Abstract
We show that the 3′ boundary of the chicken β-globin locus bears striking structural similarities to the 5′ boundary. In erythroid cells a clear transition in DNase I sensitivity of chromatin at the 3′ end of the locus is observed, the location of this transition is marked by a constitutive DNase I hypersensitive site (HS), and DNA spanning this site has the enhancer-blocking capacity of an insulator. This HS contains a binding site for the transcription factor CTCF. As in the case of the 5′ insulator, the CTCF site is both necessary and sufficient for the enhancer-blocking activity of the 3′ boundary. The position of this insulator is consistent with our proposal that it may function to maintain the distinct regulatory programs of the globin genes and their closely appended 3′ neighbor, an odorant receptor gene. We conclude that both boundaries of the chicken β-globin domain are capable of playing functionally similar roles and that the same protein is a necessary component of the molecular mechanism through which these boundaries are defined.
Keywords: chromatin domains/CTCF/globin genes/insulators/odorant receptors
Introduction
The formation of independent functional domains on eukaryotic chromosomes involves specific modifications of chromatin structure. Cytological studies of Drosophila polytene chromosomes distinguish adjacent heterochromatic and euchromatic regions (Bossy et al., 1984; Rykowski et al., 1988; Friedman et al., 1991). At the molecular level, transcriptionally active genes and gene clusters are characterized by higher sensitivity to the action of nucleases (Weintraub and Groudine, 1976; Lawson et al., 1982; Jantzen et al., 1986; Farache et al., 1990) and by higher levels of core histone acetylation relative to the surrounding ‘inactive’ chromatin (Hebbes et al., 1988, 1994; O’Neill and Turner, 1995; Grunstein, 1997). The existence of active and inactive functionally distinct chromatin domains adjacent to each other raises the question of how they are kept distinct. It is known that certain DNA sequences, acting through the specific proteins bound to them, have properties that might make them suitable boundary elements. For example, the scs and scs′ elements (reviewed in Bell and Felsenfeld, 1999) are localized at the proximal and distal ends of the 87A7 heat shock locus in Drosophila. They are capable of blocking activation of a reporter gene by an enhancer when placed between enhancer and promoter, and they also protect against position effects when they surround a reporter gene (Kellum and Schedl, 1991, 1992). The gypsy retrotransposon, also found in Drosophila, has similar enhancer-blocking and position effect suppressing activities.
In erythroid cells, the chicken β-globin genes are embedded in a 33 kb chromatin domain that is sensitive to DNase I and contains hyperacetylated histones (Hebbes et al., 1988; Figure 1A). The 5′ boundary of this domain has been determined precisely and is closely associated with a ‘constitutive’ DNase I hypersensitive site (HS) (5′HS4) present in every cell type and tissue that has been examined (Reitman and Felsenfeld, 1990; Hebbes et al., 1994; Bell et al., 1999). A DNA fragment containing this HS has the properties of an insulator: it blocks enhancer action directionally and it protects a reporter gene against position effects (Chung et al., 1993, 1997; Pikaart et al., 1998; Bell et al., 1999; Recillas-Targa et al., 1999). Furthermore, we have shown that the enhancer-blocking activity of this insulator requires the DNA-binding protein CTCF (Bell et al., 1999).
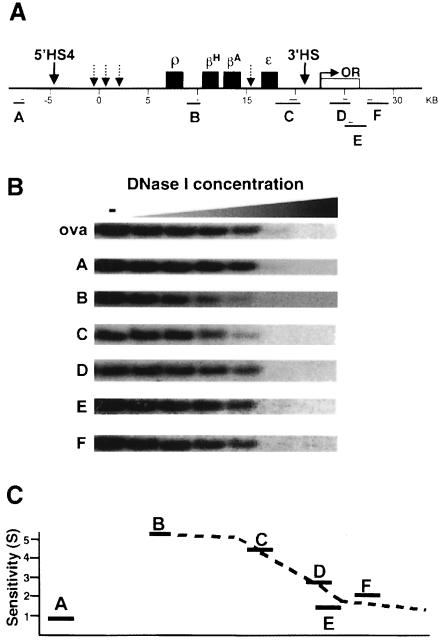
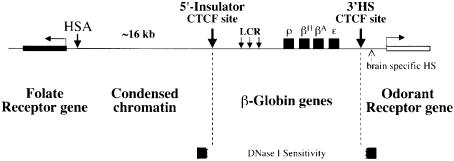
Fig. 1. A transition in DNase I sensitivity defines the 3′ boundary of the chicken β-globin domain. The 3′ boundary of generalized DNase I sensitivity is located between regions C and D. (A) Structure of the chicken β-globin locus is shown on the top. The four globin genes are indicated with black boxes. Selected HSs are shown by arrows. An open reading frame encoding the chicken odorant receptor protein (COR3′β) described in this study (Bulger et al., 1999) is shown as a white box marked ‘OR’. 5′HS4, the most upstream constitutive HS, marks the 5′ chromatin boundary and functions as a chromatin insulator. Another constitutive HS, 3′HS, was identified in this study. Other erythroid-specific HSs are shown by arrows with dotted lines. Numbers immediately below the map are map units based on the arbitrary numbering system set (Villeponteau and Martinson, 1981). Restriction fragments A–F detected in DNase I sensitivity assays in (B) are shown below the map. Probes used are indicated as thin lines. A detailed description of DNA fragments A–F and probes is given in Materials and methods. (B) Generalized DNase I sensitivity of DNA fragments A–F visualized by Southern blot hybridization. Erythrocyte nuclei isolated from 11-day-old chick embryos were treated with increasing amounts of DNase I (from right to left lanes, 0, 0.2, 0.4, 0.6, 1.0, 2.0 and 5.0 U/ml). Restriction fragments A–F were detected by Southern blot hybridizations. Relative sensitivities to DNase I correspond to the extent of loss of signal intensities of each band. DNA fragments B and C are relatively sensitive to DNase I, while fragments D–F are resistant to DNase I. A DNA fragment derived from the ovalbumin gene, which is transcriptionally inactive in this cell type, and fragment A, which is located farther upstream of the 5′ chromatin boundary, were used as DNase I resistant controls. (C) Quantitative representation of the DNase I sensitivities along the β-globin locus. Normalized DNase I sensitivity values (S) were calculated from the following equation (Pikaart et al., 1992) and plotted on a graph: S = log (GD/GU)/log (OD/OU) × T, where G and O are β-globin and ovalbumin band intensities for the undigested (U) or digested (D) samples and T is the size ratio of the ovalbumin to globin fragments. DNase I sensitivity drops significantly between C and D.
These results are consistent with the idea that the 5′HS4 insulator may help to establish and maintain the 5′ boundary of the globin domain. The need for such an element is further suggested by the presence, only 16 kb upstream of 5′HS4, of a folate receptor gene with a developmental program of expression that is distinct from that of the globin genes (Prioleau et al., 1999). We have proposed that one role of the 5′ boundary is to prevent cross-talk between the regulatory elements of these genes (Bell et al., 1999; Prioleau et al., 1999).
The existence of a 5′ boundary element immediately raised questions about the location and properties of a 3′ boundary. We have therefore examined in detail the 3′ end of the chicken β-globin domain, and find striking structural similarities with the 5′ end, although the two ends lack any long stretch of nucleotide sequence identity. A transition in the relative sensitivity of the chromatin to DNase I at the 3′ boundary is marked by a constitutive HS. Its position maps precisely to a binding site for CTCF and a DNA fragment containing this site has directional enhancer-blocking activity, which depends upon its ability to bind CTCF. Only 2 kb downstream of the 3′ boundary lies a gene that encodes an odorant receptor (OR) (Bulger et al., 1999; this paper). We suggest that the enhancer-blocking activity of this boundary is a necessary component of the mechanism that maintains the distinct regulatory programs of these neighboring genes. Furthermore, the presence of a second element at the 3′ end of the β-globin locus with such similarity to the one at the 5′ end is strong evidence that both these elements are involved with the establishment or maintenance of chromatin domain boundaries.
Results
Chromatin sensitivity to DNase I defines the 3′ chicken β-globin domain boundary
In order to identify the 3′ chromatin boundary of the chicken β-globin locus, we first surveyed the general DNase I sensitivity of a 14 kb region downstream of the embryonic ε-globin gene (Figure 1A). In these experiments, nuclei isolated from the red blood cells (RBCs) of 11-day-old chick embryos were incubated with increasing concentrations of DNase I for a fixed amount of time. The rate of disappearance of individual restriction fragments with increasing DNase I concentration served as a measure of the accessibility of the chromatin in that region to the enzyme (Weintraub and Groudine, 1976; Lawson et al., 1982; Jantzen et al., 1986; Farache et al., 1990). We found several considerations to be essential in accurate assessment of the relative sensitivities of these fragments. First, we carefully eliminated from our analysis any DNA fragments derived from regions containing sites that are hypersensitive to DNase I digestion (data not shown). Secondly, since the kinetics of digestion are dependent upon the length of the fragment examined, to avoid making large corrections for size, we chose restriction fragments of roughly equivalent length. A representative example of the data obtained by Southern blotting of such fragments is shown in Figure 1B. DNA fragments B and C are digested away with a lower concentration of DNase I relative to fragments D, E and F. When these results are quantified and normalized for length (see analysis in Figure 1 legend), a clear transition in the general DNase I sensitivity in this region is apparent (Figure 1C). The 3′ end of fragment C is 4.8 kb downstream of the ε-gene, while the 5′ end of fragment D is 7.0 kb downstream of the ε-gene. Thus, these results identify a 3′ chromatin boundary located between 4.8 and 7.0 kb downstream of the ε-gene. This position is consistent with the 3′ boundary determined previously at lower resolution by both general DNase I sensitivity and histone acetylation pattern (Hebbes et al., 1994).
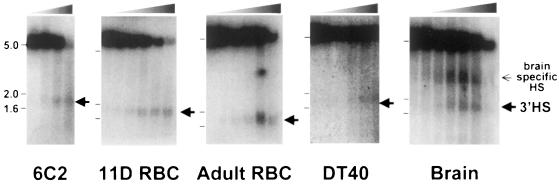
A further examination of the DNase I sensitivity in the region defined above revealed an HS (not present in the fragments examined in Figure 1) that we designated as 3′HS. This site appears to be constitutive: it is present in all of the cell types we tested, including erythroid cells at early and late stages of erythropoiesis (6C2 cells, 11-day-old embryonic and adult RBCs) and non-erythroid cells such as DT40 and embryonic brain cells (Figure 2). No additional HSs were observed within a 10 kb region downstream of the 3′HS in RBCs from 11-day-old chick embryos.
Fig. 2. A constitutive hypersensitive site (3′HS) within the 3′ chromatin boundary. Nuclei from various types of chicken cells were treated with increasing amounts of DNase I (from left to right lanes: 0, 0.06, 0.1, 0.2, 0.4 and 0.6 U/ml for RBCs and brain nuclei; 0, 8.0, 10, 20, 40 and 60 U/ml for DT40 and 6C2 cell nuclei). Genomic DNA was extracted and digested with KpnI. In addition to a parental fragment, a hypersensitive site, 3′HS, was detected as marked by arrowheads, in all cell types tested. Cells tested are a chicken erythroid precursor derived cell line (6C2), erythroid cells from either 11-day-old embryo (11D RBC) or adult blood (Adult RBC), brain from 11-day-old embryos (Brain) and a lymphoma-cell derived cell line (DT40).
A DNA fragment including the 3′HS hasenhancer-blocking activity
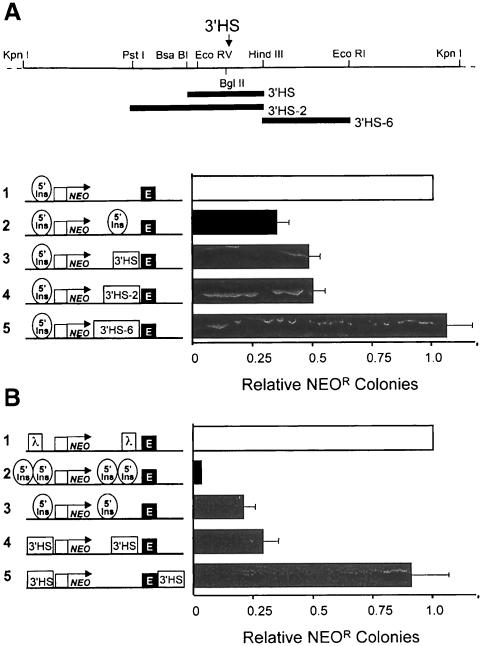
We asked whether a fragment of DNA containing the 3′HS would have one of the functions of an insulator: the capacity to block an enhancer. To examine this, we made use of the same assay for enhancer-blocking activity previously employed to study the 5′HS4 insulator (Chung et al., 1993, 1997; Bell et al., 1999). In these assays, constructs carrying the neomycin resistance gene and an enhancer are stably transformed into the human erythroleukemia line K562; enhancer action is reflected in the number of neomycin-resistant colonies obtained in stably transfected K562 cells. Two DNA fragments spanning the 3′HS and one immediately adjacent to it were tested for enhancer-blocking activity in this assay (Figure 3A). Insertion of either of the fragments that contains the HS resulted in a reduction in colony number relative to controls. This reduction in colony number was roughly equivalent to that obtained with the 5′ insulator and was not attributable to insert size as insertions of either a larger λ DNA fragment or a similarly sized fragment 3′ to the boundary (3′HS-6) had no effect on colony number. The enhancer-blocking activities of the 3′HS and 3′HS-2 fragments were independent of the orientation in which these fragments were inserted (data not shown).
Fig. 3. Directional enhancer-blocking activity of the 3′HS. (A) The human erythroleukemic cell line K562 was stably transfected with the constructs shown on the left. Each construct has the neomycin resistance gene (NEO) driven by a human βA-globin promoter with mouse β-globin HS2 as an enhancer. The DNA fragments 3′HS and 3′HS-2 include the DNase I HS 3′HS. 3′HS-6 does not contain the HS. For each construct, the 1.2 kb chromatin insulator fragment (5′Ins) including the 5′HS4 was placed upstream of the promoter in order to block influence from regulatory elements at the site of integration. The level of expression of each construct was measured as the number of neomycin-resistant colonies. Colony numbers obtained from construct 1, which does not have a DNA fragment between the promoter and the enhancer, were set at 100. Relative numbers of neomycin-resistant colonies are shown in the bar graph. We present the mean of five independent experiments. Enhancer-blocking activity resides in a DNA fragment containing the 3′HS. (B) Enhancer-blocking assays were performed using constructs shown to the left. In construct 1, a 2.3 kb fragment of λ DNA was inserted, as a spacer control, between the enhancer and the reporter. Thick bars show means of at least four independent experiments.
The enhancer-blocking activity of the 5′ insulator is positional, i.e. to block an enhancer the insulator must be inserted between the enhancer and the promoter. As the results in Figure 3B show, in these assays, enhancer blocking by the 3′HS fragment was also positional: when the 3′HS fragment flanks the enhancer and promoter, it has no effect (Figure 3B, construct 5). Thus, the 3′HS blocks enhancers in a directional manner in agreement with the properties of an insulator.
Identification of an adjacent gene
In light of the enhancer-blocking properties of the 5′ boundary, we have proposed that this element may play a necessary role in preventing cross-talk between neighboring genes (Bell et al., 1999; Prioleau et al., 1999). Given the enhancer-blocking activity of the 3′ boundary, we searched for a gene at the 3′ end of the globin locus. As we reported in a recent paper (Bulger et al., 1999), a gene encoding an OR lies only ∼2 kb downstream of the globin 3′HS. This gene, COR3′β, contains a complete open reading frame homologous to known OR proteins. Although most of the members of the OR protein family are expressed in the olfactory sensory epithelium, some are expressed in other tissues such as the cells migrating along the olfactory nerve and tissue adjacent to the neural tube (Nef and Nef, 1997), the taste buds (Abe et al., 1993), developing heart (Drutel et al., 1995), germ line (Parmentier et al., 1992) and brain (Dreyer, 1998; Raming et al., 1998). This pattern of expression is clearly different from that of the globin genes. Consistent with this difference, we discovered a brain-specific HS 0.9 kb downstream of the globin 3′HS (Figure 2). In addition, we were able to detect COR3′β-specific poly(A)+ transcripts in 10-day-old embryonic chick brain (data not shown).
CTCF is required for the enhancer-blocking activity of the 3′HS
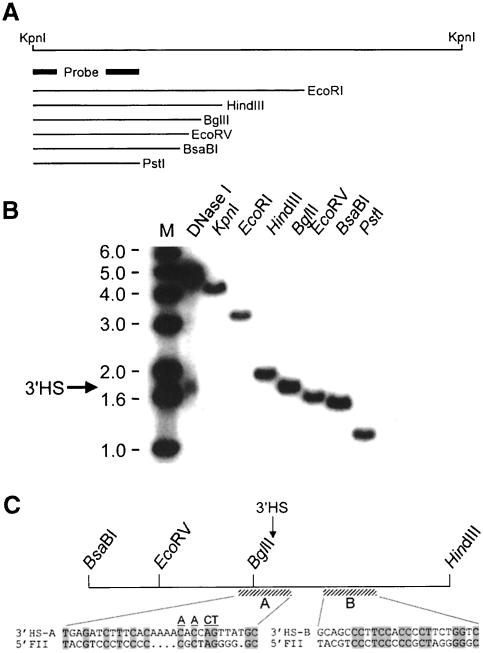
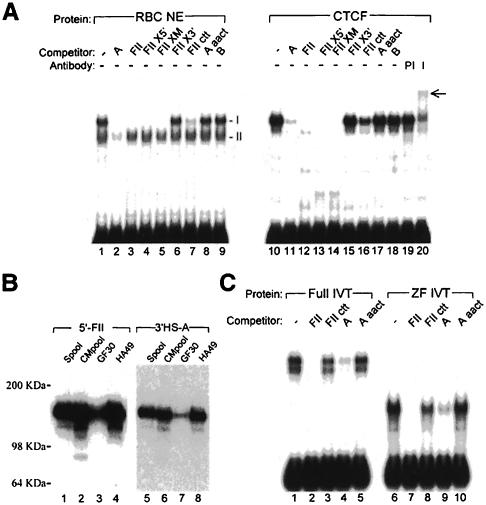
To analyze the chromatin boundary further, we mapped the position of 3′HS. Because of the availability of restriction sites conveniently located near the 3′HS, we were able to map its position with accuracy (Figure 4). The location of the HS was nearly coincident with a BglII site. Although there were no striking similarities between the sequences surrounding the 5′ and 3′ HSs, we found two relatively weak homologies between the CTCF site in the 5′ insulator (5′FII) and the 3′HS region (Figure 4C, 3′HS-A and B). The 3′HS maps directly to the 3′HS-A in this fragment (Figure 4). Although this alignment includes only a 12/30 identity (with a 4 bp insertion), a labeled 100 bp oligonucleotide containing 3′HS-A binds CTCF (Figure 5). With adult chicken RBC nuclear extracts, two slow mobility complexes were observed in a gel-shift assay with 3′HS-A (Figure 5A, lanes 1–9). The upper complex is competed by a 50-fold excess of cold 3′HS-A and 5′FII. Moreover, mutant forms of 5′FII competed to an extent that was directly proportional to their affinity for CTCF (Bell et al., 1999). This complex had an identical mobility to that observed when 5′FII was used as a labeled probe (data not shown). Partially purified CTCF gave an identical migration and competition profile, and this complex was specifically super-shifted with an antibody against CTCF (Figure 5A, lanes 10–20). Furthermore, in a Southwestern assay, both 5′FII and 3′HS-A bound specifically to a protein with an apparent mass of ∼140 kDa that had been enriched through stepwise purification of 5′FII binding protein from adult chicken RBC nuclear extract (Figure 5B). We have demonstrated by peptide sequencing that the protein responsible for this Southwestern activity is CTCF (Bell et al., 1999). Finally, the conclusion that 3′HS-A is a bona fide binding site for CTCF was confirmed by the observation that in vitro translated CTCF, and a peptide spanning its zinc finger domain, bound 3′HS-A with a specificity identical to that observed with native CTCF (Figure 5C).
Fig. 4. Sequences homologous to the 5′ insulator element of the chicken β-globin locus are found at the site of the 3′HS. The position of the 3′HS was measured by the indirect end-labeling method and the strategy is shown in (A). Nuclei from 11-day-old chick embryos were treated with 0.4 U/ml DNase I, from which genomic DNA was extracted and digested with KpnI. In (B), the position of the 3′HS (arrow) was compared with the migration of genomic fragments of known length. The 3′HS hypersensitive fragment co-migrates with a fragment derived from BglII digestion. (C) Sequences homologous to FII from 5′HS4 (Chung et al., 1997; Bell et al., 1999) are found at or close to the sites of 3′HS, 3′HS-A and 3′HS-B, respectively. Alignment of the sequences 3′HS-A and 3′HS-B with the sequences of the 5′FII is shown. Conserved bases are shaded. Bases altered to generate a mutant site are underlined.
Fig. 5. Sequences derived from the 3′HS are bound specifically by CTCF. (A) Gel retardation analysis of complexes formed with the 3′HS-A sequence. Duplex 100mer oligonucleotides containing the 3′HS-A site were incubated with adult chicken RBC nuclear extracts (lanes 1–9) or partially purified CTCF (lanes 10–20; Bell et al., 1999). Two complexes observed following incubation with nuclear extracts are labeled I and II. Complexes were competed with 50-fold excesses of unlabeled competitor oligonucleotides containing either the 3′HS-A (lanes 2 and 11), 5′FII (lanes 3 and 12), FIIX5′-mutant (lanes 4 and 13), FIIXM-mutant (lanes 5 and 14), FIIX3′-mutant (lanes 6 and 15), FII ctt-mutant (lanes 7 and 16; Bell et al., 1999), 3′HS-A aact-mutant (lanes 8 and 17) or 3′HS-B (lanes 9 and 18) sites. Pre-immune antibodies (lane 19) or anti-CTCF antibodies (lane 20) were added to the binding reactions, and supershifted material is indicated by the arrow at the right. (B) Southwestern analysis of oligonucleotide duplexes binding to immobilized fractions of purified CTCF. Fractions of increased CTCF purity were separated by polyacrylamide gel electrophoresis and immobilized on PVDF membrane. The positions of molecular weight markers are indicated. A single gel blot was cut in two and probed with 32P-labeled oligonucleotide duplexes containing the 5′FII (lanes 1–4) and 3′HS-A (lanes 5–8) sites. Following washing, probed blots were exposed to autoradiographic film for 3 min and 2 h, respectively. (C) Gel retardation analysis of complexes formed between the 3′HS sequence and in vitro translated CTCF proteins. Duplex 100mer oligonucleotides containing the 3′HS-A site were incubated with in vitro translated proteins representing the full-length (lanes 1–5) or the zinc finger domain (lanes 6–10) of CTCF. Complexes were competed with 50-fold excesses of unlabeled competitor as indicated.
Within 5′FII, base pairs at the 3′ end of the fragment are essential for CTCF binding (Bell et al., 1999). In that site, a 12 bp transversion at the 3′ end was sufficient to ablate CTCF binding in vitro and enhancer blocking in vivo completely (FIIX3′ in Figure 5). Moreover, nearly all of the binding to CTCF was eliminated by a more minimal 3 bp transversion in this same region of 5′FII (FII ctt in Figure 5). In the context of the 3′HS-A site, we substituted each of the base pairs that is identical to the 3′ region of the 5′FII site with the corresponding base transversions (A aact in Figure 5). These mutations of 3′HS-A were sufficient to eliminate CTCF binding. Although 5′FII has a more uniform similarity to 3′HS-B than to 3′HS-A, 3′HS-B showed no detectable CTCF binding (Figure 5A).
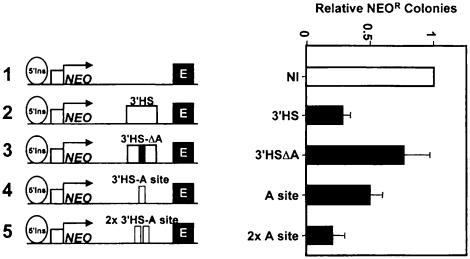
We have shown that a fragment containing the 3′HS has the enhancer-blocking properties of an insulator (Figure 3). Deletion of 3′HS-A from this fragment resulted in an essentially complete loss in enhancer-blocking activity (Figure 6). Furthermore, the 3′HS-A site was independently capable of blocking an enhancer in our assay (Figure 6).
Fig. 6. The CTCF binding site found in the 3′HS is sufficient and necessary for enhancer-blocking activity. The enhancer-blocking activities of the 3′HS-A variants indicated were measured in the colony assay. In construct 3, a 56 bp fragment spanning the A-site has been deleted from the 400 bp 3′HS fragment and inserted into construct 1. One (construct 4) and two (construct 5) copies of the 3′HS-A site alone confer enhancer-blocking activity.
Discussion
In this study, we analyzed the 3′ chromatin boundary of the chicken β-globin locus and found striking similarities with the 5′ chromatin boundary. Previous attempts to locate the 3′ boundary have provided ambiguous results. One study suggested that the 3′ chromatin boundary is located between 5 and 9 kb downstream of the ε-globin gene (Hebbes et al., 1994), another concluded that it lies at least 7 kb downstream of the ε-globin gene (Stalder et al., 1980), and a third located the 3′ chromatin boundary much further downstream (Verreault and Thomas, 1993). Some of these earlier experiments tested the general DNase I sensitivity of DNA fragments now known to contain DNase I HSs. The presence of such sites results in rapid loss of target restriction fragments in the assay, which can be mistaken for general nuclease sensitivity over the entire region. We show here that a transition in general DNase I sensitivity occurs in the region from 4.8 to 7.0 kb downstream of the embryonic β-globin gene.
We found a constitutive HS at the 3′ end of the domain similar to what is observed at the 5′ boundary. Further downstream we recently identified a complete open reading frame encoding an OR protein (COR3′β; Bulger et al., 1999) and there is another putative OR gene still further downstream (Staines and Thomas, 1999). Here we also show (Figure 2) that there is a brain-specific HS just upstream of COR3′β. The constitutive 3′HS lies between the globin genes and COR3′β. Since this was reminiscent of the properties and location of the 5′ β-globin insulator element, we analyzed the 3′ boundary region for insulator activity. We found that a DNA fragment containing the 3′HS was able to block enhancer activity in a position-dependent manner in stably transfected K562 cells. Sequence comparisons between this HS and that of the 5′ insulator element (5′HS4) revealed, at the position of the 3′HS, a sequence of moderate similarity. A direct comparison of their DNA binding activities in gel-shift experiments showed that these sequences bind the same protein, CTCF.
Comparison of the 3′ and 5′ boundary elements
As noted above, the DNA sequences around the 5′ and 3′ boundaries of the β-globin locus share many properties. Most significant is the ability of both to block enhancer action in a directional manner. An equally important and related property is the ability to bind in vitro the same protein, CTCF. We have recently shown (Bell et al., 1999) that the ability to bind CTCF is necessary and sufficient to confer enhancer-blocking activity on region 5′FII of the β-globin 5′HS4. CTCF is an 11 zinc finger protein that has been implicated in both activation (Klenova et al., 1993; Vostrov and Quitschke, 1997) and repression of expression in other systems (Arnold et al., 1996). Moreover, binding sites for CTCF are found in other reported enhancer-blocking elements associated with insulator function at non-erythroid loci (Bell et al., 1999).
Wide sequence variation among CTCF binding sites has been noted in other cases (Burcin et al., 1997). Although the 3′ and 5′ sites both bind CTCF, their affinities are different (Figure 5). Nonetheless, the enhancer-blocking activities of the two sites are similar (Figure 3). CTCF has been shown to use different subsets of fingers at different binding sites (Filippova et al., 1996). This may explain the relatively weak homology between 5′FII and the 3′ site shown in Figure 4C. For the same reason, CTCF might engage essential cofactors differently at the two sites, which could affect the strength of enhancer blocking.
There are some differences between the 3′ sequence element we describe here and the 1.2 kb DNA fragment containing 5′HS4: the 5′ element contains additional distinct sequences that protect against position effects (F.Recillas-Targa, M.J.Pikaart, B.Burgess-Beusse, A.C. Bell and G.Felsenfeld, unpublished). The 400 bp fragment containing the 3′HS that we describe here does not protect against position effects (data not shown). It may be that the requirements for an effective 3′ boundary element are somewhat different from those for the 5′ element, since at the 5′ end the folate receptor gene is separated from the globin locus by ∼16 kb of condensed chromatin. Furthermore, the 3′ condensed chromatin region must certainly be disrupted in those olfactory cells where the OR gene is expressed. In contrast, the 5′ condensed region contains no coding sequences and is probably never in an active chromatin conformation.
Biological significance of enhancer-blocking activity
The position of 5′HS4 coincides with the 5′ boundary between the active and inactive chromatin domains of the β-globin locus (Chung et al., 1993; Hebbes et al., 1994). It has seemed reasonable to speculate that 5′HS4 in some way determines the boundary, and that enhancer-blocking activity plays a role in its establishment. The results presented here show that both ends of the β-globin locus possess such elements, and thus provide a persuasive argument that these are indeed functional elements in the establishment and/or maintenance of boundaries. We note that in the case of the scs/scs′ elements of Drosophila, the proteins binding to the boundaries are quite distinct (Zhao et al., 1995; Gaszner et al., 1999). It appears that in contrast the two globin boundaries have at least one common binding component, although as noted above other components may differ.
We have speculated that the 5′ boundary serves to limit the influence of folate receptor-specific and globin-specific regulatory elements to their respective loci (Prioleau et al., 1999). Just as in the case of the 5′ boundary, which lies between the β-globin locus and an upstream folate receptor gene with a different developmental pattern of expression, the 3′ boundary lies between the nearby strong regulatory elements of the β-globin locus and the putative promoter for an OR gene, each with a distinct program of tissue-specific expression (Figure 7). The 3′HS may perform similar functions, preventing cross-talk between the downstream regulatory elements in the β-globin locus (the strong β/ε enhancer and the ε promoter) and the OR regulatory elements, which we suggest may be located at the brain-specific HS 0.9 kb downstream of the globin 3′HS (∼1.1 kb upstream of the coding sequence of COR3′β; Figure 2). Tissue-specific HSs often mark enhancer or promoter elements responsible for the tissue specificity (Reitman and Felsenfeld, 1990). The existence of the brain-specific HS therefore suggests that COR3′β is active in brain cells. Our observation of a COR3′β transcript in embryonic brain tissue (data not shown) is direct evidence for such activity. Consistent with this observation, transcripts of the mouse homologs MOR3′β1 and MOR3′β2 have been observed in olfactory epithelium (Bulger et al., 1999).
Fig. 7. Summary of the chromatin structure at the boundaries of the chicken β-globin locus in erythroid cells. A schematic view of the region extending from the chicken β-globin locus is shown. CTCF binds to both the 5′ and 3′ chromatin boundaries (Bell et al., 1999; this study), and may be involved in shielding the β-globin locus from regulatory effects from neighboring genes: the upstream folate receptor gene (Prioleau et al., 1999) and the downstream odorant receptor gene (Bulger et al., 1999; this study), respectively.
The observed insensitivity to DNase I (Figure 1) over COR3′β in erythroid cells indicates that at least in most of these cells the chromatin of this OR gene is in an inactive condensed conformation. This is confirmed by the observation that histone acetylation levels are low over the COR3′β promoter region (M.Litt and G.Felsenfeld, unpublished observations). RT–PCR measurements (data not shown) do detect a low level of RNA from this gene in 5- and 10-day-old erythroid cells, suggesting that at least in a few cells of the population the gene might be in an active conformation. We note that low levels of transcripts have been detected from at least one OR gene (HPFH1OR) near the human β-globin locus and two (MOR3′β2 and 3) near the mouse locus in some, but not all erythroid cells (Feingold et al., 1999).
The identification of the 3′ boundary element is consistent with tentative reports of enhancer-blocking activity in other globin gene systems. Recent studies in transgenic mice of the human β-globin locus (Tanimoto et al., 1999) show that inversion of the locus control region (LCR) results in greatly reduced expression of all β-like globin genes. One possible explanation of this result is that the inversion places 5′HS5 of the LCR between the other LCR elements and the globin locus. It has been reported (Chung et al., 1993; Li and Stamatoyannopoulos, 1994) that 5′HS5 has enhancer-blocking activity. In support of this view is the further observation (Tanimoto et al., 1999) that a tagged ε-globin gene placed upstream of HS5 in a normally oriented LCR is not expressed, whereas one placed immediately downstream of the LCR is active.
The mechanism by which boundaries are established is still not clear. It has been suggested (Pikaart et al., 1998; Bell and Felsenfeld, 1999) that the suppression of position effects observed with the 5′ β-globin insulating element may reflect either an ability to protect the reporter from the action of histone deacetylases, or an ability to sequester histone acetylases within the protected region. Either of these actions would help to preserve the transcriptionally active status of the domain against outside influences from nearby endogenous regulatory elements or condensed chromatin. The structure of such a domain remains to be investigated. Among other possibilities, the proteins bound at the 3′ and 5′ boundaries may interact with each other or with the nuclear architecture to create physically defined domains. Now that the location and identity of the boundaries are known, this proposition can be tested.
Materials and methods
DNase I sensitive assay
Preparation of nuclei. Embryonated White Leghorn chicken eggs (11-day-old) and adult chicken blood were obtained from Truslow Farms, Chestertown, MD. Erythrocytes were harvested from a dozen eggs, collected in phosphate-buffered saline (PBS) containing 5 mM EDTA and washed in 50 ml of buffer A [10 mM Tris–HCl pH 7.5, 80 mM NaCl, 6 mM MgCl2, 0.2 M phenylmethylsulfonyl fluoride (PMSF) and 10 µg/ml aprotinin]. Cells were lysed in buffer A with 0.1% Triton X-100. Nuclei were pelleted, washed in the same buffer three times, and resuspended in RSB buffer (10 mM Tris–HCl pH 7.5, 10 mM NaCl, 3 mM MgCl2) at a concentration of 20 A260. Nuclei from 6C2 cells and DT40 cells were prepared following the same protocol. Nuclei were digested with varying concentrations of DNase I (Worthington), typically 0–5 U/ml for 10 min. DNase I-treated DNA from chicken brain nuclei was kindly provided by Dr M.-N.Prioleau. Each DNA sample (10 µg) was digested overnight with restriction enzymes and 8 µg of DNA/lane were electrophoresed through a 0.8% agarose gel. Southern hybridizations were performed at 65°C using QuickHyb (Stratagene). After a final wash in 0.2× SSC–0.1% SDS at 65°C, filters were either autoradiographed or exposed to a Molecular Dynamics PhosphorImager to measure intensities of hybridized signals.
Hybridization probes. DNA fragments in Figure 1 were detected as follows: A, a 1.4 kb BamHI fragment detected with probe P1 (450 bp; NcoI–BamHI at –7.9 to –7.5 map units); B, a 2.1 kb BamHI–HindIII fragment detected with probe i116 (450 bp; PstI–BamHI at 9.5–9.9 map units); C, a 2.4 kb BglII–BsaBI fragment detected with probe i112 (1000 bp; KpnI–PstI at 23.3–24.3 map units); D, a 2.3 kb BglII–BsaBI fragment detected with probe P9 (500 bp; BamHI–EcoRI at 28.1–28.6 map units); E, a 3.1 kb BamHI–BamHI fragment detected with probe P9; F, a 3.2 kb BamHI–BamHI fragment detected with probe i87 (500 bp; NcoI–NcoI at 31.6–32.1 map units); Ova, a 1.4 kb BamHI–HindIII fragment detected with a probe i104 (500 bp; EcoRI–SmaI derived from a clone OV1.8; Woo et al., 1981), provided by Dr Marc Reitman. The DNase I HS, 3′HS, was detected by digestion of nuclei with increasing amounts of DNase I as above followed by digestion of extracted DNA with KpnI and probing with i112 (1000 bp; KpnI–PstI at 23.3–24.3 map units).
Plasmid constructions
Constructs used in Figure 3A: construct 1 was pNI (Chung et al., 1997) and construct 2 was pJC5-4 (Chung et al., 1993). In order to create constructs 3 (pJC3′HS), 4 (pJC3′HS-2) and 5 (pJC3′HS-6), DNA fragments ‘3′HS’, ‘3′HS-2’ and ‘3′HS-6’ as indicated in Figure 3 were PCR amplified from cosmid +ENH (Barton et al., 1990). Primers used were ns11 (5′-AGGCGCGCCAGATTTTCATCACCTCTTCCCCTGC-3′) and ns12 (5′-AGGCGCGCCAAGCTTAGCTATGTGCCCAAAAGGATGAGGA-3′) for ‘3′HS’, ns12 and ns14 (5′-AGGCGCGCCTTCCTGTGAAAACATCACCTAGCCA-3′) for ‘3′HS-2’, and ns13 (5′-AGGCGCGCCTTCCTGTGAAAACATCACCTAGCCAT-3′) and ns20 (5′-AGGCGCGCCGGGTTGGGCTGAGTGGGCTT-3′) for ‘3′HS-6’. Each amplified fragment was inserted in the AscI site of pNI.
Constructs used in Figure 3B: the constructs 1, 2 and 3 were pJC3-4, pJC13-1 and pJC5-4 (Chung et al., 1993), respectively. Construct 4 (pJC3′double) was derived from pJC3′HS (see above). The 1.2 kb of 5′Ins sequence at XbaI site was removed and replaced with the PCR-amplified 3′HS DNA fragment. The construct 5 (3′pJC3′) was derived from the plasmid pJC3′double. ‘3′HS’ at the AscI site was removed, and inserted at the NdeI site.
Constructs used in Figure 6: the constructs 1 and 2 were pNI (Chung et al., 1997) and pJC3′HS (see above). Constructs 3–7 were derived from pNI. 3′HS-ΔA was generated by two-step overlapping PCR using the cosmid +ENH (Barton et al., 1990) as the first template and two sets of primers ns11 and ns77 (5′-TTCAGAGGCATAGCAGACGCAGTTCTCCGAGTCTGACTTT), and ns12 and ns76 (5′-AAAGTCAGACTCGGAGAACTGCGTCTGCTATGCCTCTGAA). The product of that reaction was used as the template for the second round of PCR using primers ns11 and ns12. 3′HS-A and 2×∼3′HS-A were created by inserting, in one and two copies, respectively, duplexed oligomers identical to those used in DNA-binding assays into the AscI site of pNI.
Colony assay by stable transfection of K562 cells
Colony assays were performed as described previously (Chung et al., 1993). Briefly, exponentially growing K562 cells were harvested, washed in PBS, and resuspended in PBS at 2 × 107 cells/ml. Linearized plasmid (250 ng) was transfected by electroporation with a Bio-Rad Gene Pulser at 200 V and 960 µF. The cells were transferred to 20 ml of growth medium and cultured for 48 h. For drug selection, 3 ml of the cell suspension were mixed with 27 ml of growth media complemented with 0.3% cell culture agar and 750 µg/ml active G418 (Gibco). Colonies grown in the presence of G418 were counted 2–3 weeks after selection.
DNA sequencing and analysis
Plasmids used as sequencing templates were p556 and p932, originally derived from the cosmid p918, kindly provided by Dr Marc Reitman (Barton et al., 1990). In this study, nucleotide sequences were determined from 21.0 to 28.2 map units of the chicken β-globin locus (DDBJ/EMBL/GenBank accession No. L17432).
Gel mobility shift assays
Oligonucleotides used as templates and competitors were synthesized with an Applied Biosystems Synthesizer, and purified on urea–polyacrylamide gels prior to use (Sambrook et al., 1989). Each upper strand was labeled at the 5′ end with polynucleotide kinase (New England Biolab) and [γ-32P]ATP. The reaction was terminated by adding EDTA at 10 mM and heating at 90°C for 10 min. To prepare duplex DNA, 1.5× molar excess of the corresponding lower strand was added, heated at 94°C for 10 min, followed by slow cooling to room temperature. To remove salt and unincorporated nucleotides, annealed products were spun through a Sephadex G50 spin column (Pharmacia). To prepare cold competitors, strands were annealed in TEN buffer (10 mM Tris–HCl pH 7.9, 1 mM EDTA, 50 mM NaCl) by heating at 94°C for 10 min, and then slowly cooling to room temperature. In vitro translated CTCF, nuclear extract and partially purified CTCF were prepared as previously described (Bell et al., 1999). For gel mobility shift assays, 20 fmol of radiolabeled probe, ∼1 µl of nuclear extract (3 µg/µl) or S-pool fraction (∼1 µg/µl), 1 µg of poly dG–dC, 1 mM dithiothreitol (DTT), 0.5% Triton X-100 and 1 pmol of competitor oligonucleotide, where indicated, were mixed in 1× GS buffer (20 mM HEPES pH 7.9, 150 mM KCl, 5 mM MgCl2, 5% glycerol) at room temperature for 15 min. Samples were electrophoresed on 5% acrylamide gels in 0.5× TBE (45 mM Tris–borate pH 7.5, 1 mM EDTA) at room temperature. Southwestern blotting was performed as previously described (Bell et al., 1999).
Acknowledgments
Acknowledgements
We would like to acknowledge Dr Marc Reitman for fruitful discussions and for supplying plasmid clones used in this study. We thank Dr M.Krause for helping with DNA sequencing, and Drs Catherine Farrell and Pascale Nony for providing helpful advice and reagents. We also thank Drs Robert Martin and Marc Reitman for critical reading of this manuscript. N.S. was supported by JSPS (Japan Society for Promotion of Science).
References
- Abe K., Kusakabe,Y., Tanemura,K., Emori,Y. and Arai,S. (1993) Multiple genes for G protein-coupled receptors and their expression in lingual epithelia. FEBS Lett., 316, 253–256. [DOI] [PubMed] [Google Scholar]
- Arnold R., Burcin,M., Kaiser,B., Muller,M. and Renkawitz,R. (1996) DNA bending by the silencer protein NeP1 is modulated by TR and RXR. Nucleic Acids Res., 24, 2640–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton M.C., Hoekstra,M.F. and Emerson,B.M. (1990) Site-directed, recombination-mediated mutagenesis of a complex gene locus. Nucleic Acids Res., 18, 7349–7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell A.C. and Felsenfeld,G. (1999) Stopped at the border: boundaries and insulators. Curr. Opin. Genet. Dev., 9, 191–198. [DOI] [PubMed] [Google Scholar]
- Bell A.C., West,A.G. and Felsenfeld,G. (1999) The protein CTCF is required for the enhancer-blocking activity of vertebrate insulators. Cell, 98, 387–396. [DOI] [PubMed] [Google Scholar]
- Bossy B., Hall,L.M.C. and Spierer,P. (1984) Genetic activity along 315 kb of the Drosophila chromosome. EMBO J., 3, 2537–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M., von Doorninck,J.H., Saitoh,N., Telling,A., Farrell,C., Bender,M.A., Felsenfeld,G., Axel,R. and Groudine,M. (1999) Conservation of sequence and structure flanking the mouse and human β-globin loci: the β-globin genes are embedded within an array of odorant receptor genes. Proc. Natl Acad. Sci. USA, 96, 5129–5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burcin M., Arnold,R., Lutz,M., Kaiser,B., Runge,D., Lottspeich,F., Filippova,G.N., Lobanenkov,V.V. and Renkawitz,R. (1997) Negative protein 1, which is required for function of the chicken lysozyme gene silencer in conjunction with hormone receptors, is identical to the multivalent zinc finger repressor CTCF. Mol. Cell. Biol., 17, 1281–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J.H., Whiteley,M. and Felsenfeld,G. (1993) A 5′ element of the chicken β-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell, 74, 505–514. [DOI] [PubMed] [Google Scholar]
- Chung J.H., Bell,A.C. and Felsenfeld,G. (1997) Characterization of the chicken β-globin insulator. Proc. Natl Acad. Sci. USA, 94, 575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyer W.J. (1998) The area code hypothesis revisited: olfactory receptors and other related transmembrane receptors may function as the last digits in a cell surface code for assembling embryos. Proc. Natl Acad. Sci. USA, 95, 9072–9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drutel G., Arrang,J.M., Diaz,J., Wisnewsky,C., Schwartz,K. and Schwartz,J.C. (1995) Cloning of OL1, a putative olfactory receptor and its expression in the developing rat heart. Receptors Channels, 3, 33–40. [PubMed] [Google Scholar]
- Farache G., Razin,S.V., Recillas-Targa,F. and Scherrer,K. (1990) Organization of the 3′-boundary of the chicken α globin gene domain and characterization of a CR1-specific protein binding site. Nucleic Acids Res., 18, 401–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feingold E.A., Penny,L.A., Nienhuis,A.W. and Forget,B.G. (1999) An olfactory receptor gene is located in the extended human β-globin gene cluster and is expressed in erythroid cells. Genomics, 61, 15–23. [DOI] [PubMed] [Google Scholar]
- Filippova G.N., Fagerlie,S., Klenova,E.M., Myers,C., Dehner,Y., Goodwin,G., Neiman,P.E., Collins,S.J. and Lobanenkov,V.V. (1996) An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol. Cell. Biol., 16, 2802–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman T.B., Owens,K.N., Burnett,J.B., Saura,A.O. and Wallrath,L.L. (1991) The faint band/interband region 28C2 to 28C4-5(-) of the Drosophila melanogaster salivary gland polytene chromosomes is rich in transcripts. Mol. Gen. Genet., 226, 81–87. [DOI] [PubMed] [Google Scholar]
- Gaszner M., Vazquez,J. and Schedl,P. (1999) The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer-promoter interaction. Genes Dev., 13, 2098–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M. (1997) Histone acetylation in chromatin structure and transcription. Nature, 389, 349–352. [DOI] [PubMed] [Google Scholar]
- Hebbes T.R., Thorne,A.W. and Crane-Robinson,C. (1988) A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J., 7, 1395–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbes T.R., Clayton,A.L., Thorne,A.W. and Crane-Robinson C. (1994) Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken β-globin chromosomal domain. EMBO J., 13, 1823–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantzen K., Fritton,H. and Igo-Kemenes,T. (1986) The DNase I sensitive domain of the chicken lysozyme gene spans 24 kb. Nucleic Acids Res., 14, 6085–6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellum R. and Schedl,P. (1991) A position-effect assay for boundaries of higher order chromosomal domains. Cell, 64, 941–950. [DOI] [PubMed] [Google Scholar]
- Kellum R. and Schedl,P. (1992) A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol. Cell. Biol., 12, 2424–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenova E.M., Nicolas,R.H., Paterson,H.F., Carne,A.F., Heath,C.M., Goodwin,G.H., Neiman,P.E. and Lobanenkov,V.V. (1993) CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol. Cell. Biol., 13, 7612–7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson G.M., Knoll,B.J., March,C.J., Woo,S.L., Tsai,M.J. and O’Malley,B.W. (1982) Definition of 5′ and 3′ structural boundaries of the chromatin domain containing the ovalbumin multigene family. J. Biol. Chem., 257, 1501–1507. [PubMed] [Google Scholar]
- Li Q. and Stamatoyannopoulos,G. (1994) Hypersensitive site 5 of the human β locus control region functions as a chromatin insulator. Blood, 84, 1399–1401. [PubMed] [Google Scholar]
- Nef S. and Nef,P. (1997) Olfaction: transient expression of a putative odorant receptor in the avian notochord. Proc. Natl Acad. Sci. USA, 94, 4766–4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill L.P. and Turner,B.M. (1995) Histone H4 acetylation distinguishes coding regions of the human genome from heterochromatin in a differentiation-dependent but transcription-independent manner. EMBO J., 14, 3946–3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmentier M., Libert,F., Schurmans,S., Schiffmann,S., Lefort,A., Eggerickx,D., Ledent,C., Mollereau,C., Gerard,C. and Perret,J. (1992) Expression of members of the putative olfactory receptor gene family in mammalian germ cells. Nature, 355, 453–455. [DOI] [PubMed] [Google Scholar]
- Pikaart M.J., Feng,J. and Villeponteau,B. (1992) The polyomavirus enhancer activates chromatin accessibility on integration into the HPRT gene. Mol. Cell. Biol., 12, 5785–5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikaart M.J., Recillas-Targa,F. and Felsenfeld,G. (1998) Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev., 12, 2852–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prioleau M.N., Nony,P., Simpson,M. and Felsenfeld,G. (1999) An insulator element and condensed chromatin region separate the chicken β-globin locus from an independently regulated erythroid-specific folate receptor gene. EMBO J., 18, 4035–4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raming K., Konzelmann,S. and Breer,H. (1998) Identification of a novel G-protein coupled receptor expressed in distinct brain regions and a defined olfactory zone. Receptors Channels, 6, 141–151. [PubMed] [Google Scholar]
- Recillas-Targa F., Bell,A.C. and Felsenfeld,G. (1999) Positional enhancer-blocking activity of the chicken β-globin insulator in transiently transfected cells. Proc. Natl Acad. Sci. USA, 96, 14354–14359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitman M. and Felsenfeld,G. (1990) Developmental regulation of topoisomerase II sites and DNase I-hypersensitive sites in the chicken β-globin locus. Mol. Cell. Biol., 10, 2774–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rykowski M.C., Parmelee,S.J., Agard,D.A. and Sedat,J.W. (1988) Precise determination of the molecular limits of a polytene chromosome band: regulatory sequences for the Notch gene are in the interband. Cell, 54, 461–472. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. 2nd edn, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Staines D.M. and Thomas,J.O. (1999) A sequence with homology to human HPFH- linked enhancer elements and to a family of G-protein linked membrane receptor genes is located downstream of the chicken β-globin locus. Gene, 234, 345–352. [DOI] [PubMed] [Google Scholar]
- Stalder J., Larsen,A., Engel,J.D., Dolan,M., Groudine,M. and Weintraub,H. (1980) Tissue-specific DNA cleavages in the globin chromatin domain introduced by DNase I. Cell, 20, 451–460. [DOI] [PubMed] [Google Scholar]
- Tanimoto K., Liu,Q., Bungert,J. and Engel,J.D. (1999) Effects of altered gene order or orientation of the locus control region on human β-globin gene expression in mice. Nature, 398, 344–348. [DOI] [PubMed] [Google Scholar]
- Verreault A. and Thomas,J.O. (1993) Chromatin structure of the β-globin chromosomal domain in adult chicken erythrocytes. Cold Spring Harb. Symp. Quant. Biol., 58, 15–24. [DOI] [PubMed] [Google Scholar]
- Villeponteau B. and Martinson,H. (1981) Isolation and characterization of the complete chicken β-globin gene region: frequent deletion of the adult β-globin genes in λ. Nucleic Acids Res., 9, 3731–3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vostrov A.A. and Quitschke,W.W. (1997) The zinc finger protein CTCF binds to the APBb domain of the amyloid β-protein precursor promoter. Evidence for a role in transcriptional activation. J. Biol. Chem., 272, 33353–33359. [DOI] [PubMed] [Google Scholar]
- Weintraub H. and Groudine,M. (1976) Chromosomal subunits in active genes have an altered conformation. Science, 193, 848–856. [DOI] [PubMed] [Google Scholar]
- Woo S.L., Beattie,W.G., Catterall,J.F., Dugaiczyk,A., Staden,R., Brownlee,G.G. and O’Malley,B.W. (1981) Complete nucleotide sequence of the chicken chromosomal ovalbumin gene and its biological significance. Biochemistry, 20, 6437–6446. [DOI] [PubMed] [Google Scholar]
- Zhao K., Hart,C.M. and Laemmli,U.K. (1995) Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell, 16, 879–889. [DOI] [PubMed] [Google Scholar]