Abstract
Trypanosoma cruzi, the causative agent of Chagas disease, is subdivided into six discrete typing units (DTUs; TcI–TcVI) of which TcI is ubiquitous and genetically highly variable. While clonality is the dominant mode of propagation, recombinant events play a significant evolutive role. Recently, foci of wild Triatoma infestans have been described in Bolivia, mainly infected by TcI. Hence, for the first time, we evaluated the level of genetic exchange within TcI natural potentially panmictic populations (single DTU, host, area and sampling time).
Seventy-nine TcI stocks from wild T. infestans, belonging to six populations were characterized at eight microsatellite loci. For each population, Hardy-Weinberg equilibrium (HWE), linkage disequilibrium (LD), and presence of repeated multilocus genotypes (MLG) were analyzed by using a total of seven statistics, to test the null hypothesis of panmixia (H0).
For three populations, none of the seven statistics allowed to rejecting H0; for another one the low size did not allow us to conclude, and for the two others the tests have given contradictory results. Interestingly, apparent panmixia was only observed in very restricted areas, and was not observed when grouping populations distant of only two kilometers or more. Nevertheless it is worth stressing that for the statistic tests of "HWE", in order to minimize the type I error (i. e. incorrect rejection of a true H0), we used the Bonferroni correction (BC) known to considerably increase the type II error ( i. e. failure to reject a false H0). For the other tests (LD and MLG), we did not use BC and the risk of type II error in these cases was acceptable. Thus, these results should be considered as a good indicator of the existence of panmixia in wild environment but this must be confirmed on larger samples to reduce the risk of type II error.
Introduction
Trypanosoma cruzi is the causative agent of Chagas disease, which affects about eight million people in Latin America, of whom 30–40% either suffers or will develop cardiomyopathy, digestive megasyndromes, or both. Moreover, Chagas disease is becoming an emerging health problem in nonendemic areas because of the increasing number of migrants from endemic areas [1]. The T. cruzi species exhibits a very high genetic variability similar to that observed within different species of other kinetoplastidae such as Leishmania [2]. Consensual taxonomy recognized six discrete typing units (DTUs named TcI–TcVI) [3] and one additional group only found in bats (Tcbat) [4] within T. cruzi [5]; TcI is the most genetically diversified and ubiquitous of them, spreading from the United States to Argentina, and present in both sylvatic and domestic biotopes. As a result of the dominant clonal multiplication, identical multilocus genotypes (MLGs) have been sampled over several years and over large geographical distances, leading to considering the species as multiclonal [6]. The long-term clonal evolution is involved in the current important genetic diversity of the species, but more and more “genetic exchange” events are being described. Scarce hybridization events are the source of two hybrid DTUs [7–9], mitochondrial introgression events have been detected [10,11], and different levels of gene recombination have been described [12–14]. In addition, high genome plasticity is also a source of variability. Aneuploidy is suspected [15], occurrence of allele loss is possible during genetic exchanges, the mitochondrial genome is probably more complex than previously described, and maxicircle gene recombination occurs as well as intragenic recombination [14]; heteroplasmy has also been reported [16]. Several of these genetic exchange mechanisms have been triggered in vitro [17] and are still hotly debated in the field. As previously stated [18]: “From an epidemiological and medical point of view, the important parameter to evaluate is the stability of the genetic clones in space and time.” This stability directly depends on the level of genetic exchanges (in the broad sense). Indeed, within a strict clonal framework the clones are stable in space and time, and they convey similar biological characteristics that can be crucial for epidemiological and medical features generation after generation. In contrast, with more or less frequent recombination, such correlations are not necessarily expected, hence the importance of studying genetic exchanges between stocks.
In general terms, to test panmixia, two prerequisites are needed: (i) the use of an appropriate genetic marker not subjected to selection and with a sufficient level of polymorphism and (ii) populations isolated in restricted areas where parasites are assumed to be in sympatry. Our previous work showed that microsatellite markers are relevant for studying the population genetics of T. cruzi at the DTU level [19]. Moreover, abundant and accessible foci of wild Triatoma infestans vectors mainly infected by TcI have been recently described in Bolivia [20,21]; hence, in the present work it was possible to evaluate the level of genetic exchanges in potentially panmictic T. cruzi TcI populations isolated from sylvatic T. infestans in Bolivia.
Materials and Methods
Parasite stocks and multilocus microsatellite typing (MLMT)
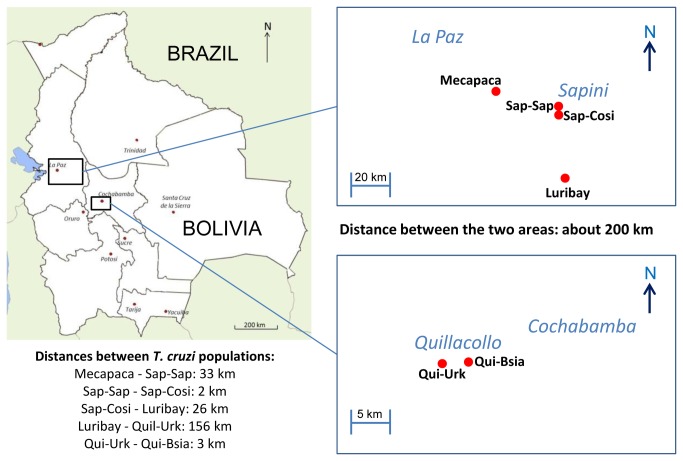
Seventy-nine T. cruzi stocks, previously assigned to the DTU TcI using the multiplex miniexon PCR method [22] and isolated from six potentially panmictic Bolivian sylvatic T. infestans populations (see Figure 1) were compared to 21 TcI sylvatic reference stocks ranging from the United States to South America (see Table 1). These populations were defined in small geographic areas in which we believe that the T. infestans vector can move freely (maximum distance between two stocks less than five hundred meters). Four of them are located in La Paz department (namely, Luribay, central sampling point at 17°3'54.90"S / 67°39'53.85"W; Mecapaca, 16°42'45.90"S / 67°59'27.13"W; Sap-Sap, 16°48'47.23"S / 67°42'9.83"W; and Sap-Cosi, 16°49'50.00"S / 67°42'22.20"W), while the other two populations are located in Cochabamba department (namely, Qui-Urk, 17°25'29.00"S / 66°17'45.20"W and Qui-Bsia, 17°25'28.81"S / 66°15'52.75"W). The distances between the populations are given in Figure 1. The stocks directly isolated from wild triatomines, all captured with mice bait Noireau’s traps, were cultured in LIT medium supplemented with 10% fetal calf serum. DNA was extracted with a conventional CTBA 2% method and the solutions diluted to 20 ng/µl before use. Eight previously described microsatellite loci were used, namely MCLE01, SCLE10, SCLE11, MCLF10, A427, MCLG10, C875, and MCLE08 [17,23] using the same PCR conditions [19]. Electrophoreses of fluorescent-labeled PCR products, diluted and denatured in 20 µl of HiDi formamide, were carried out on a ABI3130xL Genetic Analyzer (Applied Biosystems, Carlsbad, CA, USA), with Genescan 500 LIZ as the internal size standard. GeneMapper® software (Applied Biosystems, Carlsbad, CA, USA) was used to characterize the alleles.
Figure 1. Map of Bolivia: localization of the six populations of Trypanosoma cruzi under study isolated from sylvatic Triatoma infestans and distances between populations.
Table 1. Codes, locations, genotypes at each locus and reference numbers of each multilocus genotype (MLG) of the 79 Trypanosoma cruzi TcI stocks isolated from six potentially panmictic populations and of the 21 T. cruzi TcI reference strains.
| Stock code | Location* | MCLE01 |
SCLE10 |
SCLE11 |
MCLF10 |
A427 |
MCLG10 |
C875 |
MCLE08 |
MLG | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LaPaz / Mecapaca / Tun1 / Mecapaca |
||||||||||||||||||
| MEC095 | id. | 128 | 128 | 250 | 254 | 138 | 138 | 188 | 188 | 185 | 185 | 155 | 155 | 191 | 191 | 117 | 117 | 66 |
| MEC099 | id. | 128 | 128 | 250 | 254 | 138 | 138 | 188 | 188 | 186 | 186 | 155 | 155 | 183 | 187 | 117 | 117 | 74 |
| MEC101 | id. | 128 | 128 | 250 | 254 | 138 | 138 | 188 | 188 | 186 | 186 | 155 | 155 | 183 | 187 | 117 | 119 | 73 |
| MEC102 | id. | 128 | 128 | 250 | 254 | 138 | 138 | 184 | 188 | 186 | 186 | 155 | 155 | 183 | 187 | 117 | 119 | 71 |
| MEC103 | id. | 128 | 129 | 250 | 254 | 139 | 139 | 184 | 188 | 186 | 186 | 155 | 155 | 191 | 191 | 117 | 117 | 76 |
| MEC107 | id. | 128 | 128 | 254 | 254 | 138 | 138 | 184 | 188 | 186 | 186 | 155 | 155 | 191 | 191 | 117 | 119 | 70 |
| MEC161 | id. | 128 | 128 | 250 | 254 | 139 | 139 | 188 | 188 | 186 | 186 | 155 | 155 | 187 | 191 | 119 | 119 | 86 |
| MEC166 | id. | 128 | 128 | 250 | 254 | 139 | 139 | 188 | 188 | 186 | 186 | 155 | 155 | 187 | 191 | 119 | 119 | 86 |
| MEC170 | id. | 128 | 128 | 250 | 254 | 139 | 139 | 188 | 188 | 186 | 186 | 155 | 155 | 183 | 187 | 119 | 119 | 85 |
| MEC171 | id. | 127 | 127 | 250 | 250 | 138 | 138 | 184 | 188 | 178 | 184 | 155 | 155 | 183 | 191 | 117 | 119 | 58 |
| MEC173 | id. | 128 | 128 | 250 | 250 | 139 | 141 | 184 | 188 | 177 | 184 | 155 | 155 | 183 | 191 | 119 | 119 | 59 |
| LaPaz / Luribay / Luribay / Luribay | ||||||||||||||||||
| LUR229 | id. | 128 | 128 | 238 | 250 | 138 | 138 | 184 | 184 | 178 | 186 | 155 | 155 | 189 | 189 | 117 | 117 | 67 |
| LUR237 | id. | 128 | 128 | 250 | 250 | 138 | 138 | 184 | 184 | 185 | 185 | 155 | 155 | 189 | 189 | 117 | 117 | 68 |
| LUR245 | id. | 120 | 128 | 250 | 254 | 138 | 138 | 184 | 188 | 186 | 186 | 146 | 155 | 189 | 189 | 117 | 117 | 64 |
| LUR250 | id. | 128 | 128 | 250 | 254 | 138 | 138 | 184 | 188 | 185 | 185 | 155 | 155 | 189 | 189 | 0 | 0 | 65 |
| LUR258 | id. | 128 | 128 | 250 | 250 | 138 | 140 | 184 | 184 | 179 | 186 | 155 | 155 | 189 | 189 | 117 | 117 | 69 |
| LUR265 | id. | 128 | 128 | 250 | 250 | 138 | 138 | 184 | 184 | 177 | 187 | 155 | 155 | 187 | 187 | 0 | 0 | 60 |
| LaPaz / Murillo / Sapini / Sap-Sap | ||||||||||||||||||
| SAP203 | id. | 129 | 129 | 250 | 254 | 138 | 138 | 184 | 188 | 175 | 178 | 155 | 155 | 187 | 189 | 117 | 119 | 21 |
| SAP207 | id. | 128 | 128 | 250 | 254 | 138 | 138 | 188 | 188 | 178 | 178 | 155 | 155 | 187 | 189 | 117 | 119 | 48 |
| SAP223 | id. | 120 | 128 | 250 | 254 | 138 | 138 | 184 | 188 | 175 | 178 | 155 | 155 | 187 | 189 | 117 | 119 | 53 |
| SAP233 | id. | 129 | 129 | 250 | 254 | 138 | 138 | 184 | 188 | 175 | 178 | 155 | 155 | 189 | 191 | 117 | 119 | 22 |
| SAP241 | id. | 117 | 128 | 250 | 254 | 138 | 138 | 184 | 188 | 177 | 177 | 146 | 146 | 187 | 189 | 117 | 119 | 50 |
| SAP242 | id. | 129 | 129 | 250 | 254 | 138 | 138 | 184 | 188 | 175 | 178 | 155 | 155 | 187 | 189 | 117 | 119 | 21 |
| SAP242b | id. | 129 | 129 | 250 | 254 | 138 | 138 | 184 | 188 | 178 | 178 | 146 | 155 | 187 | 189 | 117 | 119 | 24 |
| SAP243 | id. | 129 | 129 | 250 | 250 | 138 | 140 | 188 | 188 | 178 | 178 | 155 | 155 | 187 | 189 | 117 | 119 | 35 |
| SAP256 | id. | 128 | 128 | 250 | 254 | 138 | 138 | 182 | 186 | 175 | 178 | 146 | 146 | 187 | 189 | 117 | 119 | 51 |
| SAP259 | id. | 128 | 128 | 250 | 254 | 139 | 139 | 184 | 188 | 175 | 178 | 146 | 155 | 187 | 189 | 117 | 119 | 52 |
| SAP260 | id. | 129 | 129 | 250 | 254 | 138 | 138 | 184 | 188 | 175 | 175 | 146 | 146 | 187 | 189 | 117 | 119 | 26 |
| SAP261 | id. | 129 | 129 | 250 | 254 | 138 | 138 | 184 | 188 | 175 | 175 | 146 | 155 | 187 | 189 | 117 | 119 | 25 |
| SAP263 | id. | 129 | 129 | 250 | 254 | 138 | 138 | 184 | 188 | 175 | 175 | 155 | 155 | 189 | 191 | 117 | 119 | 23 |
| SAP264 | id. | 129 | 129 | 250 | 254 | 138 | 138 | 184 | 184 | 178 | 178 | 155 | 155 | 189 | 189 | 119 | 119 | 19 |
| SAP265 | id. | 129 | 129 | 250 | 254 | 138 | 138 | 184 | 188 | 175 | 184 | 155 | 155 | 187 | 187 | 117 | 119 | 30 |
| SAP266 | id. | 129 | 129 | 254 | 254 | 138 | 138 | 188 | 188 | 178 | 178 | 155 | 155 | 189 | 189 | 117 | 119 | 17 |
| SAP267 | id. | 120 | 128 | 250 | 254 | 138 | 138 | 184 | 188 | 175 | 178 | 146 | 155 | 187 | 189 | 117 | 119 | 54 |
| SAP270 | id. | 128 | 128 | 250 | 254 | 138 | 138 | 184 | 188 | 175 | 178 | 155 | 155 | 187 | 189 | 117 | 119 | 55 |
| SAP271 | id. | 129 | 129 | 250 | 250 | 138 | 140 | 184 | 188 | 186 | 186 | 146 | 155 | 187 | 189 | 117 | 119 | 39 |
| SAP272 | id. | 129 | 129 | 250 | 254 | 138 | 138 | 184 | 188 | 175 | 186 | 146 | 146 | 187 | 189 | 117 | 119 | 27 |
| SAP391 | id. | 120 | 128 | 250 | 250 | 138 | 138 | 184 | 184 | 177 | 177 | 155 | 155 | 187 | 187 | 119 | 119 | 62 |
| SAP404 | id. | 128 | 128 | 250 | 254 | 138 | 138 | 184 | 188 | 175 | 175 | 155 | 155 | 187 | 189 | 117 | 119 | 56 |
| SAP405 | id. | 117 | 128 | 250 | 250 | 138 | 138 | 184 | 184 | 177 | 177 | 155 | 155 | 187 | 187 | 119 | 119 | 63 |
| SAP445 | id. | 120 | 128 | 250 | 254 | 138 | 138 | 184 | 184 | 177 | 177 | 155 | 155 | 187 | 187 | 119 | 119 | 61 |
| SAP491 | id. | 120 | 128 | 250 | 254 | 138 | 138 | 184 | 188 | 175 | 175 | 155 | 155 | 187 | 189 | 117 | 119 | 57 |
| SAP492 | id. | 120 | 128 | 250 | 254 | 138 | 138 | 184 | 188 | 175 | 175 | 155 | 155 | 187 | 189 | 117 | 119 | 57 |
| SAP500 | id. | 128 | 128 | 250 | 254 | 138 | 138 | 184 | 188 | 175 | 175 | 155 | 155 | 187 | 189 | 117 | 119 | 56 |
| LaPaz / Loayza / Cosiraya / Sap-Cosi | ||||||||||||||||||
| SAP302 | id. | 129 | 129 | 250 | 250 | 137 | 137 | 188 | 188 | 178 | 186 | 155 | 155 | 187 | 187 | 117 | 119 | 37 |
| SAP303 | id. | 128 | 128 | 250 | 254 | 137 | 137 | 188 | 188 | 178 | 178 | 155 | 155 | 187 | 187 | 117 | 119 | 49 |
| SAP304 | id. | 129 | 129 | 250 | 250 | 138 | 138 | 184 | 188 | 178 | 186 | 155 | 155 | 187 | 189 | 117 | 119 | 34 |
| SAP310 | id. | 129 | 129 | 250 | 254 | 138 | 138 | 184 | 188 | 186 | 186 | 146 | 155 | 187 | 187 | 117 | 119 | 28 |
| SAP312 | id. | 129 | 129 | 254 | 254 | 138 | 138 | 188 | 188 | 186 | 186 | 146 | 146 | 189 | 189 | 117 | 119 | 18 |
| SAP313 | id. | 129 | 129 | 250 | 254 | 138 | 138 | 184 | 188 | 178 | 186 | 155 | 155 | 187 | 187 | 117 | 119 | 31 |
| SAP318 | id. | 129 | 129 | 250 | 250 | 138 | 138 | 188 | 188 | 178 | 186 | 146 | 146 | 187 | 187 | 117 | 119 | 46 |
| SAP319 | id. | 129 | 129 | 250 | 250 | 138 | 138 | 188 | 188 | 178 | 178 | 146 | 146 | 187 | 187 | 117 | 119 | 45 |
| SAP321 | id. | 129 | 129 | 250 | 250 | 138 | 140 | 178 | 188 | 179 | 186 | 146 | 155 | 187 | 187 | 117 | 119 | 40 |
| SAP323 | id. | 129 | 129 | 250 | 254 | 138 | 138 | 184 | 188 | 179 | 186 | 155 | 155 | 187 | 187 | 117 | 117 | 32 |
| SAP334 | id. | 129 | 129 | 250 | 250 | 138 | 140 | 178 | 178 | 186 | 186 | 155 | 155 | 187 | 187 | 111 | 117 | 41 |
| SAP336 | id. | 129 | 129 | 250 | 250 | 138 | 138 | 178 | 178 | 178 | 186 | 146 | 155 | 187 | 187 | 117 | 119 | 43 |
| SAP337 | id. | 129 | 129 | 250 | 250 | 138 | 138 | 178 | 178 | 178 | 186 | 146 | 146 | 187 | 187 | 117 | 119 | 44 |
| SAP346 | id. | 129 | 129 | 250 | 250 | 138 | 138 | 184 | 188 | 179 | 186 | 155 | 155 | 187 | 187 | 117 | 117 | 33 |
| SAP347 | id. | 129 | 129 | 250 | 250 | 138 | 138 | 188 | 188 | 178 | 186 | 155 | 155 | 187 | 189 | 117 | 119 | 36 |
| SAP348 | id. | 129 | 129 | 250 | 250 | 137 | 137 | 188 | 188 | 178 | 186 | 146 | 155 | 187 | 189 | 117 | 119 | 38 |
| SAP349 | id. | 129 | 129 | 250 | 250 | 138 | 138 | 188 | 188 | 178 | 186 | 146 | 146 | 187 | 187 | 117 | 117 | 47 |
| SAP372 | id. | 129 | 129 | 250 | 250 | 138 | 138 | 184 | 184 | 178 | 186 | 146 | 155 | 189 | 189 | 117 | 119 | 20 |
| SAP374 | id. | 129 | 129 | 250 | 250 | 138 | 138 | 184 | 188 | 179 | 179 | 146 | 155 | 187 | 187 | 117 | 119 | 42 |
| Cochabamba / Quillacollo / VillaUrkipiña / Qui-Urk | ||||||||||||||||||
| QUI755 | id. | 129 | 129 | 250 | 254 | 138 | 138 | 188 | 188 | 186 | 186 | 155 | 155 | 187 | 187 | 117 | 119 | 29 |
| QUI757 | id. | 129 | 129 | 250 | 254 | 139 | 139 | 188 | 188 | 186 | 186 | 155 | 155 | 187 | 187 | 119 | 119 | 88 |
| QUI762 | id. | 120 | 128 | 250 | 254 | 139 | 139 | 188 | 188 | 186 | 186 | 155 | 155 | 187 | 187 | 119 | 119 | 84 |
| QUI763 | id. | 129 | 129 | 250 | 254 | 139 | 139 | 188 | 188 | 186 | 186 | 155 | 155 | 187 | 187 | 117 | 119 | 89 |
| QUI766 | id. | 129 | 129 | 250 | 254 | 139 | 139 | 188 | 188 | 186 | 186 | 155 | 155 | 187 | 187 | 117 | 119 | 89 |
| QUI768 | id. | 129 | 129 | 250 | 254 | 138 | 138 | 188 | 188 | 186 | 186 | 155 | 155 | 187 | 187 | 117 | 119 | 29 |
| QUI769 | id. | 120 | 128 | 250 | 250 | 139 | 139 | 188 | 188 | 186 | 186 | 155 | 155 | 187 | 187 | 117 | 119 | 83 |
| QUI774 | id. | 129 | 129 | 250 | 254 | 139 | 139 | 188 | 188 | 186 | 186 | 155 | 155 | 187 | 187 | 117 | 119 | 89 |
| QUI775 | id. | 129 | 129 | 250 | 254 | 139 | 139 | 188 | 188 | 186 | 186 | 155 | 155 | 187 | 187 | 117 | 119 | 89 |
| QUI907 | id. | 129 | 129 | 250 | 254 | 139 | 139 | 188 | 188 | 186 | 186 | 155 | 155 | 187 | 187 | 117 | 119 | 89 |
| QUI913 | id. | 129 | 129 | 250 | 254 | 139 | 139 | 188 | 188 | 186 | 186 | 155 | 155 | 187 | 187 | 119 | 119 | 88 |
| QUI916 | id. | 129 | 129 | 250 | 254 | 139 | 139 | 188 | 188 | 186 | 186 | 155 | 155 | 187 | 187 | 117 | 117 | 87 |
| Cochabamba / Quillacollo / BSIA14T1 / Qui-Bsia | ||||||||||||||||||
| QUI026 | id. | 128 | 128 | 250 | 254 | 138 | 138 | 188 | 188 | 186 | 186 | 155 | 155 | 187 | 187 | 117 | 117 | 75 |
| QUI027 | id. | 128 | 128 | 250 | 254 | 138 | 138 | 188 | 188 | 186 | 186 | 155 | 155 | 187 | 187 | 117 | 117 | 75 |
| QUI053 | id. | 131 | 131 | 250 | 254 | 136 | 136 | 188 | 188 | 186 | 186 | 146 | 155 | 187 | 187 | 117 | 117 | 80 |
| QUI054 | id. | 117 | 125 | 250 | 254 | 130 | 140 | 188 | 188 | 186 | 186 | 155 | 155 | 187 | 187 | 117 | 117 | 81 |
| Countries of reference strains | ||||||||||||||||||
| 361-TA | Colombia | 131 | 133 | 252 | 254 | 135 | 138 | 186 | 186 | 174 | 174 | 157 | 157 | 176 | 182 | 119 | 119 | 7 |
| 458 | Colombia | 131 | 131 | 248 | 251 | 138 | 140 | 186 | 186 | 178 | 178 | 153 | 155 | 172 | 176 | 114 | 119 | 2 |
| 85/818 | Bolivia | 123 | 141 | 251 | 251 | 138 | 138 | 174 | 174 | 177 | 181 | 146 | 155 | 174 | 174 | 119 | 119 | 1 |
| 93041401P | USA | 135 | 141 | 255 | 255 | 135 | 135 | 174 | 184 | 188 | 188 | 157 | 157 | 165 | 165 | 114 | 114 | 11 |
| 93070103P | USA | 135 | 141 | 255 | 255 | 135 | 135 | 174 | 174 | 188 | 188 | 157 | 157 | 165 | 165 | 114 | 117 | 12 |
| A269 | Guiana | 143 | 145 | 251 | 251 | 140 | 140 | 184 | 184 | 179 | 179 | 155 | 157 | 178 | 184 | 114 | 114 | 4 |
| Cuicacl1 | Brazil | 128 | 128 | 250 | 254 | 139 | 139 | 188 | 188 | 186 | 186 | 146 | 155 | 187 | 187 | 117 | 119 | 82 |
| Cutiacl1 | Brazil | 129 | 129 | 254 | 254 | 139 | 139 | 178 | 188 | 173 | 186 | 155 | 155 | 165 | 165 | 117 | 117 | 77 |
| FX18 | Colombia | 127 | 131 | 255 | 255 | 136 | 136 | 178 | 178 | 173 | 173 | 159 | 161 | 166 | 185 | 114 | 114 | 5 |
| G-38-1 | Brazil | 129 | 129 | 254 | 254 | 138 | 138 | 182 | 182 | 177 | 177 | 153 | 153 | 172 | 172 | 117 | 117 | 15 |
| H10 | Mexico | 135 | 137 | 252 | 255 | 135 | 135 | 176 | 176 | 173 | 173 | 157 | 157 | 165 | 165 | 117 | 119 | 9 |
| OPS21cl11 | Venezuela | 135 | 135 | 252 | 255 | 135 | 135 | 184 | 186 | 173 | 186 | 157 | 157 | 165 | 165 | 117 | 119 | 10 |
| P209cl93 | Bolivia | 129 | 129 | 238 | 254 | 138 | 138 | 178 | 178 | 186 | 186 | 155 | 155 | 165 | 165 | 117 | 119 | 16 |
| PB3cl2 | Bolivia | 127 | 155 | 252 | 254 | 138 | 140 | 186 | 188 | 178 | 178 | 155 | 155 | 170 | 170 | 114 | 117 | 3 |
| PERU | Peru | 127 | 127 | 252 | 255 | 128 | 128 | 184 | 186 | 182 | 182 | 149 | 157 | 165 | 165 | 114 | 119 | 8 |
| SABP3 | Peru | 128 | 128 | 250 | 254 | 138 | 138 | 188 | 188 | 186 | 186 | 155 | 155 | 187 | 187 | 119 | 119 | 72 |
| Saimiri4A | Venezuela | 142 | 144 | 251 | 251 | 136 | 136 | 191 | 191 | 178 | 178 | 157 | 157 | 185 | 189 | 111 | 117 | 6 |
| SP31 | Chile | 128 | 128 | 254 | 254 | 139 | 139 | 184 | 188 | 173 | 186 | 155 | 155 | 165 | 165 | 119 | 119 | 78 |
| T.cruzi#1 | Honduras | 134 | 134 | 252 | 252 | 135 | 135 | 184 | 184 | 0 | 0 | 157 | 157 | 165 | 165 | 119 | 119 | 13 |
| V120 | Chile | 128 | 128 | 254 | 254 | 139 | 139 | 188 | 188 | 173 | 186 | 155 | 155 | 165 | 169 | 114 | 119 | 79 |
| Z17 | Mexico | 137 | 137 | 255 | 255 | 135 | 135 | 184 | 184 | 175 | 175 | 157 | 157 | 165 | 165 | 119 | 119 | 14 |
* Department / Municipality / Area / Population
Data analysis
“For a majority of pathogens, including the Trypanosomatidae family, the reproductive strategy was mainly deduced from population genetics analysis” [24]. Here, the analyses were focused on two kinds of events involved in sexual exchanges: allelic segregation and genetic recombination. Allelic segregation was explored through Hardy-Weinberg equilibrium (HWE) or F is, while genetic recombination was explored through linkage disequilibrium analysis (LD, nonrandom association between genotypes at independent loci) and the presence / absence of repeated multilocus genotypes (MLG). A previous study, based on simulations and aiming to estimate the level of clonal reproduction in diploids [25] advised the simultaneous use of F is (mean and variance) and LD estimators.
F is is a measure of inbreeding of individuals within a subsample; it also represents the deviation from random union of gametes and varies from −1 (fixed heterozygous) to +1 (fixed homozygous) via F is = 0 (Hardy-Weinberg equilibrium). This Wright F-statistic [26] was estimated with Weir and Cockerham’s unbiased estimators [27] called f. Negative values of F is (excess heterozygosity) can be caused by accumulation of mutations in an ancient clonal lineage, a phenomenon called the Meselson effect [28], and are generally regarded as a mark of clonality as observed in Bdelloid rotifers [29]. Positive values of F is correspond to inbreeding within the sample, a particular case being the Wahlund effect, when the sample comes from heterogeneous and structured populations. It is worth noting that if the mean F is values are good estimators of HWE, low F is values associated with substantial variance of F is among loci (with some loci displaying an extreme heterozygote deficit and others an extreme excess) can reveal very low levels of sex (cryptic sex) [30]. All statistical tests were based on randomization: data sets fitting the null hypothesis (H0 = panmixia) were generated by randomizing the relevant unit (allele, genotype, etc.). Here, to test HWE within the subsamples, the alleles were permuted among individuals within each subsample and F is was used as a HWE estimator, while for testing the overall HWE, alleles were permuted among subsamples and F it was used as an estimator. Moreover, since the presence of null alleles artificially increases F is estimations, we tested the impact of null alleles on the increased F is values.
Linkage disequilibrium (LD) is another measure of deviation from panmixia. Here it was estimated in three different ways: (i) by the classical index IA [31], which has the disadvantage of increasing with the number of loci, so we also used a slightly modified index (ȓd) which is independent of the number of loci [32]; (ii) by the log-likelihood ratio G-statistics [33]; the P-value of this test is obtained as follows: genotypes at the 2 loci are associated at random a number of times and the statistic is recalculated on the randomized data set; the P-value is estimated as the proportion of statistics from randomized data sets that are larger or equal to the observed and (iii) by comparing the observed number of MLGs and the frequency of the most frequent MLG to the expected ones in simulated panmixia. As for F is, all the LD statistical tests are based on H0 = panmixia (i.e., the genotypes at the two loci are associated at random a number of times depending on the sample size and the statistics are recalculated on the randomized data set).
LD and HWE tests are based on multiple comparisons, so the Bonferroni corrections should be applied; this consists in dividing the p-value (or α, generally 5%), which is the threshold for rejecting H0, by the number of comparisons. For example, testing eight loci within seven different populations leads to 56 comparisons and theoretically α (0.05) would become α′ = α / 56 = 0.00089. Nevertheless, the Bonferroni correction entails a high risk of falsely accepting H0 (bias towards Type II error) and therefore masking real deviations from panmixia. Teriokhin et al. [34] suggested that a high test power can be preserved by using the binomial test instead of the Bonferroni correction in order to check whether the proportion of tests found significant at the 5% level was significantly above 0.05: if this is true, the test is significant and H0 is rejected, and if it is not true, H0 is not rejected ; for example here with 8 loci, to test the genotype association at two loci by using G-statistics there are 26 comparisons and hence 26 values of G-Statistics: if 3 of them are below 0.05 the binomial test (written in R “binom.test (3, 26, p=0.05)”) give a no significant P-value of 0.1386, meaning that 3 values under 0.05 out of 26 are not sufficient to reject Ho; in reality we need 4 values below 0.05 out of 26 to reject significantly H0 (P-value of the binomial test in this case is 0.03874). Because rejecting or accepting the null hypothesis is crucial here, we chose to use and discuss all the p-values (with or without the Bonferroni corrections and the p-values given by the exact binomial tests).
To test F is, LD, and MLG, we examined nine subsamples: the six populations under study (Luribay, Mecapaca, Sap-Sap, Sap-Cosi, Qui-Urk, and Qui-Bsia); the subsample “overall Sapini,” which clusters the two populations from Sapini (Sap-Sap + Sap-Cosi); the subsample “overall Quillacollo,” which clusters the two populations from Quillacollo (Qui-Urk + Qui-Bsia); and the “overall” sample including all stocks (N = 79). The different indices and p-values were associated with their level of significance (NS, not significant; * significant at 5% and ** significant at 1%). As several tests were applied for F is, LD, and repeated MLG, a decision about accepting or rejecting H0 is proposed in each case, namely “reject H0” or “not reject H0” when all tests are congruent, and “ambiguous” when at least one of the tests gave a discordant result.
To process the data, different programs were used: (i) the HierFstat package [35] in R [36] to compute the 95% confidence intervals of F is, (ii) the “binom.test” function in R to test the null hypothesis about the probability of success in Bernoulli’s experiments, (iii) MicroChecker v.2.2.3 [37] to test the load of null alleles, (iv) Multilocus v1.3b [32] for IA and ȓd indices and to test the probabilities of repeated MLG and different MLG, (v) Populations (v.1.2.30© 1999, Olivier Langella, CNRS UPR9034) to build a general clustering analysis between all stocks using the Cavalli-Sforza and Edwards’ chord genetic distances [38], and (vi) Fstat [39] for all other tests.
Results
Genetic diversity of the six populations under study
Genetic diversity was explored within the six local wild T. cruzi TcI populations (79 stocks) and within the 21 reference strains. Details of the origin and allelic microsatellite composition of each stock studied are listed in Table 1.
Null alleles: Only two stocks from the Luribay population did not amplify at locus MCL08 and one reference stock at locus A427. Analyzing the six potentially panmictic populations with MicroChecker, 43 null alleles were expected at loci presenting high F is over 1264 alleles, hence 3.40%, which is already very low. The proportion of observed null alleles in this sample (n = 4, hence 0.32%) is lower than expected (exact binomial test, p = 4e-14). Thus, the role of null alleles in inflated F is may be considered here as negligible.
Overall polymorphism: The main indices of genetic diversity as well as observed and expected heterozygotes and F is by locus and by population are listed in Table 2. It is worth noting that, as expected, the subsample of the reference strains (n = 21) is by far the most polymorphic. Moreover, 42 alleles out of 82 (51.2% of the total number of alleles) were specific to reference strains (see Table 1). Eighty-nine different multilocus genotypes (MLGs) were observed among the 100 stocks (including references) versus only 68 MLGs among the 79 stocks under study (without references). The most repeated MLG (no. 89, repeated five times) was identified in a single population, Qui-Urk, in the Cochabamba valley (Table 1). The number of alleles per locus ranged from 4 to 18 and from 2 to 8 with and without references, respectively. Similarly, the mean allelic richness by locus systematically decreased when reference strains were removed. For the six local populations, the F is values per locus and per population showed high variance, ranging from −1.00 (fixed heterozygosity for locus SCLE10 in Qui-Bsia population) to 1.00 (fixed homozygosity for loci MCLE01 in Sap-Cosi, SCLE11 in Qui-Urk, and C875 in Luribay), while only positive F is values were observed for the reference population (ranging from 0.30 to 0.82) as is expected when pooling differentiated reproductive units within a single subpopulation [25]. The mean allelic richness in local populations was weakly variable, ranging from 1.49 (Qui-Urk) to 2.27 (Sap-Sap) and higher within the reference strains (4.49). The clustering analysis (NJ tree not shown) of all the stocks using the Cavalli-Sforza and Edwards distance method showed that six of the reference strains, namely P209cl93, SABP3, Cutiacl1, SP31, V120, and Cuicacl1, were closely related to some of the wild stocks under study, the other reference strains forming a separate group not supported by a significant bootstrap value. The analysis of genetic distances between each of the 21 reference strains and the 79 wild stocks (mean of pairwise distances) showed that the three reference strains closest to the Bolivian wild stocks were SABP3 from Peru, Cuicacl1 from Brazil, and P209cl93 from Bolivia, with genetic distances of 0.36, 0.41, and 0.50, respectively; the three reference strains farthest from the wild stocks were FX18 from Colombia and 93041401P and 93070103P from the US, with mean genetic distances of 0.89, 0.88, and 0.87, respectively.
Table 2. Main indices of genetic diversity and Hardy-Weinberg equilibrium by locus (vertical) and by population (horizontal) of the 79 Trypanosoma cruzi TcI stocks isolated from six potentially panmictic populations and of the 21 T. cruzi TcI reference strains.
| Population | MCLE01 | SCLE10 | SCLE11 | MCLF10 | A427 | MCLG10 | C875 | MCLE08 | Overall | |
|---|---|---|---|---|---|---|---|---|---|---|
| Luribay | N/no. all/all. rich. | 6/2/1.67 | 6/3/2.58 | 6/2/1.67 | 6/2/1.91 | 6/6/4.66 | 6/2/1.67 | 6/2/1.91 | 4/1/1.00 | 2.13* |
| Ho/He/F is | 0.17/0.17/0.00 | 0.50/0.44/-0.15 | 0.17/0.17/0.00 | 0.33/0.30/-0.11 | 0.50/0.82/0.41 | 0.17/0.17/0.00 | 0.00/0.30/1.00 | NA | 0.23/0.30/0.24 | |
| MLG | - | - | - | - | - | - | - | - | 6 | |
| Mecapaca | N/no. all/all. rich. | 11/3/1.97 | 11/2/2.00 | 11/3/2.36 | 11/2/1.92 | 11/5/2.94 | 11/1/1.00 | 11/3/2.92 | 11/2/2.00 | 2.14* |
| Ho/He/F is | 0.09/0.25/0.65 | 0.73/0.52/-0.43 | 0.09/0.56/0.84 | 0.45/0.37/-0.25 | 0.18/0.47/0.63 | NA | 0.73/0.67/-0.08 | 0.36/0.52/0.31 | 0.33/0.42/0.23 | |
| MLG | - | - | - | - | - | - | - | - | 10 | |
| Qui-Urk | N/no. all/all. rich. | 12/3/2.13 | 12/2/2.00 | 12/2/1.83 | 12/1/1.00 | 12/1/1.00 | 12/1/1.00 | 12/1/1.00 | 12/2/1.97 | 1.49* |
| Ho/He/F is | 0.17/0.30/0.46 | 0.92/0.52/-0.83 | 0.00/0.29/1.00 | NA | NA | NA | NA | 0.67/0.50/-0.33 | 0.22/0.20/-0.08 | |
| MLG | - | - | - | - | - | - | - | - | 6 | |
| Qui-BSIA | N/no. all/all. rich. | 4/4/4.00 | 4/2/2.00 | 4/4/4.00 | 4/1/1.00 | 4/1/1.00 | 4/2/2.00 | 4/1/1.00 | 4/1/1.00 | 2.00* |
| Ho/He/F is | 0.25/0.75/0.70 | 1.00/0.57/-1.00 | 0.25/0.75/0.70 | NA | NA | 0.25/0.25/0.00 | NA | NA | 0.22/0.29/0.28 | |
| MLG | - | - | - | - | - | - | - | - | 3 | |
| Sap-Sap | N/no. all/all. rich. | 27/4/2.89 | 27/2/1.99 | 27/3/1.55 | 27/4/2.29 | 27/5/3.25 | 27/2/1.91 | 27/3/2.27 | 27/2/1.99 | 2.27* |
| Ho/He/F is | 0.30/0.63/0.53 | 0.81/0.50/-0.64 | 0.07/0.14/0.48 | 0.74/0.54/-0.37 | 0.37/0.68/0.46 | 0.18/0.37/0.51 | 0.78/0.54/-0.44 | 0.85/0.50/-0.73 | 0.51/0.49/-0.05 | |
| MLG | - | - | - | - | - | - | - | - | 24 | |
| Sap-Cosi | N/no. all/all. rich. | 19/2/1.38 | 19/2/1.78 | 19/3/2.17 | 19/3/2.72 | 19/3/2.70 | 19/2/1.99 | 19/2/1.84 | 19/3/2.20 | 2.10* |
| Ho/He/F is | 0.00/0.10/1.00 | 0.21/0.27/0.23 | 0.10/0.36/0.71 | 0.37/0.57/0.36 | 0.68/0.61/-0.12 | 0.31/0.50/0.38 | 0.16/0.31/0.49 | 0.84/0.52/-0.64 | 0.33/0.41/0.18 | |
| MLG | - | - | - | - | - | - | - | - | 19 | |
| REF | N/no. all/all. rich. | 21/15/6.07 | 21/7/4.23 | 21/6/4.26 | 21/8/4.98 | 20/10/5.31 | 21/7/3.38 | 21/13/4.60 | 21/4/3.05 | 4.49* |
| Ho/He/F is | 0.43/0.91/0.54 | 0.43/0.80/0.47 | 0.14/0.80/0.82 | 0.29/0.85/0.67 | 0.25/0.87/0.72 | 0.29/0.68/0.58 | 0.29/0.74/0.62 | 0.48/0.67/0.30 | 0.32/0.79/0.60 | |
| MLG | - | - | - | - | - | - | - | - | 21 | |
| Overall with reference strains (N = 100) / without reference strains (N = 79) | ||||||||||
| no. all | 18/7 | 7/3 | 9/7 | 8/5 | 13/8 | 7/2 | 15/4 | 4/3 | - | |
| Size range** | 117-155 | 238-255 | 128-141 | 174-191 | 173-188 | 146-161 | 165-191 | 111-119 | - | |
| mean all. rich. | 3.64/2.75 | 2.96/2.02 | 3.19/2.58 | 3.08/2.37 | 4.24/3.75 | 2.54/1.83 | 3.70/2.67 | 2.44/2.04 | - | |
| F is | 0.56/0.57 | -0.17/-0.49 | 0.76/0.72 | 0.20/-0.07 | 0.44/0.31 | 0.48/0.41 | 0.17/-0.07 | -0.24/-0.46 | - | |
| Hs | 0.46/0.38 | 0.51/0.45 | 0.45/0.39 | 0.39/0.30 | 0.51/0.44 | 0.29/0.22 | 0.38/0.31 | 0.39/0.34 | - | |
N, size of the sample; no. all, number of alleles; all. rich., allelic richness; Ho, observed heterozygotes; He, expected heterozygotes; MLG, number of different multilocus genotypes in the subsample; REF, reference stocks; Hs, Nei's gene diversity; *, mean allelic richness; **Size range of alleles calculated with all stocks (including references); NA, not available because the locus is monomorphic.
Panmixia tests within the six populations under study
F is among populations: F is values per population and their 95% confidence intervals are shown in Figure 2. The F is values were also examined by grouping the most adjacent populations, Sap-Sap with Sap-Cosi (1.9 km apart), Qui-Urk with Qui-Bsia (3.3 km apart), and all the populations (overall). F is varied from −0.08 (Qui-Urk population) to 0.29 (overall). Considering the significance using the Bonferroni correction (BC), none of the F is were significant (H0 not rejected, see Table 3) except for the overall sample. As we know that BC may falsely accept H0, we also considered the p-values without BC: here H0 is rejected with α = 1% within the “overall” sample and for only one sample grouping two local populations “overall Sapini” and was not rejected in all the local populations. Consequently, the decisions about panmixia were rejection for the “overall” sample, ambiguous for “overall Sapini,” and no rejection for all local populations (Table 3).
Figure 2. Observed F is of the six Trypanosoma cruzi populations under study and three artificial clusters (all stocks from Sapini, all stocks from Quillacollo, and all TcI Bolivian stocks) and their 95% confidence intervals.
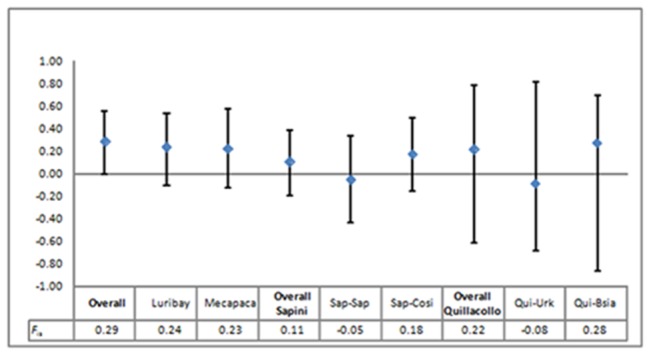
Table 3. Analysis of F is, disequilibrium linkage (LD) and repeated multilocus genotypes (MLGs) of the 79 Trypanosoma cruzi strains isolated from six potentially panmictic populations.
| Populations | Overall | Luribay | Mecapaca | Overall Sapini | Sap-Sap | Sap-Cosi | Overall Quillacollo | Qui-Urk | Qui-Bsia |
|---|---|---|---|---|---|---|---|---|---|
| Sample size | 79 | 6 | 11 | 46 | 27 | 19 | 16 | 12 | 4 |
| Statistical tests of HWE based on Fis statistics | |||||||||
| Real p-value without BC(1) | 0.006** | 0.0371* | 0.0182* | 0.0075** | 0.2243NS | 0.0148* | 0.0178* | 0.3954NS | 0.1096NS |
| Signification with BC(2) | ** | NS | NS | NS | NS | NS | NS | NS | NS |
| Decision about H0 | reject H0 | no reject H0 | no reject H0 | ambiguous | no reject H0 | no reject H0 | no reject H0 | no reject H0 | no reject H0 |
| Statistical tests of LD | |||||||||
| Ratio signif. / total(3) | 16/28** | 0/21NS | 2/21NS | 11/28** | 7/28** | 1/28NS | 1/10NS | 0/6NS | 0/3NS |
| IA (4) | 0.25** | -0.05NS | 0.39* | 0.31** | 0.56** | 0.002NS | 0.63** | -0.08NS | 1.31NS |
| ȓd (5) | 0.04** | -0.009NS | 0.07* | 0.05** | 0.08** | 0.0003NS | 0.20** | -0.03NS | 0.68NS |
| Decision about H0 | reject H0 | no reject H0 | ambiguous | reject H0 | reject H0 | no reject H0 | ambiguous | no reject H0 | no reject H0 |
| Statistical tests of repeated MLG | |||||||||
| No. of different MLGs | 68** | 6NS | 10NS | 43* | 24* | 19NS | 9** | 6NS | 3NS |
| Maximum frequency of MLG | 5** | 1NS | 2NS | 2NS | 2NS | 1NS | 5NS | 5NS | 2NS |
| Decision about H0 | reject H0 | no reject H0 | no reject H0 | ambiguous | ambiguous | no reject H0 | ambiguous | no reject H0 | no reject H0 |
Results of statistical tests and decisions about H0 (reject or not reject panmixia). For all tests: NS or NS = not significant; * = significant at 5% risk; ** = significant at 1% risk (1) p-value for F is within samples without Bonferroni correction (BC); (2) significance of the test with BC; (3) Ratio: significant loci pairwise comparisons / total comparisons, tested by the binomial test with R program; (4) Value of index of association; (5) Value of ȓd index.
Linkage disequilibrium (LD): three parameters were tested: (i) the proportion of significant LD tests over the total number of comparisons by pairs of loci, using the binomial test, (ii) the association index (IA), a direct measure of LD, and (iii) a special index (ȓd) derived from IA. These indices and their associated significance are given in Table 3. Of the six local populations under study, H0 was not rejected in four of them (Luribay, Sap-Cosi, Qui-Urk and Qui-Bsia); two results were ambiguous (Mecapaca and overall Quillacollo) and three rejected H0 (Overall, Overall Sapini and Sap-Sap).
Repeated multilocus genotypes: We tested two parameters, the number of different MLGs and the maximum frequency of the most repeated MLG. The results showed (Table 3) that H0 is rejected in only one sample (Overall), not rejected in five populations (Luribay, Mecapaca, Sap-Cosi, Qui-Urk, and Qui-Bsia) and ambiguous in three populations (Overall Sapini, Sap-Sap, and Overall Quillacollo).
Considering only the six potentially panmictic populations under study, in four of them (Luribay, Sap-Cosi, Qui-Urk, and Qui-Bsia) the decisions for F is, LD, and MLG were “no rejecting H0” , while in the two others (Mecapaca and Sap-Sap) contradictory results were observed between the different tests of panmixia. Nevertheless, for the only F is tests within the populations from Luribay and Sap-Cosi, there is a potential risk of type II error
Discussion
Likely panmixia in several T. cruzi populations isolated from wild T. infestans
As previously recommended [25], we used three classes of classical population genetics parameters to study the mode of reproduction (i.e., Hardy-Weinberg equilibrium, linkage equilibrium, and presence of repeated MLG) and we showed that in four out of six potentially panmictic T. cruzi populations (Luribay, Sap-Cosi, Qui-Urk, and Qui-Bsia) sampled in restricted areas, true panmixia cannot be excluded. In the case of Luribay and Sap-Cosi, the F IS tests required the Bonferroni correction to lead to the decision “no rejecting Ho”, carrying a high risk of Type II error. However, as Qui-Bsia has a small size (N = 4) and that we cannot rule out a statistical type II error in this case, we must consider only three panmictic populations Luribay, Sap-Cosi, and Qui-Urk. For the two other populations, the tests gave contradictory results in Sap-Sap and ambiguous results for LD tests in Mecapaca, which appears “more panmictic” than Sap-Sap. In this case we could infer a lack of power of the tests to explain these results; nevertheless, and except for Qui-Bsia where the sample size is very small, a high β error value (type II error) is unlikely because for comparable population sizes the tests can reject or not reject H0. Moreover, multiplying the tests decreases the probability of type II error and increases the power of the test. Hence we can consider that no rejection of H0 is equivalent to accepting panmixia, possibly except for Qui-Bsia. Moreover, it is interesting to note that the tests were very sensitive to the Wahlund effect (sampling from heterogeneous populations): when we grouped all six populations (overall), all the tests became highly significant, proving the heterogeneity between populations at the regional level. This was true to a lesser extent for overall Sapini and overall Quillacollo, showing a genetic structure at a very low geographic scale (a few kilometers).
Role of sympatry and sampling design
To test panmixia, the first condition is natural sympatry; indeed, a nonsympatric sample may lead to genetic structuring and generate a Wahlund effect and consequently a false rejection of H0. As nobody knows precisely what sympatry means for this parasite, we picked up the populations within a very small area, not more than 1 ha, in which the triatomes and mammal hosts are assumed to move enough to allow parasite transmission from one host to another and hence generate opportunities for genetic exchanges; we named these populations “potentially panmictic” and tested them. Consequently, in such populations, when H0 is not rejected, and excluding a type II error discussed above, we can consider, a posteriori, that these populations were truly sympatric. Inversely, when H0 is rejected by some tests, as is the case for the Sap-Sap population and to a lesser extent for Mecapaca, a Wahlund effect due to a hidden genetic structure (itself possibly due to a lack of sympatry) could be inferred. Interestingly, when we analyzed the microsatellite data by the software Structure [40], we showed the presence of two distinct genomes in only Sap-Sap and Mecapaca, hence a hidden genetic structure, which can explain the rejection of H0 for some tests within these two populations (data not shown). Meanwhile for these two populations, choosing between the two alternative hypotheses (i.e., lack of sympatry or presence of some extent of clonality) is almost impossible. Sampling in areas that are not actually sympatric may therefore result in falsely rejecting H0. Inversely, as previously stated by others [41], selecting only one individual per subpopulation and pooling each of them into an artificial population generates misleading patterns and false conclusions regarding the mode of reproduction, in particular a significant reduction of LD and modified HW equilibrium, sometimes giving an erroneous picture of the recombining organism despite a high level of clonality. Obviously, our sampling method did not fit this pattern and consequently absence of H0 rejection cannot be attributed to this sampling bias. All these remarks emphasize the importance of sampling design to test the hypothesis under study, for example here, to test panmixia, we need potential sympatric areas, not allopatric areas.
Clonality versus recombination in T. cruzi species
Since the pioneering studies using isoenzymes [6], T. cruzi has been considered by most authors to have a basically clonal population structure, with occasional bouts of genetic exchange or hybridization. These facts were confirmed on many occasions with other genetic markers and a clonal theory of parasitic protozoa was proposed [2,42] with the notable exception of Plasmodium falciparum in which sex occurs [43]; the theory was reaffirmed with both Trypanosoma and Leishmania genera [44] and extended to fungi bacteria and viruses in a recent review [45]. The question of determining whether sex occurs or not in T. cruzi is not trivial, nor needless. Because of a reduced or absent gene flow, clonality must have a major impact on the biological and medical properties of the parasites, which has been explored [46,47]. On the other hand, genetic exchanges can take different forms, the best known being hybridization that has been provoked in vitro [17] and has naturally occurred, playing a crucial role in T. cruzi evolution (generating new DTUs). It is generally admitted that two hybridization events have defined the population structure of T. cruzi [7], the first one very ancient, between TcI and TcII, leading to TcIII and TcIV, and the second one, recent, between TcII and TcIII, leading to TcV and TcVI. The in vitro hybrids showed a fusion of parental genotypes, loss of alleles, homologous recombination, and uniparental inheritance of kinetoplast maxicircle DNA [17], and it is accepted that natural hybridization might occur in a similar but contrasted way [48]. In addition to hybridization, many authors have reported incongruence between phylogenetic trees, which is generally a sign of recombination: for example 13,49, mitochondrial introgression [10,11] and even mitochondrial heteroplasmy (heterogeneous mitochondrial genomes in an individual cell) was demonstrated recently [16] using the promising mtMLST method (mitochondrial multilocus sequence typing), itself derived from the MLST method using nuclear genes [50]. The last way of genetic exchanges might be conventional recombination mechanisms, as in sexual diploids, which can be detected by the usual tools of population genetics (F IS, LD, etc., like here). Because we do not know the cytological mechanisms involved, we named these events “recombination-like” in order to differentiate them from the known genetic exchanges involving meiosis in sexual diploids. One of the first studies regarding this event [51], reported at one isoenzyme locus (phosphoglucomutase), observed homozygous and heterozygous frequencies almost identical to those predicted by the theoretical Hardy-Weinberg distribution in sylvatic TcI. Later, using microsatellites, some recombinations were suggested in a general clonal framework in sylvatic TcI over the endemic area [52], TcI in Ecuador [53], and TcIII [54]: in the latter, the authors could not effectively discriminate a recombination from a high genome-wide frequency of gene conversion. Finally, three recent studies emphasize the role of genetic exchanges and the extraordinary genome plasticity of T. cruzi, (i) using genomic CNV (copy number variation) [15]; (ii) another team [14] reported gross incongruence in Colombian TcI between nuclear and mitochondrial markers, mosaic maxicircle sequences, and the genetic resorting mechanism; (iii) other authors [55] showed that hybrid stocks contain haplotypes that are mosaics probably originating from intragenic recombination. In all these examples, it is worth noting that hybridization or introgression may occur between distant DTUs, whereas “recombination-like” events generally are intra-DTU, as shown in the present study. The “clonality or genetic exchanges” duality for T. cruzi has definitively became obsolete; this species obviously has used both mechanisms to evolve and probably to adapt to its multiple hosts, associated with an extraordinarily plastic genome shaped by clonal evolution and several kinds of genetic exchanges. The mode of reproduction of T. cruzi could oscillate between clonality and sexuality and the true questions are why, when, how, and to what extent T. cruzi recombines? Nevertheless, we agree with Tibayrenc and Ayala’s [45] definition of clonality as “restrained recombination on an evolutionary scale,” which has already been observed in T. cruzi since the same MLGs can be sampled at different times and in distant regions. The same authors stated that “recombination seems easier between closely related genotypes pertaining to the same near-clade in both fungi and parasitic protozoa”; this probably constitutes the most parsimonious explanation for the co-occurrence of recombination at restricted space / time levels and of clonality at larger space / time scales. Interestingly, in bacteria “the probability of acceptance of a recombination event decreases exponentially with genetic distance between the donor and recipient DNA” [56], which is an effect of sexual isolation in bacteria [57]; this could be true for T. cruzi and should be further investigated.
Conclusion, limitations and warning
For the first time we report panmixia, notably through linkage disequilibrium statistics, in T. cruzi TcI populations isolated from wild T. infestans in Bolivia. In absence of additional studies involving other sylvatic vectors, it is not possible to associate panmixia with the sylvatic biotopes; further studies of panmixia should be conducted in other biotopes where parasites should be sympatric. As previously mentioned, “mixed clonal / sexual reproduction is nearly indistinguishable from strict sexual reproduction as long as the proportion of clonal reproduction is not strongly predominant” [30], so, although unlikely, we cannot exclude a certain level of clonality in these populations, even when all tests did not reject the panmixia hypothesis. Moreover, it is worth noting that the parasite strains used here were not cloned and some artifacts due to multiple infections could be a possible explanation for some contradictory results between the different tests. The Leishmania genome is aneuploid [58], every chromosome in every cell may be present in different ploidy states (monosomic, disomic, or trisomic). If this is the case for T. cruzi, as suspected [15], there could be a serious bias with all the codominant nuclear markers, particularly in the studies involving microsatellites: artificially decreasing F is in the trisomic state (excess heterozygosity) and artificially increasing F is in the monosomic state (excess homozygosity). Hence, all the F is results should be interpreted with caution, especially when there is a substantial variance of F is between loci. Moreover, F is is not linearly related to the rate of clonal reproduction [59]. As stated above, the sampling strategy is crucial to confirm or reject these results in other natural contexts, avoiding sampling stocks that have a foreign origin because of passive transport by humans. For this purpose (to specify the mating system at the local scale), we recommend starting with a reduced time and space scale in order to avoid the Wahlund bias as much as possible, which does not hamper the opposite strategy previously proposed [45], “taking a birds-eye view of genetic variability over years and continents, from different hosts and ecosystems” to look at the evolution of the species over space and time.
Acknowledgments
We are particularly grateful to the leaders of the Inlasa (Instituto de Laboratorios de Salud, La Paz Bolivia), Dr. Walter Agreda for having hosted this work in the Department of Entomology directed by Dr. Tamara Chavez.
Funding Statement
This work was funded by the IRD, the National Agency for Research (ANR No 3624, France) and the European Commission Framework Program Project “Comparative epidemiology of genetic lineages of Trypanosoma cruzi” ChagasEpiNet, Contract No. 223034. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Rassi A Jr., Rassi A, Marin-Neto JA (2010) Chagas disease. Lancet 375: 1388-1402. doi: 10.1016/S0140-6736(10)60061-X. PubMed: 20399979. [DOI] [PubMed] [Google Scholar]
- 2. Tibayrenc M, Kjellberg F, Ayala FJ (1990) A clonal theory of parasitic protozoa: the population structures of Entamoeba, Giardia, Leishmania, Naegleria, Plasmodium, Trichomonas, and Trypanosoma and their medical and taxonomical consequences. Proc Natl Acad Sci USA 87: 2414-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brisse S, Barnabé C, Tibayrenc M (2000) Identification of six Trypanosoma cruzi phylogenetic lineages by random amplified polymorphic DNA and multilocus enzyme electrophoresis. Int J Parasitol 30: 35-44. doi: 10.1016/S0020-7519(99)00168-X. PubMed: 10675742. [DOI] [PubMed] [Google Scholar]
- 4. Marcili A, Lima L, Cavazzana M, Junqueira AC, Veludo HH et al. (2009) A new genotype of Trypanosoma cruzi associated with bats evidenced by phylogenetic analyses using SSU rDNA, cytochrome b and Histone H2B genes and genotyping based on ITS1 rDNA. Parasitology 136: 641-655. doi: 10.1017/S0031182009005861. PubMed: 19368741. [DOI] [PubMed] [Google Scholar]
- 5. Zingales B, Miles MA, Campbell DA, Tibayrenc M, Macedo AM et al. (2012) The revised Trypanosoma cruzi subspecific nomenclature: Rationale, epidemiological relevance and research applications. Infect Genet Evol 12: 240–253. doi: 10.1016/j.meegid.2011.12.009. PubMed: 22226704. [DOI] [PubMed] [Google Scholar]
- 6. Tibayrenc M, Ward P, Moya A, Ayala FJ (1986) Natural populations of Trypanosoma cruzi, the agent of Chagas disease, have a complex multiclonal structure. Proc Natl Acad Sci U S A 83: 115-119. doi: 10.1073/pnas.83.1.115. PubMed: 3510428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Westenberger SJ, Barnabé C, Campbell DA, Sturm NR (2005) Two hybridization events define the population structure of Trypanosoma cruzi . Genetics 171: 527-543. doi: 10.1534/genetics.104.038745. PubMed: 15998728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lewis MD, Llewellyn MS, Yeo M, Acosta N, Gaunt MW et al. (2011) Recent, independent and anthropogenic origins of Trypanosoma cruzi hybrids. PLoS Negl Trop. Drosophila Inf Service 5: e1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sturm NR, Vargas NS, Westenberger SJ, Zingales B, Campbell DA (2003) Evidence for multiple hybrid groups in Trypanosoma cruzi . Int J Parasitol 33: 269-279. doi: 10.1016/S0020-7519(02)00264-3. PubMed: 12670512. [DOI] [PubMed] [Google Scholar]
- 10. Barnabé C, Brenière SF (2012) Scarce events of mitochondrial introgression in Trypanosoma cruzi: New case with a Bolivian strain. Infect Genet Evol 12: 1879-1883. doi: 10.1016/j.meegid.2012.08.018. PubMed: 22982157. [DOI] [PubMed] [Google Scholar]
- 11. Machado CA, Ayala FJ (2001) Nucleotide sequences provide evidence of genetic exchange among distantly related lineages of Trypanosoma cruzi . Proc Natl Acad Sci U S A 98: 7396-7401. doi: 10.1073/pnas.121187198. PubMed: 11416213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bogliolo AR, Lauria-Pires L, Gibson WC (1996) Polymorphisms in Trypanosoma cruzi: Evidence of genetic recombination. Acta Trop (Basel) 61: 31-40. doi: 10.1016/0001-706X(95)00138-5. PubMed: 9133162. [DOI] [PubMed] [Google Scholar]
- 13. Brisse S, Henriksson J, Barnabé C, Douzery EJ, Berkvens D et al. (2003) Evidence for genetic exchange and hybridization in Trypanosoma cruzi based on nucleotide sequences and molecular karyotype. Infect Genet Evol 2: 173-183. doi: 10.1016/S1567-1348(02)00097-7. PubMed: 12797979. [DOI] [PubMed] [Google Scholar]
- 14. Ramírez JD, Guhl F, Messenger LA, Lewis MD, Montilla M et al. (2012) Contemporary cryptic sexuality in Trypanosoma cruzi . Mol Ecol 21: 4216-4226. doi: 10.1111/j.1365-294X.2012.05699.x. PubMed: 22774844. [DOI] [PubMed] [Google Scholar]
- 15. Minning TA, Weatherly DB, Flibotte S, Tarleton RL (2011) Widespread, focal copy number variations (CNV) and whole chromosome aneuploidies in Trypanosoma cruzi strains revealed by array comparative genomic hybridization. BMC Genomics 12: 139. doi: 10.1186/1471-2164-12-139. PubMed: 21385342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Messenger LA, Llewellyn MS, Bhattacharyya T, Franzen O, Lewis MD et al. (2012) Multiple mitochondrial introgression events and heteroplasmy in Trypanosoma cruzi revealed by maxicircle MLST and next generation sequencing. PLoS Negl Trop. Drosophila Inf Service 6: e1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gaunt MW, Yeo M, Frame IA, Stothard JR, Carrasco HJ et al. (2003) Mechanism of genetic exchange in American trypanosomes. Nature 421: 936-939. doi: 10.1038/nature01438. PubMed: 12606999. [DOI] [PubMed] [Google Scholar]
- 18. Tibayrenc M, Barnabé C, Telleria J (2010) Reticulate Evolution in Trypanosoma cruzi: Medical and epidemiological implications. In: Telleria J, Tibayrenc M. American trypanosomiasis Chagas disease One hundred years of research. Burlington, USA: Elsevier; pp. 475-488. [Google Scholar]
- 19. Barnabé C, De Meeûs T, Noireau F, Bosseno MF, Monje EM et al. (2011) Trypanosoma cruzi discrete typing units (DTUs): Microsatellite loci and population genetics of DTUs TcV and TcI in Bolivia and Peru. Infect Genet Evol 11: 1752-1760. doi: 10.1016/j.meegid.2011.07.011. PubMed: 21801854. [DOI] [PubMed] [Google Scholar]
- 20. Brenière SF, Aliaga C, Waleckx E, Buitrago R, Salas R et al. (2012) Genetic characterization of Trypanosoma cruzi DTUs in wild Triatoma infestans from Bolivia: predominance of TcI. PLoS Negl Trop. Drosophila Inf Service 6: e1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Buitrago R, Waleckx E, Bosseno MF, Zoveda F, Vidaurre P et al. (2010) First report of widespread wild populations of Triatoma infestans (Reduviidae, Triatominae) in the valleys of La Paz, Bolivia. Am J Trop Med Hyg 82: 574-579. doi: 10.4269/ajtmh.2010.09-0325. PubMed: 20348501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Aliaga C, Brenière SF, Barnabé C (2011) Further interest of miniexon multiplex PCR for a rapid typing of Trypanosoma cruzi DTU groups. Infect Genet Evol 11: 1155-1158. doi: 10.1016/j.meegid.2010.11.013. PubMed: 21255686. [DOI] [PubMed] [Google Scholar]
- 23. Oliveira RP, Broude NE, Macedo AM, Cantor CR, Smith CL et al. (1998) Probing the genetic population structure of Trypanosoma cruzi with polymorphic microsatellites. Proc Natl Acad Sci U S A 95: 3776-3780. doi: 10.1073/pnas.95.7.3776. PubMed: 9520443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rougeron V, De Meeus T, Kako Ouraga S, Hide M, Banuls AL (2010) “Everything you always wanted to know about sex (but were afraid to ask)” in Leishmania after two decades of laboratory and field analyses. PLOS Pathog 6: e1001004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. de Meeûs T, Balloux F (2004) Clonal reproduction and linkage disequilibrium in diploids: a simulation study. Infect Genet Evol 4: 345-351. doi: 10.1016/j.meegid.2004.05.002. PubMed: 15374532. [DOI] [PubMed] [Google Scholar]
- 26. Wright S (1965) The Interpretation of Population Structure by F-Statistics with Special Regard to Systems of Mating. Evolution 19: 395-420. doi: 10.2307/2406450. [DOI] [Google Scholar]
- 27. Weir BS, Cockerham CC (1984) Estimating F-Statistics for the Analysis of Population-Structure. Evolution 38: 1358-1370. doi: 10.2307/2408641. [DOI] [PubMed] [Google Scholar]
- 28. Judson OP, Normark BB (1996) Ancient asexual scandals. Trends Ecol Evolut 11: A41-A46. doi: 10.1016/0169-5347(96)81040-8. PubMed: 21237759. [DOI] [PubMed] [Google Scholar]
- 29. Mark Welch DB, Meselson M (2000) Evidence for the evolution of Bdelloid rotifers without sexual reproduction or genetic exchange. Science 288: 1211-1215. doi: 10.1126/science.288.5469.1211. PubMed: 10817991. [DOI] [PubMed] [Google Scholar]
- 30. Balloux F, Lehmann L, de Meeûs T (2003) The population genetics of clonal and partially clonal diploids. Genetics 164: 1635-1644. PubMed: 12930767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brown AHD, Feldman MW, Nevo E (1980) Multilocus structure of natural populations of Hordeum spontaneum . Genetics 96: 523-536. PubMed: 17249067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Agapow PM, Burt A (2001) Indices of multilocus linkage disequilibrium. Mol Ecol Notes 1: 101-102. doi: 10.1046/j.1471-8278.2000.00014.x. [DOI] [Google Scholar]
- 33. Goudet J, Raymond M, de Meeüs T, Rousset F (1996) Testing differentiation in diploid populations. Genetics 144: 1933-1940. PubMed: 8978076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Teriokhin AT, De Meeûs T, Guégan JF (2007) On the power of some binomial modifications of the Bonferroni multiple test. Zh Obshch Biol 68: 332-340. PubMed: 18038646. [PubMed] [Google Scholar]
- 35. Goudet J (2005) hierfstat, a package for R to compute and test hierarchical F-statistics. Mol Ecol Notes 5: 184-186. doi: 10.1111/j.1471-8286.2004.00828.x. [DOI] [Google Scholar]
- 36. R Development Core Team (2008) R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 37. Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4: 535-538. doi: 10.1111/j.1471-8286.2004.00684.x. [DOI] [Google Scholar]
- 38. Cavalli-Sforza LL, Edwards AW (1967) Phylogenetic analysis. Models and estimation procedures. Am J Hum Genet 19: 233-257. PubMed: 6026583. [PMC free article] [PubMed] [Google Scholar]
- 39. Goudet J (1995) FSTAT (Version 1.2): A Computer Program to Calculate F-Statistics. J Hered 86: 485-486. [Google Scholar]
- 40. Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155: 945-959. PubMed: 10835412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Prugnolle F, De Meeus T (2010) Apparent high recombination rates in clonal parasitic organisms due to inappropriate sampling design. Heredity (Edinb) 104: 135-140. doi: 10.1038/hdy.2009.128. PubMed: 19812614. [DOI] [PubMed] [Google Scholar]
- 42. Tibayrenc M, Ayala FJ (1991) Towards a population genetics of microorganisms: the clonal theory of parasitic protozoa. Parasitol Today 7: 228-232. doi: 10.1016/0169-4758(91)90234-F. PubMed: 15463504. [DOI] [PubMed] [Google Scholar]
- 43. Tibayrenc M, Kjellberg F, Arnaud J, Oury B, Brénière SF et al. (1991) Are eukaryotic microorganisms clonal or sexual? A population genetics vantage. Proc Natl Acad Sci U S A 88: 5129-5133. doi: 10.1073/pnas.88.12.5129. PubMed: 1675793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tibayrenc M, Ayala FJ (1999) Evolutionary genetics of Trypanosoma and Leishmania . Microbes Infect 1: 465-472. doi: 10.1016/S1286-4579(99)80050-1. PubMed: 10602679. [DOI] [PubMed] [Google Scholar]
- 45. Tibayrenc M, Ayala FJ (2012) Reproductive clonality of pathogens: A perspective on pathogenic viruses, bacteria, fungi, and parasitic protozoa. Proc Natl Acad Sci USA. PubMed: 22949662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Revollo S, Oury B, Laurent JP, Barnabé C, Quesney V et al. (1998) Trypanosoma cruzi: impact of clonal evolution of the parasite on its biological and medical properties. Exp Parasitol 89: 30-39. doi: 10.1006/expr.1998.4216. PubMed: 9603486. [DOI] [PubMed] [Google Scholar]
- 47. Laurent JP, Barnabe C, Quesney V, Noel S, Tibayrenc M (1997) Impact of clonal evolution on the biological diversity of Trypanosoma cruzi . Parasitology 114: 213-218. doi: 10.1017/S0031182096008414. PubMed: 9075341. [DOI] [PubMed] [Google Scholar]
- 48. Lewis MD, Llewellyn MS, Gaunt MW, Yeo M, Carrasco HJ et al. (2009) Flow cytometric analysis and microsatellite genotyping reveal extensive DNA content variation in Trypanosoma cruzi populations and expose contrasts between natural and experimental hybrids. Int J Parasitol 39: 1305-1317. doi: 10.1016/j.ijpara.2009.04.001. PubMed: 19393242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Higo H, Miura S, Agatsuma T, Mimori T, Yanagi T et al. (2007) Identification of Trypanosoma cruzi sublineages by the simple method of single-stranded conformation DNA polymorphism (SSCP). Parasitol Res 100: 1023-1031. doi: 10.1007/s00436-006-0376-8. PubMed: 17171567. [DOI] [PubMed] [Google Scholar]
- 50. Yeo M, Mauricio IL, Messenger LA, Lewis MD, Llewellyn MS et al. (2011) Multilocus sequence typing (MLST) for lineage assignment and high resolution diversity studies in Trypanosoma cruzi. PLoS Negl Trop. Drosophila Inf Service 5: e1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Carrasco HJ, Frame IA, Valente SA, Miles MA (1996) Genetic exchange as a possible source of genomic diversity in sylvatic populations of Trypanosoma cruzi . Am J Trop Med Hyg 54: 418-424. PubMed: 8615458. [DOI] [PubMed] [Google Scholar]
- 52. Llewellyn MS, Miles MA, Carrasco HJ, Lewis MD, Yeo M et al. (2009) Genome-scale multilocus microsatellite typing of Trypanosoma cruzi discrete typing unit I reveals phylogeographic structure and specific genotypes linked to human infection. PLoS Pathog 5: e1000410 PubMed: 19412340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ocana-Mayorga S, Llewellyn MS, Costales JA, Miles MA, Grijalva MJ (2010) Sex, subdivision, and domestic dispersal of Trypanosoma cruzi lineage I in southern Ecuador. PLoS Negl Trop. Drosophila Inf Service 4: e915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Llewellyn MS, Lewis MD, Acosta N, Yeo M, Carrasco HJ et al. (2009) Trypanosoma cruzi IIc: Phylogenetic and phylogeographic insights from sequence and microsatellite analysis and potential impact on emergent Chagas disease. PLoS Negl Trop. Drosophila Inf Service 3: e510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ferreira RC, Briones MR (2012) Phylogenetic evidence based on Trypanosoma cruzi nuclear gene sequences and information entropy suggest that inter-strain intragenic recombination is a basic mechanism underlying the allele diversity of hybrid strains. Infect Genet Evol 12: 1064-1071. doi: 10.1016/j.meegid.2012.03.010. PubMed: 22449773. [DOI] [PubMed] [Google Scholar]
- 56. Didelot X, Maiden MC (2010) Impact of recombination on bacterial evolution. Trends Microbiol 18: 315-322. doi: 10.1016/j.tim.2010.04.002. PubMed: 20452218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Majewski J (2001) Sexual isolation in bacteria. FEMS Microbiol Lett 199: 161-169. doi: 10.1111/j.1574-6968.2001.tb10668.x. PubMed: 11377861. [DOI] [PubMed] [Google Scholar]
- 58. Sterkers Y, Lachaud L, Bourgeois N, Crobu L, Bastien P et al. (2012) Novel insights into genome plasticity in Eukaryotes: mosaic aneuploidy in Leishmania . Mol Microbiol, 86: 15–23. PubMed: 22857263. [DOI] [PubMed] [Google Scholar]
- 59. De Meeûs T, Lehmann L, Balloux F (2006) Molecular epidemiology of clonal diploids: a quick overview and a short DIY (do it yourself) notice. Infect Genet Evol 6: 163-170. doi: 10.1016/j.meegid.2005.02.004. PubMed: 16290062. [DOI] [PubMed] [Google Scholar]