Abstract
Transcriptional regulation is one of the most important steps in control of cell identity, growth, differentiation and development. Many signaling pathways controlling these processes ultimately target the core transcription machinery that, for protein coding genes, consists of RNA polymerase II (Pol II) and the general transcription factors (GTFs). New studies on the structure and mechanism of the core assembly and how it interfaces with promoter DNA and coactivator complexes have given tremendous insight into early steps in the initiation process, genome-wide binding, and mechanisms conserved for all nuclear and archaeal Pols. Here we review recent developments in dissecting the architecture of the Pol II core machinery with a focus on early and regulated steps in transcription initiation.
The transcription initiation pathway
None of the multi subunit RNA polymerases (Pols) specifically initiate transcription at promoter sequences – all require accessory factors. Although bacterial Pols use a family of sigma factors to recognize core promoters and interface with many transcriptional regulators [1], archaeal and eukaryotic Pols use a complex set of general transcription factors (GTFs) that function in promoter recognition, Pol recruitment, interaction with regulatory factors, DNA unwinding, and transcription start site (TSS) recognition [2–4]. The earliest intermediate, and a frequently regulated step for expression of protein coding genes, is formation of the preinitiation complex (PIC), containing Pol II, TFIIA, TATA binding protein (TBP), TFIIB, TFIIF, TFIIE, and TFIIH. In the PIC, a network of DNA-GTF interactions anchors Pol II to double stranded promoter DNA that lies outside the Pol II active site cleft. Next, TFIIH promotes ATP-dependent unwinding of about 10 base pairs of promoter DNA, leading to the open complex state (OC). Pol II then locates the TSS and eventually transitions into a processive elongation mode. In metazoans, this transition frequently involves pausing of Pol II at many developmentally regulated genes and is a commonly regulated step [5, 6].
It’s important to note that nearly all biochemical and structural studies have examined these steps at only TATA-containing promoters. Although TATA promoters account for only 20–30% of eukaryotic promoters [7–9], they are typically the most highly regulated. By contrast, TATA-less promoters often direct transcription of housekeeping genes and have heterogeneous TSSs. The biochemical study of initiation at TATA-less promoters has been hampered by difficulties in PIC assembly and low initiation activity. Here, we review recent advances in understanding the function of the GTFs, the architecture of the initiation complex, and new insights into the mechanism of transcription initiation and its regulation.
Architecture of the preinitiation complex (PIC)
After the GTFs were identified, genes cloned, and structures of Pol II and some individual GTFs were solved, the next challenges were to understand how these factors worked together to promote initiation and interface with regulatory factors and promoter DNA. Pioneering studies utilizing DNA-protein crosslinking of human PICs showed that Pol II is at the center of the PIC, covering over 60 bp of promoter DNA [10, 11]. TBP, TFIIB, and TFIIF primarily contact DNA upstream of the TSS. TFIIE is positioned to overlap the TSS. The TFIIH ATPase subunit Ssl2/XPB, required for promoter opening, is located at the downstream end of the PIC. Over the past ten years, a combination of protein biochemistry [12–16], biophysical measurements [17], x-ray structures [18–20] and, most recently, cryo EM of the PIC and intermediates in the assembly pathway [21] have resulted in tremendous advances in understanding the architecture of the PIC and the role of the GTFs. This work has set the stage for probing the mechanism of initiation and understanding how coactivators and other regulators interact with the core transcription machinery.
The role of TFIIB in PIC architecture and allosteric regulation of Pol II activity
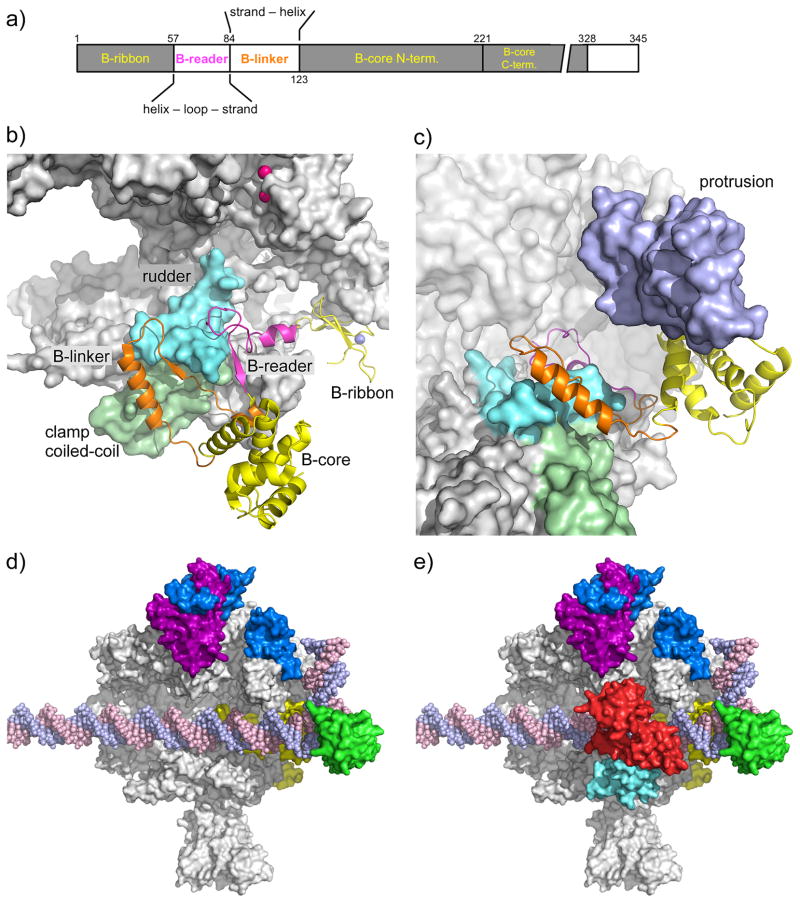
TBP binding to TATA bends DNA by ~90 degrees, forming an asymmetric platform for PIC assembly [22, 23]. The GTF TFIIA binds TBP-DNA, interacting with DNA upstream of TATA, stabilizing the protein DNA complex, and blocking regulators such as Mot1 and NC2 from binding TBP [24, 25]. TFIIB, a factor that bridges TBP and Pol II, contains 4 functional domains termed B-ribbon, reader, linker, and core (comprised of two cyclin-like repeats) [18, 26, 27] and all of these domains interact with Pol II (Fig 1a,b). From x-ray and biochemical analysis, the B-ribbon is strongly anchored to the Pol II dock domain. The most striking advances in understanding TFIIB-Pol II interactions are recent x-ray structures of Pol II-TFIIB complexes and cryo EM of PIC intermediates. Early structural work reported that the B-reader and linker regions formed a hairpin, termed the B-finger, positioned within the Pol II cleft and close to the active site [28], however, later work has so far failed to support this model. Instead, the B-reader helix enters the Pol II active site cleft through the RNA exit channel where it positions the reader B-loop on the Pol II rudder domain and Rpb2 fork loop 1 [18–20] (Fig 1b). These interactions locate the B-loop distant from the Pol II catalytic site, meaning that TFIIB must exert indirect effects on initial Pol II catalysis. Consistent with this model, TFIIB binding leads to ordering of the Pol II protrusion domain, with which it only peripherally interacts, and induces a second Mg atom to bind the Pol II active site [19] (Fig 1b,c). The findings that the TFIIB reader stimulates transcription in several systems, despite its location away from the active site, is further evidence for the allosteric model [18, 29]. The Blinker binds to the coil-coil region of the Pol II clamp where it connects to the TFIIB N-terminal cyclin repeat located on the Pol II wall [18–20]. As the B-core cyclin repeats also bind TBP-DNA, its location on the wall dictates the DNA path across the opening of the Pol II cleft [30].
Fig. 1. Location of the general factors in the Pol II PIC.
a) TFIIB domain organization. The domain color code (colored lettering) is used in panels a–c.
b) Pol II-TFIIB complex [19]. TFIIB is shown as a cartoon, Pol II as a grey surface. Rpb2 is removed for clarity. The Rpb1 rudder (cyan) and clamp coiled-coil (light green) are highlighted. The TFIIB Zn-atom is shown as blue sphere, the two Mg-atoms in the Pol II active site are shown as pink spheres.
c) TFIIB binding to Pol II results in ordering of the Rpb2 protrusion. To visualize the protrusion (light purple), Rpb2 (partially transparent) has been added and Pol II has been rotated upwards by ~40 degrees compared to panel 1b.
d) TFIIF binding to the PIC. As positioned by site-specific protein cleavage and crosslinking assays [13, 15], dimerization domains of TFIIF subunits Tfg1 (deep purple) and Tfg2 (blue) are located on the Pol II lobe. A C-terminal winged helix (WH) domain in Tfg2 resides on top of the Pol II protrusion domain, close to upstream promoter DNA. In contrast, a cryo EM structure suggested that the human TFIIF WH lies just downstream from the TFIIB core domain (not shown). TFIIB is shown in yellow, TBP in green. EM showed that promoter DNA is slightly bent over the cleft by ~ 18 degrees and the Pol II clamp is in an open state to accommodate dsDNA for the initial step in OC formation [21].
e) A triple WH structure of TFIIE spans the Pol II cleft. As positioned by site-specific protein cleavage assays [16], Tfa1 WH domain (cyan) anchors TFIIE on the Pol II clamp and dimerizes with the Tfa2 tandem WH domain (red) that encircles promoter DNA at positions 18 to 8 relative to the human TSS at +1.
TFIIF may modulate opening of the Pol II clamp
TFIIF enters the PIC along with Pol II [31] and consists of two conserved polypeptides (yeast Tfg1/Tfg2; human Rap74/30), each containing an N-terminal dimerization domain and C-terminal winged helix (WH). Both yeast and human TFIIF have unrelated insertions within the dimerization domain [15]; the human insertion is termed the TFIIF arm [32]. Biochemical probes and crosslinking have shown that the TFIIF dimerization domain binds to the lobe domain of the Pol II subunit Rpb2 [13, 15]. The Tfg2 WH domain binds to the Pol II protrusion with the linker connecting the two TFIIF domains on top of the protrusion and cleft [16] (Fig 1d). Strikingly, these two locations are far from the active site, suggesting that TFIIF somehow exerts an indirect effect on Pol II activity. It was reported in the human system that once TFIIF promotes PIC formation, it could be removed with little or no loss of transcription activity [33], suggesting an allosteric role during PIC assembly.
A new EM structure suggests a striking and unexpected allosteric role for TFIIF: opening of the Pol II clamp [21]. X-ray analysis has captured various forms of Pol II in either the clamp open or closed conformations; in the open state, the clamp is wide enough for double stranded DNA to enter, but is only wide enough for single stranded DNA in the closed state. Until now, only Pol II lacking the subunits Rpb4/7 (comprising the stalk domain) has been observed in the open state. In contrast, all previous structures of complete 12 subunit Pol II are in the closed state and it was suggested that subunits Rpb4/7 act as a wedge to close the clamp [34]. Whether the clamp is open or closed has important implications for the initiation mechanism: If closed in the PIC, DNA melting must occur outside of the Pol II cleft, with single stranded DNA entering after unwinding. In bacteria, recent single molecule studies have shown that, in the closed complex (analogous to the PIC), the clamp is opened and that DNA unwinding eventually causes clamp-closure [35]. Strikingly, cryo EM analysis indicates that TFIIF addition results in clamp opening and ordering of promoter DNA over the cleft. This is completely unexpected because TFIIF is situated apart from the clamp and downstream DNA. How this could occur is a puzzle, but if borne out in future studies, will be a central advance in understanding TFIIF function.
EM also showed that the path of DNA is smoothly bent over the cleft by ~18 degrees, a trajectory that is preserved in the complete PIC. Another unexpected finding from the EM analysis is the location of the TFIIF small subunit WH domain, located near DNA adjacent to the TFIIB N-terminal cyclin-like repeat. This location is in contrast to protein-specific cleavage assays and protein-DNA crosslinking in fully assembled PICs, which showed the WH on top of the protrusion domain [15], at a location where Tfg2 can interact with DNA both upstream and downstream of the B-core domain [10, 12]. Further experiments are required to resolve these differences.
TFIIE and open complex (OC) formation
TFIIE is comprised of two subunits (yeast Tfa1/2; human TFIIEα/β) with Tfa1 containing an N-terminal WH and central Zn-ribbon, and Tfa2 containing tandem WH domains. The Tfa1 WH and ribbon are essential and conserved in archaea, while the Tfa2 tandem WH domains are functionally redundant and conserved only in eukaryotes. TFIIE (and TFIIH; see below) function primarily in OC formation as pre-opening the DNA eliminates the requirement for these factors in basal transcription [29, 36]. TFIIEβ binds single stranded DNA [37], leading to the proposal that, in the unwound state, TFIIE functions in part to bind and stabilize the non-template DNA strand. TFIIE is also thought to directly interact with TFIIH, as TFIIH is recruited to the PIC only after TFIIE binds. Recent work using site-specific protein cleavage reagents indicated that the three TFIIE WH domains form a triple WH structure that lies adjacent to the TFIIB core and spans the Pol II cleft from the clamp to the protrusion [16] (Fig 1e). After DNA unwinding, either of the Tfa2 WH domains are in position to bind single stranded non-template DNA. In agreement with these results, cryo EM showed a TFIIE-dependent density in this location [21]. An additional TFIIE-dependent density was observed near the Rpb4/7 subunits and this agrees with studies in archaea showing that the ribbon domain from the orthologous factor TFE lies near the Pol stalk in an OC [38].
TFIIH architecture and its role in DNA unwinding
TFIIH, the most complex GTF, contains 10 subunits, three of which contain ATP-dependent enzymatic activities [39]: (i) CDK7, the Pol II CTD kinase, (ii) Rad3/XPD, a DNA helicase, and (iii) Ssl2/XPB, likely a DNA translocase. TFIIH functions in both general and transcription-coupled DNA excision repair (NER), where it unwinds DNA in preparation for excision of the damaged DNA strand [40]. TFIIH is also a component of the PIC where it functions in DNA unwinding during OC formation and phosphorylation of the Pol II CTD at Ser5 during initiation. Unraveling the functions of TFIIH in NER and transcription initiation is of fundamental importance, as defects in TFIIH DNA repair function lead to cancer and the human diseases Xeroderma Pigmentosum and Cocayne Syndrome [40]. More severe mutations that also disrupt transcription activity can lead to Tricothiodistrophy. Rad3/XPD helicase activity functions exclusively in repair, whereas Ssl2/XPB functions in DNA unwinding during OC formation with an additional role in DNA repair. TFIIH is comprised of several modules: core, kinase (CAK), and Ssl2, all of which can dissociate from each other during purification under non-optimal conditions.
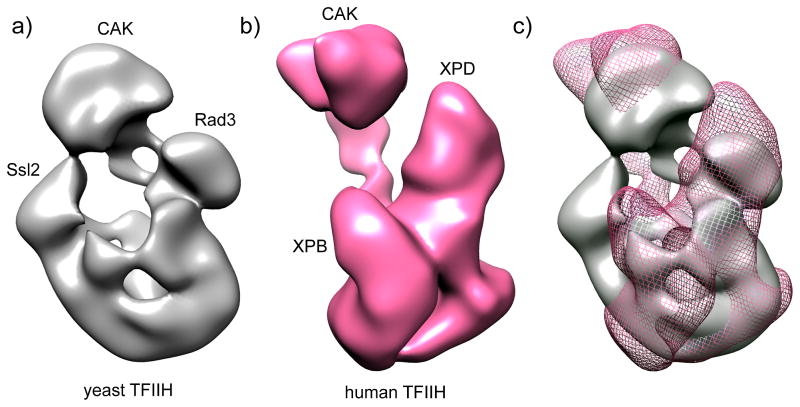
Progress in determining TFIIH architecture and mechanism has been hampered because of difficulty in biochemical purification, stability, and inherent flexibility. However, new purification methods for yeast TFIIH [29, 41], and the discovery that eliminating the Tfb6 protein increases Ssl2 stability within TFIIH, have produced quantities of TFIIH sufficient for biochemical and higher resolution structural studies. In addition, the structures of many TFIIH subunit subdomains as well as archaeal homologs of XPB and XPD were previously determined (summarized in [41]). In a recent study, holo TFIIH and several partial complexes were purified, negatively stained and examined by EM [41] (Fig 2a). This was a significant advance, as it conclusively assigned the positions of Ssl2, Rad3, and the kinase module. An EM structure of human TFIIH was also recently determined [21] (Fig 2b). Although the yeast and human structures have similar shapes, there are several significant differences (Fig 2c). Whether these are due to yeast/human differences, subunits that are invisible in the EM density, or alternate TFIIH conformations remains to be determined. Although these new structures are a leap in understanding TFIIH architecture, there is not yet sufficient resolution or other information to conclusively dock known x-ray/NMR structures into these EM densities. An important future priority is obtaining new information that will allow definitive placement of known high-resolution structures within the EM volume.
Fig. 2. Structure of TFIIH.
The modules containing the cyclin-dependent kinase (CDK)-activating kinase complex (CAK), Ssl2/XPB, and Rad3/XPD are labeled.
a) Negative stain EM image of yeast TFIIH [41]. The locations of subunits Ssl2, Rad3 and the CAK module (identified by EM analysis of TFIIH lacking these subunits or by gold labeling) are indicated.
b) Negative stain EM image of human TFIIH [21].
c) Alignment of both densities, showing yTFIIH in grey and hTFIIH as pink mesh. EM data suggest a similar general TFIIH shape with several significant differences.
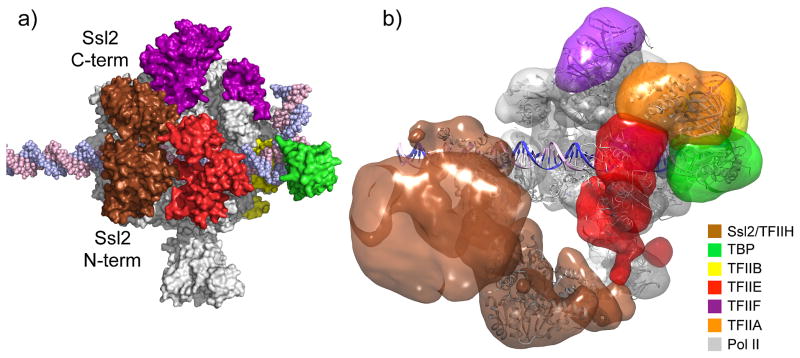
A second key question concerning TFIIH is how it fits into the PIC. Site-specific cleavage probes on TFIIE showed that the Ssl2/XPB subunit is positioned adjacent to TFIIE at the downstream edge of an isolated PIC [16] (Fig 3a), as predicted from earlier protein-DNA crosslinking and cleavage assays [10, 12]. With this fixed reference point, where is the rest of TFIIH? A recent negatively stained EM structure suggests that the TFIIH core is pointed away from the PIC and DNA, whereas Rad3/XPD and potentially the kinase module (not visible in the structure) are pointing toward the Pol II stalk [21] (Fig 3b). As mentioned below, TFIIH also contacts the coactivator Mediator, which was absent in the latest EM structure. It is likely that Mediator, and possibly other regulatory factors, will “lock down” the position of TFIIH resulting in stable structures that may be amenable to structural analysis. A 3D view of PIC assembly and overall architecture of the PIC is shown in movie S1 and 3D coordinates of the latest PIC and OC models are available as a supplemental file.
Fig. 3. Architecture of the basal PIC.
Components of the PIC are colored according to the color scheme shown in the color chart. 3D coordinates of the model and a movie are available as supplemental material.
a) PIC model based on crystal structures and biochemical mapping. Note that the two Ssl2 RecA-like domains (labeled Ssl2 C-term and Ssl2 N-term) abut TFIIE, encircling promoter DNA at positions −2 to +6 regarding the human transcription start site at +1.
b) Negative stain structure of the human PIC, containing all GTFs and Pol II [21]. EM density corresponding to the TFIIH core complex is shown in brown. Crystal structures and homology model of PIC components were docked into the EM density, suggesting Ssl2 binding to downstream promoter DNA at positions +10 to +20. Note that models in panels a and b show different downstream locations for Ssl2, either adjacent to TFIIE or in a more downstream location.
The first step in the transition from the PIC to the OC state is the ATP-dependent DNA unwinding promoted by Ssl2/XPB. It was first thought that this factor uses DNA helicase activity to unwind DNA, due to its sequence and structural similarity to DNA helicases, as well as weak 3′ to 5′ helicase activity detected with the purified XPB subunit [42]. Contrary to this model is the finding that 3′ to 5′ helicase activity has not been observed with holo TFIIH [43] and the fact that Ssl2/XPB binds downstream from the unwound DNA [10]. New mapping studies suggested that Ssl2 functions instead as an ATP-dependent DNA translocase, inserting ~ 15 bp double stranded DNA into the Pol II active site, leading to DNA unwinding [16] (Box 1). Although Pol II transcription in vitro is notoriously inefficient, with typically only a few percent of templates generating RNA transcripts, DNA unwinding is reportedly a very efficient step [44]. If borne out by future experiments, this would indicate that inefficiency in a later step of the pathway, such as transition to the elongation state, accounts for the inherent inefficiency observed in vitro.
Box 1. Ssl2/XPB translocase model and the mechanism of DNA unwinding.
The function of the TFIIH subunit Ssl2/XPB has been a key unanswered question in the mechanism of Pol II transcription. The ATPase activity of this subunit is required for Pol II promoter opening and transition to the open complex (OC). It was first presumed that Ssl2/XPB was a DNA helicase because of its sequence homology to helicases and the 3′-5′ helicase activity of the isolated human subunit [42] and its archaeal homolog [68]. However, XPB was reported to have no DNA helicase activity when it is incorporated into TFIIH [43]. Thus, the ATP-dependent mechanism by which Ssl2 drives transcription bubble formation remained obscure.
Protein-DNA crosslinking of human PICs showed that XPB binds downstream from the site of DNA opening, first suggesting that XPB does not work by a traditional helicase mechanism [10]. These studies suggested that XPB was contacting essentially the same promoter location before and after ATP addition, implying that XPB induces torsional stress by rotation, leading to DNA unwinding. However, biochemical mapping of Ssl2 position in the yeast PIC [16], combined with a model for the OC [18] and supported by recent EM of human PICs [21], has suggested a DNA translocase model for DNA opening. Comparison of the PIC and OC models shows that in addition to the opening of the promoter DNA, ~15 bp of double stranded DNA are inserted into the Pol II cleft. This suggested that Ssl2 acts as a DNA translocase, screwing ~15 bp of DNA into the cleft during OC formation (Figure I). Both DNA helicases and translocases (e.g, ATP-dependent chromatin remodeling enzymes and type I restriction enzymes) move directionally along the DNA using the energy of ATP hydrolysis [69]. The difference is that helicases unzip DNA strands as they move, whereas double stranded DNA translocases are thought to track along one strand of the DNA backbone without unwinding..
In the PIC, upstream DNA is fixed through TBP-TFIIB-Pol II binding, while the TFIIE triple winged helix structure and possibly other protein-protein interactions prevent physical movement of Ssl2. Thus, as Ssl2 tracks along the one strand of the DNA helix, it feeds DNA into the Pol II cleft through a right-handed threading of DNA. This movement leads to opening of the DNA strands to relieve the resulting tortional stress. DNA rotation resulting from translocation by the ATP-dependent remodeling enzymes has also been proposed as the basis for nucleosome movement [70]
Key experiments to test this model for promoter unwinding are to demonstrate Ssl2/XPB has double stranded DNA translocase activity and that it causes directional movement of promoter DNA during PIC formation. Confirming translocase activity and deciphering how Ssl2/XPB drives promoter opening will also be important for understanding the function of this factor in DNA repair and the molecular defects that lead to various TFIIH-related diseases. It is still unknown as to why Pol II requires this activity for OC formation whereas all other multi-subunit Pols spontaneously form OCs.
Figure I.
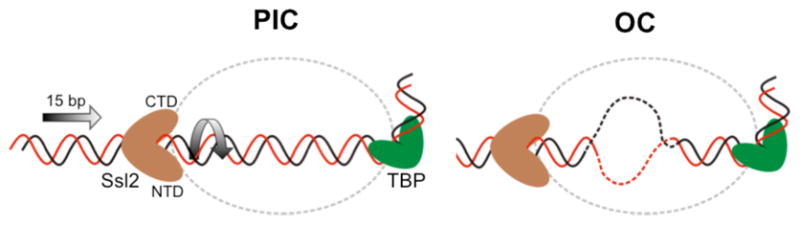
Diagram of Ssl2 function in OC formation. In the left panel, upstream promoter DNA is in a fixed position due to binding by TBP-TFIIB-Pol II. Ssl2 lies on downstream DNA at a fixed position due to interaction with TFIIE. Ssl2 feeds DNA into the cleft by a right-handed threading mechanism, rotating and unwinding DNA. Figure reused with permission from [16].
Open complex formation can be regulated in eukaryotes
OC formation in bacteria is a regulated step [1], but it was only recently found that OC formation is regulated in eukaryotes [45, 46]. In resting lymphocytes, Pol II is preloaded on most genes that are induced upon lymphocyte activation, but DNA melting at the promoter is observed only upon lymphocyte activation. This, coupled with the finding that levels of TFIIH are limiting in resting cells, suggests that transcription is stimulated upon activation by promoting OC formation. It remains to be seen whether this is a commonly regulated step in higher eukaryotes.
Architecture of the open complex and implications for the initiation mechanism
Two important questions are: what is the architecture of the OC state and, how does this structure poise Pol II for initiation? The most striking molecular details on the OC structure come from x-ray analysis of a Pol II-TFIIB complex containing a mimic of an early transcription intermediate with a short DNA template and 6 bases of RNA:DNA hybrid [19]. This structure showed that the B-reader helix contacts the DNA template strand 7–8 nucleotides upstream from the catalytic site. From this location, the B-reader is proposed to stabilize the nascent RNA:DNA hybrid and to orient template DNA and the RNA:DNA hybrid in an active, non-tilted position. From the structure of this complex, it seems that once the nascent RNA has grown to ~6 bases, it will clash with the B-linker helix, causing separation of the RNA:DNA hybrid. Further growth of the RNA chain to 12–13 bases will clash with the B-reader and B-ribbon, leading to dissociation of TFIIB from Pol II, a prediction in agreement with earlier biochemical studies [47]. Extrapolating to the OC state before RNA synthesis is initiated, the B-reader is predicted to contact single stranded template DNA 7–8 bases upstream of the initiation site. This is consistent with the base at this position being part of the yeast TSS recognition sequence. Finally, the B-linker helix is positioned near the upstream junction between single and double stranded DNA in the OC, suggesting its involvement in DNA opening or stability of the open state. Consistent with this, mutation of either the B-linker or reader causes a severe transcription defect that is restored by preopening the DNA via a heteroduplex bubble [18, 29].
Cryo EM analysis of an OC mimic also supports these models for OC architecture. This complex contained an arrested Pol II early elongation complex with a 9 base RNA:DNA hybrid and 6 bases of backtracked RNA (where the RNA 3′ end has slipped backwards past the enzyme catalytic site), as well as all GTFs [21]. The positions of the GTFs had not moved with respect to the PIC, the Pol II clamp was in the closed position as predicted, and the DNA path seemed identical to that in a Pol II elongating complex. A surprising feature observed was a new density spanning the cleft from the Pol II clamp to the TFIIF dimerization domain. This was interpreted as the TFIIF arm domain interacting with the Pol II rudder to prevent reannealing of melted DNA. However, the identity and function of this new density is not clear, as the TFIIF arm domain is not conserved and there is not yet data to show it performs an important function.
Scanning for the transcription start site (TSS)
Another unsolved problem is how Pol II locates the TSS. Whereas mammalian Pol II at TATA-containing promoters initiates from a site within the initial 10 bp of unwound DNA, initiation by human Pol II at TATA-less promoters can be distributed over a range of ~50 bp [48, 49]. In the best-studied example, transcription from yeast TATA-containing promoters typically occurs from ~50–100 bp downstream from TATA [50]. It’s thought that after initial DNA opening, Pol II scans for an appropriate TSS, perhaps because initiation at the mammalian location is blocked. Consistent with the scanning model, yeast OCs formed using heteroduplex DNA bubbles have striking flexibility, and are active when the bubbles are located 30–80 bp downstream from TATA [29]. Mutations in GTFs and Pol II can alter the position of the yeast TSS with some mutations promoting upstream initiation and others promoting initiation downstream from the normal starts [50]. What mechanisms drive the scanning, and is this a fundamentally different mechanism than occurs in mammalian systems? One possible mechanism for scanning is that movement of DNA within the Pol II active site is driven by the Ssl2 translocase – feeding DNA into the cleft in a process analogous to DNA scrunching by bacterial Pol, where Pol pulls downstream DNA into itself and past its active center during initiation to create a stressed intermediate [16, 21]. An alternative model is that movement is driven by Pol II synthesis of short abortive RNAs [51]. Answering this question will undoubtedly involve single molecule methods to visualize Pol II movement and its dependence on Ssl2/ATP vs. RNA synthesis.
The current theory is that the TSS is recognized at least in part by the B-reader sequence interacting with upstream DNA; however, other factors including Pol II likely play a role. For example, initiation on heteroduplex bubble promoters is greatly stimulated by TFIIB and TBP, although the positions of initiation do not change [29]. This suggests that Pol II itself has affinity for initiation at particular sequences. Mammalian transcription often starts at Inr sequences, and these elements play a dual role as recognition sequences for coactivator TFIID binding and TSS recognition.
Is recognition of the TSS different at TATA-less promoters?
Although biochemical methods have not been very successful in the study of TATA-less promoters, a new genome-wide method termed ChIP-exo has allowed high resolution mapping of all yeast PICs [9]. It was found that nearly all TATA-less PICs contained TBP, within the context of the coactivator TFIID, situated at a TATA-like sequence, containing one or two mismatches from the consensus TATA. TFIID in these PICs was often situated adjacent to the +1 nucleosome, suggesting a possible role of this nucleosome in PIC positioning. Constraining Pol II scanning by the +1 nucleosome was postulated to play a role in TSS selection. Consistent with this hypothesis, TSSs at TATA-less promoters are typically found at the upstream border of the +1 nucleosome [9]. Future work on interaction of the core machinery and chromatin will be very informative in analyzing the initiation and regulatory mechanisms at these promoters.
Interaction of coactivators with the core machinery
PIC formation is regulated by a series of gene-specific activators and repressors, working though mechanisms such as recruitment of the GTFs, modification of chromatin, and regulation of Pol II pausing. Coactivators, complexes that are recruited by activators to modulate transcription, have two broad types of activities: modification of chromatin and/or direct interaction with the core transcription machinery. Here, we briefly discuss several key coactivators that directly interact with the core machinery: Mediator, TFIID, and SAGA. Structural studies are beginning to unravel the architecture of these coactivators. The next challenge will be to determine how these coactivators interface with the core transcription machinery and modulate its activity.
Mediator, TFIID and SAGA architecture and interaction with the core transcription machinery
Mediator is a conserved coactivator and a common activator target [52]. The complex is comprised of four separate modules: head, middle, kinase, and tail, and structures of the head module have recently been elucidated [53–55]. Mediator was genetically identified from mutations in several subunits that can suppress truncations in the Pol II CTD and it has long been thought that CTD-Mediator contacts are a key mechanism for connection of Mediator to Pol II. Other biochemical studies have suggested that the middle and head modules interact with Pol II and a number of studies have given conflicting results. However, recent EM studies provided an explanation for these discrepancies [56]. Pol II and only the unphosphorylated CTD polypeptide, were found to strongly bind to the Mediator middle module and, to a lesser and variable extent, the head module. Consistent with biochemical and genetic results, the kinase module binds at two distinct locations within the Mediator middle module where it competes with Pol II binding. The head-Pol II interaction is consistent with in vivo crosslinks between Med17 (a head subunit) and the Pol II subunit Rpb3 [57] as well as a recent structure [54].
These recent results indicate that, in the absence of other factors and DNA, Mediator-Pol II connections are not “locked down” into a single conformation. This suggests that Mediator interactions with additional components of the core machinery are important for the final architecture of the complex. For example, TFIIF was reported to influence Pol II-Mediator binding [51, 58, 59] and Med11/22 (head subunits) are thought to interact with the TFIIH subunit Rad3 [60].
TFIID contains TBP and 12–13 TBP-associated factors (TAFs) and binds transcription activators and promoter elements, interacts with covalently modified nucleosomes, and is reported to have other enzymatic activities [61]. Several tissue-specific forms of TFIID are found in addition to canonical TFIID. In yeast, TFIID is typically utilized at TATA-less promoters [62], but it is used at a wider variety of promoters in metazoans. The large size and flexible nature of TFIID have so far prevented high resolution structures of the entire complex, but structures of a number of subdomains have been determined, as well as recent cryo EM structures of holo TFIID [63] and a key core TFIID subcomplex upon which the remaining TAFs and TBP are assembled [64]. TFIID is a three lobed complex where the position of lobe A exists in two alternate locations, differing by 100Å [63]. DNA binding and the GTF TFIIA were found to induce large structural rearrangements in the location of TFIID lobe A and the position of TATA, INR and the DPE promoter elements were mapped in this complex. Additional data on the position of TBP within TFIID and connections of TAFs to other GTFs should allow docking of TFIID into the PIC model to predict the overall shape of the complex. Important questions to be answered include whether TFIID influences the activity of the core machinery apart from recruiting TBP to the promoter, and how TFIID interaction with activators or other regulators influences TFIID conformation and its interactions with the core machinery and chromatin.
SAGA is another large coactivator complex that has several important roles: histone acetyl transferase, ubiquitin protease, transcription activator and TBP-binding [50]. SAGA is assembled around a subset of TAFs that are shared with TFIID, although a low resolution EM structure of SAGA shows that TFIID and SAGA have very different shapes [65]. SAGA is responsible for regulation of ~10% of yeast genes, typically the highly regulated TATA-containing promoters [62]. In vivo studies in yeast have shown that SAGA plays a role in TBP recruitment [66], whereas studies in mammalian cells have indicated that SAGA plays a role after PIC recruitment [67]; the reason for these differences has yet to be determined. Studies on the architecture of SAGA are at an early stage, so determining connections between the SAGA modules and the overall architecture of the complex are important areas for future work. Although genetic studies have indicated that SAGA-TBP interaction is an important aspect of function, biochemical studies have shown that this is a very weak protein-protein interaction. How this weak interaction translates into gene regulation is another important question. Finally, how SAGA associates with the PIC is unknown and, as with TFIID and Mediator, important insights into gene regulation will be obtained from learning how SAGA fits into the PIC.
Concluding remarks
Although tremendous progress has recently been made on understanding the architecture and activity of the core transcription machinery, there is much to be learned. For example, we still do not understand important aspects of how some GTFs function and their activities are modulated. In particular the structure, function, and docking of TFIIH within the PIC is poorly understood. How PICs form on TATA-less promoters – the major form of Pol II promoters in cells – is a complete mystery. Another important area for future work is to understand how coactivator complexes dock with the PIC and modulate its activity. Finally, recent work has raised the question: which factors are in PICs apart from the core machinery? Mass spectrometry analysis of mammalian PICs unexpectedly found that many chromatin remodeling complexes and elongation factors appeared to be stable PIC components and many may enter via interaction with Mediator and/or activators [67]. Is the association of these factors promoter-specific and how is their recruitment and activity regulated? The coming years promise more striking advances in understanding the mechanisms of gene regulation, insight into defects leading to human disease, and illuminating specific steps in the transcription pathway that can be targeted to overcome defects caused by mutations or epigenetic aberrations.
Supplementary Material
Box 2. Nuclear and archaeal Pols share a functionally and structurally conserved core initiation complex.
All eukaryotic and archaeal Pols share a 10 subunit catalytic core which binds additional subunits forming the heterodimeric Pol stalk and other Pol-specific subcomplexes (A49/34.5 for Pol I; C37/53 and C82/34/31 for Pol III) [71]. Counterparts of the Pol II GTFs except TFIIA and TFIIH exist for Pol I and III, whereas archaea also lack TFIIF and the small subunit ortholog of TFIIE. For Pol I and III, some of these GTF orthologs are stably associated Pol subunits, rather than separable GTFs, however, they are presumed to have similar functions as their Pol II counterparts. For example, the TFIIF dimerization domain binds the Pol II lobe and the same binding locations were found for the homologous dimerization domains of Pol I subunits A49/34.5 and Pol III subunits C37/53 [72, 73]. Similarly, the tandem WH domains in Pol III subunit C34 and Pol I subunit A49 span the Pol cleft in the same approximate position as the tandem WH domain of TFIIE, suggesting a conserved function [72, 74, 75]. All eukaryotic and archaeal systems contain a TFIIB-like factor [76–78]. Protein swapping experiments have shown that the Blinker regions from TFIIB and its Pol III ortholog Brf1 can be substituted for the analogous regions in the Pol I TFIIB-like factor Rrn7 [77]. Similarly, the Zn ribbon domain of Brf1 can be substituted within Rrn7. These experiments suggest that the TFIIB-like factors play mechanistic roles within the Pol cleft similar to that of TFIIB. By contrast, other details of PIC assembly may be distinct in the different systems. For example, Brf1 binds TBP most strongly through a Pol III-specific C-terminal domain [79]. Similarly, Rrn6, a subunit of the Pol I GTF, termed Core Factor, reportedly binds TBP more strongly than the TFIIB-like factor Rrn7 [80]. Structural and biochemical studies in the next few years will likely give fascinating insights into the architecture and mechanisms used in all these transcription systems.
Highlights.
Models for the architecture of the RNA Pol II preinitiation complex and open complex are discussed.
General factors TFIIB and TFIIF function in part through allosteric regulation of Pol II.
A DNA translocase activity is proposed to open DNA strands during initiation.
Eukaryotic Pols share a conserved core initiation complex.
New progress in understanding the interface between coactivators and the core transcription machinery is described.
Acknowledgments
We thank Valentin Grünberg for his help in editing the supplementary movie. Supported by NIH grants GM053451 and GM075114 to SH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Saecker RM, et al. Mechanism of bacterial transcription initiation: RNA polymerase - promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. Journal of molecular biology. 2011;412:754–771. doi: 10.1016/j.jmb.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hahn S. Structure and mechanism of the RNA polymerase II transcription machinery. Nature structural & molecular biology. 2004;11:394–403. doi: 10.1038/nsmb763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 4.Cheung AC, Cramer P. A movie of RNA polymerase II transcription. Cell. 2012;149:1431–1437. doi: 10.1016/j.cell.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nature reviews Genetics. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuda NJ, et al. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohler U, et al. Computational analysis of core promoters in the Drosophila genome. Genome Biol. 2002;3:RESEARCH0087. doi: 10.1186/gb-2002-3-12-research0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang C, et al. Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters. Gene. 2007;389:52–65. doi: 10.1016/j.gene.2006.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rhee HS, Pugh BF. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature. 2012;483:295–301. doi: 10.1038/nature10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim TK, et al. Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science. 2000;288:1418–1422. doi: 10.1126/science.288.5470.1418. [DOI] [PubMed] [Google Scholar]
- 11.Forget D, et al. RAP74 indices promoter contacts by RNA Pol II upstream and downstream of a DNA bend centered on the TATA box. Proc Natl Acad Sci USA. 1997;94:7150–7155. doi: 10.1073/pnas.94.14.7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller G, Hahn S. A DNA-tethered cleavage probe reveals the path for promoter DNA in the yeast preinitiation complex. Nature structural & molecular biology. 2006;13:603–610. doi: 10.1038/nsmb1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen ZA, et al. Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. The EMBO journal. 2010;29:717–726. doi: 10.1038/emboj.2009.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen HT, et al. The positions of TFIIF and TFIIE in the RNA polymerase II transcription preinitiation complex. Nature structural & molecular biology. 2007;14:696–703. doi: 10.1038/nsmb1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eichner J, et al. Position of the general transcription factor TFIIF within the RNA polymerase II transcription preinitiation complex. The EMBO journal. 2010;29:706–716. doi: 10.1038/emboj.2009.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grunberg S, et al. Architecture of the RNA polymerase II preinitiation complex and mechanism of ATP-dependent promoter opening. Nature structural & molecular biology. 2012;19:788–796. doi: 10.1038/nsmb.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Treutlein B, et al. Dynamic architecture of a minimal RNA polymerase II open promoter complex. Molecular cell. 2012;46:136–146. doi: 10.1016/j.molcel.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 18.Kostrewa D, et al. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature. 2009;462:323–330. doi: 10.1038/nature08548. [DOI] [PubMed] [Google Scholar]
- 19.Sainsbury S, et al. Structure and function of the initially transcribing RNA polymerase II-TFIIB complex. Nature. 2013;493:437–440. doi: 10.1038/nature11715. [DOI] [PubMed] [Google Scholar]
- 20.Liu X, et al. Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism. Science. 2010;327:206–209. doi: 10.1126/science.1182015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He Y, et al. Structural visualization of key steps in human transcription initiation. Nature. 2013;495:481–486. doi: 10.1038/nature11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim Y, et al. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 23.Kim JL, et al. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 24.Geiger JH, et al. Crystal structure of the yeast TFIIA/TBP/DNA complex. Science. 1996;272:830–836. doi: 10.1126/science.272.5263.830. [DOI] [PubMed] [Google Scholar]
- 25.Tan S, et al. Crystal structure of a yeast TFIIA/TBP/DNA complex. Nature. 1996;381:127–134. doi: 10.1038/381127a0. [DOI] [PubMed] [Google Scholar]
- 26.Chen HT, et al. Structure of a (Cys3His) zinc ribbon, a ubiquitous motif in archaeal and eucaryal transcription. Protein Sci. 2000;9:1743–1752. doi: 10.1110/ps.9.9.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikolov DB, et al. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature. 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- 28.Bushnell DA, et al. Structural basis of transcription: an RNA polymerase II-TFIIB cocrystal at 4.5 Angstroms. Science. 2004;303:983–988. doi: 10.1126/science.1090838. [DOI] [PubMed] [Google Scholar]
- 29.Fishburn J, Hahn S. Architecture of the yeast RNA polymerase II open complex and regulation of activity by TFIIF. Molecular and cellular biology. 2012;32:12–25. doi: 10.1128/MCB.06242-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen HT, Hahn S. Mapping the location of TFIIB within the RNA polymerase II transcription preinitiation complex: a model for the structure of the PIC. Cell. 2004;119:169–180. doi: 10.1016/j.cell.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 31.Roeder RG. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 32.Gaiser F, et al. Novel dimerization fold of RAP30/RAP74 in human TFIIF at 1.7 A resolution. Journal of molecular biology. 2000;302:1119–1127. doi: 10.1006/jmbi.2000.4110. [DOI] [PubMed] [Google Scholar]
- 33.Cabart P, et al. Transcription factor TFIIF is not required for initiation by RNA polymerase II, but it is essential to stabilize transcription factor TFIIB in early elongation complexes. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:15786–15791. doi: 10.1073/pnas.1104591108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Armache KJ, et al. Architecture of initiation-competent 12-subunit RNA polymerase II. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:6964–6968. doi: 10.1073/pnas.1030608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chakraborty A, et al. Opening and closing of the bacterial RNA polymerase clamp. Science. 2012;337:591–595. doi: 10.1126/science.1218716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan G, Greenblatt J. Initiation of transcription by RNA Polymerase II is limited by melting of the promoter DNA in the region immediately upstream of the initiation site. J Biol Chem. 1994;269:30101–30104. [PubMed] [Google Scholar]
- 37.Okamoto T, et al. Analysis of the role of TFIIE in transcriptional regulation through structure-function studies of the TFIIEbeta subunit. The Journal of biological chemistry. 1998;273:19866–19876. doi: 10.1074/jbc.273.31.19866. [DOI] [PubMed] [Google Scholar]
- 38.Grohmann D, et al. The initiation factor TFE and the elongation factor Spt4/5 compete for the RNAP clamp during transcription initiation and elongation. Molecular cell. 2011;43:263–274. doi: 10.1016/j.molcel.2011.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Egly JM, Coin F. A history of TFIIH: two decades of molecular biology on a pivotal transcription/repair factor. DNA repair. 2011;10:714–721. doi: 10.1016/j.dnarep.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 40.Compe E, Egly JM. TFIIH: when transcription met DNA repair. Nat Rev Mol Cell Bio. 2012;13:343–354. doi: 10.1038/nrm3350. [DOI] [PubMed] [Google Scholar]
- 41.Gibbons BJ, et al. Subunit architecture of general transcription factor TFIIH. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:1949–1954. doi: 10.1073/pnas.1105266109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guzder SN, et al. RAD25 is a DNA helicase required for DNA repair and RNA polymerase II transcription. Nature. 1994;369:578–581. doi: 10.1038/369578a0. [DOI] [PubMed] [Google Scholar]
- 43.Coin F, et al. Mutations in the XPD helicase gene result in XP and TTD phenotypes, preventing interaction between XPD and the p44 subunit of TFIIH. Nature genetics. 1998;20:184–188. doi: 10.1038/2491. [DOI] [PubMed] [Google Scholar]
- 44.Murakami K, et al. Formation and fate of a complete 31-protein RNA polymerase II transcription preinitiation complex. The Journal of biological chemistry. 2013;288:6325–6332. doi: 10.1074/jbc.M112.433623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kouzine F, et al. Global regulation of promoter melting in naive lymphocytes. Cell. 2013;153:988–999. doi: 10.1016/j.cell.2013.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li JLY, Rhee HS, Ghosh SK, Bai L, Pugh BF, Gilmour DS. Kinetic Competition between Elongation Rate and Binding of NELF Controls Promoter-Proximal Pausing. Molecular cell. 2013;50:711–722. doi: 10.1016/j.molcel.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pal M, et al. The role of the transcription bubble and TFIIB in promoter clearance by RNA polymerase II. Molecular cell. 2005;19:101–110. doi: 10.1016/j.molcel.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 48.Sandelin A, et al. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nature reviews Genetics. 2007;8:424–436. doi: 10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
- 49.Valen E, Sandelin A. Genomic and chromatin signals underlying transcription start-site selection. Trends Genet. 2011;27:475–485. doi: 10.1016/j.tig.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 50.Hahn S, Young ET. Transcriptional regulation in Saccharomyces cerevisiae: transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics. 2011;189:705–736. doi: 10.1534/genetics.111.127019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaplan CD, et al. Dissection of Pol II trigger loop function and Pol II activity-dependent control of start site selection in vivo. PLoS genetics. 2012;8:e1002627. doi: 10.1371/journal.pgen.1002627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ansari SA, Morse RH. Mechanisms of Mediator complex action in transcriptional activation. Cellular and molecular life sciences: CMLS. 2013 doi: 10.1007/s00018-013-1265-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Imasaki T, et al. Architecture of the Mediator head module. Nature. 2011;475:240–243. doi: 10.1038/nature10162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robinson PJ, et al. Structure of the mediator head module bound to the carboxy-terminal domain of RNA polymerase II. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:17931–17935. doi: 10.1073/pnas.1215241109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lariviere L, et al. Structure of the Mediator head module. Nature. 2012;492:448–451. doi: 10.1038/nature11670. [DOI] [PubMed] [Google Scholar]
- 56.Tsai KL, et al. A conserved Mediator-CDK8 kinase module association regulates Mediator-RNA polymerase II interaction. Nature structural & molecular biology. 2013;20:611–619. doi: 10.1038/nsmb.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soutourina J, et al. Direct interaction of RNA polymerase II and mediator required for transcription in vivo. Science. 2011;331:1451–1454. doi: 10.1126/science.1200188. [DOI] [PubMed] [Google Scholar]
- 58.Bernecky C, et al. Molecular architecture of the human Mediator-RNA polymerase II-TFIIF assembly. PLoS biology. 2011;9:e1000603. doi: 10.1371/journal.pbio.1000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takagi Y, et al. Head module control of mediator interactions. Molecular cell. 2006;23:355–364. doi: 10.1016/j.molcel.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 60.Esnault C, et al. Mediator-dependent recruitment of TFIIH modules in preinitiation complex. Molecular cell. 2008;31:337–346. doi: 10.1016/j.molcel.2008.06.021. [DOI] [PubMed] [Google Scholar]
- 61.Cler E, et al. Recent advances in understanding the structure and function of general transcription factor TFIID. Cellular and molecular life sciences: CMLS. 2009;66:2123–2134. doi: 10.1007/s00018-009-0009-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Molecular cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- 63.Cianfrocco MA, et al. Human TFIID binds to core promoter DNA in a reorganized structural state. Cell. 2013;152:120–131. doi: 10.1016/j.cell.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bieniossek C, et al. The architecture of human general transcription factor TFIID core complex. Nature. 2013;493:699–702. doi: 10.1038/nature11791. [DOI] [PubMed] [Google Scholar]
- 65.Wu PY, et al. Molecular Architecture of the S. cerevisiae SAGA Complex. Molecular cell. 2004;15:199–208. doi: 10.1016/j.molcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 66.Mohibullah N, Hahn S. Site-specific cross-linking of TBP in vivo and in vitro reveals a direct functional interaction with the SAGA subunit Spt3. Genes & development. 2008;22:2994–3006. doi: 10.1101/gad.1724408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen XF, et al. Mediator and SAGA have distinct roles in Pol II preinitiation complex assembly and function. Cell reports. 2012;2:1061–1067. doi: 10.1016/j.celrep.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fan L, et al. Conserved XPB core structure and motifs for DNA unwinding: implications for pathway selection of transcription or excision repair. Molecular cell. 2006;22:27–37. doi: 10.1016/j.molcel.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 69.Singleton MR, et al. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 70.Durr H, et al. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell. 2005;121:363–373. doi: 10.1016/j.cell.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 71.Vannini A, Cramer P. Conservation between the RNA Polymerase I, II, and III Transcription Initiation Machineries. Molecular cell. 2012;45:439–446. doi: 10.1016/j.molcel.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 72.Vannini A, et al. Molecular basis of RNA polymerase III transcription repression by Maf1. Cell. 2010;143:59–70. doi: 10.1016/j.cell.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 73.Wu CC, et al. The TFIIF-like Rpc37/53 dimer lies at the center of a protein network to connect TFIIIC, Bdp1, and the RNA polymerase III active center. Molecular and cellular biology. 2011;31:2715–2728. doi: 10.1128/MCB.05151-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu CC, et al. RNA polymerase III subunit architecture and implications for open promoter complex formation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:19232–19237. doi: 10.1073/pnas.1211665109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jennebach S, et al. Crosslinking-MS analysis reveals RNA polymerase I domain architecture and basis of rRNA cleavage. Nucleic acids research. 2012;40:5591–5601. doi: 10.1093/nar/gks220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Knutson BA, Hahn S. TFIIB-related factors in RNA polymerase I transcription. Biochimica et biophysica acta. 2013;1829:265–273. doi: 10.1016/j.bbagrm.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Knutson BA, Hahn S. Yeast Rrn7 and human TAF1B are TFIIB-related RNA polymerase I general transcription factors. Science. 2011;333:1637–1640. doi: 10.1126/science.1207699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Naidu S, et al. TAF1B Is a TFIIB-Like Component of the Basal Transcription Machinery for RNA Polymerase I. Science. 2011;333:1640–1642. doi: 10.1126/science.1207656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Juo ZS, et al. Crystal structure of a transcription factor IIIB core interface ternary complex. Nature. 2003;422:534–539. doi: 10.1038/nature01534. [DOI] [PubMed] [Google Scholar]
- 80.Lalo D, et al. RRN11 encodes the third subunit of the complex containing Rrn6p and Rrn7p that is essential for the initiation of rDNA transcription by yeast RNA polymerase I. The Journal of biological chemistry. 1996;271:21062–21067. doi: 10.1074/jbc.271.35.21062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.