Abstract
Colorectal carcinoma RKO cells expressing reduced levels of the RNA-binding protein HuR (ASHuR) displayed markedly reduced growth. In synchronous RKO populations, HuR was almost exclusively nuclear during early G1, increasing in the cytoplasm during late G1, S and G2. The expression and half-life of mRNAs encoding cyclins A and B1 similarly increased during S and G2, then declined, indicating that mRNA stabilization contributed to their cell cycle-regulated expression. In gel-shift assays using radiolabeled cyclin RNA transcripts and RKO protein extracts, only those transcripts corresponding to the 3′-untranslated regions of cyclins A and B1 formed RNA–protein complexes in a cell cycle-dependent fashion. HuR directly bound mRNAs encoding cyclins A and B1, as anti-HuR antibodies supershifted such RNA–protein complexes. Importantly, the expression and half-life of mRNAs encoding cyclins A and B1 were reduced in ASHuR RKO cells. Our results indicate that HuR may play a critical role in cell proliferation, at least in part by mediating cell cycle-dependent stabilization of mRNAs encoding cyclins A and B1.
Keywords: cyclin A/cyclin B1/HuR/proliferation/mRNA stability
Introduction
The mamalian cell division cycle is governed through the orchestrated activation and inactivation of cyclin-dependent kinases (cdks), whose activity is modulated by a host of regulatory events (Johnson and Walker, 1999). Negative regulators of cdk include cdk-inhibitory proteins, and positive regulators include cyclins, their catalytic partners. During the G1 phase, cyclins D1, D2 and D3 form complexes with cdk4 or cdk6, and cyclin E with cdk2; these complexes function as Rb kinases, modulating the activity of E2F and hence the expression of proliferative genes. Cyclin A associates with cdk2 during the S phase, and with cdc2 (cdk1) at the S–G2 boundary and into G2. Progression through G2, culminating in mitosis, further requires that cdc2 form complexes with cyclins B1 and B2.
Given the central role of cyclins in controlling cell cycle progression, their expression during the cell division cycle is tightly regulated. The transcriptional regulation of cyclins D1 and E, which are induced after mitogenic stimulation, has been extensively documented (Ohtani et al., 1995; Guttridge et al., 1999). Likewise, some studies have reported cell cycle-dependent transcriptional regulation of cyclin A (Henglein et al., 1994; Zwicker et al., 1995) and of cyclin B1 (Hwang et al., 1995, 1998; Piaggio et al., 1995). Post-transcriptional events controlling cyclin expression include their rapid and specific proteolysis by the ubiquitin–proteasome pathway (King et al., 1996; Koepp et al., 1999).
However, it has long been recognized that the expression of cyclins and cell cycle-related proteins is also critically regulated by altering the stability of their mRNAs. Such regulatory mechanisms have been proposed for cyclin D1 (Lebwohl et al., 1994; Hashemolhosseini et al., 1998; Hosokawa et al., 1998), cyclin E (Oda et al., 1995), cyclin A (Howe et al., 1995; Maity et al., 1997), cyclin B1 (Trembley et al., 1994; Maity et al., 1995), p21 (Schwaller et al., 1995; Esposito et al., 1997; Gorospe et al., 1998), p27 (Baghdassarian et al., 1999), cdk2 (Oda et al., 1995), cdc2 and cdc25 (Datta et al., 1992), among other cell cycle-regulatory genes. Indeed, the stability of some cyclin mRNAs varies as a function of the cell cycle (Maity et al., 1995, 1997).
The process of mRNA turnover is increasingly recognized as critical for regulating the expression of many genes during processes such as immune-cell activation, response to mitogens, differentiation, and cell cycle regulation (Malter and Hong, 1991; Schiavi et al., 1992; Ross, 1995; Liebhaber, 1997). Many mRNAs subject to message turnover identified to date bear AU-rich elements (AREs) in their 3′-untranslated regions (3′ UTRs), which often also contain the pentamer AUUUA (Caput et al., 1986; Shaw and Kamen, 1986; Shyu et al., 1989; Lagnado et al., 1994; Chen and Shyu, 1995; Xu et al., 1997). These sequences have been shown to participate in rapid changes in the steady-state levels of mRNAs encoding critical growth-response genes (Treisman et al., 1985), cytokines (Wodnar-Filipowicz and Moroni, 1990; Henics et al., 1994) and cell-cycle regulatory proteins (Datta et al., 1992; Lebwohl et al., 1994; Markiewicz et al., 1994; Nemer and Stuebing, 1996).
Among the best characterized ARE-binding proteins are the Elav/Hu family of RNA-binding proteins, composed of the ubiquitously expressed HuR (HuA) and the neuronal specific Hel-N1 (HuB), HuC and HuD (Levine et al., 1993; Good, 1995; Chung et al., 1996; Ma et al., 1996; Antic and Keene, 1997). Hu proteins were first identified as specific tumor antigens in cancers (particularly lung carcinomas) of individuals with paraneoplastic neurological disorder (Dalmau et al., 1990; Szabo et al., 1991). Patients developed autoantibodies against Hu proteins, which penetrated the blood–brain barrier and led to neuronal degeneration. The features of paraneoplastic disease suggest that Hu proteins may play a pivotal role in controlling the expression of growth-regulatory genes. Indeed, Hu proteins have been found to bind to critical ARE-containing mRNAs and either stabilize them, enhance their translation, or both (Fan and Steitz, 1998a; Peng et al., 1998; Ford et al., 1999). For example, there is evidence for binding, stabilization and/or enhanced translation of GLUT-1, neurofilament M and c-myc mRNAs by Hel-N1 (Levine et al., 1993; Jain et al., 1997; Antic et al., 1999); N-myc, GAP-43 and tau mRNAs by HuD (Chung et al., 1997; Aranda-Abreu et al., 1999; Lazarova et al., 1999); and c-fos, PAI-2, VEGF, p21 and c-myc mRNAs by HuR (Joseph et al., 1998; Levy et al., 1998; Peng et al., 1998; Maurer et al., 1999; Wang et al., 2000). In addition, the intracellular distribution of Elav proteins varies (Antic and Keene, 1997; Keene, 1999). HuR, which bears the recently described nuclear shuttling sequence HNS (Fan and Steitz, 1998b), is primarily nuclear, but can redistribute to the cytoplasm (Atasoy et al., 1998; Fan and Steitz, 1998b; Peng et al., 1998). While it is becoming apparent that the neuronal members (Hel-N1, HuC and HuD) function in the terminal differentiation of neurons (Aranda-Abreu et al., 1999), the role of HuR in cell proliferation or differentiation has not been directly addressed.
We recently demonstrated that HuR’s cytoplasmic localization increased under conditions of stress and further showed that this redistribution was coupled with its ability to bind to and stabilize the mRNA encoding the cdk inhibitor p21 (Wang et al., 2000). During the course of those studies, we generated cells whose HuR levels were downregulated through expression of an antisense (AS) HuR transcript and observed that ASHuR cells exhibited markedly reduced proliferation rates. In the present study we have directly examined the influence of HuR on cell proliferation. Our observations reveal the cell cycle dependency of (i) the levels of cytoplasmic HuR, (ii) the formation of RNA–protein complexes with 3′ UTR transcripts of cyclins A and B1 and (iii) the stabilization of mRNAs encoding cyclins A and B1. Importantly, ASHuR-expressing cells exhibited decreased binding to their respective mRNAs, significantly reduced half-lives for mRNAs encoding cyclins A and B1, lower levels of cyclins A and B1, and, consequently, reduced proliferation rates. Our observations strongly support HuR’s function in controlling the expression of cyclins A and cyclin B1 through upregulation of the half-life of their mRNAs. Thus, our observations not only illustrate the importance of HuR in cell proliferation, but also yield novel insight into the post-transcriptional regulation of cyclin A and cyclin B1 expression.
Results
RKO cells expressing reduced HuR levels exhibit slower cell proliferation
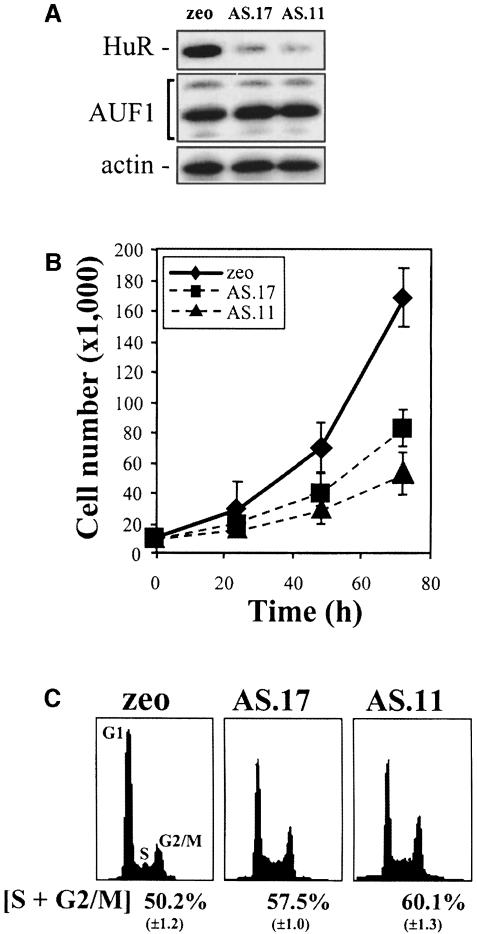
Several RKO clonal lines expressing lower levels of HuR by transfection with an antisense transcript complementary to HuR mRNA were generated in our laboratory during the course of earlier studies (Wang et al., 2000). HuR expression in two such ASHuR clones, AS.17 and AS.11 (Figure 1A), was 75% (3-fold) and 80% (4-fold) lower than that seen in the control zeo population. This reduction was specific for HuR as the expression of AUF1, another RNA-binding protein (Zhang et al., 1993), remained unchanged (Figure 1A). AS.17 and AS.11, as well as other ASHuR clonal isolates, exhibited markedly decreased proliferation rates and displayed greater S and G2/M compartments (Figure 1B and C). The differences in population growth were unlikely to result from enhanced death of ASHuR cells, since Trypan blue-exclusion assays revealed <2% non-viable cells in all lines. Instead, we hypothesized that HuR may affect the expression of genes involved in cell cycle progression and sought to investigate this function of HuR further.
Fig. 1. RKO cells expressing reduced HuR show reduced growth rates. (A) Western blot analysis depicting HuR and AUF1 expression in RKO cells transfected with the empty pSVzeo vector (zeo), and in two RKO clonal lines transfected with pSVzeo(–) HuR to express an antisense HuR transcript (AS.17 and AS.11). Blots were sequentially stripped and rehybridized to assess the expression of AUF1 and actin. (B) Proliferation rates in RKO zeo, AS.17 and AS.11 cells, measured after seeding 10 000 cells and counting at 24 h intervals using a hemacytometer. (C) Representative FACS histograms of RKO zeo, AS.17 and AS.11 cultures undergoing logarithmic growth. Data represent the mean ± SEM of four independent experiments.
Cell cycle-dependent cytoplasmic localization of HuR
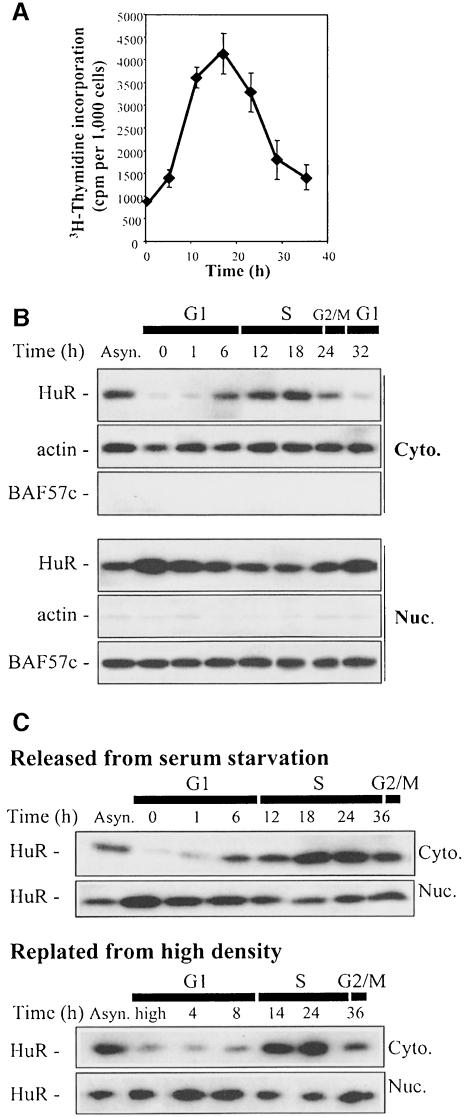
To explore the potential role of HuR in cell proliferation, we first examined the expression of HuR throughout the cell division cycle. To this end, RKO cells were subjected to a synchronization regimen consisting of 3-day culture in serum-free medium that markedly enriched the G1-cell compartment, from 30–35% in asynchronous cultures to >75% after serum starvation, as described previously (Gorospe and Holbrook, 1996). G1-enriched, growth-inhibited cultures were released from arrest by the addition of serum (final 10%) and cell cycle progression was monitored by FACS analysis (described in Gorospe and Holbrook, 1996) and by measuring incorporation of [3H]thymidine (Figure 2A). As observed, the highest [3H]thymidine incorporation was seen between 12 and 24 h. It was estimated that, under these conditions, RKO cells required ∼28 h to complete the entire first division cycle; synchrony was rapidly lost in subsequent rounds of division.
Fig. 2. Cell cycle-dependent cytoplasmic localization of HuR. (A) Thymidine incorporation in synchronous RKO zeo populations after serum addition. Data represent the mean ± SEM of four independent experiments. (B) Western blot analysis of HuR (34 kDa) levels in the cytoplasmic (Cyto.) and nuclear (Nuc.) fractions of RKO cells that were either serum starved for 3 days, then serum stimulated for the times indicated, or asynchronously growing (Asyn.). To monitor the quality of the fractionation procedure and the evenness in loading and transfer among samples, membranes were stripped and rehybridized to detect either actin (cytoplasmic, 43 kDa) or BAF57c (nuclear, 57 kDa). (C) Western blot analysis of HuR levels in the cytoplasmic and nuclear fractions of synchronous mouse embryo fibroblast populations that were either serum starved for 3 days, then stimulated by addition of 10% serum, or grown to high density (high), then replated at low density.
By Western blot analysis, whole-cell protein lysates revealed no substantial cell cycle-dependent differences in HuR expression (not shown). However, several recent reports have provided evidence that HuR is transported between the nucleus and the cytoplasm (Atasoy et al., 1998; Fan and Steitz, 1998b; Peng et al., 1998; Keene, 1999; Wang et al., 2000). As these translocation events may be linked to HuR’s function, we directly examined the subcellular localization of HuR as a function of the cell cycle stage. Interestingly, HuR was almost entirely nuclear during the first hours after serum addition (early G1), but by 6–12 h, when cells begin to replicate their DNA and progress through S phase, HuR’s presence in the cytoplasm increased dramatically. A concomitant reduction, though moderate, was observed in nuclear HuR (Figure 2B). Approximately at the time when cells are completing G2, HuR’s presence in the cytoplasm decreased (24 h time point), becoming lower again by the following G1 phase (32 h). Subsequent divisions occurred with less synchrony and cultures rapidly approached confluence.
Serum addition profoundly alters many cellular processes. To rule out the possibility that HuR’s translocation was merely due to such ‘serum effects’ and was due, instead, to the cells’ progression through their division cycle, we monitored the distribution of HuR in cultures that were synchronized by alternative means. Thus, we employed mouse embryo fibroblasts that could be synchronized either by growth to high density in medium containing 10% serum (91% G1 cells by 48 h after reaching confluence), a synchronization regimen to which RKO cells are not amenable, or serum deprivation for 3 days (87% G1 cells). Release by addition of serum or replating at low density, respectively, led to cell cycle progression, as monitored by FACS analysis (not shown), with one division cycle requiring ∼36 h. Despite differences in cell cycle duration, HuR essentially displayed the same subcellular distribution: its abundance in the cytoplasm was low during early and mid G1, but subsequently increased, reaching a maximum level during the time period corresponding to the S and G2 phases; it then decreased again at the onset of the following G1 (Figure 2C). In conclusion, HuR’s presence in the cytoplasm varied in a cell cycle-dependent manner and was highest during the S and G2 phases.
Cyclin mRNA expression and stability during cell cycle progression
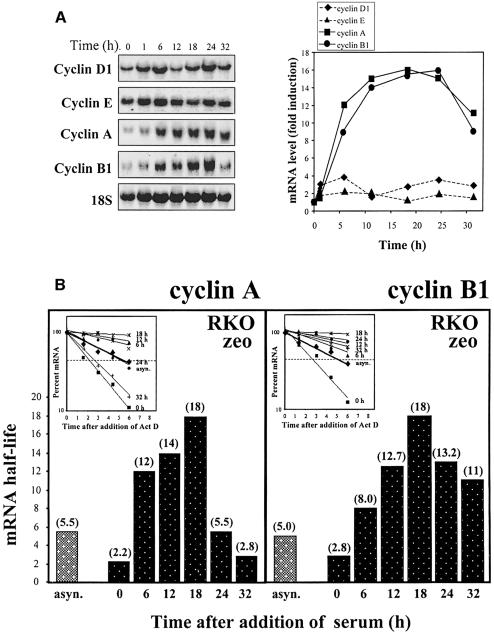
We next examined the expression of cyclins in RKO cells following release from serum starvation. Shown in Figure 3A are Northern blot analyses of the expression and relative induction of mRNAs encoding cyclins D1, E, A and B1. Throughout the time period studied, only a moderate fluctuation in the expression of mRNAs encoding cyclins D1 and E was observed, with maximal inductions of 4- and 2-fold, respectively, over the levels seen at time 0. Expression of cyclins D2 and D3 was unchanged (not shown). In contrast, mRNAs encoding cyclins A and B1 were greatly induced following serum addition: expression of each transcript between 10 and 20 h was 14- to 16-fold higher than that seen before serum induction.
Fig. 3. Cyclin mRNA expression and stability during the cell division cycle. (A) Left: representative Northern blot analysis of the expression of cyclins D1, E, A and B1. After a 3 day synchronization period, cells were serum stimulated for the times shown, whereupon RNA was extracted for analysis. Blots were sequentially stripped and rehybridized to detect each of the transcripts shown; 18S rRNA signals served to quantitate differences in loading and transfer among samples. Right: quantitation of the levels of each cyclin mRNA from three independent experiments. Data are represented as fold induction in mRNA levels relative to those at time 0 (before serum addition). (B) The half-lives of cyclin A and cyclin B1 mRNAs in either asynchronous (asyn.) or synchronous populations (serum stimulated for the times indicated) were calculated after adding 2 µg/ml actinomycin D after each serum stimulation period, preparing RNA at the times indicated thereafter, measuring the remaining signals of cyclin A and cyclin B1 mRNAs by Northern blot analysis, normalizing them to 18S rRNA, and plotting them on a logarithmic scale (inset). mRNA half-lives at each time following serum stimulation are indicated in parentheses.
Such increases in the steady-state levels of mRNAs encoding cylins A and B1 were due, at least in part, to changes in the turnover rates of each transcript. When the half-life of each transcript was calculated (by standard actinomycin D-chase analysis) at different times after serum addition, they were found to vary significantly (Figure 3B). Both for cyclin A and cyclin B1 mRNAs, half-lives were lowest (2.2 and 2.8 h, respectively) before serum addition (0 h), when most cells were in early G1. After serum addition half-lives progressively increased, with the longest half-lives of >12 h seen by 18 h (S-phase peak), and decreased afterwards, as cells exited G2 (Figure 3B).
HuR complexes with cyclin A and cyclin B1 transcripts in vitro and in vivo. Cell cycle-dependent binding to cyclin A and cyclin B1 3 ′ UTRs
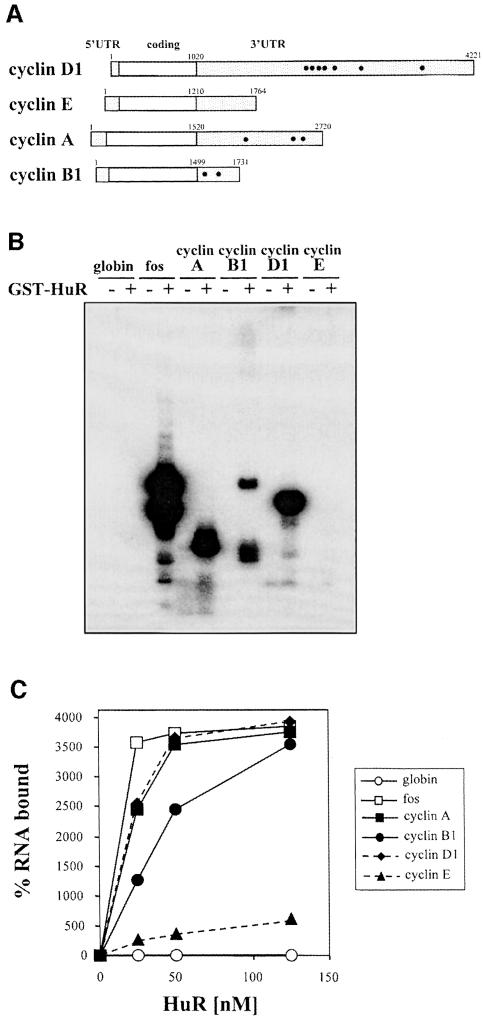
Based on the cell cycle-dependent regulation of HuR localization (Figure 2B) and the changes in cyclin mRNA levels and stability (Figure 3), we hypothesized that mRNAs encoding cyclins A and B1 were possible targets of HuR, mediating its growth-promoting function. To investigate this possibility directly, we tested whether mRNAs encoding cyclins A, B1, D1 and E (Figure 4A) contained binding sites for HuR by incubating radiolabeled transcripts corresponding to their respective 3′ UTRs with purified recombinant HuR. RNase T1 selection assays showed that cyclin A, cyclin B1 and cyclin D1 harbored HuR-binding sites, as demonstrated by the selection of specific RNA oligonucleotides (Figure 4B). A nitrocellulose-binding assay using recombinant HuR further revealed that HuR has high affinity for transcripts encoding cyclins A, B1 and D1, but is unlikely to bind to cyclin E (Figure 4C).
Fig. 4. HuR binds transcripts encoding cyclins A, B1 and D1. (A) Schematic representation of mRNAs encoding cyclins D1, E, A and B1. Small circles represent AUUUA elements. (B) RNase T1 selection assay. Radiolabeled RNAs were incubated in the presence (+) or absence (–) of 10 nM GST–HuR, then digested with RNase T1. (C) Nitrocellulose filter binding assays were performed after incubating radiolabeled transcripts with the indicated concentrations of GST–HuR; percentages of bound RNA are shown.
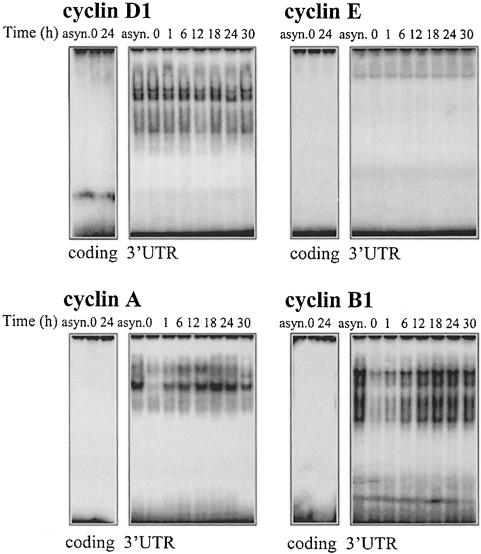
Binding to these transcripts was then investigated using lysates prepared from synchronous RKO cultures at the times indicated after serum addition. Lysates were incubated with radiolabeled transcripts and the formation of protein–RNA complexes examined. As shown (Figure 5), incubation of cytoplasmic fractions with the coding region of each of the mRNAs studied revealed no protein–RNA complexes. In contrast, 3′ UTR transcripts corresponding to cyclins A, B1 and D1 exhibited abundant complex formation. While complexes forming with the cyclin D1 3′ UTR transcript remained essentially unchanged throughout the time course studied, binding to 3′ UTR transcripts of cyclins A and B1 was largely dependent on the cell cycle stage. The most abundant binding was seen between 12 and 24 h, a time period coinciding with the cells’ progression through the S and G2/M phases, when the half-life of mRNAs encoding cyclins A and B1 is the longest (Figure 3B). Little complex formation was observed with the cyclin E 3′ UTR. When nuclear lysates were assayed, all transcripts formed complexes, but no time dependence of binding was seen (not shown). In conclusion, the 3′ UTR of mRNAs encoding cyclins A and B1 forms complexes through binding to RKO cytoplasmic proteins in a cell cycle-dependent manner.
Fig. 5. Cell cycle-dependent binding to transcripts encoding cyclins A and cyclin B1. Binding activity of radiolabeled RNAs corresponding to either the coding regions or 3′ UTRs of cyclins D1, E, A and B1, and proteins present in cytoplasmic lysates of asynchronous (asyn.) or synchronous RKO cultures after addition of serum for the times indicated is shown.
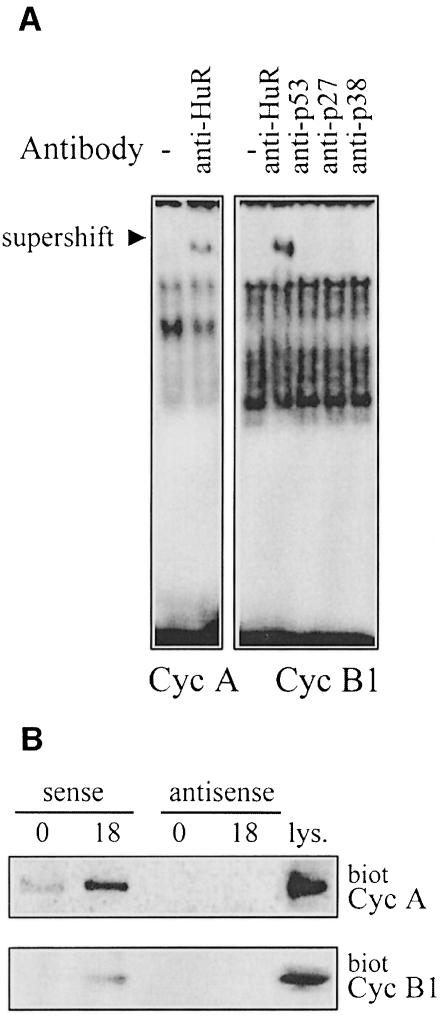
To test directly whether endogenous HuR bound the 3′ UTRs of cyclins A and B1, we carried out supershift analyses in which the migration of HuR–RNA complexes (but not that of other protein–mRNA complexes) is altered by the binding of a monoclonal antibody (Wang et al., 2000). Using this approach, we observed that both HuR–cyclin A 3′ UTR and HuR–cyclin B1 3′ UTR complexes formed with lysates from serum-stimulated RKO cells (Figure 6A); no supershifted complexes were observed with non-specific antibodies. Likewise, no supershifted complexes were seen when using 3′ UTR transcripts from cyclins D1 or E (not shown), suggesting that, under these experimental conditions, HuR does not bind cyclin D1 (a somewhat unexpected observation given its many AUUUA motifs and its binding to recombinant HuR in vitro) or cyclin E mRNAs. Of the bands that routinely formed with the UTRs of cyclins A and B1, some were more clearly depleted by the HuR antibody, suggesting that HuR forms part of those particular complexes. The bands that are not supershifted by the HuR antibody may contain other RNA-binding proteins also recognizing their 3′ UTRs. Alternatively, they may represent post-translational modifications of HuR, or associations of HuR with other proteins that mask the HuR epitope recognized by this monoclonal antibody (although it was generated against the N-terminus to avoid this problem). Finally, after incubation with RKO cell lysates, biotinylated RNAs that encompassed the 3′ UTRs of cyclins A and B1 effectively ‘pulled down’ HuR (described in Materials and methods), as detected by Western blot analysis. Irrelevant biotinylated transcripts, such as RNAs complementary to cyclin A and B1 3′ UTRs (Figure 6B), or RNAs encompassing each coding region (not shown), did not pull down HuR.
Fig. 6. HuR forms complexes with cyclin A and cyclin B1 3′ UTR transcripts. (A) Complexes forming with lysates from synchronous RKO zeo cells (18 h serum stimulated) and radiolabeled 3′ UTR transcripts from cyclins A and B1 were assayed for their ability to be supershifted by the indicated antibodies. (B) Biotinylated transcripts (sense, 3′ UTR; antisense, complementary to each 3′ UTR) corresponding to the 3′ UTRs of either cyclin A or cyclin B1 were incubated with cytoplasmic RKO lysates (prepared 0 or 18 h after serum stimulation). Complexes were pulled down using streptavidin-conjugated magnetic beads and analyzed by Western blotting to assess HuR levels. lys., 35 µg of RKO zeo whole-cell lysate.
RKO ASHuR cells exhibit reduced binding to transcripts from cyclins A and B1 and decreased cyclin A and B1 mRNA stability, mRNA levels and protein expression
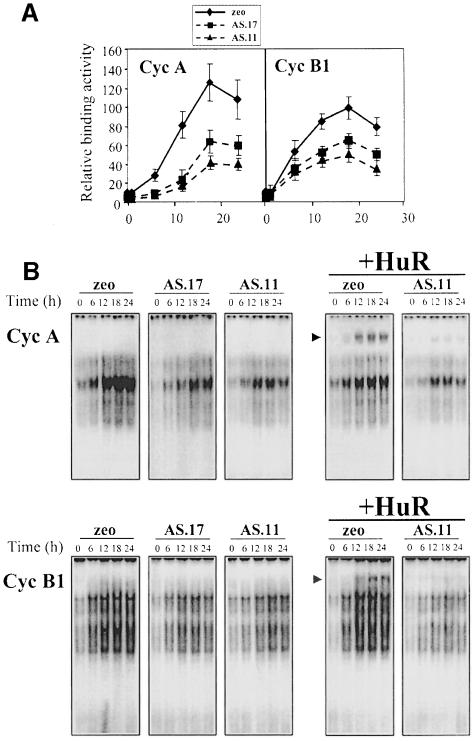
To examine further the significance of HuR’s binding to the 3′ UTR of cyclins A and B1, complexes forming in ASHuR-expressing cells were studied. As shown, the cell cycle-dependent increase in binding to the 3′ UTRs of either cyclin A or cyclin B1 was greatly diminished when assaying lysates from AS.17 and AS.11 (quantitated in Figure 7A). Supershifting of complexes was markedly reduced in ASHuR cells (Figure 7B, +HuR). In summary, cells with reduced HuR expression exhibit substantially reduced binding to transcripts of cyclins A and B1 as the cell progresses through its proliferative cycle.
Fig. 7. Binding of RKO ASHuR cells to transcripts from cyclins A and B1. (A) Quantitation of radioactive signals (sum of all bands, represented as the mean ± SEM from three independent experiments) from binding experiments similar to those shown in (B). (B) Repre sentative complexes forming between cytoplasmic lysates from synchronous populations of RKO zeo or ASHuR cells (prepared at the times indicated after serum stimulation), and radiolabeled cyclin A 3′ UTR (top) or cyclin B1 3′ UTR (bottom). Supershift analysis was carried out where indicated (+HuR); the arrowhead shows supershifted complexes.
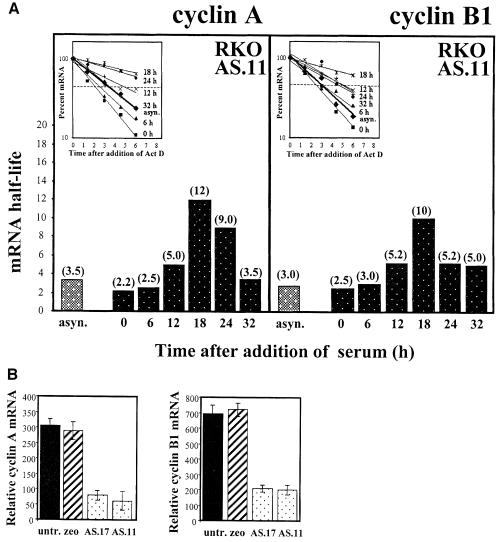
A number of studies support the notion that HuR plays a protective role in mRNA stability and enhances mRNA half-life (Jain et al., 1997; Fan and Steitz, 1998a; Wang et al., 2000). To investigate whether HuR also had a protective influence on mRNA turnover in the cell cycle progression paradigm discussed here, we measured the half-lives of mRNAs encoding cyclins A and B1 in RKO ASHuR cells. As observed, the half-life values measured in AS.11 cells (Figure 8A) were significantly lower than those measured in RKO zeo cells (Figure 3B). For example, mRNA half-lives for cyclins A and B1 in asynchronous cultures were 5.5 and 5 h, respectively, in RKO zeo, while they were 3.5 and 3 h, respectively, in AS.11 cells. Likewise, at almost all time points after serum stimulation examined, cyclins A and B1 in RKO zeo cells had longer lived mRNAs than did AS.11 cells. Thus, the half-life of cyclin A mRNA in RKO zeo and AS.11 cells was, respectively, 12 versus 2.5 h (6 h time point), 14 versus 5 h (12 h time point) and 18 versus 12 h (18 h time point). The half-life of cyclin B1 mRNA was also longer in RKO zeo than AS.11 cells at all time points examined: 8 versus 3 h (6 h time point), 12.7 versus 5.2 h (12 h time point), 18 versus 10 h (18 h time point), 13.2 versus 5.2 h (24 h time point) and 11 versus 5 h (32 h time point). The cyclin A mRNA half-life was somewhat shorter in RKO zeo cells at late time points after serum release, possibly due to additional compensatory mRNA-stabilizing factors upregulated in AS.11 during the late stages of the cell’s proliferative cycle.
Fig. 8. Stability and levels of mRNAs encoding cyclins A and B1. (A) Half-lives of mRNAs encoding cyclins A and B1 in AS.11 populations were calculated as described in Figure 3B. mRNA half-lives at each time point following serum stimulation are indicated in parentheses. (B) Relative steady-state levels of mRNAs encoding cyclins A and B1 in RKO untransfected (untr.), zeo, AS.17 and AS.11 cells. Data represent the mean ± SEM from eight independent Northern blots.
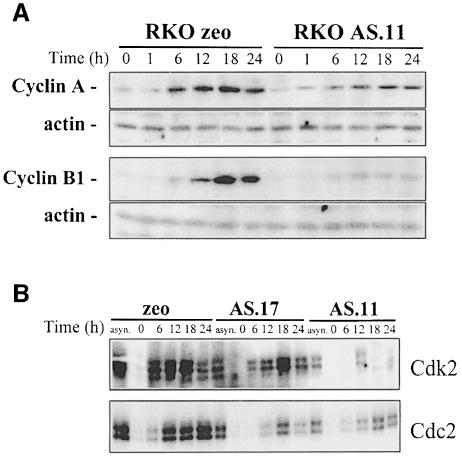
These changes in mRNA stability, in turn, influenced the steady-state levels of mRNAs encoding cyclins A and B1 (Figure 8B), as they were reduced overall in asynchronous populations of AS.l7 and AS.11 cells (and other ASHuR lines examined; not shown). Finally, cell cycle-dependent expression of cyclins A and B1 was also diminished (Figure 9A), and consequently the kinase activity associated with either cdk2 or cdc2 decreased in cells with reduced HuR expression (Figure 9B).
Fig. 9. Cyclin A and B1 protein levels and cdk2- and cdc2-associated kinase activity in ASHuR cells. (A) Representative Western blots of cyclin A and cyclin B1 in whole-cell lysates prepared from synchronous RKO zeo and AS.11 cells at the times indicated after serum stimulation. Actin signals served to monitor the equality of sample loading and transfer. (B) Representative cdk2 and cdc2 kinase activities in RKO zeo and ASHuR cells after serum stimulation for the times indicated. Phosphorylated histone H1 is shown.
In summary, cells with markedly reduced HuR display lower steady-state levels of cyclin A mRNA and cyclin B1 mRNA, decreased levels of each protein, and reduced activity of the cyclin-dependent kinases regulated by cyclins A and B1. These alterations are likely to contribute directly to their diminished proliferation rates. In conclusion, our studies strongly support a model whereby HuR stabilizes mRNAs encoding cyclins A and B1 during cellular proliferation.
Discussion
In the present study we provide evidence supporting the hypothesis that the ARE-binding protein HuR participates in the regulation of cyclin A and cyclin B1 expression by stabilizing their mRNAs. Using two different synchronization paradigms, we show that HuR is mostly nuclear during G1, is mobilized to the cytoplasm through late G1, S and G2 phases, and becomes mostly nuclear again by the next early G1 phase. Concomitant with S-phase entry, the half-lives of mRNAs encoding cyclins A and B1, and hence the levels of cyclin A and cyclin B1 mRNA and protein (but not those of cyclins D1, D2, D3 and E), increase markedly. We further show that, in a cell cycle-dependent fashion, RNA-binding proteins present in the cytoplasm of RKO cells recognize and form complexes with 3′ UTR transcripts of cyclins A and B1, and by supershift analysis and biotinylated RNA pull-down assays we provide evidence that HuR forms part of those associations. Cells expressing markedly reduced levels of HuR through an antisense approach exhibit a reduction in cyclin A and cyclin B1 mRNA steady-state levels and half-life, decreased binding to their 3′ UTR transcripts and slower proliferation rates. Taken together, our results strongly support the notion that HuR regulates the expression of cyclins A and B1 by enhancing their stability during the cell’s proliferative cycle.
The nature of the transport mechanism(s) regulating HuR’s proliferation-dependent subcellular localization is presently unclear. It is plausible that the recently described HNS sequence (Fan and Steitz, 1998b) plays a role, but this possibility has not been tested directly. Atasoy et al. (1998) also described serum-induced and T-cell activation-induced changes in HuR’s subcellular localization, although they report HuR to be cytoplasmic during G1, returning to a nuclear localization during S phase. The reasons for the discrepancy between their results and ours are presently unclear. That cyclin B1 mRNA expression might be regulated by HuR was also postulated previously by this group, based on the parallel regulation of cyclin B1 expression and the observed subcellular localization of HuR (Atasoy et al., 1998).
While both cyclins A and B1 have distinct AREs in their 3′ UTRs, the precise HuR-binding region remains to be mapped. Delineating these binding sites may be important since our unpublished observations using lysates from RKO and other carcinoma lines reveal that another RNA-binding protein, AUF1, also binds to the 3′ UTRs of cyclins A and B1. Since binding of AUF1 to target mRNAs has been linked to their enhanced turnover (Brewer, 1991; DeMaria and Brewer, 1996; Loflin et al., 1999), it will be of interest to examine whether AUF1 and HuR bind to the same sequences on the mRNAs encoding cyclins A and B1. If this is the case, we may envision a scenario in which two RNA-binding proteins, one promoting stabilization and one promoting decay, compete for binding to the same target mRNA. Of note, neither AUF1 levels nor its subcellular localization varied throughout the cell cycle (not shown); interestingly, however, AUF1 binding to 3′ UTRs of cyclins A and B1 increased dramatically under conditions of stress that are known to downregulate cyclin A and cyclin B1 expression rapidly (manuscript in preparation). Binding to the cyclin D1 transcript may constitute another example of the competition between the relative affinities of different RNA-binding proteins as endogenous AUF1, but not endogenous HuR, binds to it efficiently (S.Lin, W.Wang, G.M.Wilson, G.Brewer, N.J.Holbrook and M.Gorospe, submitted), while recombinant GST–HuR binds to it in vitro, in the absence of competing RNA-binding proteins.
As described here, HuR exerts a regulatory influence on the expression of cyclins A and B1, two proliferation-associated genes that are upregulated in many cancers (Wang et al., 1997; Dobashi et al., 1998; Huuhtanen et al., 1999; for reviews, see Hunter and Pines, 1991; MacLachlan et al., 1995). It is, therefore, tempting to hypothesize that the enhanced expression of Elav/Hu proteins in SCLC and Hu syndrome-related cancers may be linked to the enhanced half-life of cell cycle-related and proliferative genes. In support of this hypothesis are also our unpublished observations that HuR is overexpressed in virtually all tumors examined and is largely cytoplasmic. Furthermore, while a clear role of HuR or other RNA-binding proteins in cancer has yet to be established, it is noteworthy that many oncogenes and tumor suppressor genes have ARE sequences in their 3′ UTR; mechanisms of coordinate regulation of the half-life of the cell cycle and growth-regulatory genes have been postulated (Shyu et al., 1989; Brewer, 1991; Datta et al., 1992).
HuR has also been shown to play a role in enhancing mRNA translation, as shown for GLUT-1 and NF-M, (Jain et al., 1997; Antic and Keene, 1998; Antic et al., 1999). Whether or not HuR, in addition to stabilizing mRNAs encoding cyclins A and B1, also regulates their translation remains to be determined. In this regard, the 3′ UTRs of cyclins A and B1 also contain cytoplasmic polyadenylation elements (CPEs) that regulate their translation (Standart et al., 1990; de Moor and Richter, 1999; Minshall et al., 1999); the existence of a functional link between mRNA turnover and translational control by CPE-binding proteins remains an intriguing possibility that awaits investigation. It also remains to be established whether HuR regulates the expression of additional cell cycle control genes with labile mRNAs, since they may contribute to the profound effects on proliferation of ASHuR cells described here. In this regard, our recent study (Wang et al., 2000), showing that HuR stabilizes the p21 mRNA following UVC irradiation, supports this notion, although our unpublished results suggest that HuR does not mediate the cell cycle-dependent expression of p21. These observations illustrate the existence of additional levels of complexity in the mechanisms regulating the turnover of target mRNAs. Finally, our study highlights the importance of post-transcriptional regulatory events such as those elicited by HuR, which, coupled with transcriptional and proteolytic mechanisms, cooperate in the regulation of two important cyclins, cyclin A and cyclin B1, and participate in the control of cell proliferation.
Materials and methods
Cell culture, treatments, synchronization and transfection
Human colorectal carcinoma RKO cells (Kessis et al., 1993) were cultured in minimum essential medium (Gibco BRL, Gaithersburg, MD) and mouse embryo fibroblasts in Dulbecco’s modified essential medium, each supplemented with 10% fetal bovine serum (Hyclone, Logan, UT) and antibiotics. Actinomycin D was from Sigma (St Louis, MO). To establish lines expressing ASHuR mRNA constitutively, RKO cells were transfected with either pZeoSV2(zeo) or pZeoSV2(–) HuR (ASHuR) by calcium phosphate precipitation, and selected in 600 µg/ml zeocin (Invitrogen, Carlsbad, CA). Only 1/10 of transfected clones expressed adequately reduced HuR (≤75% than that expressed in zeo transfectants); these clones were stored as frozen aliquots and used within 3 weeks. For synchronization studies, RKO cultures were maintained in serum-free medium for 3 days, then released by serum addition; under this synchronization protocol the G1 phase compartment, which generally constitutes 30–35% of the total population, was considerably enriched, reaching >75% (Gorospe and Holbrook, 1996). For [3H]thymidine incorporation studies (performed using standard methods), RKO cells were cultured in 24-well plates, synchronized by serum starvation and collected after addition of serum for the times indicated; 10 µCi of [3H]thymidine were used per well.
Subcellular fractionation and Northern and Western blot analysis
Northern blot analysis was carried out as described (Gorospe et al., 1998). For detection of mRNAs encoding cyclins A, B1, D1 and E, corresponding PCR fragments encompassing each coding region (obtained as indicated below) were random primer labeled with [α-32P]dATP; 18S rRNA was detected as described (Gorospe et al., 1998). Incorporation of 32P was visualized and quantitated using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA). Cytoplasmic and nuclear fractions were prepared as described previously (Wang et al., 2000). For Western blot analysis, whole-cell (20 µg), cytoplasmic (40 µg) or nuclear (10 µg) lysates were size-fractionated by SDS–PAGE and transferred onto PVDF membranes. HuR was detected with the monoclonal antibody 19F12 (Wang et al., 2000), BAF57c and AUF1 with polyclonal antibodies (Zhang et al., 1993; Wang et al., 1998), and actin, cyclin A and cyclin B1 were detected using monoclonal antibodies from Santa Cruz (Santa Cruz, CA).
Preparation of transcripts
Complementary DNA, prepared from RKO cell RNA using a kit from Invitrogen, was used as a template for PCR amplification of different mRNA regions of cyclins A, B1, E and D1. All 5′ oligonucleotides contained the T7 RNA polymerase promoter sequence CCAAGCTTCTAATACG ACTCACTATAGGGAGA (T7). The following oligonucleotides were used to prepare the cyclin coding region and 3′ UTR templates.
Cyclin A coding region and 3′ UTR templates, respectively:
(T7)GAGCAGTGATGTTGGGCAAC and CAGATTTAGTGTCTCTGGTG (region 214–1516);
(T7)CCAGAGACACTAAATCTGTAAC and GGTAACAAATTTCTGGTTTATTTC (region 1499–2718).
Cyclin B1 coding and 3′ UTR templates, respectively:
(T7)CTGCCTGGTGAAGAGGAAGC and GAGTGCTGATCTTAGCATGC (region 158–1416);
(T7)GTCAAGAACAAGTATGCCA and CTGAAGTGGGAGCGGAAAAG (region 1369–1702).
Cyclin E coding and 3′ UTR templates, respectively:
(T7)GGACACCATGAAGGAGGACG and GTGGTCAGGCCATTTCCGGC (region 18–1217);
(T7)CACAGAGCGGTAAGAAGCAG and GGATAGATATAGCAGCACTTAC (region 1169–1714).
Cyclin D1 coding and 3′ UTR templates, respectively:
(T7)TAGCAGCGAGCAGCAGAG and CTCAGATGTCCACGTCCCG (region 2–1030);
(T7) ACATCTGAGGGCGCCAGG and CCACCTCCCTTCAACACTTC (region 1022–2870).
Corresponding RNAs were synthesized from PCR-generated DNA fragments as described (Gorospe and Baglioni, 1994) and used at 100 000 c.p.m./µl (2–10 fmol/µl). The preparation of c-fos and globin transcripts is described in Chung et al. (1996). For the preparation of biotinylated transcripts, PCR fragments encompassing the 3′ UTRs of cyclins A and B1 were synthesized bearing T7 on either the 5′ (for sense transcripts) or the 3′ (for antisense transcripts) ends, then transcribed in vitro using biotinylated CTP [(Sigma), 1/10 of total CTP].
RNA–protein binding, supershift, RNase T1 selection and nitrocellulose filter binding assays
For the analysis of complexes forming with radiolabeled RNAs, binding reactions and native gel electrophoresis were carried out as described (Wang et al., 2000). For supershift assays, antibodies were incubated with lysates for 1 h on ice before addition of radiolabeled RNA; all subsequent steps were as described above. Antibodies used in supershift assays were from PharMingen (San Diego, CA) except those recognizing HuR (Wang et al., 2000). RNase T1 selection assays and nitrocellulose filter binding assays were carried out as described (Joseph et al., 1998). Binding of proteins to biotinylated transcripts was performed using 140 µg of cytoplasmic lysate supplemented with RNase inhibitor (5′→3′, Boulder, CO) and protease inhibitor cocktail (Sigma), and 2 µg of biotinylated transcript for 30 min at room temperature. Complexes were isolated with paramagnetic streptavidin Dynabeads (Dynal, Oslo, Norway), washed with phosphate-buffered saline and subjected to Western blot analysis to detect HuR.
Immunoprecipitation and kinase assays to monitor cdk2 and cdc2 kinase activity
Immunoprecipitation and kinase assays were carried out as described previously for cdk2 (Gorospe et al., 1996). cdk2 and cdc2 were immunoprecipitated from 200-µg aliquots of lysate after incubation with either anti-cdk2 (PharMingen) or anti-cyclin B1 antibodies (Santa Cruz), respectively. Kinase activities associated with cdk2- and cdc2-containing immunoprecipitates were assayed using histone H1 (Ambion, Austin, TX) as substrate.
Acknowledgments
Acknowledgements
We thank M.B.Kastan for the RKO cells, W.Wang for the BAF57c antibody and G.Brewer for the AUF1 antibody. We are also grateful to our colleagues N.J.Holbrook and D.L.Longo for critical reading of the manuscript.
References
- Antic D. and Keene,J.D. (1997) Embryonic lethal abnormal visual RNA-binding proteins involved in growth, differentiation and posttranscriptional gene expression. Am. J. Hum. Genet., 61, 273–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antic D. and Keene,J.D. (1998) Messenger ribonucleoprotein complexes containing human ELAV proteins: interactions with cytoskeleton and translational apparatus. J. Cell Sci., 111, 183–197. [DOI] [PubMed] [Google Scholar]
- Antic D., Lu,N. and Keene,J.D. (1999) ELAV tumor antigen, hel-N1, increases translation of neurofilament M mRNA and induces formation of neurites in human teratocarcinoma cells. Genes Dev., 13, 449–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda-Abreu G.E., Behar,L., Chung,S., Furneaux,H. and Ginzburg,I. (1999) Embryonic lethal abnormal vision-like RNA-binding proteins regulate neurite outgrowth and τ expression in PC12 cells. J. Neurosci., 19, 6907–6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atasoy U., Watson,J., Patel,D. and Keene,J.D. (1998) ELAV protein HuA (HuR) can redistribute between nucleus and cytoplasm and is upregulated during serum stimulation and T cell activation. J. Cell Sci., 111, 3145–3156. [DOI] [PubMed] [Google Scholar]
- Baghdassarian N., Peiretti,A., Devaux,E., Bryon,P.A. and Ffrench,M. (1999) Involvement of p27Kip1 in the G1- and S/G2-phase lengthening mediated by glucocorticoids in normal human lymphocytes. Cell Growth Differ., 10, 405–412. [PubMed] [Google Scholar]
- Brewer G. (1991) An A+U-rich element RNA binding factor regulates c-myc mRNA stability in vitro. Mol. Cell. Biol., 11, 2460–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caput D., Beutler,B., Hartog,K., Thayer,R., Brown-Shimer,S. and Cerami,A. (1986) Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc. Natl Acad. Sci. USA, 83, 1670–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Y. and Shyu,A.B. (1995) AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci., 20, 465–470. [DOI] [PubMed] [Google Scholar]
- Chung S., Jiang,L., Cheng,S. and Furneaux,H. (1996) Purification and properties of HuD, a neuronal RNA-binding protein. J. Biol. Chem., 271, 11518–11524. [DOI] [PubMed] [Google Scholar]
- Chung S., Eckrich,M., Perrone-Bizzozero,N., Kohn,D.T., Furneaux,H. (1997) The Elav-like proteins bind to a conserved regulatory element in the 3′-untranslated region of GAP-43 mRNA. J. Biol. Chem., 272, 6593–6598. [DOI] [PubMed] [Google Scholar]
- Dalmau J., Furneaux,H.M., Gralla,R.J., Kris,M.G. and Posner,J.B. (1990) Detection of the anti-Hu antibody in the serum of patients with small cell lung cancer—a quantitative western blot analysis. Ann. Neurol., 27, 544–552. [DOI] [PubMed] [Google Scholar]
- Datta R., Hass,R., Gunji,H., Weichselbaum,R. and Kufe,D. (1992) Down-regulation of cell cycle control genes by ionizing radiation. Cell Growth Differ., 3, 637–644. [PubMed] [Google Scholar]
- DeMaria C.T. and Brewer,G. (1996) AUF1 binding affinity to A+U-rich elements correlates with rapid mRNA degradation. J. Biol. Chem., 271, 12179–12184. [DOI] [PubMed] [Google Scholar]
- de Moor C.H. and Richter,J.D. (1999) Cytoplasmic polyadenylation elements mediate masking and unmasking of cyclin B1 mRNA. EMBO J., 18, 2294–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobashi Y., Shoji,M., Jiang,S.X., Kobayashi,M., Kawakubo,Y. and Kameya,T. (1998) Active cyclin A–CDK2 complex, a possible critical factor for cell proliferation in human primary lung carcinomas. Am. J. Pathol., 153, 963–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito F., Cuccovillo,F., Vanoni,M., Cimino,F., Anderson,C.W., Appella,E. and Russo,T. (1997) Redox-mediated regulation of p21 (waf1/cip1) expression involves a post-transcriptional mechanism and activation of the mitogen-activated protein kinase pathway. Eur. J. Biochem., 245, 730–737. [DOI] [PubMed] [Google Scholar]
- Fan X.C. and Steitz,J.A. (1998a) Overexpression of HuR, a nuclear–cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J., 17, 3448–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X.C. and Steitz,J.A. (1998b) HNS, a nuclear–cytoplasmic shuttling sequence in HuR. Proc. Natl Acad. Sci. USA, 95, 15293–15298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford L.P., Watson,J., Keene,J.D. and Wilusz,J. (1999) ELAV proteins stabilize deadenylated intermediates in a novel in vitro mRNA deadenylation/degradation system. Genes Dev., 13, 188–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good P.J. (1995) A conserved family of Elav-like genes in vertebrates. Proc. Natl Acad. Sci. USA, 89, 4557–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorospe M. and Baglioni,C. (1994) Degradation of unstable interleukin-1α mRNA in a rabbit reticulocyte cell-free system. Localization of an instability determinant to a cluster of AUUUA motifs. J. Biol. Chem., 269, 11845–11851. [PubMed] [Google Scholar]
- Gorospe M. and Holbrook,N.J. (1996) Role of p21 in prostaglandin A2-mediated cellular arrest and death. Cancer Res., 56, 475–479. [PubMed] [Google Scholar]
- Gorospe M., Liu,Y., Xu,Q., Chrest,F.J. and Holbrook,N.J. (1996) Inhibition of G1 cyclin-dependent kinase activity during growth arrest of human breast carcinoma cells by prostaglandin A2. Mol. Cell. Biol., 16, 762–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorospe M., Wang,X. and Holbrook,N.J. (1998) p53-dependent elevation of p21Waf1 expression by UV light is mediated through mRNA stabilization and involves a vanadate-sensitive regulatory system. Mol. Cell. Biol., 18, 1400–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttridge D.C., Albanese,C., Reuther,J.Y., Pestell,R.G. and Baldwin,A.S. (1999) NF-κB controls cell growth and differentiation through transcriptional regulation of cyclin D1. Mol. Cell. Biol., 19, 5785–5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashemolhosseini S., Nagamine,Y., Morley,S.J., Desrivieres,S., Mercep,L. and Ferrari,S. (1998) Rapamycin inhibition of the G1 to S transition is mediated by effects on cyclin D1 mRNA and protein stability. J. Biol. Chem., 273, 14424–14429. [DOI] [PubMed] [Google Scholar]
- Henglein B., Chenivesse,X., Wang,J., Eick,D. and Brechot,C. (1994) Structure and cell cycle-regulated transcription of the human cyclin A gene. Proc. Natl Acad. Sci. USA, 91, 5490–5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henics T., Sanfridson,A., Hamilton,B.J., Nagy,E. and Rigby,W.F. (1994) Enhanced stability of interleukin-2 mRNA in MLA 144 cells. Possible role of cytoplasmic AU-rich sequence-binding proteins. J. Biol. Chem., 269, 5377–5383. [PubMed] [Google Scholar]
- Hosokawa Y., Suzuki,R., Joh,T., Maeda,Y., Nakamura,S., Kodera,Y., Arnold,A. and Seto,M. (1998) A small deletion in the 3′-untranslated region of the cyclin D1/PRAD1/bcl-1 oncogene in a patient with chronic lymphocytic leukemia. Int. J. Cancer, 76, 791–796. [DOI] [PubMed] [Google Scholar]
- Howe J.A., Howell,M., Hunt,T. and Newport,J.W. (1995) Identification of a developmental timer regulating the stability of embryonic cyclin A and a new somatic A-type cyclin at gastrulation. Genes Dev., 9, 1164–1176. [DOI] [PubMed] [Google Scholar]
- Hunter T. and Pines,J. (1991) Cyclins and cancer. Cell, 66, 1071–1074. [DOI] [PubMed] [Google Scholar]
- Huuhtanen R.L., Blomqvist,C.P., Bohling,T.O., Wiklund,T.A., Tukiainen,E.J., Virolainen,M., Tribukait,B. and Andersson,L.C. (1999) Expression of cyclin A in soft tissue sarcomas correlates with tumor aggressiveness. Cancer Res., 59, 2885–2890. [PubMed] [Google Scholar]
- Hwang A., Maity,A., McKenna,W.G. and Muschel,R.J. (1995) Cell cycle-dependent regulation of the cyclin B1 promoter. J. Biol. Chem., 270, 28419–28424. [DOI] [PubMed] [Google Scholar]
- Hwang A., McKenna,W.G. and Muschel,R.J. (1998) Cell cycle-dependent usage of transcriptional start sites. A novel mechanism for regulation of cyclin B1. J. Biol. Chem., 273, 31505–31509. [DOI] [PubMed] [Google Scholar]
- Jain R.G., Andrews,L.G., McGowan,K.M., Pekala,P.H. and Keene,J.D. (1997) Ectopic expression of Hel-N1, an RNA-binding protein, increases glucose transporter (GLUT1) expression in 3T3-L1 adipocytes. Mol. Cell. Biol., 17, 954–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D.G. and Walker,C.L. (1999) Cyclins and cell cycle checkpoints. Annu. Rev. Pharmacol. Toxicol., 39, 295–312. [DOI] [PubMed] [Google Scholar]
- Joseph B., Orlian,M. and Furneaux,H. (1998) p21waf1 mRNA contains a conserved element in its 3′-untranslated region that is bound by the Elav-like mRNA-stabilizing proteins. J. Biol. Chem., 273, 20511–20516. [DOI] [PubMed] [Google Scholar]
- Keene J.D. (1999) Why is Hu where? Shuttling of early-response-gene messenger RNA subsets. Proc. Natl Acad. Sci. USA, 96, 5–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessis T.D., Slebos,R.J., Nelson,W.D., Kastan,M.B., Plunkett,B.S., Han,S.M., Lorincz,A.T., Hedrick,L. and Cho.K.R. (1993) Human papilloma virus 16 E6 expression disrupts the p53-mediated cellular response to DNA damage. Proc. Natl Acad. Sci. USA, 90, 3988–3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King R.W., Deshaies,R.J., Peters,J.M. and Kirschner,M.W. (1996) How proteolysis drives the cell cycle. Science, 274, 1652–1659. [DOI] [PubMed] [Google Scholar]
- Koepp D.M., Harper,J.W. and Elledge,S.J. (1999) How the cyclin became a cyclin: regulated proteolysis in the cell cycle. Cell, 97, 431–434. [DOI] [PubMed] [Google Scholar]
- Lagnado C.A., Brown,C.Y. and Goodall,G.J. (1994) AUUUA is not sufficient to promote poly(A) shortening and degradation of an mRNA: the functional sequence within AU-rich elements may be UUAUUUA(U/A) (U/A). Mol. Cell. Biol., 14, 7984–7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarova D.L., Spengler,B.A., Biedler,J.L. and Ross,R.A. (1999) HuD, a neuronal-specific RNA-binding protein, is a putative regulator of N-myc pre-mRNA processing/stability in malignant human neuroblasts. Oncogene, 18, 2703–2710. [DOI] [PubMed] [Google Scholar]
- Lebwohl D.E., Muise-Helmericks,R., Sepp-Lorenzino,L., Serve,S., Timaul,M., Bol,R., Borgen,P. and Rosen,N. (1994) A truncated cyclin D1 gene encodes a stable mRNA in a human breast cancer cell line. Oncogene, 9, 1925–1929. [PubMed] [Google Scholar]
- Levine T.D., Gao,F., King,P.H., Andrews,L.G. and Keene,J.D. (1993) Hel-N1: an autoimmune RNA-binding protein with specificity for 3′ uridylate-rich untranslated regions of growth factor mRNAs. Mol. Cell. Biol., 13, 3494–3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy N.S., Chung,S., Furneaux,H. and Levy,A.P. (1998) Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J. Biol. Chem., 273, 6417–6423. [DOI] [PubMed] [Google Scholar]
- Liebhaber S.A. (1997) mRNA stability and the control of gene expression. Nucleic Acids Symp. Ser., 36, 29–32. [PubMed] [Google Scholar]
- Loflin P., Chen,C.-Y.A. and Shyu,A.-B. (1999) Unraveling a cytoplasmic role for hnRNPD in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev., 13, 1884–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W.J., Cheng,S., Campbell,C., Wright,A. and Furneaux,H. (1996) Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem., 271, 8144–8151. [DOI] [PubMed] [Google Scholar]
- MacLachlan T.K., Sang,N. and Giordano,A. (1995) Cyclins, cyclin-dependent kinases and cdk inhibitors: implications in cell cycle control and cancer. Crit. Rev. Eukaryot. Gene Expr., 5, 127–156. [DOI] [PubMed] [Google Scholar]
- Maity A., McKenna,W.G. and Muschel,R.J. (1995) Evidence for post-transcriptional regulation of cyclin B1 mRNA in the cell cycle and following irradiation in HeLa cells. EMBO J., 14, 603–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maity A., McKenna,W.G. and Muschel,R.J. (1997) Cyclin A message stability varies with the cell cycle. Cell Growth Differ., 8, 311–318. [PubMed] [Google Scholar]
- Malter J.S. and Hong,Y. (1991) A redox switch and phosphorylation are involved in the post- translational up-regulation of the adenosine-uridine binding factor by phorbol ester and ionophore. J. Biol. Chem., 266, 3167–3171. [PubMed] [Google Scholar]
- Markiewicz D.A., McKenna,W.G., Flick,M.B., Maity,A. and Muschel,R.J. (1994) The effects of radiation on the expression of a newly cloned and characterized rat cyclin B mRNA. Int. J. Radiat. Oncol. Biol. Phys., 28, 135–144. [DOI] [PubMed] [Google Scholar]
- Maurer F., Tierney,M. and Medcalf,R.L. (1999) An AU-rich sequence in the 3′-UTR of plasminogen activator inhibitor type 2 (PAI-2) mRNA promotes PAI-2 mRNA decay and provides a binding site for nuclear HuR. Nucleic Acids Res., 27, 1664–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshall N., Walker,J., Dale,M. and Standart,N. (1999) Dual roles of p82, the clam CPEB homolog, in cytoplasmic polyadenylation and translational masking. RNA, 5, 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemer M. and Stuebing,E.W. (1996) WEE1-like CDK tyrosine kinase mRNA level is regulated temporally and spatially in sea urchin embryos. Mech. Dev., 58, 75–88. [DOI] [PubMed] [Google Scholar]
- Oda S., Nishida,J., Nakabeppu,Y. and Sekiguchi,M. (1995) Stabilization of cyclin E and cdk2 mRNAs at G1/S transition in Rat-1A cells emerging from the G0 state. Oncogene, 10, 1343–1351. [PubMed] [Google Scholar]
- Ohtani K., DeGregori,J. and Nevins,J.R. (1995) Regulation of the cyclin E gene by transcription factor E2F1. Proc. Natl Acad. Sci. USA, 92, 12146–12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S.S., Chen,C.Y., Xu,N. and Shyu,A.B. (1998) RNA stabilization by the AU-rich element binding protein HuR, an ELAV protein. EMBO J., 17, 3461–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piaggio G., Farina,A., Perrotti,D., Manni,I., Fuschi,P., Sacchi,A. and Gaetano,C. (1995) Structure and growth-dependent regulation of the human cyclin B1 promoter. Exp. Cell Res., 216, 396–402. [DOI] [PubMed] [Google Scholar]
- Ross J. (1995) mRNA stability in mammalian cells. Microbiol. Rev., 59, 423–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavi S.C., Belasco,J.G. and Greenberg,M.E. (1992) Regulation of proto-oncogene mRNA stability. Biochim. Biophys. Acta, 1114, 95–106. [DOI] [PubMed] [Google Scholar]
- Schwaller J., Koeffler,H.P., Niklaus,G., Loetscher,P., Nagel,S., Fey, M.F. and Tobler,A. (1995) Posttranscriptional stabilization underlies p53-independent induction of p21WAF1/CIP1/SDI1 in differentiating human leukemic cells. J. Clin. Invest., 95, 973–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw G. and Kamen,R. (1986) A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell, 46, 659–667. [DOI] [PubMed] [Google Scholar]
- Shyu A.B., Greenberg,M.E. and Belasco,J.G. (1989) The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes Dev., 3, 60–72. [DOI] [PubMed] [Google Scholar]
- Standart N., Dale,M., Stewart,E. and Hunt,T. (1990) Maternal mRNA from clam oocytes can be specifically unmasked in vitro by antisense RNA complementary to the 3′-untranslated region. Genes Dev., 4, 2157–2168. [DOI] [PubMed] [Google Scholar]
- Szabo A., Dalmau,J., Manley,G., Rosenfeld,M.R., Wong,E., Henson,J., Posner,J.B. and Furneaux,H. (1991) HuD, a paraneoplastic encephalomyelitis antigen, contains RNA-binding domains and is homologous to Elav and Sex-lethal. Cell, 67, 325–333. [DOI] [PubMed] [Google Scholar]
- Treisman R. (1985) Transient accumulation of c-fos RNA following serum stimulation requires a conserved 5′ element and c-fos 3′ sequences. Cell, 42, 889–902. [DOI] [PubMed] [Google Scholar]
- Trembley J.H., Kren,B.T. and Steer,C.J. (1994) Posttranscriptional regulation of cyclin B messenger RNA expression in the regenerating rat liver. Cell Growth Differ., 5, 99–108. [PubMed] [Google Scholar]
- Wang A., Yoshimi,N., Ino,N., Tanaka,T. and Mori,H. (1997) Overexpression of cyclin B1 in human colorectal cancers. J. Cancer Res. Clin. Oncol., 123, 124–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Chi,T., Xue,Y., Kuo,A., Zhou,S. and Crabtree,G. (1998) Architectural DNA binding by a high-mobility-group/kinesin-like subunit in mammalian SWI/SNF-related complexes. Proc. Natl Acad. Sci. USA, 95, 492–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Furneaux,H., Cheng,H., Caldwell,M.C., Hutter,D., Liu,Y., Holbrook,N. and Gorospe,M. (2000) HuR regulates p21 mRNA stabilization by ultraviolet light. Mol. Cell. Biol., 20, 760–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodnar-Filipowicz A. and Moroni,C. (1990) Regulation of interleukin 3 mRNA expression in mast cells occurs at the posttranscriptional level and is mediated by calcium ions. Proc. Natl Acad. Sci. USA, 87, 777–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N., Chen,C.-Y.A. and Shyu,A.-B. (1997) Modulation of the fate of cytoplasmic mRNA by AU-rich elements: key sequence features controlling mRNA deadenylation and decay. Mol. Cell. Biol., 17, 4611–4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Wagner,B.J., Ehrenman,K., Schaefer,A.W., DeMaria,C.T., Crater,D., DeHaven,K., Long,L. and Brewer,G. (1993) Purification, characterization and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol. Cell. Biol., 13, 7652–7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwicker J., Lucibello,F.C., Wolfraim,L.A., Gross,C., Truss,M., Engeland,K. and Muller,R. (1995) Cell cycle regulation of the cyclin A, cdc25C and cdc2 genes is based on a common mechanism of transcriptional repression. EMBO J., 14, 4514–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]