 (leucine) | – | 31 (LeuAMS) |
| No. of solvent molecules | 127 water, 4 sulfate | 267 water, 4 sulfate | 518 water, 2 sulfate |
| No. of metal atoms | 2 Zn | 1 Zn, 1 Hg (Cys128) | 2 Zn |
| <B> protein | 45.6 | 39.1 | 29.9 |
| <B> solvent | 48.2 | 42.4 | 40.1 |
| <B> substrate | 49.1 | – | 20.0 |
| R.m.s.d. bonds (Å) | 0.0065 | 0.0060 | 0.0065 |
| R.m.s.d. angles (degrees) | 1.30 | 1.22 | 1.30 |
| Ramachandran plot | |||
| favourable % | 90.8 | 92.0 | 93.1 |
| additional % | 8.7 | 7.7 | 6.3 |
| generous % | 0.3 | 0.1 | 0.4 |
| disallowed % | 0.1 (1, Ala 8) | 0.1 (1, Ala 8) | 0.1 (1, Ala 8) |
Concluding remarks
In common with the other two very large class 1a synthetases with editing activity, leucyl-tRNA synthetase has a very complex modular architecture. Three distinct insertion modules, the editing domain, ZN-1 domain and leucyl-specific domain, are flexibly linked to the enzyme core by β-ribbon structures and can probably make large rigid-body motions with respect to each other and the central catalytic domain, as already observed by comparison of the two structures of IleRS (Nureki et al., 1998; Silvian et al., 1999). As demonstrated here, other elements forming the synthetic active site are also mobile and respond particularly to the binding of the adenosine moiety of the adenylate and presumably ATP. The large magnitude of the conformational changes expected means that in structural studies on such a system one has to be careful to crystallize pre-formed, well defined functional states. In this way one should avoid artifacts arising from substrate soaking experiments, which due to crystal constraints may not be successful at all or give only partial conformational transitions. It is also clear that the complexity of the system means that many different substrate–complex structures, combined with biochemical and mutagenesis studies, will be required to understand fully the mechanism of action of LeuRS in aminoacylation, editing and eventually splicing.
Materials and methods
Crystals and data collection
Wild-type leucyl-tRNA synthetase was purified from T.thermophilus strain HB-27 (LeuRSTT). The corresponding gene was cloned and codes for a protein subunit of 878 residues and 101 kDa (M.Tukalo, S.Cusack and A.Yaremchuk, unpublished results). Wild-type and seleno-methionated proteins were expressed in E.coli (Yaremchuk et al., 2000). Single crystals of LeuRSTT were grown by the hanging drop method using ammonium sulfate as precipitate (Yaremchuk et al., 2000). Some crystals were grown in the presence of 5′-O-[N-(leucyl)-sulfamoyl]adenosine. This compound was synthesized by a method analogous to that described for the alanyl compound (Ueda et al., 1991) except that 2′,3′-O-isopropylidene-5′-O-sulfamoyladenosine was reacted with t-butoxycarbonyl-l-leucine-N-hydroxysuccinimide ester (Yaremchuk et al., 2000). Other crystals were soaked or co-crystallized with l-leucine, dl-norvaline, dl-homocysteine and various heavy metal compounds. The crystals are of space group C2221 (#20) with one monomer in the asymmetric unit and a solvent content of ∼65%. They grow as very thin plates, exceptionally to a maximum size of 1800 × 300 × 60 µm3 and diffract to ∼1.9 Å resolution. Diffraction data on frozen crystals cryoprotected with 30% glycerol were collected on beamline ID14 at the ESRF and integrated using MOSFLM (Leslie, 1992). Details of the data collections used for structure solution and refinement are given in Table II. All subsequent analysis was carried out with the CCP4 package (CCP4, 1994), except where indicated. The incompleteness of the data at high resolution reflects a certain anisotropy of the data (B11 = 4.79, B22 = 8.26, B33 = 3.47 Å2).
Table II. X-ray crystallography data collection statistics.
| Components co-crystallized | LeuRS + leucine + norvaline | LeuRS + LeuAMS + norvaline | LeuRS + leucine + norvaline + KAuCl4 | LeuRS + homocysteine + Hg acetate | LeuRS:SeMet + Leu peak | LeuRS:SeMet + Leu inflection | LeuRS:SeMet + Leu remote |
|---|---|---|---|---|---|---|---|
| Beamline/detector | ID14-EH4/ADSC | ID14-EH4/ADSC | ID14-EH4/ADSC | ID14-EH2/MarCCD | ID14-EH4/ADSC | ID14-EH4/ADSC | ID14-EH4/ADSC |
| Wavelength (Å) | 0.979 | 0.979 | 0.979 | 0.933 | 0.9794 | 0.9795 | 0.9301 |
| Exposure/image | 6 s/0.5° | 5 s/0.5° | 5 s/0.5° | 12 s/0.5° | 7 s/0.5° | 7 s/0.5° | 2 s/0.5° |
| Cell dimensions (Å) | a = 102.4 | a = 102.4 | a = 103.3 | a = 102.3 | a = 102.2 | a = 102.2 | a = 102.2 |
| b = 154.1 | b = 155.6 | b = 152.6 | b = 153.7 | b = 154.0 | b = 154.0 | b = 154.0 | |
| c = 174.3 | c = 176.3 | c = 174.4 | c = 173.5 | c = 174.6 | c = 174.6 | c = 174.6 | |
| Resolution (Å) | 20–2.25 | 20–2.0 | 20–2.3 | 30–2.0 | 20–3.2 | 20–3.2 | 20–2.7 |
| Unique reflections | 57 966 | 88 328 | 55 640 | 86 260 | 22 864 | 22 846 | 37 780 |
| Average redundancy | 3.2 | 4.1 | 3.0 | 2.7 | 3.3 | 3.5 | 3.4 |
| Completeness (%) (highest bin) | 88.7 (47.8) | 90.4 (60.2) | 90.6 (53.6) | 93.6 (59.1) | 99.0 (99.1) | 99.0 (99.7) | 99.0 (99.5) |
| Rmerge (highest bin) | 0.056 (0.261) | 0.077 (0.39) | 0.054 (0.184) | 0.073 (0.372) | 0.075 (0.227) | 0.069 (0.205) | 0.074 (0.310) |
Structure determination and refinement
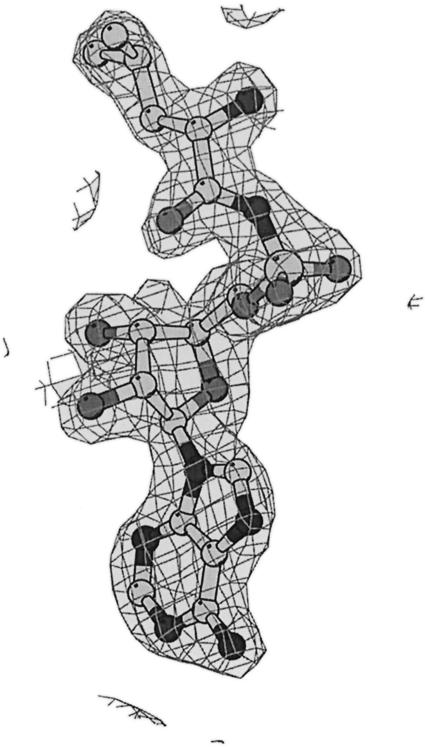
The structure was solved initially at 3.5 Å resolution using data from a three-wavelength MAD experiment on a single crystal of seleno-methionated LeuRSTT. Fifteen out of the 24 possible selenium sites were found by SOLVE giving a mean figure of merit <m> = 0.59 and Z-score = 32.8 (Terwilliger and Berendzen, 1999). Phases from the MAD experiment were used to locate heavy atom sites in the gold (two sites), platinum (four sites) and mercury (one site) derivatives and three additional selenium sites. Improved phases were then calculated to 2.7 Å resolution with MLPHARE using the three heavy atom derivatives and the peak and remote selenium data, giving <m> = 0.51 for 37 781 reflections. A partial model (651 residues) was built into the electron density map after solvent flattening with DM and then transferred by molecular replacement into the non-isomorphous crystal form co-crystallized with the LeuAMS. The higher resolution (2.0 Å) enabled WarpNtrace automatically to build and refine 750/814 visible residues including side chain positions (Perrakis et al., 1999). The C-terminal residues 815–878 are invisible in the electron density. Subsequent refinement was performed with CNS using standard protocols including maximum likelihood target, solvent correction and anisotropic temperature factor (Brünger et al., 1998). Refinement statistics are reported in Table I for the LeuAMS complex, the leucine complex and a mercury derivative with no substrate. In the corresponding crystals, either norvaline or homocysteine was present, but no clear density could be assigned to these amino acids. Electron density for the LeuAMS is shown in Figure 5.
Fig. 5. Final electron density at 2.0 Å resolution for LeuAMS in the active site of LeuRSTT contoured at 2 σ.
Acknowledgments
Acknowledgements
The authors thank the ESRF–EMBL Joint Structural Biology Group for access to ESRF synchrotron beamline facilitites. We are particularly indebted to Morton Grøtli for synthesis of LeuAMS, to Sean McSweeney and Raimond Ravelli for the success of the MAD measurements on ID14-EH4, and to Anastassis Perrakis for help with the automatic model building. Figures of structures were prepared with BOBSCRIPT (Esnouf, 1997).
References
- Asahara H., Himeno,H., Tamura,K., Hasegawa,T., Watanabe,K. and Shimizu,M. (1993) Recognition nucleotides of Escherichia coli tRNALeu and its elements facilitating discrimination from tRNASer and tRNATyr. J. Mol. Biol., 231, 219–229. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallographic and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Carter C.W. Jr (1993) Cognition, mechanism and evolutionary relationships in aminoacyl-tRNA synthetases. Annu. Rev. Biochem., 62, 715–748. [DOI] [PubMed] [Google Scholar]
- Cavarelli J., Delagoutte,B., Eriani,G., Gangloff,J. and Moras,D. (1998) l-arginine recognition by yeast arginyl-tRNA synthetase. EMBO J., 17, 5438–5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Cusack S., Yaremchuk,A. and Tukalo,M. (1996) The crystal structure of the ternary complex of T.thermophilus seryl-tRNA synthetase with tRNAser and a seryl-adenylate analogue reveals a conformational switch in the active site. EMBO J., 15, 2834–2842. [PMC free article] [PubMed] [Google Scholar]
- Deckert G. et al. (1998) The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature, 392, 353–358. [DOI] [PubMed] [Google Scholar]
- Dujardin G. and Herbert,C.J. (1997) Aminoacyl tRNA synthetases involved in group I intron splicing. In Green,R. and Schroeder,R. (eds), Ribosomal RNA and Group I Introns. Landes Bioscience, Austin, TX, pp. 179–198. [Google Scholar]
- Englisch S., Englisch,U., von der Haar,F. and Cramer,F. (1986) The proofreading of hydroxy analogues of leucine and isoleucine by leucyl-tRNA synthetases from E.coli and yeast. Nucleic Acids Res., 14, 7529–7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnouf R.M. (1997) An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph., 15, 133–138. [DOI] [PubMed] [Google Scholar]
- Herbert C.J., Labouesse,M., Dujardin,G. and Slonimski,P.P. (1988) The NAM2 proteins from S.cerevisiae and S.douglasii are mitochondrial leucyl-tRNA synthetases and are involved in mRNA splicing. EMBO J., 7, 473–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowski H. (1995) Editing of homocysteine by aminoacyl-tRNA synthetases in Eschericha coli. J. Biol. Chem., 270, 17672–17673. [PubMed] [Google Scholar]
- Jakubowski H. and Goldman,E. (1992) Editing of errors in selection of amino acids for protein synthesis. Microbiol. Rev., 56, 412–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. and Sander,C. (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers, 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- Ilyin V.A., Temple,B., Hu,M., Li,G., Yin,Y., Vachette,P. and Carter,C.W. (2000) 2.9Å crystal structure of ligand-free tryptophanyl-tRNA synthetase: domain movements fragment the adenine nucleotide binding site. Protein Sci., 9, 218–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labouesse M., Dujardin,G. and Slonimski,P.P. (1985) The yeast nuclear gene NAM2 is essential for mitochondrial DNA integrity and can cure a mitochondrial RNA-maturase deficiency. Cell, 41, 133–143. [DOI] [PubMed] [Google Scholar]
- Landro J.A., Schmidt,E., Schimmel,P., Tierney,D.L. and Penner-Hahn,J.E. (1994) Thiol ligation of two zinc atoms to a class 1 tRNA synthetase: evidence for unshared thiols and role in amino acid binding and utilization. Biochemistry, 33, 14213–14220. [DOI] [PubMed] [Google Scholar]
- Leslie A.G.W. (1994) Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography, No. 26. Daresbury Laboratory, Warrington, UK. [Google Scholar]
- Mechulam Y., Schmitt,E., Maveyraud,L., Zelwer,C., Nureki,O., Yokoyama,S., Konno,M. and Blanquet,S. (1999) Crystal structure of E.coli methionyl-tRNA synthetase highlights species-specific features. J. Mol. Biol., 294, 1287–1297. [DOI] [PubMed] [Google Scholar]
- Nomanbhoy T.K., Hendricjson,T.L. and Schimmel,P. (1999) Transfer RNA-dependent translocation of misactivated amino acids to prevent errors in protein synthesis. Mol. Cell, 4, 519–528. [DOI] [PubMed] [Google Scholar]
- Nureki O., Kohno,T., Sakamoto,K., Miyazawa,T. and Yokoyama,S. (1993) Chemical modification and mutagenesis studies on zinc binding of aminoacyl-tRNA synthetases. J. Biol. Chem., 268, 15368–15373. [PubMed] [Google Scholar]
- Nureki O. et al. (1998) Enzyme structure with two catalytic sites for double-sieve selection of substrate. Science, 280, 578–582. [DOI] [PubMed] [Google Scholar]
- Nureki O. et al. (1999) Proofreading by isoleucyl-transfer RNA synthetase: response. Science, 283, 459a. [Google Scholar]
- Pauling L. (1958) The Probability of Errors in the Process of Synthesis of Protein Molecules. Festschrift Arthur Stoll. Birkhauser Verlag, Basel, Switzerland, pp. 597. [Google Scholar]
- Perrakis A., Morris,R.J. and Lamzin,V.S. (1999) Automated protein model building combined with iterative structure refinement. Nature Struct. Biol., 6, 458–463. [DOI] [PubMed] [Google Scholar]
- Silvian L.F., Wang,J. and Steitz,T.A. (1999) Insights into editing from an Ile-tRNA synthetase structures with tRNAile and mupirocin. Science, 285, 1074–1077. [PubMed] [Google Scholar]
- Soma A., Uchiyama,K., Sakamoto,T., Maeda,M. and Himeno,H. (1999) Unique recognition style of tRNAleu by Haloferax volcanii leucyl-tRNA synthetase. J. Mol. Biol., 293, 1029–1038. [DOI] [PubMed] [Google Scholar]
- Starzyk R.M., Webster,T.A. and Schimmel,P. (1987) Evidence for dispensable sequences inserted into a nucleotide fold. Science, 237, 1614–1618. [DOI] [PubMed] [Google Scholar]
- Sugiura I. et al. (2000) The 2.0Å structure of T.thermophilus methionyl-tRNA synthetase reveals two RNA-binding modules present in other class-I enzymes. Structure, 8, 197–208. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen,J. (1999) Automated structure solution for MIR and MAD. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H., Shoku,Y., Hayashi,N., Mitsunaga,J., In,Y., Doi,M., Inoue,M. and Ishida,T. (1991) X-ray crystallographic conformational study of 5′-O-[N-(l-alanyl)-sulfamoyl]adenosine, a substrate analogue for alanyl-tRNA synthetase. Biochim. Biophys. Acta, 1080, 126–134. [DOI] [PubMed] [Google Scholar]
- Yaremchuk A., Cusack,S., Gudzera,O., Grøtli,M. and Tukalo,M. (2000) Crystallisation and preliminary analysis of Thermus themophilus leucyl-tRNA synthetase and its complexes with leucine and non-hydrolysable leucyl-adenylate analogue. Acta Crystallogr. D, in press. [DOI] [PubMed] [Google Scholar]