Abstract
Purpose: Methods for quantification of breast density on MRI using semiautomatic approaches are commonly used. In this study, the authors report on a fully automatic chest template-based method.
Methods: Nonfat-suppressed breast MR images from 31 healthy women were analyzed. Among them, one case was randomly selected and used as the template, and the remaining 30 cases were used for testing. Unlike most model-based breast segmentation methods that use the breast region as the template, the chest body region on a middle slice was used as the template. Within the chest template, three body landmarks (thoracic spine and bilateral boundary of the pectoral muscle) were identified for performing the initial V-shape cut to determine the posterior lateral boundary of the breast. The chest template was mapped to each subject's image space to obtain a subject-specific chest model for exclusion. On the remaining image, the chest wall muscle was identified and excluded to obtain clean breast segmentation. The chest and muscle boundaries determined on the middle slice were used as the reference for the segmentation of adjacent slices, and the process continued superiorly and inferiorly until all 3D slices were segmented. The segmentation results were evaluated by an experienced radiologist to mark voxels that were wrongly included or excluded for error analysis.
Results: The breast volumes measured by the proposed algorithm were very close to the radiologist's corrected volumes, showing a % difference ranging from 0.01% to 3.04% in 30 tested subjects with a mean of 0.86% ± 0.72%. The total error was calculated by adding the inclusion and the exclusion errors (so they did not cancel each other out), which ranged from 0.05% to 6.75% with a mean of 3.05% ± 1.93%. The fibroglandular tissue segmented within the breast region determined by the algorithm and the radiologist were also very close, showing a % difference ranging from 0.02% to 2.52% with a mean of 1.03% ± 1.03%. The total error by adding the inclusion and exclusion errors ranged from 0.16% to 11.8%, with a mean of 2.89% ± 2.55%.
Conclusions: The automatic chest template-based breast MRI segmentation method worked well for cases with different body and breast shapes and different density patterns. Compared to the radiologist-established truth, the mean difference in segmented breast volume was approximately 1%, and the total error by considering the additive inclusion and exclusion errors was approximately 3%. This method may provide a reliable tool for MRI-based segmentation of breast density.
Keywords: model-based breast segmentation, chest body template, breast MRI, breast density, demons algorithm
INTRODUCTION
Breast MRI is a well-established clinical imaging modality for the management of breast diseases. The current clinical indications include screening, detection, diagnosis, preoperative staging, therapy response monitoring, and surveillance. In addition, since MRI acquires a set of three-dimensional (3D) images with a strong contrast between fibroglandular tissue (or, generally described as “breast density”) and fatty tissue, it is the optimal imaging modality for measurement of volumetric breast density. Breast density has been proven as an independent risk factor associated with development of breast cancer,1, 2 and there is a strong interest to develop reliable quantitative methods that can be used to evaluate density on different imaging modalities. Since MRI acquires many 3D images to cover the whole breast, applying computer algorithm-based segmentation methods is the only feasible approach. An initial segmentation of the breast area on MR images is the first required step. Then, the chest wall (pectoralis) muscle needs to be identified and excluded. Within the segmented breast, the fibroglandular tissue, fatty tissue, and the skin are further separated to measure the volume of fibroglandular tissue and the percent density by normalizing to the breast volume.
Breasts have a large variety in shapes and density patterns, and it is difficult to develop an automatic algorithm-based segmentation method that can work for all types of breasts. The major task is to define the lateral posterior boundary using a consistent criterion, and to delineate the chest wall muscle. For cases with dense breasts, the fibroglandular tissue may be very close to the chest wall muscle, which increases the technical difficulty to exclude the muscle while preserving the dense tissue.
Currently, most of the reported methods in the literature are based on semiautomated methods that require some operator interventions.3, 4, 5, 6, 7, 8 They are time-consuming and subject to variations from an operator's personal judgment. Lee et al.6 used edge detection to determine the breast-air interface and manually outlined the breast-chest wall interface. Khazen et al.4 used a thresholding operation to limit the extent of breast volume and manually defined a straight line anterior to the chest wall muscle at each axial slice. In our previous work, we used body landmarks of each individual woman, e.g., using the thoracic spine and lateral boundary of pectoral muscle for a V-shape cut7 or using the sternum for a horizontal cut3 to standardize the initial segmentation and minimize an operator's variation.
Some studies have attempted to develop automatic breast segmentation methods either for density analysis or for computer-aided-diagnosis of breast cancer. Active contour based methods9, 10 have been applied to detect the boundary of chest wall muscle. Wang et al.11 used Hessian filtering to locate the potential location of chest wall muscle and further used the morphological operations to automatically delineate the actual boundary. However, these methods still required an initial segmentation of the breast region, and also they were not integrated into a complete framework for analysis of breast density. Template-based registration methods may be applied for automatic segmentation. This approach is commonly used for the brain, but it is much more challenging for the breast. Gallego and Martel12 proposed an automated model-based 3D segmentation method for the breast. The subject's breast boundary was identified using edge detection to generate the surface meshes, and they were matched to the surface meshes of the template for segmentation. Gubern-Mérida et al.13 proposed a probabilistic atlas which included both body and breast tissues for breast segmentation in MRI. Reed et al.14 proposed a 3D model-based method for breast segmentation in CT using nonrigid registration. A cellular neural network has also been applied to extract the breast region from MR images.15
So far, most of the model-based segmentation methods mentioned above used the whole breast as the template. However, because of the large variability in the shape of breasts, simply using one universal template may not be robust enough to segment all types of breasts. On the other hand, in most breast MR scans, the chest region including the lung and the heart can be detected at similar locations with similar shape and intensity features. In this study, we present a new automatic template-based method using the chest body model for breast segmentation. The analysis starts from the middle slice of the 3D image set. The chest template is coregistered to each subject's chest region to obtain a subject-specific chest model on the middle slice that is excluded. On the remaining image, a chest wall muscle detection algorithm with Bezier curve fitting is developed to exclude the muscle and obtain a clean segmentation of the breast. The boundary of the chest and the muscle determined on the middle slice is then used as the reference for segmentation on the adjacent slices, and the process continues until all slices are segmented. The obtained segmentation results using this method were evaluated by an experienced radiologist to calculate the error percentage. The resulting error in the segmented fibroglandular tissue was also evaluated.
MATERIALS AND METHODS
The step-by-step procedures of our template-based breast segmentation method are described in this section. The segmentation starts from the middle slice of the acquired 3D imaging slab (Sec. 2B), and then the segmented chest region on this slice is used in the adjacent images until all images are segmented (Sec. 2C). Evaluation of this method is in Sec. 2D.
Image data
T1-weighted MR scans from 31 healthy female volunteers (age range 22–54 y/o, mean 42 y/o) without any symptom of breast diseases were used in this study. Imaging was performed on a Philips Achieva 3.0T scanner (Eindhoven, Netherlands) using the turbo spin echo pulse sequence without fat suppression. A total of 90 image slices with 2 mm thickness were acquired to cover the entire breast. Other imaging parameters were: TR/TE = 645/9.0 ms, echo train = 5, slice gap = 0, phase encoding R-L, bandwidth per pixel = 174 Hz, field of view = 330 mm, imaging matrix = 328 × 384, and parallel imaging with SENSE factor = 2. These 31 cases presented a variety of breast sizes, shapes, and fibroglandular tissue patterns. One case was selected to generate the chest region template, and the remaining 30 cases were used for testing and evaluation.
Breast segmentation on the middle 2D MR slice
For each case, the starting slice number and the ending slice number of the breasts were determined by a radiologist, and the middle slice was selected for the initial segmentation. The generation of the chest region template with three body landmarks (thoracic spine and bilateral boundary of the pectoral muscle) for a V-shape cut to determine the posterior boundary of the breast is described in Sec. 2B1. The registration of the template to the chest region of an individual subject for initial segmentation using the V-shape cut is described in Sec. 2B2. The identification and exclusion of the chest wall muscle to obtain the final segmented breast is described in Sec. 2B3.
Generation of the chest region template
The 3D breast MR scans of the 31 subjects covered a large region to ensure that both breasts were entirely contained in the imaging slab. In the middle of this imaging slab, the chest region always included the lung and the heart, and showed similar shape and signal intensity features. Therefore, the center slice was chosen as the starting point to ensure the presence of these similar tissue components (heart, lung, thoracic spine, pectoral muscles). Of the 31 cases, one case was randomly selected to generate the chest template (Fig. 1). An approximate contour of the chest body region was roughly outlined manually along the posterior chest wall muscle and the outer boundary of the lung [the red contour in Fig. 1a]. We used three body landmarks: the thoracic spine and the lateral margins of the bilateral pectoralis muscles [the highlighted points in Fig. 1a], to perform an initial V-shape cut to define the posterior boundary of the breast. These three points were manually marked. Within this roughly drawn region, the k-means clustering algorithm was used to identify the dark signal intensity coming from the lung tissue for refining the contour as the chest template [cyan contour in Fig. 1b]. After the chest region was defined, it was excluded by setting the signal intensity to zero. The image was then smoothed, normalized, and cropped to generate a “SNC image” used for registration. In order to have the registration process focus on the chest region, the image was cropped at 8 mm above the chest model contour. An ideal SNC image needed to keep some tissues above the chestwall muscle, and with a flat anterior boundary to avoid complications during the registration. A thickness of 8 mm was chosen to keep sufficient tissues above and also to make sure that the boundary was flat. The remaining image was normalized using the k-means clustering algorithm followed by Gaussian smoothing. The normalization and smoothing was required to achieve the optimal quality in registration. The final SNC image of the generated chest region template is shown in Fig. 1c, and was registered to other subjects’ SNC images to generate subject-specific chest models. Detailed methods to generate the SNC image, including the reasons for choosing the cluster number in k-means clustering, will be described in Sec. 2B2.
Figure 1.
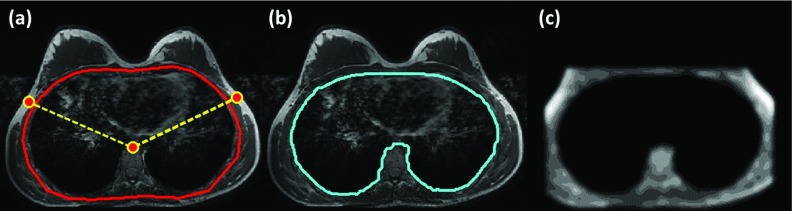
Generation of the chest template using one arbitrarily selected case. (a) An approximate contour of the chest body region is roughly outlined manually along the posterior chest wall muscle and the outer boundary of the lung. Three body landmarks: the thoracic spine and the lateral margin of the bilateral pectoralis muscles are also manually marked. (b) Within this roughly drawn region, the k-means clustering algorithm is used to identify the dark signal intensity coming from the lung tissue for refining the contour as the chest template. (c) The image is cropped at 8 mm above the chest model contour, and after excluding the chest determined in (b) the remaining image is normalized into ten clusters using k-means clustering algorithm followed by Gaussian smoothing to generate the Smoothed, Normalized, and Cropped “SNC image” for registration.
Registration between chest region template and subject's chest region
The chest region on the middle slice includes the lungs and heart (and the band of artifacts associated with the heart motion) which have different signal intensities. Figure 2 illustrates the step-by-step procedures for estimating the air region (lung tissue) inside the chest to generate the chest contour, similar to that shown in Fig. 1b, so they can be coregistered. The original breast MR image with background noise (e.g., the ghosting artifact coming from the heart motion outside the breast) is shown in Fig. 2a. This outside noise is removed by using Otsu's automated thresholding algorithm16 followed by morphological operations of erosion and open [Fig. 2b]. Since the lung (mainly containing air) is much darker compared to other tissues, an initial k-means clustering with (k = 2) is applied to locate the dark region [Fig. 2c], which may also include fibroglandular tissue and chest wall muscle. Within this contour [Fig. 2d], k-means clustering (k = 10) is applied again to identify the area with the lowest intensity (cluster#1). The center line is manually drawn based on the center point between the left and the right breasts along with the thoracic spine. The largest area of cluster#1 in the left side and the right side is detected and the center of mass in each side is used as the seed point for region growing. The intensity step for region growing is set as α · max (I) with α = 0.1. The region growing results from these two seed points are merged as the initial air region [Fig. 2e]. We have tried different cluster numbers and found that by using a total number of N = 10 clusters the lowest cluster identified the darkest tissue with a sufficient number of pixels to define a center of mass as the seed for region growing.
Figure 2.
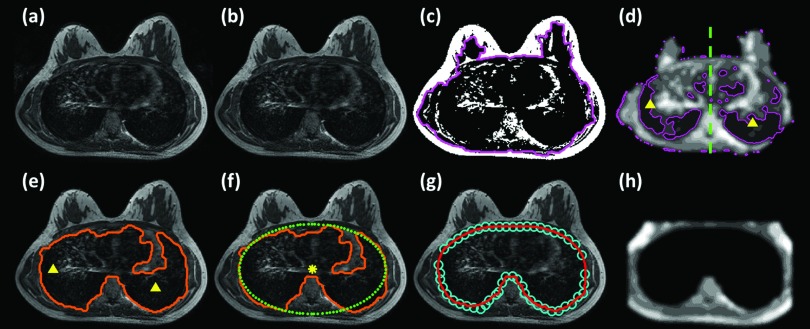
The step-by-step procedures for estimating the air (lung region) in each subject to generate the chest model for coregistration with the template. (a) The original breast MR image with background noise (e.g., the ghosting artifact coming from the heart motion outside the breast). (b) This outside noise is removed by using Otsu's automated thresholding algorithm followed by morphological operations of erosion and open. (c) An initial k-means clustering with (k = 2) is applied to locate the dark region, which may also include fibroglandular tissue and chest wall muscle. (d) Within this contour, k-means clustering (k = 10) is applied again to identify the area with the lowest intensity (cluster#1). (e) The largest area of cluster#1 in the left side and the right side is detected and the center of mass in each side is used as the seed point for region growing. (f) Based on the center of the initial air region (the highlighted dot), an ellipse that has the best fit to the initial air region is then generated. For the chest region above the center, the boundary of the initial air region and the estimated ellipse is averaged and smoothed to obtain a smooth boundary. For the chest region below the center, the boundary estimated by region growing is directly used. (g) The resulted contour is indicated by circles. Bezier curve fitting is then applied to the circles to obtain a smooth contour of the estimated chest region. (h) Finally, the SNC image is generated (cropped at 8 mm above the chest model contour, and then normalized into 10 clusters and smoothed).
As shown in Fig. 2e, the upper boundary is not smooth, and it is not the correct upper chest boundary. Based on the center of the initial air region [the highlighted dot in Fig. 2f], an ellipse that has the best fit to the initial air region is then generated. For the chest region above the center, the boundary of the initial air region, and the estimated ellipse is averaged and smoothed to obtain a smooth boundary [circles in Fig. 2g]. For the chest region below the center, the boundary estimated by region growing is directly used. Bezier curve fitting17 is then applied to the circles to obtain a smooth contour of the estimated chest region [Fig. 2g]. Finally, the SNC image is generated based on the description in Sec. 2B1 for registration [cropped at 8 mm above the anterior point of the chest model contour, and then normalized into ten clusters and smoothed, Fig. 2h]. The SNC images of the subject and the template are co-registered using the non-rigid demons algorithm.18, 19
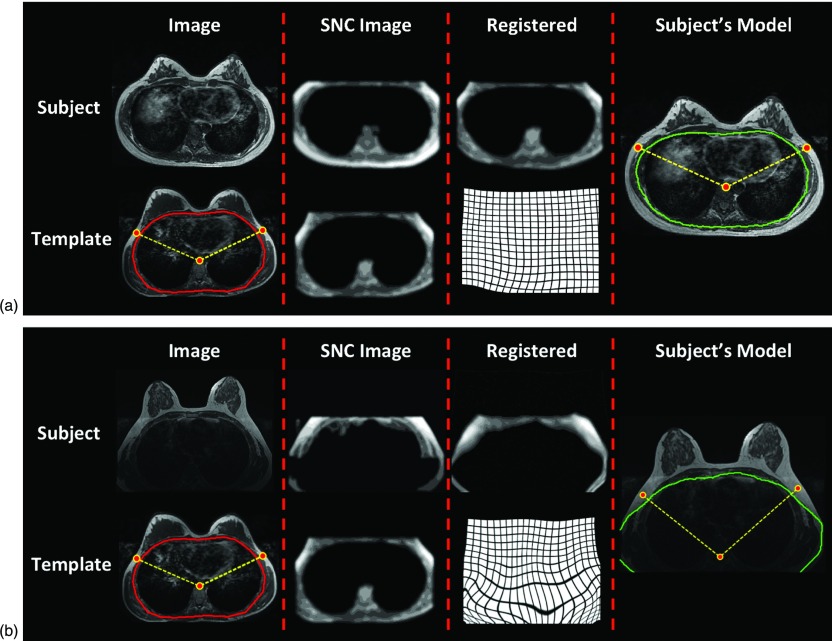
To demonstrate the robustness of this chest-template based segmentation method, we chose two subjects who have different breast and body shapes compared to that of the template case for illustrations, shown in Figs. 3a, 3b, respectively. The subject in Fig. 3a had a complete chest covered within the field of view, showing relatively small breasts in a large body with more subcutaneous body fat. The subject in Fig. 3b had an incomplete chest covered within the field of view, with the bottom of the chest cut-off and showing very dark signal intensity. As noted in Fig. 3b, although part of chest was cut-out, the registration in the anterior part of the chest still worked well and the landmarks used for v-shape cut were mapped to reasonable locations. After applying the v-shape cut, the lateral boundary of the breast was well defined and the remaining image showed decent signal intensity for the next chest wall muscle exclusion procedure.
Figure 3.
The coregistration process to obtain subject-specific chest model. (a) This subject has a complete chest covered within the field of view, and has relatively small breasts in a large body with more subcutaneous body fat. (b) This subject has an incomplete chest covered within the field of view, with the bottom of the chest cut-out and showing very dark signal intensity. The original images of the subject and the template case are shown in the first column. The subject SNC and the template SNC images are shown in the second column. The template SNC is coregistered to the subject's SNC image using the Demons algorithm to obtain the deformation matrix, shown in the third column. The last column shows the obtained chest model superimposed on the original subject image. The three body landmarks manually marked on the template are also transformed to the subject's space for performing the V-Shape cut to define the posterior extent of the bilateral breasts. As noted in (b), although part of chest is cut-out, the registration in the anterior part of the chest still works well and the landmarks used for v-shape cut are mapped to reasonable locations.
Exclusion of residual chest wall muscle
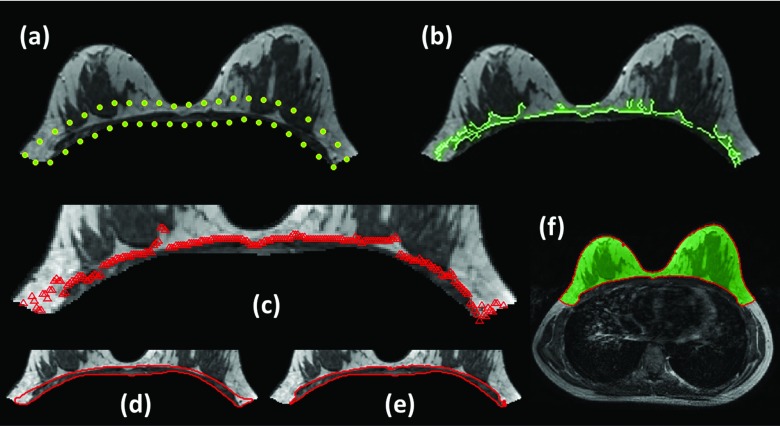
After applying the V-shape cut and excluding the chest area using the subject's specific chest model (the last column images shown in Fig. 3), one example of the residual breast image is shown in Fig. 4a. Since the chest template was mainly used to exclude the lung tissue, the chest wall muscle still remained on the resulting image and needed to be removed. The search of the chest wall muscle was confined within 15 mm from the posterior boundary [the highlighted area in Fig. 4a]. This thickness was decided based on the consideration to cover the entire muscle, and not cover excessive breast tissue that can lead to increased segmentation difficulty. The choice of 15 mm worked well for all analyzed cases.
Figure 4.
Procedures to define and exclude chest wall muscle. (a) The residual image after applying the V-shape cut and excluding the chest area using the subject's specific chest model. (b) Canny edge detection method is applied to search the edge of chest wall muscle in a confined area within 15 mm from the posterior breast boundary. (c) Bezier curve fitting is then applied to find a smooth chest wall muscle boundary. (d) The area between this fitted curve and the posterior boundary is used to segment the chest wall muscle by using the k-means clustering (k = 3). (e) The two lower intensity clusters (including cluster#1 and cluster#2) are selected as the chest wall muscle. (f) The layer of the skin is excluded by using gradient-based skin detection method to obtain a clean segmented breast.
In general, there is a strong contrast between the chest wall muscle and the adjacent fatty tissues, and edge detection is able to detect such intensity contrast. In the present study, the Canny edge detection method20 with the threshold of 0.2 was applied [Fig. 4b]. As shown in the figure, some edge points belong to the breast dense tissue and not the chest wall muscle, so Bezier curving fitting was then applied to find a smooth chest wall muscle boundary [Fig. 4c]. The area between this fitted curve and the posterior boundary [Fig. 4d] was used to segment the chest wall muscle by using the k-means clustering (k = 3). The two lower intensity clusters (including cluster#1 and cluster#2) were selected as the chest wall muscle [Fig. 4e] and excluded. Since the covered area was small, the choice of three clusters was the most commonly used setting. Finally, the layer of the skin was excluded by using our published gradient-based skin detection method21 to obtain a clean segmented breast [Fig. 4f].
3D segmentation framework
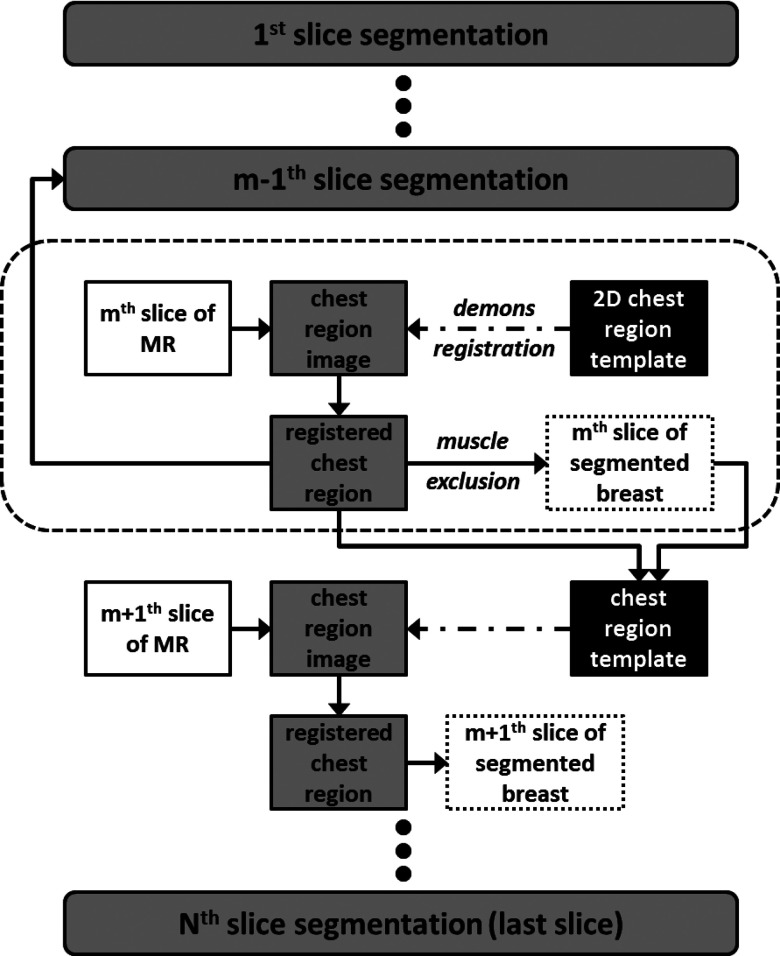
The scheme for segmentation of all MR slices based on the central slice is shown in Fig. 5. After the breast region in the center slice (the mth slice) is first segmented, the chest model for the mth slice is then used as reference for the m − 1th slice and the m + 1th slice. The boundary for m + 1th is used for m + 2th; the boundary for m − 1th is used for m − 2th, etc. The process continues superiorly and inferiorly until all slices are segmented. The rationale is based on the fact that there is often a high similarity and correlation between two adjacent MR slices. Each slice is only 2 mm thick.
Figure 5.
The scheme for segmentation of all MR slices based on the central slice. After the breast region in the center slice (the mth slice) is first segmented, the chest model for the mth slice is then used as reference for the m − 1th slice and the m + 1th slice. The boundary for m + 1th is used for m + 2th; the boundary for m − 1th is used for m − 2th, etc. The process continues superiorly and inferiorly until all slices are segmented.
Figure 6 shows the segmentation results in the adjacent images in the superior (slice-44) and inferior (slice-46) direction based on the central slice (slice-45) as the template. As shown in the figure, the differences between adjacent slices are relatively small. Especially in the chest body region, the location of the chest wall muscle can be assumed to have a small shift compared to the one in the neighboring slice. Figure 6 also shows the results in the far superior (slice-20) and far inferior (slice-74) directions. It can be seen that as slices move on in both directions, the images become very different, yet by starting from the middle slice and going one slice by one slice, the method works well for the entire set of 3D breast images.
Figure 6.
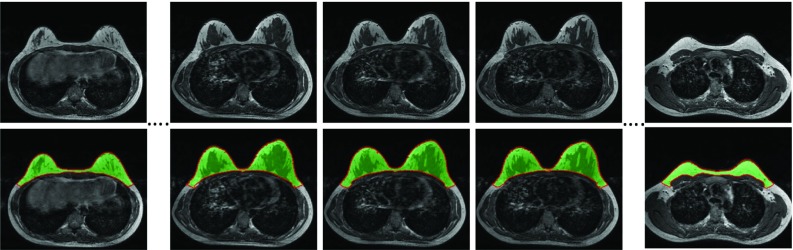
The segmentation results in the adjacent images in the superior (slice-44) and inferior (slice-46) direction based on the central slice (slice-45). As slices move on in both directions, the images become very different, yet the method can still yield excellent segmentation results in the far superior (slice-20) and far inferior (slice-74) directions. The images from left to right are from slice-20, 44, 45, 46, and 74, respectively.
After the segmentation of the breast region was completed for all slices, the k-means clustering method was used to segment the fibroglandular tissue. The detailed procedures have been published in our previous work.22 Briefly, a bias-field correction algorithm combining N3 and FCM was applied to correct for the signal intensity inhomogeneity, and k-means clustering (k = 6) was used to separate the fibroglandular tissue (the lower three intensity clusters) and the fatty tissue (the higher three intensity clusters). From our experience, since the image contrast between fat and fibroglandular tissue was very strong, the typical setting was to use a total cluster of N = 6 (3 for fat, and 3 for fibroglandular tissue); or use a total cluster of N = 5 (2 or 3 for fat, and 3 or 2 for fibroglandular tissue).3, 7, 22
Evaluation metrics
An experienced radiologist evaluated the accuracy of the segmented breast and made corrections manually by using a graphical-user-interface (GUI) program developed inhouse. Two types of corrections were made: (1) inclusion: to select the breast tissues that were missed by the algorithm; (2) exclusion: to remove the tissues that were wrongly included as part of the breast. The radiologist only needed to drag the wrongly segmented boundary near the correct location, and the program would perform fitting to find the correct boundary. The total voxels that were marked as inclusion and exclusion corrections were independently recorded. The % difference between the algorithm-measured volume and the radiologist-corrected volume was calculated. In addition, in order not to have the inclusion and exclusion errors cancel each other out, the total error percentage was calculated as the total corrected voxels (inclusion + exclusion) divided by the total number of breast voxels from the true breast region determined by the radiologist.
Since one main purpose for developing the automated breast segmentation methods is for density analysis, the total fibroglandular tissue volume analyzed based on the breast region segmented by the computer algorithm and the ground truth corrected by the radiologist was quantitatively compared. Similarly, the inclusion and exclusion errors were determined. The % difference of the fibroglandular tissue volume measured within the algorithm-measured and radiologist-measured breasts were calculated, and the total error by considering the additive effect of inclusion error and exclusion error was calculated.
RESULTS
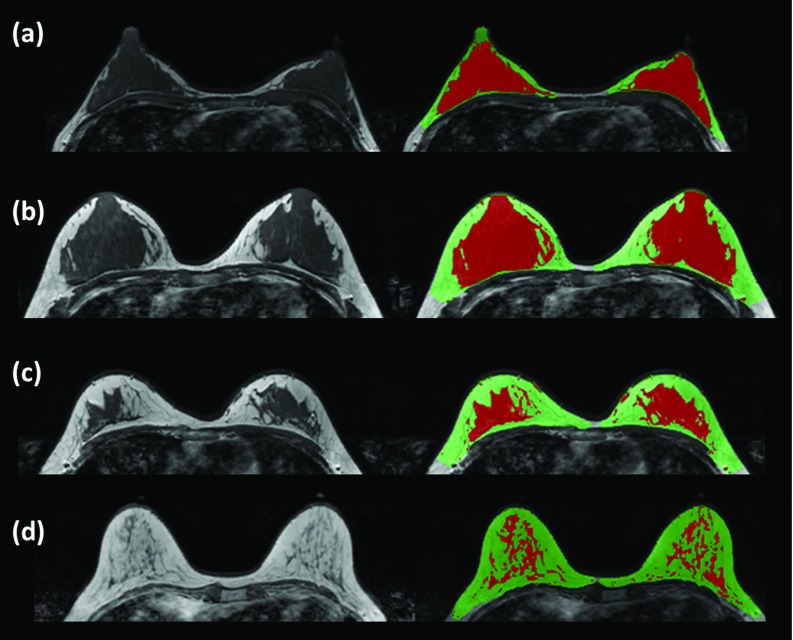
Figure 7 shows the segmentation results in four cases with different breast shapes and density patterns using the proposed method. The two cases shown in Figs. 7a, 7b have a high density, with fibroglandular tissue close to the chest wall muscle. Figure 7c shows one case with moderately dense breast and Fig. 7d shows one case with a low density. The results demonstrate that our breast segmentation method worked well, and that the chest template randomly selected from one of these 31 subjects could be used to detect the breast region in all other subjects with different breast and body shapes. The results also show that our chest wall muscle detection method worked well to exclude the muscle while preserving fibroglandular tissues nearby.
Figure 7.
The segmentation results in four cases with different breast shapes and density patterns. (a) and (b) Two cases with a high density and fibroglandular tissue close to the chest wall muscle; (c) one case with moderately dense breast; (d) one case with low density.
The proposed method was implemented in MATLAB and run on a laptop computer with the Intel Core i7 2.5 Ghz CPU and 8 GB RAM. After the template was generated, all other procedures were fully automatic without the need of any operator intervention. The execution time for analyzing cases with the resolution of 384 × 384 × 90 ranged from 2 to 4 min. The proposed method can be further speeded up by at least 3–4 times with GPU implementation on C/C++.
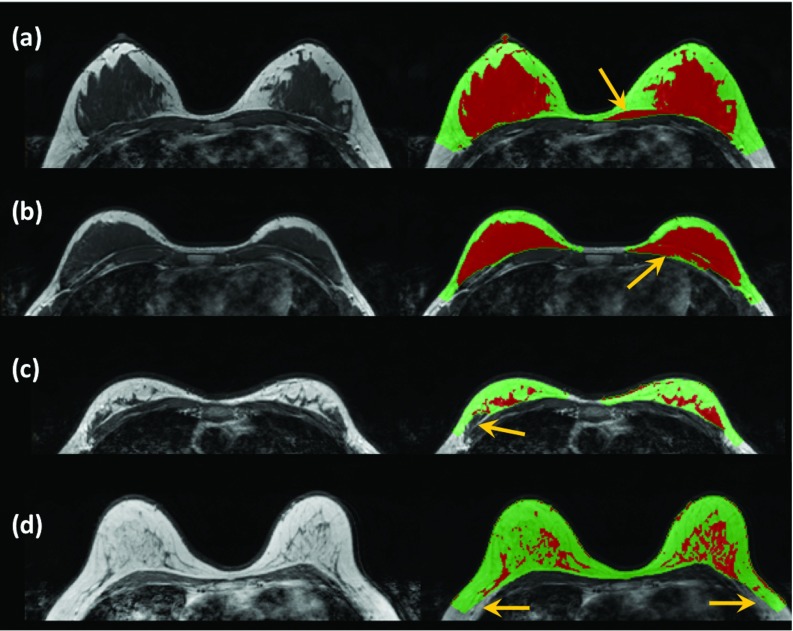
Figure 8 shows four examples of the imaging slices with wrong segmentation that need correction. Figures 8a, 8b have the fibroglandular tissues closely connected to the chest wall muscle. As indicated by the arrows, although the fitted chest wall muscle boundary presents a smooth contour, some part of the chest wall muscle was mistakenly included into the breast region. These required “exclusion correction,” and the radiologist needed to exclude them using the correction program. Figure 8c shows a case that needed “inclusion correction.” One part of the fibroglandular tissue connected to the chest wall muscle was identified as chest wall muscle and mistakenly excluded. The radiologist needed to include it into the breast region using the correction program. In Fig. 8d, some parts of fatty tissue on both breasts were wrongly excluded by the chest model. They also required “inclusion correction” and the radiologist needed to include them back to the breast region using the correction program. Since this correction only involved fatty tissue, it does not affect the accuracy in the segmentation of fibroglandular tissue.
Figure 8.
Illustration of the corrections done in four examples using the GUI contour modification program by an experienced radiologist. (a) and (b) Two cases that have the fibroglandular tissues closely connected to the chest wall muscle. Some part of the chest wall muscle is mistakenly included into the breast region in both cases and they need to be excluded, as “exclusion error.” (c) One part of the fibroglandular tissue connected to the chest wall muscle is identified as chest wall muscle and mistakenly excluded. It needs to be included back into the breast, as “inclusion error.” (d) Parts of fatty tissue on bilateral breasts are wrongly excluded by the chest model. They need to be included back into the breast region, as “inclusion error.”
Table 1 shows the results of the breast volume segmented using the template-based method and the results after the radiologist's correction. The whole breast volume (including both sides, after radiologist's correction) ranges from 376 to 1590 cm3 with a mean of 834 ± 293 cm3. The breast volume measured by the proposed algorithm is very close to the radiologist's corrected volume, with the % difference ranging from 0.01%–3.04% and a mean of 0.86% ± 0.72%. The volume of exclusion correction ranges from 0.28 to 46.11 cm3 with a mean of 15.16 ± 11.35 cm3. The volume of inclusion correction ranges from 0 to 35.87 cm3 with a mean of 10.11 ± 8.86 cm3. The total error percentage by considering both inclusion and exclusion pixels ranges from 0.05% to 6.75% with a mean of 3.05% ± 1.93%.
Table 1.
The breast volume segmented by using the proposed template-based algorithm, the corrected volume by an experienced radiologist, the % difference, and the inclusion, exclusion and total % errors.
| Breast | Breast | Inclusion | Exclusion | |||
|---|---|---|---|---|---|---|
| vol (cm3) | vol (cm3) | Difference | correction | correction | Total | |
| Cases | algorithm | radiologist | % | vol (cm3) | vol (cm3) | error % |
| 1 | 704 | 706 | −0.22% | 14.1 | 12.6 | 3.78% |
| 2 | 1292 | 1287 | 0.36% | 13.8 | 18.5 | 2.51% |
| 3 | 1545 | 1540 | 0.30% | 3.08 | 7.74 | 0.70% |
| 4 | 1087 | 1083 | 0.35% | 14.3 | 18.2 | 3.00% |
| 5 | 730 | 718 | 1.65% | 8.24 | 20.1 | 3.95% |
| 6 | 561 | 550 | 1.88% | 2.33 | 12.7 | 2.72% |
| 7 | 747 | 739 | 1.07% | 2.41 | 10.3 | 1.73% |
| 8 | 898 | 881 | 1.89% | 16.9 | 33.5 | 5.72% |
| 9 | 802 | 801 | 0.11% | 12.7 | 13.5 | 3.27% |
| 10 | 912 | 905 | 0.83% | 24.2 | 31.6 | 6.17% |
| 11 | 934 | 932 | 0.29% | 19.7 | 22.3 | 4.51% |
| 12 | 817 | 810 | 0.89% | 6.98 | 14.2 | 2.61% |
| 13 | 784 | 779 | 0.53% | 24.2 | 28.4 | 6.75% |
| 14 | 343 | 341 | 0.54% | 6.32 | 8.16 | 4.25% |
| 15 | 1600 | 1590 | 0.64% | 35.9 | 46.1 | 5.16% |
| 16 | 1066 | 1061 | 0.39% | 21.2 | 25.4 | 4.39% |
| 17 | 734 | 736 | −0.19% | 11.0 | 9.65 | 2.81% |
| 18 | 423 | 418 | 1.05% | 10.6 | 15.0 | 6.13% |
| 19 | 717 | 702 | 2.11% | 11.9 | 26.7 | 5.48% |
| 20 | 960 | 953 | 0.77% | 5.98 | 13.3 | 2.03% |
| 21 | 380 | 376 | 1.09% | 1.42 | 5.52 | 1.83% |
| 22 | 877 | 890 | −1.49% | 17.8 | 4.54 | 2.55% |
| 23 | 949 | 949 | 0.01% | 0.23 | 0.28 | 0.05% |
| 24 | 912 | 899 | 1.41% | 0 | 12.7 | 1.39% |
| 25 | 623 | 624 | −0.16% | 1.68 | 0.67 | 0.38% |
| 26 | 441 | 439 | 0.61% | 0 | 2.66 | 0.60% |
| 27 | 752 | 759 | −1.00% | 8.16 | 0.57 | 1.16% |
| 28 | 1033 | 1033 | −0.02% | 3.26 | 3.09 | 0.61% |
| 29 | 868 | 842 | 3.04% | 5.11 | 30.8 | 4.13% |
| 30 | 686 | 680 | 0.88% | 0 | 5.96 | 0.87% |
| Mean | 839 | 834 | 0.86%1 | 10.11 | 15.16 | 3.05% |
| Stdev | 294 | 293 | 0.72%1 | 8.86 | 11.35 | 1.93% |
The mean % difference was calculated by considering the absolute values of the % difference from all cases, so the undermeasured and overmeasured cases would not have a cancellation effect.
Table 2 shows the results of the fibroglandular tissue volume measured based on the breast segmented using the automatic template-based method and the radiologist's corrected breast. The volume of the fibroglandular tissues (including both sides, within radiologist-measured breasts) ranges from 41 to 513 cm3 with a mean of 180 ± 116 cm3. The fibroglandular tissue segmented within the breast region determined by the algorithm and the radiologist are also very close, with the differences ranging from 0.02% to 2.52% and a mean of 1.03% ± 1.03%. The exclusion volume ranges from 0.18 to 27.35 cm3 with a mean of 3.48 ± 5.17 cm3. The inclusion volume ranges from 0 to 9.35 cm3 with a mean of 1.51 ± 1.93 cm3. The total error percentage by adding the inclusion and exclusion pixels ranges from 0.16% to 11.8% with a mean of 2.89% ± 2.55%. The largest total error of 11.8% is seen in case #22 (in Table 2), but interestingly, the% difference between the two methods was only −0.73%. In this case, the inclusion error was 9.35 cm3 and exclusion error was 8.26 cm3, and they canceled each other out leading to a close measurement of the fibroglandular volume with a very small error of 0.73%. This case illustrates why we need to consider both inclusion and exclusion errors separately for a detailed understanding about the sources of errors.
Table 2.
The fibroglandular tissue volume segmented within the breast region obtained by using the proposed template-based algorithms and within the breast region corrected by the radiologist, the % difference, and the inclusion, exclusion, and total % errors.
| Fibrogland | Fibrogland | Inclusion | Exclusion | |||
|---|---|---|---|---|---|---|
| vol (cm3) | vol (cm3) | Difference | correction | correction | Total | |
| Cases | algorithm | radiologist | error % | vol (cm3) | vol (cm3) | error % |
| 1 | 134.4 | 134.0 | 0.29% | 0.42 | 0.81 | 0.92% |
| 2 | 129.6 | 128.5 | 0.90% | 1.22 | 2.38 | 2.80% |
| 3 | 94.69 | 93.53 | 1.24% | 0.47 | 1.63 | 2.25% |
| 4 | 518.0 | 513.0 | 0.98% | 2.14 | 7.15 | 1.81% |
| 5 | 248.2 | 246.9 | 0.55% | 0.12 | 1.48 | 0.65% |
| 6 | 104.9 | 103.4 | 1.49% | 0.62 | 2.16 | 2.69% |
| 7 | 243.7 | 241.8 | 0.77% | 1.47 | 3.32 | 1.98% |
| 8 | 163.7 | 162.5 | 0.70% | 0.41 | 1.54 | 1.20% |
| 9 | 41.39 | 41.24 | 0.36% | 0.47 | 0.62 | 2.64% |
| 10 | 231.0 | 228.4 | 1.13% | 1.54 | 4.12 | 2.48% |
| 11 | 197.2 | 196.6 | 0.32% | 0.21 | 0.84 | 0.53% |
| 12 | 97.11 | 96.24 | 0.90% | 1.54 | 2.41 | 4.10% |
| 13 | 88.03 | 87.48 | 0.63% | 0.68 | 1.23 | 2.18% |
| 14 | 48.48 | 48.04 | 0.92% | 1.25 | 1.69 | 6.12% |
| 15 | 49.22 | 48.01 | 2.52% | 1.02 | 2.23 | 6.77% |
| 16 | 107.2 | 105.2 | 1.83% | 0.65 | 2.58 | 3.07% |
| 17 | 102.0 | 101.7 | 0.31% | 1.02 | 1.34 | 2.32% |
| 18 | 108.1 | 109.1 | −0.88% | 1.85 | 0.89 | 2.51% |
| 19 | 62.43 | 61.94 | 0.79% | 0 | 0.49 | 0.79% |
| 20 | 136.5 | 135.5 | 0.72% | 1.45 | 2.43 | 2.86% |
| 21 | 209.7 | 207.5 | 1.09% | 2.12 | 4.39 | 3.14% |
| 22 | 148.0 | 149.0 | −0.73% | 9.35 | 8.26 | 11.8% |
| 23 | 274.7 | 274.8 | −0.02% | 0.25 | 0.18 | 0.16% |
| 24 | 232.5 | 223.4 | 4.07% | 2.69 | 11.79 | 6.48% |
| 25 | 157.4 | 157.1 | 0.20% | 0.23 | 0.54 | 0.49% |
| 26 | 151.5 | 150.7 | 0.58% | 1.43 | 2.3 | 2.48% |
| 27 | 200.0 | 200.1 | −0.05% | 0.91 | 0.81 | 0.86% |
| 28 | 360.9 | 359.4 | 0.42% | 1.39 | 2.89 | 1.19% |
| 29 | 474.5 | 453.9 | 4.54% | 6.77 | 27.35 | 7.52% |
| 30 | 334.9 | 332.0 | 0.87% | 1.57 | 4.45 | 1.81% |
| Mean | 182 | 180 | 1.03%1 | 1.51 | 3.48 | 2.89% |
| Stdev | 118 | 116 | 1.03%1 | 1.93 | 5.17 | 2.55% |
The mean % difference was calculated by considering the absolute values of the % difference from all cases, so the undermeasured and overmeasured cases would not have a cancellation effect.
The percent density was calculated as the volume of the fibroglandular tissue divided by the volume of the breast (×100%, using radiologist-corrected results), which ranged from 3.02% to 55.2%, with a mean of 23.35% ± 14.33%. The percent density calculated using algorithm-analyzed results were very close: 23.40% ± 14.39%. The subjects analyzed in this study were healthy Asian women with a mean of 42 years old, and they have a higher mean percent density compared to our previously reported results from normal breasts of diagnostic patients in our institute, e.g., 17% reported in Chang et al.3
DISCUSSION
In this study, we report on an automatic chest template-based method for segmentation of the breast on MRI. Most model-based breast segmentation methods reported in the literature use the breast as the template to initialize the segmentation.12, 14 The breasts often have a large variability in shape and density pattern, and it is difficult to define a universal breast template that works well for all types of breast. The chest body region, on the other hand, often presents an ellipse shape on breast MR images and it is much easier to fit its contour for initial exclusion to facilitate further breast segmentation. Although the entire chest body region may vary substantially among individual subjects, the shape is similar in the middle slice and the demons algorithm is robust enough to achieve a good registration quality. Therefore, we propose to use the chest body region in the middle slice as the starting slice to obtain the subject-specific chest model in each case. Since the chest region from all women is similar at the middle location, this approach can be applied to most women regardless of the differences in the breast shapes and body sizes. After the chest region is defined at the middle slice, it is then used as the reference for adjacent slices. Again, based on the high similarity between neighboring slices, this approach has a high likelihood of success, as demonstrated in the illustrated case examples in this work.
Since there is no clear boundary to indicate where the lateral breast ends, the initial identification of the breast region is a challenging task. We have previously proposed to use a V-shape cut based on thoracic spine and the lateral boundary of pectoral muscle7 or a horizontal cut based on the sternum3 to determine the posterior boundaries of the bilateral breasts. In this study, since the chest region that includes the three landmarks used for V-shape cut is mapped out, they can be easily identified and transformed to the subject's image space to perform the initial V-shape cut. After the V-shape lines are determined on the middle slice, they are propagated to all image slices in the entire 3D dataset. Therefore, one important advantage of using the chest model is the ability to automatically locate these body landmarks based on registration, for each individual woman. This provides a consistent criterion for performing this initial segmentation to define the breast area that will be further analyzed, without the need of operator's subjective input. Since the purpose of the V-shape cut is to determine the lateral posterior boundary of the breast, even if the three body landmarks are not precisely located, the formed V-lines can still define the breast boundary reasonably well, as in the case illustrated in Fig. 3b.
The segmentation needs to be done slice-by-slice, which may be time-consuming even using algorithms. The nonrigid registration methods are often criticized as a time-consuming process; therefore, we only apply the demons algorithm to the middle slice. As shown in Fig. 6, since the slice thickness is only 2 mm, the chest region has high similarity in neighboring slices, and we can use the segmented chest and the muscle for mth slice as the reference for adjacent m + 1th and m − 1th slices. The boundary for m + 1th is used for m + 2th; the boundary for m − 1th is used for m − 2th, etc. The chestwall muscle detection method is based on the signal intensity contrast between the muscle and the nearby fatty tissue right above the muscle. On nonfat-sat images, the muscle is dark and the fat is bright, so the contrast is very strong to help determine a precise boundary. For MR images that are acquired using different scan protocols and parameters, the method needs to be modified. For example, if fat suppression is used, the fatty tissue will appear much darker than in nonfat-sat images analyzed in this study, and it will be much more difficult to differentiate them from the chest wall muscle. The edge detection method used in this study may not work well on fat-sat images.
For studies that use a smaller field of view or only contain part of the chest region, the template used in this study may not be valid any more. However, as shown in Fig. 3b case, as long as a major part of chest is present, since the segmentation process is mainly focused in the anterior part of the chest region, our method is still applicable. We have applied this method and the template used in this study to several nonfat-sat image datasets that were acquired using different imaging protocols and different MR systems (GE, Siemens, and other Philips scanners), and obtained excellent results without the need of further modifying the program. This method is based on excluding the chest region; so one critical factor that is needed for this method to work is a good contrast between the chest (dark intensity lung tissue), chest wall muscle (medium intensity), and the fat breast tissues (bright intensity). The typical nonfat-suppressed breast image has a strong contrast to differentiate these three different tissues, so the method is robust and the results are not dependent on the choice of the template or the patient's body shape and sizes. For other datasets that use a rectangle field of view and only cover a very small part of the chest, it may be possible to generate an appropriate study-specific template by only including a very small part of the chest. In this case, some modifications in the program, e.g., putting more weighting in the anterior part of the chest, may also help, but this needs to be further investigated.
We did a careful evaluation of the segmentation quality by using the slice-by-slice correction done by an experienced radiologist. As shown in the case examples in Figs. 678, our framework by incorporating various algorithms provides a fully automatic, robust, and fast method for breast segmentation in the analyzed datasets. Specifically, we evaluated the inclusion error and exclusion error separately to better characterize the source of errors. When only the segmented volumes were concerned, the inclusion and exclusion errors canceled each other out, and that led to a very good estimate of volume, with a mean of <1% error in segmented breast volume and approximately 1% error in the fibroglandular tissue volume. When comparing the inclusion and exclusion errors, the exclusion error was seen more often with more pixels [e.g., in Figs. 8a, 8b, part of muscle was wrongly included into the breast and needed to be excluded]. In breast segmentation results summarized in Table 1, of the total of 30 cases, the algorithm-estimated breast volume was greater than radiologist's corrected volume in 26 cases. The volume of exclusion correction ranged from 0.28 to 46.11 cm3 with a mean of 15.16 ± 11.35 cm3, which was greater compared to the inclusion correction ranging from 0 to 35.87 cm3 with a mean of 10.11 ± 8.86 cm3. The total error percentage by considering both inclusion and exclusion pixels ranged from 0.05% to 6.75% with a mean of 3.05% ± 1.93%. This total error of 3% was still very small. Most errors were minor and happened in the posterior breast regions where the tissue contrast was lower. The three main source of errors were: part of chest wall muscle was included into breast region [Figs. 8a, 8b], dense tissue very close to the muscle was excluded [Fig. 8c], or fatty tissue near the chest boundary was excluded [Fig. 8d].
Since we did a slice-by-slice correction, the errors could be carefully inspected for developing more sophisticated correction algorithms in the future. The wrong segmentation in Figs. 8a, 8b came from the noise in chest wall muscle (the bright tissues inside the chest wall muscle). In this method, the potential boundary points of the chest wall muscle were selected using edge detection, and this might not work well when there were artifacts inside the chest wall muscle. In order to solve this problem, the potential boundary points can be selected from the longest continuously connected edge.23 The segmentation error in Figs. 8c, 8d came from the wrong chest region exclusion. For the middle slice, this came from the registration error to the template. For all other slices, it might come from the suboptimal reference of the chest region built based on previous slices. Different solutions are needed for solving different problems, but their pros and cons need to be carefully evaluated to find optimal correction methods.
CONCLUSION
In summary, we demonstrate a new automatic chest template-based breast segmentation method for nonfat-sat breast MR images in this study. Unlike most of the model-based breast segmentation methods that use the breast region as the template, we used the chest body region as the template. Since these reported methods were developed for different studies that were acquired using different MR imaging protocols with different tissue contrasts, it was difficult to make a direct comparison about their performances. The bottom line is: before a particular method is applied, the reliability needs to be evaluated through careful error analysis. The method proposed in this work is fully automatic, and can achieve satisfactory results with small errors in cases with different types of breast shapes and different breast density patterns. This method may provide a great tool for reliable volumetric measurement of breast density based on nonfat-sat MRI. For the fat-sat images, since the contrast near the chest wall muscle is not as clear, the program may not work well and will need further modification.
ACKNOWLEDGMENTS
This work was supported in part by NIH R01 CA127927, R21 CA170955, R03 CA136071, California BCRP #16GB-0056, and the National Science Council of Taiwan NSC-98-2221-E-039-009.
References
- Boyd N. F., Guo H., Martin L. J., Sun L., Stone J., Fishell E., Jong R. A., Hislop G., Chiarelli A., Minkin S., and Yaffe M. J., “Mammographic density and the risk and detection of breast cancer,” N. Engl. J. Med. 356, 227–236 (2007). 10.1056/NEJMoa062790 [DOI] [PubMed] [Google Scholar]
- McCormack V. A. and Silva I. D. S., “Breast density and parenchymal patterns as markers of breast cancer risk: A meta-analysis,” Cancer. Epidem. Biomar. 15, 1159–1169 (2006). 10.1158/1055-9965.EPI-06-0034 [DOI] [PubMed] [Google Scholar]
- Chang D. H., Chen J. H., Lin M., Bahri S., Yu H. J., Mehta R. S., Nie K., Hsiang D. J., Nalcioglu O., and Su M. Y., “Comparison of breast density measured on MR images acquired using fat-suppressed versus nonfat-suppressed sequences,” Med. Phys. 38, 5961–5968 (2011). 10.1118/1.3646756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khazen M., Warren R. M., Boggis C. R., Bryant E. C., Reed S., Warsi I., Pointon L. J., Kwan-Lim G. E., Thompson D., Eeles R., Easton D., Evans D. G., and Leach M. O., “A pilot study of compositional analysis of the breast and estimation of breast mammographic density using three-dimensional T1-weighted magnetic resonance imaging,” Cancer Epidemiol. Biomarkers Prev. 17, 2268–2274 (2008). 10.1158/1055-9965.EPI-07-2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klifa C., Carballido-Gamio J., Wilmes L., Laprie A., Lobo C., Demicco E., Watkins M., Shepherd J., Gibbs J., and Hylton N., “Quantification of breast tissue index from MR data using fuzzy clustering,” Conf. Proc. IEEE Eng. Med. Biol. Soc. 3, 1667–1670 (2004). [DOI] [PubMed] [Google Scholar]
- Lee N. A., Rusinek H., Weinreb J., Chandra R., Toth H., Singer C., and Newstead G., “Fatty and fibroglandular tissue volumes in the breasts of women 20-83 years old: Comparison of x-ray mammography and computer-assisted MR imaging,” AJR, Am. J. Roentgenol. 168, 501–506 (1997). 10.2214/ajr.168.2.9016235 [DOI] [PubMed] [Google Scholar]
- Nie K., Chen J. H., Chan S., Chau M. K., Yu H. J., Bahri S., Tseng T., Nalcioglu O., and Su M. Y., “Development of a quantitative method for analysis of breast density based on three-dimensional breast MRI,” Med. Phys. 35, 5253–5262 (2008). 10.1118/1.3002306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Engeland S., Snoeren P. R., Huisman H., Boetes C., and Karssemeijer N., “Volumetric breast density estimation from full-field digital mammograms,” IEEE Trans. Med. Imaging 25, 273–282 (2006). 10.1109/TMI.2005.862741 [DOI] [PubMed] [Google Scholar]
- Lu W., Yao J., Lu C., Prindiville S., and Chow C., “DCE-MRI segmentation and motion correction based on active contour model and forward mapping,” in Proceedings of the Seventh ACIS International Conference on Software Engineering, Artificial Intelligence, Networking, and Parallel/Distributed Computing (IEEE Computer Society, 2006), pp. 208–212.
- Wang Q., Ding H., and Wang G., “Multistage processing procedure for 4D breast MRI segmentation,” Conf. Proc. IEEE Eng. Med. Biol. Soc. 3036–3039 (2008). [DOI] [PubMed] [Google Scholar]
- Wang L., Filippatos K., Friman O., and Hahn H. K., “Fully automated segmentation of the pectoralis muscle boundary in breast MR images,” Proc. SPIE 7963, 796309 (2011). 10.1117/12.877645 [DOI] [Google Scholar]
- Gallego C. and Martel A. L., “Automatic model-based 3D segmentation of the breast in MRI,” Proc. SPIE 7962, 796215 (2011). 10.1117/12.877712 [DOI] [Google Scholar]
- Gubern-Mérida A., Kallenberg M., Marti R., and Karssemeijer N., “Fully automatic fibroglandular tissue segmentation in breast MRI: An atlas-based approach,” in MICCAI Workshop: Breast Image Analysis, Toronto, Canada, 2011.
- Reed V. K., Woodward W. A., Zhang L., Strom E. A., Perkins G. H., Tereffe W., Oh J. L., Yu T. K., Bedrosian I., Whitman G. J., Buchholz T. A., and Dong L., “Automatic segmentation of whole breast using atlas approach and deformable image registration,” Int. J. Radiat. Oncol., Biol., Phys. 73, 1493–1500 (2009). 10.1016/j.ijrobp.2008.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertas G., Gulcur H. O., Osman O., Ucan O. N., Tunaci M., and Dursun M., “Breast MR segmentation and lesion detection with cellular neural networks and 3D template matching,” Comput. Biol. Med. 38, 116–126 (2008). 10.1016/j.compbiomed.2007.08.001 [DOI] [PubMed] [Google Scholar]
- Otsu N., “A threshold selection method from gray-level histograms,” IEEE Trans. Syst. Man Cybern. 9, 62–66 (1979). 10.1109/TSMC.1979.4310076 [DOI] [Google Scholar]
- Hearn D. and Baker M., Computer Graphics C Version, 2nd ed. (Prentice Hall, Upper Saddle River, NJ, 1997). [Google Scholar]
- Kroon D. J. and Slump C. H., “MRI modality transformation in demon registration,” in Proceedings of the IEEE International Symposium on Biomedical Imaging (ISBI) (IEEE ISBI, 2009), pp. 963–966.
- Thirion J. P., “Image matching as a diffusion process: An analogy with Maxwell's demons,” Med. Image Anal. 2, 243–260 (1998). 10.1016/S1361-8415(98)80022-4 [DOI] [PubMed] [Google Scholar]
- Canny J., “A computational approach to edge detection,” IEEE Trans. Pattern Anal. Mach. Intell. PAMI-8, 679–698 (1986). 10.1109/TPAMI.1986.4767851 [DOI] [PubMed] [Google Scholar]
- Nie K., Chang D., Chen J.-H., Shih T.-C., Hsu C.-C., Nalcioglu O., and Su M.-Y., “Impact of skin removal on quantitative measurement of breast density using MRI,” Med. Phys. 37, 227–233 (2010). 10.1118/1.3271353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M., Chan S., Chen J. H., Chang D., Nie K., Chen S. T., Lin C. J., Shih T. C., Nalcioglu O., and Su M. Y., “A new bias field correction method combining N3 and FCM for improved segmentation of breast density on MRI,” Med. Phys. 38, 5–14 (2011). 10.1118/1.3519869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Weinstein S. P., Conant E. F., Localio A. R., Schnall M. D., and Kontos D., “Fully automated chest wall line segmentation in breast MRI by using context information,” Proc. SPIE 8315, 831507 (2012). 10.1117/12.911612 [DOI] [Google Scholar]