Abstract
Pythiosis is a rare infectious disease caused by Pythium insidiosum, which typically occurs in tropical and subtropical regions. The high mortality rate may be in consequence of the lack of diagnosis. The objective of this study was to evaluate reliability of a new single-tube nested PCR for detection of P. insidiosum DNA. A total of 78 clinical isolates of various fungi and bacteria, 106 clinical specimens and 80 simulated positive blood samples were tested. The developed primer pairs CPL6–CPR8 and YTL1–YTR1 are located on 18S subunit of the rRNA gene of P. insidiosum. The specificity, negative and positive predictive values were 100, 100 and 87.5 %, respectively, as compared with direct microscopy and cultivation. The detection limit of the single-tube nested PCR was 21 zoospores corresponding to 2.7 pg of the DNA. The results demonstrate that the new single-tube nested PCR offers a highly sensitive, specific and rapid genetic method for detecting P. insidiosum.
Keywords: Pythium insidiosum, Pythiosis, Single-tube nested PCR, 18S rRNA
Introduction
Pythiosis is an emerging and life-threatening infectious disease caused by the oomycete Pythium insidiosum, which can develop in humans and animals [1–4]. The infective stage of P. insidiosum is a biflagellate zoospore [1]. Human pythiosis occurs predominantly in tropical areas of the world and particularly in Thailand [1, 5, 6]. There, the first documented case of human pythiosis was reported in 1985 [7]. According to clinical signs, human pythiosis can be divided into four types: cutaneous/subcutaneous, vascular, ocular and disseminated. The first type is characterized by chronic swelling, painful subcutaneous granulomatous infiltration and ulceration, usually located in the face or legs [8]. Chronic arthritis in the lower extremities resulting in arterial occlusion and gangrenous ulceration of feet or legs is typical for vascular pythiosis [9]. The ocular form is usually manifested as corneal ulcers or keratitis. As a result of all these forms of infection, P. insidiosum can spread via the bloodstream to various internal organs or organ systems such as the gastrointestinal tract, brain, liver, kidney or rhinosinus [10]. Currently, the diagnosis of pythiosis is based on microscopy, culture, detection of antibodies and molecular genetic techniques [9, 11–13]. However, microscopy cannot distinguish zygomycetes because of the coenocytic form of the mycelium [7]. Culture is time-consuming, and obtaining infected tissue samples may be difficult [1]. Because of low antibody response, false-negative results frequently occur in serological tests, particularly in ocular pythiosis [11]. Nested PCR has been developed for the diagnosis of pythiosis using the internal transcribed spacer 1 (ITS1) of the gene for rRNA [14]. Although it is highly sensitive, the main problem of this PCR is a high risk of contamination as the product of the first reaction needs to be transferred into another tube for the second reaction. The purpose of this study was to solve this problem by developing a nested PCR for detecting P. insidiosum in a single tube and to evaluate its reliability using various clinical specimens, including simulated positive blood samples and clinical isolates of bacteria and fungi.
Materials and Methods
Clinical Isolates
The study comprised 34 isolates of P. insidiosum as specified in Table 1, 29 fungal isolates (Aspergillus fumigatus, Aspergillus flavus, Aspergillus niger, Basidiobolus ranarum, Candida albicans, Candida tropicalis, Cladophialophora carrionii, Curvularia spp., Exophiala jeanselmei, Filobasidiella neoformans, Fusarium spp., Hortaea werneckii, Lichtheimia spp., Microsporum gypseum, Mucor spp., Penicillium spp., Rhodotorula spp., Rhizopus spp., Saccharomyces cerevisiae, Scedosporium apiospermum, Syncephalastrum spp., Talaromyces marneffei, Trichophyton spp., Trichophyton concentricum, Trichophyton rubrum, Trichophyton schoenleinii, Trichophyton tonsurans, Trichophyton violaceum and Trichosporon spp.), 10 bacterial isolates (Burkholderia pseudomallei, Corynebacterium spp., Enterococcus spp., Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Salmonella typhi, Staphylococcus aureus, Streptococcus pyogenes and Streptococcus viridans) and 5 isolates of non-insidiosum Pythium spp. (P. aphanidermatum, P. deliense, P. grandisporangium, P. middletonii and P. ultimum). All microorganisms were obtained from patients of the Siriraj Hospital, Mahidol University, Bangkok, Thailand. All isolates were identified based on conventional microbiological methods and then stored in skim milk (Oxoid, UK) 100 mg/ml with 33 % glycerol at −20 °C.
Table 1.
Sources of P. insidiosum strains
| Reference no. | Clinical form of pythiosis | Patient information | ||
|---|---|---|---|---|
| Age/sex | Years | Residential province (Thailand) | ||
| 1. CBS 673.85 | Cutaneous | 23/M | 1985 | NA |
| 2. MCC 1 | Ocular | 20/M | 1989 | Samut Prakan |
| 3. MCC 2 | Ocular | 33/F | 1992 | Chai Nat |
| 4. MCC 3 | Cutaneous | 40/M | 1991 | Phichit |
| 5. MCC 4 | Vascular | 25/M | 1986 | Suphan Buri |
| 6. MCC 5 | Vascular | 49/F | 1993 | Bangkok |
| 7. MCC 6 | Ocular | NA | NA | Ayutthaya |
| 8. MCC 7 | Ocular | 62/M | NA | Ayutthaya |
| 9. MCC 8 | Ocular | 62/M | NA | Narathiwat |
| 10. MCC 9 | Vascular | 46/F | 1986 | Phichit |
| 11. MCC 10 | Disseminated | 12/M | 2000 | Saraburi |
| 12. MCC 11 | Ocular | 48/F | NA | Chon Buri |
| 13. MCC 29 | Disseminated | NA | NA | Ratchaburi |
| 14. SIMI 10201 | Ocular | 23/F | 1989 | Phatthalung |
| 15. SIMI 149-41 | Vascular | 14/M | 1988 | Lop Buri |
| 16. SIMI 16068 | Ocular | 26/F | 1994 | NA |
| 17. SIMI 1839-46 | Vascular | 19/F | 2003 | Nakhon Pathom |
| 18. SIMI 18093 | Ocular | 50/F | 1995 | Samut Sakhon |
| 19. SIMI 240-37 | Ocular | NA/M | 1994 | Samut Sakhon |
| 20. SIMI 283-40 | Ocular | 58/F | 1997 | Bangkok |
| 21. SIMI 2921-45 | Ocular | 75/F | 2002 | Ratchaburi |
| 22. SIMI 2989-42 | Vascular | 72/F | 1999 | Suphan Buri |
| 23. SIMI 322-37 | Ocular | 26/M | 1994 | NA |
| 24. SIMI 3306-44 | Ocular | NA/F | 2001 | Nakhon Si Thammarat |
| 25. SIMI 348-37 | Ocular | 36/M | 1994 | Bangkok |
| 26. SIMI 4523-45 | Ocular (corneal transplantation) | 37/M | 2002 | Pathum Thani |
| 27. SIMI 4763 | Cutaneous | 40/M | 2001 | Ratchaburi |
| 28. SIMI 6666 | Ocular | 42/M | 1986 | Kamphaeng Phet |
| 29. SIMI 7873 | Vascular | 31/F | 1988 | Chanthaburi |
| 30. SIMI 7874 | Vascular | 31/F | 1988 | Chanthaburi |
| 31. SIMI 8659 | Vascular | 52/M | 1988 | Suphan Buri |
| 32. SIMI 8727 | Vascular | 19/M | 1988 | Yasothon |
| 33. SIMI 9642 | Ocular | 53/F | 1989 | Yasothon |
| 34. SIMI 9743 | Ocular | 53/F | 1989 | Yasothon |
CBS Centralbureau voor Schimmelcultures, Utrecht, The Netherlands; MCC mycology culture collection at the Department of Microbiology, Mahidol University; SIMI microbial culture collection at the Siriraj Hospital Microbiology Laboratory, Mahidol University, Bangkok, Thailand; NA data not available; M male; F female
Clinical Specimens
One hundred and six clinical specimens from patients with suspected fungal infection obtained from a routine mycology laboratory in the Srinagarind Hospital, Khon Kaen University, Khon Kaen, Thailand were evaluated prospectively from May 2011 to February 2012. They included pus (n = 27), tissue biopsies (n = 19), blood (n = 10), bone marrow (n = 9), lymph nodes (n = 6), peritoneal dialysis fluid (n = 6), sputum (n = 5), bronchial washing (n = 5), tracheal secretion (n = 5), urine (n = 5), cerebrospinal fluid (n = 4), synovial fluid (n = 2), ascitic fluid (n = 2) and pleural fluid (n = 1). In addition, 80 simulated positive blood specimens were prepared from normal blood samples. For this purpose, each blood sample was mixed with zoospores of P. insidiosum so that each sample contained 1.15 × 106 zoospores ml/l.
Evaluation by Phenotypic Methods
All clinical specimens were evaluated microscopically in 20 % potassium hydroxide preparation for the presence of P. insidiosum hyphae. Culture was performed, with each specimen being inoculated on two Sabouraud dextrose agars (SDA; Oxoid, UK), two Mycosel agars (MCA; BD Diagnostics) and one blood agar (Oxoid, UK) for detection of P. insidiosum growth. One SDA and MCA each were incubated at 25 °C and the other media at 37 °C. All agars were evaluated for the P. insidiosum growth until 30 days. The suspected colonies were identified as P. insidiosum by induction of zoospores [15]. Results of these phenotypic methods were then compared with a single-tube nested PCR for sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV).
DNA Extraction for the PCR
DNA from all clinical and simulated positive specimens was extracted with the NucleoSpin Tissue kit (Macherey–Nagel, Germany) and QIAamp DNA Mini Kit (Qiagen) according to the manufacturers’ instructions. All fungal isolates were cultured in 250 ml of Sabouraud dextrose broth (Oxoid, UK) and incubated at a room temperature for 7 days with shaking (150 rpm) in a rotary shaker (PSU 2T plus, BioSan, Latvia). Fungal mycelia were filtered, washed twice with deionized water and frozen at −20 °C until used. Bacterial isolates were cultured in 3 ml of Luria broth (Oxoid, UK) and incubated at 37 °C for 16–18 h with shaking (200 rpm). Then, cultures were transferred to a 1.5 ml microtube, centrifuged and stored at 4 °C. For cell disruption, approximately 30 mg of frozen fungal mycelia and 0.14–1.21 g of the bacterial pellet were rubbed in liquid nitrogen until a fine powder. The bacterial powder was then suspended in a lysis buffer (25 mM Tris–HCl pH 8, 10 mM EDTA pH 8, 100 mM NaCl). DNA was extracted from the bacterial suspensions and powders from fungal mycelia as described by Sambrook and Russell [16]. Purity of isolated DNA was calculated using a spectrophotometer at OD 260 and 280.
Single-Tube Nested PCR
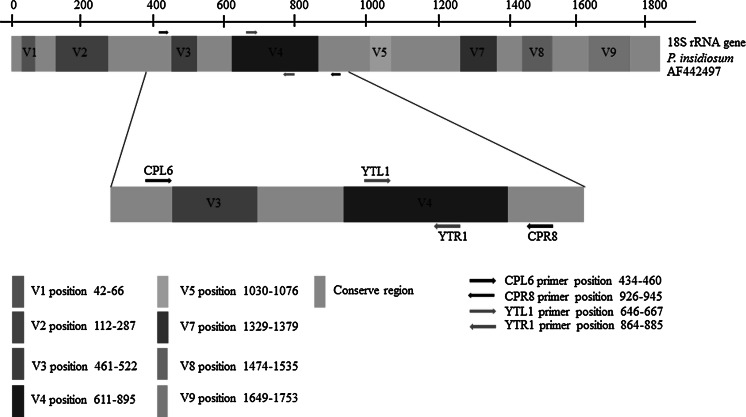
The single-tube nested PCR was performed using outer primers CPL6 (5′-GAC ACA GGG AGG TAG TGA CAA TAA ATA-3′) and CPR8 (5′-CTT GGT AAA TGC TTT CGC CT-3′), and inner primers YTL1 (5′-CTT TGA GTG TGT TGC TAG GAT G-3′) and YTR1 (5′-CTG GAA TAT GAA TAC CCC CAA C-3′) designed by ourselves using the GenBank database. The primers are located on the 18S subunit of the rRNA gene of P. insidiosum (GenBank accession no. AF442497). The position of the newly designed primer is shown in Fig. 1. The reaction mixture in a total volume of 50 μl contained 1 U of Platinum Taq DNA polymerase (Invitrogen), 0.2 mM MgCl2, 25 pmol of each primers and 0.2 mM dNTPs. Amplification was performed in a thermal cycler (Mastercycler Personal, Eppendorf International) using the following temperature cycles: initial denaturation at 95 °C for 5 min, followed by 30 cycles (denaturation at 95 °C for 1 min, annealing at 68 °C for 1 min and extension at 72 °C for 1 min), another 30 cycles (denaturation at 95 °C for 1 min, annealing at 57 °C for 1 min and extension at 72 °C for 1 min) and final extension for 10 min at 72 °C. Amplicons were visualized under UV light using a manual documentation system (InGenius3, Syngene, USA) after electrophoresis of 10 μl of the reaction solution in 2 % UltraClean agarose gel (Mo Bio Laboratories) containing the SYBR Gold nucleic acid gel stain (Invitrogen).
Fig. 1.
Position of the annealing sites of the primer pairs CPL6–CPR8 and YTL1–YTR1 in the 18S rRNA gene of P. insidiosum
Evaluation of PCR Sensitivity
To determine the lowest detection limit of our nested PCR, seven 10-fold dilutions were prepared from simulated positive blood specimen. Then, 180 μl of each dilution containing 2.07 × 105–2.07 × 10−1 zoospores was used for nested PCR. As a control, DNA of P. insidiosum hyphae was also prepared as 10-fold serial dilution from 1 ng to 1 fg, and then, it was also amplified by a single-tube nested PCR. This study was approved by the Ethical Committee on Human Experimentation in Khon Kaen University in accordance with the Declaration of Helsinki (HE 541111).
Results
If P. insidiosum DNA was presented in clinical material, four amplicons sized 512, 452, 340 and 240 bp were detected by our nested PCR.
Evaluation of PCR Specificity Within Clinical Isolates
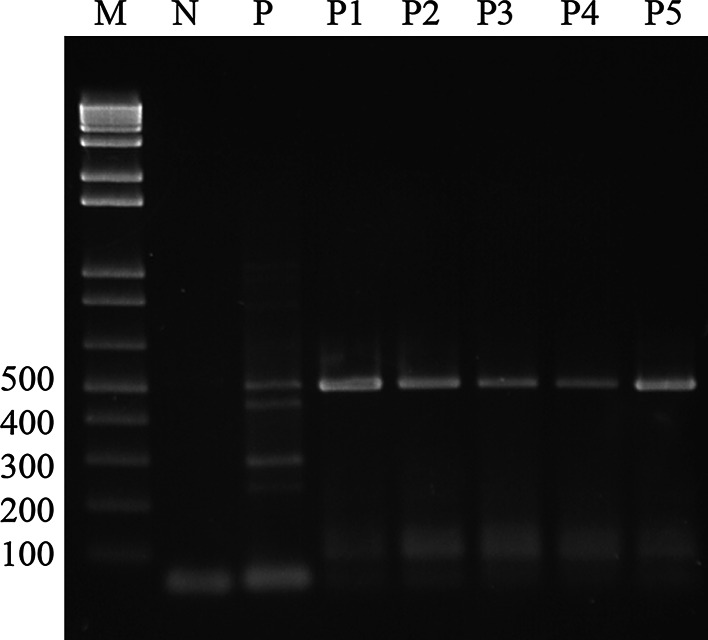
Four amplicons were detected in all 34 P. insidiosum isolates. Only one genus-specific product (512 bp) was found in five non-insidiosum Pythium spp. as demonstrated in Fig. 2. No product was found after amplification of DNA from all the remaining 29 fungal and 10 bacterial isolates. Therefore, our single-tube nested PCR had 100 % specificity within the group of clinical microorganisms tested in this study.
Fig. 2.
Specificity of the single-tube nested PCR for amplification of DNA from Pythium spp. Lane M 100 bp DNA ladder, lane N negative control, lane P P. insidiosum, lane P1 P. middletonii, lane P2 P. ultimum, lane P3 P. aphanidermatum, lane P4 P. deliense, lane P5 P. grandisporangium
Evaluation of PCR Specificity in Clinical Specimens
Based on positivity of phenotypic methods, seven pus specimens from corneal ulcers were evaluated as positive within all tested clinical specimens. From that, three specimens were microscopically negative; culture and zoospore induction were positive in all seven specimens. Moreover, as demonstrated in Table 2, one more pus specimen, also from a corneal ulcer, which was negative in both phenotypic methods, was positive in our single-tube nested PCR.
Table 2.
Comparison of P. insidiosum detection in positive clinical specimens by phenotypic methods and a single-tube nested PCR
| Specimen no. | Phenotypic method | Genotypic method | |
|---|---|---|---|
| Microscopy | Culture and zoospore induction | Single-tube nested PCR | |
| 1 | + | + | + |
| 2 | + | + | + |
| 3 | + | + | + |
| 4 | − | + | + |
| 5 | − | − | + |
| 6 | + | + | + |
| 7 | − | + | + |
| 8 | − | + | + |
All specimens were pus from corneal ulcers
Evaluation of PCR Sensitivity
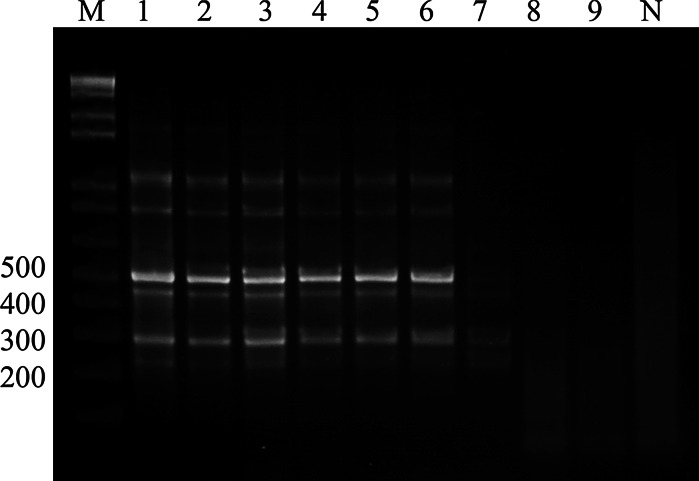
As shown in Fig. 3, the lowest detection limit in diluted simulated positive blood specimen was 21 zoospores, which corresponded to 2.7 pg of DNA. The lowest detection limit of serially diluted DNA isolated from P. insidiosum hyphae was 1 pg.
Fig. 3.
Sensitivity of the single-tube nested PCR for detection of P. insidiosum DNA from zoospores. Lane M 100 bp DNA ladder, lane N negative control, lane 1 P. insidiosum DNA (positive control), lane 2 2.07 × 105 zoospores in double distilled water (DDW), lanes 3–9 dilution of simulated positive blood specimen in DDW (lane 3 2.07 × 105 zoospores, lane 4 2.07 × 104 zoospores, lane 5 2.07 × 103 zoospores, lane 6 2.07 × 102 zoospores, lane 7 2.07 × 101 zoospores, lane 8 2.07 zoospores, lane 9 2.07 × 10−1 zoospores)
Evaluation of PPV and NPV of PCR in Clinical Specimens
These results are summarized in Table 3. Based on results of phenotypic methods, 7 specimens were evaluated as positive for P. insidiosum. Using our single-tube nested PCR, DNA of P. insidiosum was detected in 8 specimens, whereas 7 of them were positive by phenotypic methods. Therefore, NPV and PPV were 100 and 87.5 %, respectively.
Table 3.
Comparison of phenotypic test results with a single-tube nested PCR
| Phenotypic tests (microscopy in 20 % KOH and culture) | Total | ||
|---|---|---|---|
| Positive | Negative | ||
| Single-tube nested PCR | |||
| Positive | 7 | 1 | 8 |
| Negative | 0 | 98 | 98 |
| Total | 7 | 99 | 106 |
Discussion
In our set of clinical specimens, one patient was found to be positive in single-tube nested PCR, but negative in phenotypic tests. In our study, the criterion for detecting pythiosis was positivity in one of two phenotypic methods. However, Krajaejun et al. [9] defined the criteria for pythiosis as culture positivity, seropositivity or clinical manifestation. The patient who had negative results of phenotypic tests and was positive in nested PCR had symptoms of ocular pythiosis evaluated by a clinician. The reason why phenotypic methods could not confirm pythiosis in this case was probably their low sensitivity [17]. So, this finding demonstrates the superiority of PCR over phenotypic methods from the sensitivity point of view. The detection limit of our nested PCR in this study (2.7 pg of DNA in relation to 21 counted zoospores) allows us to suppose that one zoospore contains 128.57 fg of DNA. However, our experiments showed that the lowest detected amount of DNA from P. insidiosum isolated from its mycelium was 1 pg of DNA, corresponding to about eight zoospores. According to this detection limit, one zoospore could contain 48–129 fg of DNA. This is the first report which tried to estimate the amount of DNA in P. insidiosum zoospores. Hussain et al. [18] reported a detection limit of PCR for Phytophthora infestans of 0.5 pg, which corresponded to four zoospores; in that case, one zoospore should contain 125 fg of DNA. The DNA content is similar to data about plant pathogenic organisms from the order Peronosporales (Oomycota) ranging from 46 to 163 fg [19]. The newly designed, genus-specific outer set of primers CPL6 and CPR8 amplified a 512 bp fragment from a conserved part of the 18S subunit of the rRNA gene and species-specific inner primers YTL1 and YTR1 amplified a 240 bp fragment from variable region of this subunit were firstly evaluated in this study. Although finally four PCR products were seen in a single-tube nested PCR, the test showed excellent sensitivity and specificity. The fragment of 452 bp was the product of CPL6 and YTR1 primers, and the 300 bp fragment was amplified by the YTL1 and CPR8 primers. Four PCR products were detected in all 34 strains of P. insidiosum and in clinical samples from patients with pythiosis. A sensitivity of 100 % and a low detection limit allow us to recommend our single-tube nested PCR as a useful tool for laboratories, which need to detect P. insidiosum in clinical material. Pythiosis is a rare but fatal infection in humans. Krajaejun et al. [8] reported 102 cases of human pythiosis in Thailand from 1985 to 2003. The zoospore is the infective stage of P. insidiosum; therefore, the selected spiking of blood with zoospores simulates the state in really infected blood better than spiking it with pure DNA. The sensitivity of our nested PCR tested on 78 various clinical isolates and 186 clinical specimens including simulated positive blood samples was higher than with other tests [20–22]. In addition, nested PCR solves the main drawbacks of other methods for detecting pythiosis. It overcomes problems with culture and allows identification in a substantially shorter time directly in clinical specimens. Thus, the prognosis of patients can be improved because treatment can be started earlier. Moreover, performing nested PCR in a single tube markedly reduces the risk of cross-contamination. Results of this study demonstrate that our single-tube nested PCR is suitable for detecting P. insidiosum directly from clinical specimens and identification of clinical isolates as well. As this method is rapid, highly sensitive and has minimal risk of cross-contamination, it has a great potential in the diagnosis of suspected cases of pythiosis.
Acknowledgments
This work was supported by the Thailand Research Fund (TRF) through the Royal Golden Jubilee Ph.D. Program (Grant No. PHD/0071/2551), DBG 5080010 TRF Grant and by the Internal Grant Agency of Palacky University Olomouc (Grant No. LF_2012_006). The authors are grateful to the Centre for Research and Development of Medical Diagnostic Laboratories, Faculty of Associated Medical Sciences, Khon Kaen University, Khon Kaen, Thailand, for provided facilities. We also thank Professor Richard W. Titball for invaluable advice in the preparation of this manuscript.
Conflict of interest
The authors declare no conflicts of interest.
References
- 1.Mendoza L, Ajello L, McGinnis MR. Infection caused by the oomycetous pathogen Pythium insidiosum. J Mycol Med. 1996;6:151–164. [Google Scholar]
- 2.Miller RI. Gastrointestinal phycomycosis in 63 dogs. J Am Vet Med Assoc. 1985;186:473–478. [PubMed] [Google Scholar]
- 3.Miller RI, Olcott BM, Archer M. Cutaneous pythiosis in beef calves. J Am Vet Med Assoc. 1985;186:984–986. [PubMed] [Google Scholar]
- 4.Mendoza L, Alfaro AA. Equine pythiosis in Costa Rica: report of 39 cases. Mycopathologia. 1986;94:123–129. doi: 10.1007/BF00437377. [DOI] [PubMed] [Google Scholar]
- 5.De Cock AW, Mendoza L, Padhye AA, Ajello L, Kaufman L. Pythium insidiosum sp. nov., the etiologic agent of pythiosis. J Clin Microbiol. 1987;25:344–349. doi: 10.1128/jcm.25.2.344-349.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaufman L. Penicilliosis marneffei and pythiosis: emerging tropical diseases. Mycopathologia. 1998;143:3–7. doi: 10.1023/A:1006958027581. [DOI] [PubMed] [Google Scholar]
- 7.Thianprasit M, Chaiprasert A, Imwidthaya P. Human pythiosis. Curr Top Med Mycol. 1996;7:43–54. [PubMed] [Google Scholar]
- 8.Krajaejun T, Sathapatayavongs B, Pracharktam R, et al. Clinical and epidemiological analyses of human pythiosis in Thailand. Clin Infect Dis. 2006;43:569–576. doi: 10.1086/506353. [DOI] [PubMed] [Google Scholar]
- 9.Krajaejun T, Imkhieo S, Intaramat A, Ratanabanangkoon K. Development of an immunochromatographic test for rapid serodiagnosis of human pythiosis. Clin Vaccine Immunol. 2009;16:506–509. doi: 10.1128/CVI.00276-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krajaejun T, Sathapatayavongs B, Chaiprasert A, Srimuang S. Do you know human pythiosis? J Infect Dis Antimicrob Agents. 2008;25:45–51. [Google Scholar]
- 11.Krajaejun T, Pracharktam R, Wongwaisayawan S, et al. Ocular pythiosis: is it under-diagnosed? Am J Ophthalmol. 2004;137:370–372. doi: 10.1016/S0002-9394(03)00908-5. [DOI] [PubMed] [Google Scholar]
- 12.Jindayok T, Piromsontikorn S, Srimuang S, Khupulsup K, Krajaejun T. Hemagglutination test for rapid serodiagnosis of human pythiosis. Clin Vaccine Immunol. 2009;16:1047–1051. doi: 10.1128/CVI.00113-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pannanusorn S, Chaiprasert A, Prariyachatigul C, et al. Random amplified polymorphic DNA typing and phylogeny of Pythium insidiosum clinical isolates in Thailand. Southeast Asian J Trop Med Public Health. 2007;38:383–391. [PubMed] [Google Scholar]
- 14.Grooters AM, Gee MK. Development of a nested polymerase chain reaction assay for the detection and identification of Pythium insidiosum. J Vet Intern Med. 2002;16:147–152. doi: 10.1892/0891-6640(2002)016<0147:doanpc>2.3.co;2. [DOI] [PubMed] [Google Scholar]
- 15.Chaiprasert A, Samerpitak K, Wanachiwanawin W, Thasnakorn P. Induction of zoospore formation in Thai isolates of Pythium insidiosum. Mycoses. 1990;33:317–323. doi: 10.1111/myc.1990.33.6.317. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook J, Russell D. Molecular cloning: a laboratory manual. 3. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 17.Garg J, Tilak R, Garg A, et al. Rapid detection of dermatophytes from skin and hair. BMC Res Notes. 2009;18:60. doi: 10.1186/1756-0500-2-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hussain S, Lees AK, Duncan JM, Cooke DEL. Development of a species-specific and sensitive detection assay for Phytophthora infestans and its application for monitoring of inoculum in tubers and soil. Plant Pathol. 2005;54:373–382. doi: 10.1111/j.1365-3059.2005.01175.x. [DOI] [Google Scholar]
- 19.Voglmayr H, Greilhuber J. Genome size determination in peronosporales (Oomycota) by Feulgen image analysis. Fungal Genet Biol. 1998;25:181–195. doi: 10.1006/fgbi.1998.1097. [DOI] [PubMed] [Google Scholar]
- 20.Botton SA, Pereira DI, Costa MM, et al. Identification of Pythium insidiosum by nested PCR in cutaneous lesions of Brazilian horses and rabbits. Curr Microbiol. 2011;62:1225–1229. doi: 10.1007/s00284-010-9781-4. [DOI] [PubMed] [Google Scholar]
- 21.Laohapensang K, Rerkasem K, Supabandhu J, Vanittanakom N. Necrotizing arteritis due to emerging Pythium insidiosum infection in patients with thalassemia: rapid diagnosis with PCR and serological tests—case reports. Int J Angiol. 2005;14:123–128. doi: 10.1007/s00547-005-2012-3. [DOI] [Google Scholar]
- 22.Vanittanakom N, Supabandhu J, Khamwan C, et al. Identification of emerging human-pathogenic Pythium insidiosum by serological and molecular assay-based methods. J Clin Microbiol. 2004;42:3970–3974. doi: 10.1128/JCM.42.9.3970-3974.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]