Abstract
The HIV-1 viral infectivity factor (Vif) neutralizes cell-encoded antiviral APOBEC3 proteins by recruiting a cellular ElonginB (EloB)/ElonginC (EloC)/Cullin5-containing ubiquitin ligase complex, resulting in APOBEC3 ubiquitination and proteolysis. The suppressors-of-cytokine-signalling-like domain (SOCS-box) of HIV-1 Vif is essential for E3 ligase engagement, and contains a BC box as well as an unusual proline-rich motif. Here, we report the NMR solution structure of the Vif SOCS–ElonginBC (EloBC) complex. In contrast to SOCS-boxes described in other proteins, the HIV-1 Vif SOCS-box contains only one α-helical domain followed by a β-sheet fold. The SOCS-box of Vif binds primarily to EloC by hydrophobic interactions. The functionally essential proline-rich motif mediates a direct but weak interaction with residues 101–104 of EloB, inducing a conformational change from an unstructured state to a structured state. The structure of the complex and biophysical studies provide detailed insight into the function of Vif's proline-rich motif and reveal novel dynamic information on the Vif–EloBC interaction.
Keywords: HIV-1 viral infectivity factor, SOCS-box domain, ElonginBC, NMR, solution structure
2. Introduction
The apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3 (APOBEC3, A3) family of cytidine deaminases contains seven members named from A to H [1] that are constitutively present in cells and exhibit diverse physiological functions [2–4]. APOBEC3G (A3G) appears to have the most active antiviral phenotype for HIV-1, as recently described in several reviews [5–8].
The viral infectivity factor (Vif) protein of HIV-1 is an accessory protein required for virus replication in natural targets of infection [5]. Vif functions by counteracting the APOBEC3-mediated pathway of intrinsic antiviral defence by inducing the proteasomal degradation of APOBEC3 proteins, including A3DE [9], A3F [10–12], A3G [13–17] and some A3H proteins [18–20]. Degradation is achieved through the formation of an E3 ubiquitin ligase core complex composed of ElonginB (EloB), ElonginC (EloC), Cullin5 (Cul5) and RING-box protein 2 [21–25] in the presence of Vif. The core binding factor β (CBFβ), a newly discovered chaperone, is crucial for the folding and stabilizing Vif [26–28], the Vif–APOBEC3G interaction, [29,30] and regulation of host gene expression [28]. The formation of this E3 ligase complex results in ubiquitination of APOBEC3 proteins, and therefore neutralization of APOBEC-mediated antiviral activity. The interface afforded by the interaction of Vif with the cellular factors mentioned above is widely viewed as a potential target for the development of new anti-HIV drugs [5,6].
Vif possesses various motifs that bind these cellular factors that have been linked to direct interactions with the proteins mentioned above. The N-terminal half of Vif mediates the interaction with members of the APOBEC3 family [31,32]. Recent work from John Gross's group shows that the N-terminal residues 1–140 also interact with CBFβ [28]. In the middle of the Vif sequence, a zinc-binding HCCH motif has been proposed to interact with Cul5 [33–35]. C-terminal to this HCCH domain, the suppressors-of-cytokine-signalling-like domain (SOCS-box) of Vif binds to the EloB–EloC heterodimer (EloBC) along with Cul5 [22,36–38]. The SOCS-box contains a conserved SLQYLA motif (residues 144–149), called the BC-box, which interacts with EloBC. Mutation of this motif leads to the inactivation of Vif, indicating an essential role for this domain [14,22,36,39,40]. The SOCS-box also has a critical proline-rich motif (161PPLPS165, PPLPS motif) [41,42] downstream of the BC-box, whose molecular mechanism of action remains unclear. For instance, some reports suggest a role in Vif homo-multimerization [43], whereas biophysical studies indicate that the proline-rich motif interacts with the flexible C-terminus of EloB and is not required for the oligomerization [37,44].
Owing to the difficulty in overexpressing, purifying and crystallizing full-length folded soluble Vif and APOBEC3G [45,46], it remains unclear from a structural biological perspective how Vif recruits cellular factors and interacts with APOBEC3G in order to stimulate ubiquitination. Currently, the crystal structure of BC-box has been solved, but not that of the entire SOCS domain [38]. We have therefore employed a range of NMR techniques to dissect the structure and dynamics of the Vif SOCS–EloBC complex. In particular, Vif SOCS-box, EloC and EloB all experience structural changes during the SOCS–EloBC interaction, as proposed previously [37]. The solution structure of EloBC was solved by NMR in 2008 [47] in complex with the BC-box of SOCS3. Here, we present a solution structure of the Vif SOCS–EloB–EloC heterotrimer. The structure of the complex is calculated by HADDOCK combined with long-distance restraints from paramagnetic relaxation enhancement (PRE) experiments and NMR chemical shift perturbations. We find that the van der Waals surface calculated around the PPLPS motif of SOCS-box touches the van der Waals surface calculated about the EloB C-terminus, thereby supplementing the tight binding interface between the BC-box and EloC. Upon binding, the C-terminal tail of EloB experiences a structural change from a flexible state to a partially folded state. Consistent with earlier structure–function analyses, the leucine, the third proline and the adjacent serine are the most important residues in this motif [40].
3. Material and methods
3.1. Protein expression and purification
The SOCS–EloBC complex for NMR spectroscopy was prepared as described previously [37]. Essentially, EloBC dimer and SOCS-box peptide were expressed individually in the Escherichia coli BL21 (DE3) Rosetta strain in LB media or M9 minimal media supplemented with different isotopes (13C, 15N, 2H), depending on the experiments. EloBC was purified in 20 mM Tris buffer pH 7.0, 50 mM NaCl and solubility-enhancement-tagged SOCS-box peptide was purified in 20 mM Tris buffer pH 8.0, 500 mM NaCl. They were mixed at a 1 : 1 ratio after elution from the Ni-NTA column and loaded onto a Superdex 75 gel filtration column to remove unbound components. EloBC-labelled sample or SOCS-labelled sample was then used in NMR spectroscopy experiments.
3.2. NMR spectroscopy
NMR spectra were acquired at 25°C on Varian NMR 800 MHz and Bruker Avance 700 MHz spectrometers equipped with 5 mm triple-resonance z-axis gradient cryogenic probes. A 0.2 mM sample (550 μl) was prepared in 50 mM phosphate buffer pH 7.0, 10% D2O, 0.05% sodium azide with protease inhibitors (Roche) tablet. HNCO, CBCA(CO)NH, HNCA, HNCACB, HN(CA)CO, HN(CA)HA, HCCH-TOCSY, 1H-15N TROSY-HSQC and 13C-edited NOESY-HSQC were recorded on EloBC, and HNCO, HN(CA)CO, HNCA, HNCACB, CBCA(CO)NH, HN(CA)HA, HCCH-COSY, HCCH-TOCSY, 15N-edited NOESY-HSQC, 13C-edited NOESY-HSQC, 1H-15N HSQC and 1H-13C HSQC spectra were used for the SOCS-box peptide backbone and side-chain assignment. All the spectra were processed with NMRPipe [48] and analysed with CcpNmr suite [49,50]. As for the perturbation studies, each EloBC mutant sample was prepared in the same way as wild-type and divided into two aliquots. Purified SOCS-box peptide was added to one aliquot at a 1.2 : 1 ratio and the other was made up to the same volume as the first aliquot with NMR buffer. Relaxation experiments were recorded on the unbound SOCS-box peptide at 25°C at 500 and 700 MHz magnetic field, respectively.
3.3. Paramagnetic relaxation enhancement experiments
Residues G143, Q158 and R167 on SOCS-box peptide were mutated to Cys, for use in paramagnetic labelling studies. Mutant protein was mixed with an approximately fivefold excess of dithiothreitol (DTT) for 2 h after elution from the Ni-NTA column. This was followed by separation of excess DTT by gel filtration chromatography. The SOCS-box monomer sample from the size exclusion column was collected and incubated with either the diamagnetic (1-acetyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl) methanethiosulfonate or the paramagnetic (1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-methyl) methanethiosulfonate (Toronto Research Chemicals) overnight at 4°C. Each modified SOCS-box sample was dialysed against NMR buffer and mixed with 15N-labelled EloBC in NMR buffer at a 1.1 : 1 ratio. The mixed sample was then used for NMR spectral acquisition. A 1H-15N HSQC spectrum was recorded for each 50 μM sample with a 3-h acquisition. Intensity ratios were converted to distances according to an established method [51] by using the Solomon–Bloembergen equation [52]. Because of the dynamics of the labels, the diameter of the paramagnetic molecule was added to or subtracted from the calculated distance, thus obtaining the upper or the lower limitation distance between the Cys and observed 15N-labelled EloBC residues.
3.4. Structure determination
SOCS-box peptide structures in the bound state were generated by Chemical Shift ROSETTA (CS-ROSETTA) [53] by inputting NOE data and chemical shift values into the BMRB CS-ROSETTA server (condor.bmrb.wisc.edu/bbee/rosetta/). Structures were further refined according to the Rosetta refinement protocol [54]. EloB and EloC structures were generated de novo as well as on the server. Using NMR perturbation studies based on 1H-15N HSQC spectra and PRE data that provide semi-quantitative long-distance constraints, the HADDOCK approach was adopted for the structure calculation of the complex [55]. In our previous work, it has been proved by various biophysical assays that the EloB DVMK stretch interacts with the proline-rich motif [37], so in the calculation on the WeNMR web server [56], five residues in SOCS-box (Q146, A149, L163, P164 and S165), four residues in EloB (D101, V102, M103 and K104) and two residues in EloC (A82 and L86) were selected as active residues. The interfacial residues sitting between the SOCS-box proline-rich motif and the C-terminus of EloB were allowed to fully move at all stages. A file with distance restraints that are always enforced was provided. Two thousand initial complex structures were generated and the best 200 structures were chosen for explicit solvent refinement. The clustering cut-off is set to 5 Å, four structures per cluster. Default parameters excluding the settings above were always applied. The assignments and structures have been deposited to BMRB (ID 19333) and PDB (ID 2MA9), respectively.
3.5. ITC binding assays
EloBC dimer sample and SOCS-box peptide were concentrated to 0.2 and 0.02 mM, respectively. All samples were dialysed against binding buffer with 20 mM Tris pH 7.5, 250 mM NaCl and 0.05% sodium azide. ITC was performed on an ITC200 calorimeter (MicroCal, Northampton, MA). Titrations were conducted by injecting 20 aliquots of 2 μl of EloBC sample into cells containing SOCS-box peptide sample at 25°C. Fresh samples were prepared thrice in order to record ITC experiments in triplicate, and one typical set of results is presented.
4. Results
4.1. The flexibility of the unbound SOCS-box domain
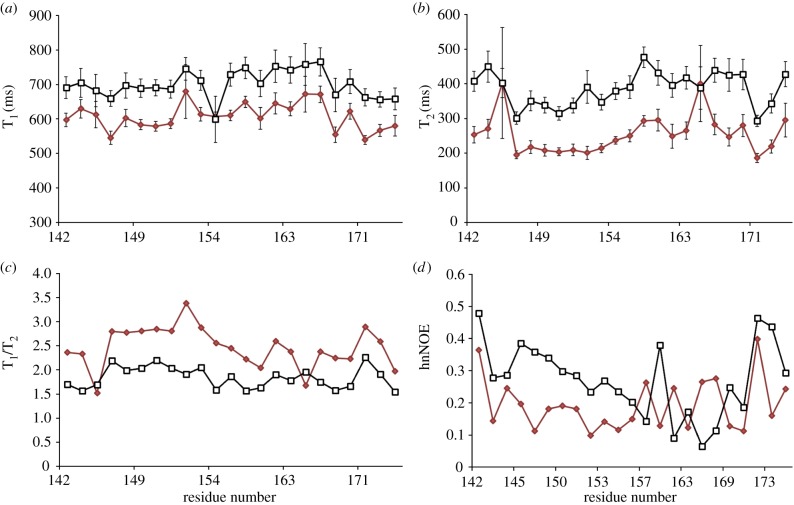
In order to address the challenges associated with Vif insolubility, we N-terminally fused the Vif SOCS-box to a solubility-enhancement tag that does not increase the molecular weight substantially and therefore is suitable for NMR studies [57]. In previous work, it was found that the unbound SOCS-box lacks secondary structure [37]. Here, the NMR relaxation experiments were recorded at two magnetic field strengths (11.75 and 16.4 T, 500 and 700 MHz at 1H frequency) in order to observe the flexibility of the SOCS-box peptide. The T1, T2, T1/T2 ratio and 15N heteronuclear nuclear Overhauser effect (hnNOE) are plotted against the residue numbers (figure 1). The fact that the T1 values of BC-box are consistently the same over the span of residues 144–154 indicates that this region is less dynamic and tumbles isotropically compared with the rest residues of the SOCS-box. However, it is of note that the N-terminal-fused tag attached to this region may also contribute to its limited motion. T2 values suggest the existence of fast motion. In addition, the variable low values of hnNOE reveal that the SOCS-box peptide possesses considerable internal motion, especially the region following the BC-box. These relaxation results show that the SOCS-box has a random coil conformation before binding to EloBC.
Figure 1.
15N relaxation rate constants at 500 MHz (red lines) and 700 MHz (black lines) magnetic fields (a) T1, (b) T2, (c) T1/T2 and (d) hnNOE plotted against residue number.
4.2. The structure of SOCS-box domain
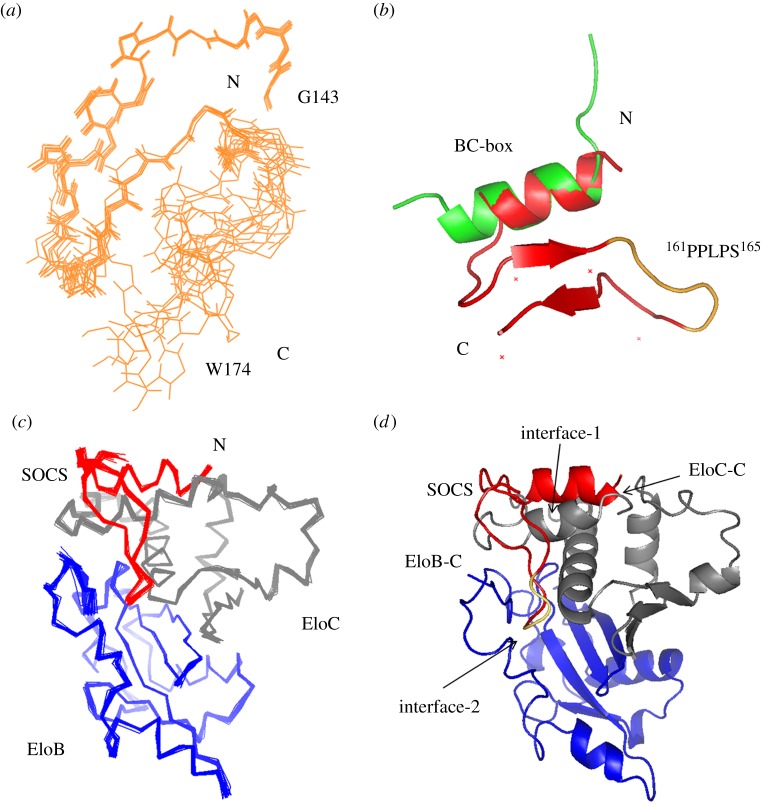
In order to solve the structure of SOCS-box, the NMR spectra (figure 2a,b) [37] were recorded on the labelled peptide produced from E. coli in the unbound state and in the complex with unlabelled EloBC heterodimer, for which the two components were co-expressed and co-purified. Although comprehensive NOE data are not available for the entire SOCS-box α-helical region, the 32-amino-acid peptide in the complex adopts a well-defined structure based on chemical shift analysis using CS-Rosetta combined with a limited set of NOE measurements (figure 2c and table 1). The RMSD between the lowest energy structure and the helical BC-box crystal structure (PDB ID: 3DCG, chain E) [38] is 0.43 Å after refinement by the Rosetta protocol. The final structure has an αββ structure in which the two β-strand-like elements are connected by the proline-rich motif loop (figure 3a,b), resulting in an exposed loop that projects into solution and is accessible for interaction with other molecules. The second β-strand appears to be flexible compared with the other regions.
Figure 2.
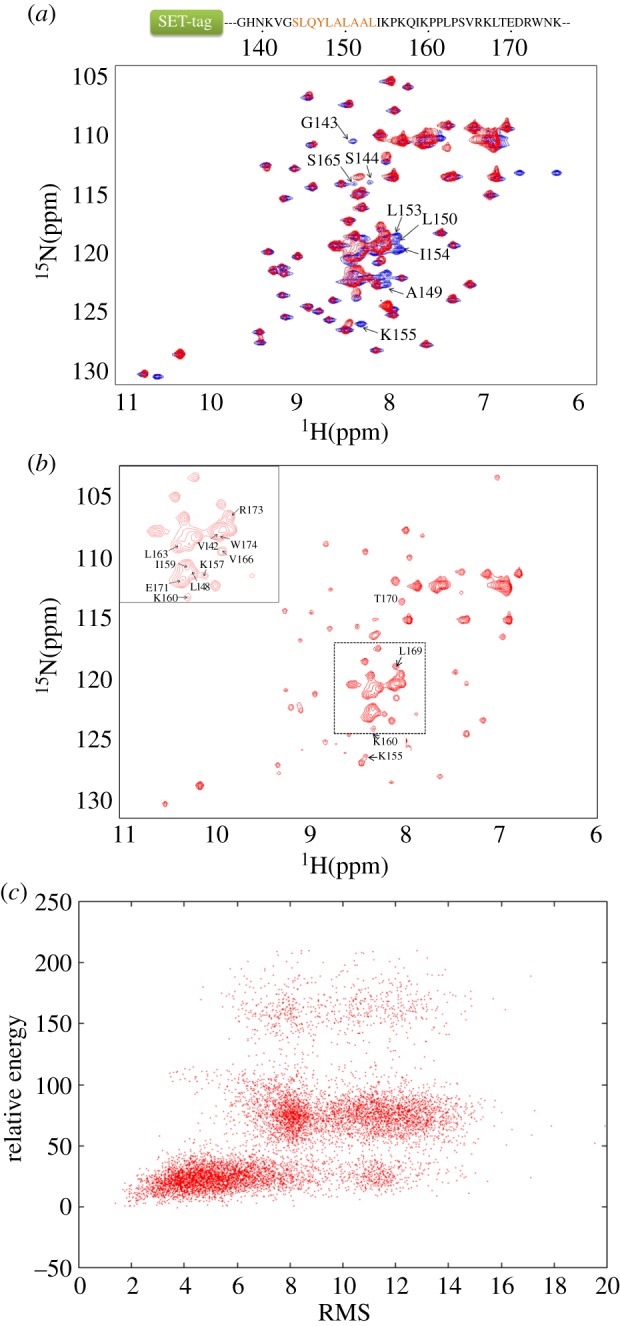
1H–15N HSQC spectra of Vif SOCS-box protein, with the sequence of the SOCS-box peptide shown on top. The 56-residue SET-tag is simplified with a rectangle, followed by the SOCS-domain peptide. The BC-box is highlighted in orange. (a) The overlay of two spectra recorded on the SOCS-box peptide in the free state (blue) and in the complex with EloBC (red). (b) Details of the spectrum recorded on the bound SOCS-box peptide. The peaks assigned to residues corresponding to the Vif SOCS-box are indicated. The rest of the peaks distributing in the spectrum are from the SET-tag. (c) The Rosetta score versus RMSD plot from CS-Rosetta structural calculation protocol. A total of 10 000 structures were generated. (a,b) are reproduced with permission from [37].
Table 1.
Structural statistics for the bound SOCS-box peptide from CS-Rosetta.
| NOE restraints comprised in calculations | |
| short range | 59 |
| medium range | 11 |
| long range | 8 |
| average RMSDs from the mean structure | |
| backbone average (Å) | 0.54 ± 0.19 |
| heavy atom average (Å) | 0.65 ± 0.22 |
| Ramachandran plot | |
| most favoured region (%) | 96.7 |
| allowed region (%) | 3.3 |
| generously allowed region (%) | 0 |
| disallowed region (%) | 0 |
Figure 3.
The structures of the HIV-1 Vif SOCS peptide and the SOCS–EloBC complex. (a) SOCS peptide alignment. Overlap of ten backbone structures for the SOCS-box after Rosetta refinement. (b) Structural comparison with the SOCS-box crystal structure (3DCG, chain E). Peptide structure is aligned against the crystal structure in green. The 165PLS167 sequence is highlighted in yellow. (c) 20 structures of the SOCS–EloBC complex with the lowest energy are aligned after HADDOCK refinement. (d) Ribbon representation of the complex. The Vif SOCS peptide is coloured in red. EloB and EloC are in blue and grey, respectively. The 165PLS167 region of SOCS is highlighted in yellow.
4.3. The solution structural determination of the SOCS-box–Elongin BC complex
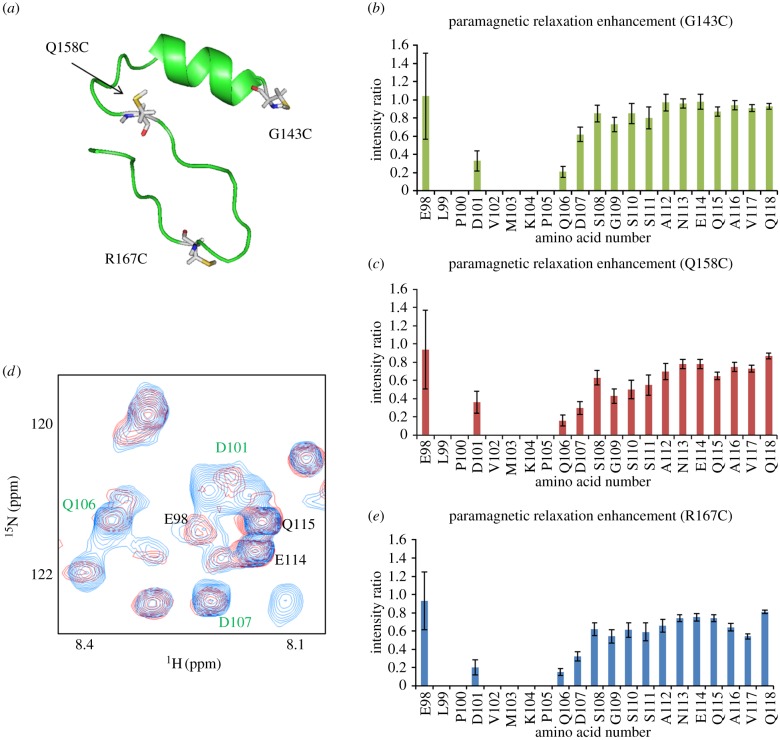
The binding of the HIV-1 Vif SOCS-box domain to EloBC forms a stable heterotrimer with a 1 : 1 : 1 stoichiometric ratio determined by gel filtration analysis [37]. In order to acquire additional intermolecular restraints among the three components, PRE experiments were performed [52]. As no cysteines are present in the SET-tag-fused SOCS-box peptide, single-point cysteine mutations could be generated in this peptide at selected locations to measure PREs within the other components of the complex, namely EloBC. In each molecule of the complex, a mutated SOCS-box peptide was engineered with a single cysteine substitution enabling the observed relaxation enhancements to be assigned to the paramagnetic moiety conjugated to the amino acid residue. Point mutations (G143C, Q158C and R167C) were placed at three widely separated sites in the SOCS peptide in order to increase the number and spread of intermolecular measurements (figure 4a). The binding of all mutants was checked by isothermal titration calorimetry (ITC) to verify that the mutation did not interfere with the binding affinity of SOCS–EloBC (see electronic supplementary material, figure S1). As the portion of the SOCS-box binding to EloBC dimer has been localized to the BC-box and the PPLPS motif, mutations were made outside these functional domains to minimize any artefactual interactions. In PRE experiments, the peak-intensity ratios measured in the EloB carboxyl terminus provide a clear indication that the DVMK stretch at the C-terminus of EloB is close in space to the labelled cysteines of the Vif SOCS-box (figure 4b,c), especially the R167C residue (figure 4d,e). The overlay of a 20-solution-structure alignment from the cluster is shown in figure 3. Structural statistics for the cluster of structures are listed in table 2.
Figure 4.
PRE experiments. (a) The three mutated sites in Vif which were distributed at widely separated positions in the peptide. G143C, Q158C and R167C are highlighted in white and the sulfur of the cysteine to be modified by label is coloured in yellow. Each predicted mutant structure is generated by Pymol in order to determine that the side chain of cysteine would not be buried inside the molecule. (b) Intensity ratio of EloB C-terminus for G143C mutant. (c) Intensity ratio of EloB C-terminus for Q158C mutant. (d) A portion of the 1H-15N HSQC spectra of EloBC in the presence of the paramagnetic molecule-labelled SOCS-box R167C peptide (red) or the diamagnetic molecule-labelled R167C peptide (blue). Peaks assigned to the EloB C-terminal tail are indicated in green (significantly broadened) or in black (not affected). (e) Intensity ratio of EloB C-terminus for R167C mutant. The intensity of peaks from residues 101–104 are broadened owing to the interaction regardless of the paramagnetic effect. The error bars show how the noise of the spectra affects the intensity ratio. Stronger peaks provide smaller deviation.
Table 2.
Structural statistics for the SOCS–EloBC complex.
| HADDOCK cluster number produced | 1 |
| average RMSDs from the mean structure | |
| backbone average (Å) | 0.26±0.03 |
| heavy atom average (Å) | 0.52±0.04 |
| HADDOCK statistics | |
| van der Waals energy (kcal mol-1) | –63.5±2.4 |
| electrostatic energy (kcal mol−1) | –574.5±15.9 |
| desolvation energy (kcal mol−1) | 3.1±3.3 |
| restraints violation energy (kcal mol−1) | 0.3±0.16 |
| buried surface area (Å2) | 2438.6±46.0 |
| experimental PRE distance restraints | |
| number | 58 |
| distance violation (Å) | 2.76±1.90 |
| Ramachandran plot | |
| most favoured region (%) | 82.3 |
| allowed region (%) | 15.2 |
| generously allowed region (%) | 1.4 |
| disallowed region (%) | 1.1 |
| deviations from ideal geometry | |
| angles (degree) | 0.6 |
| bonds (Å) | 0.003 |
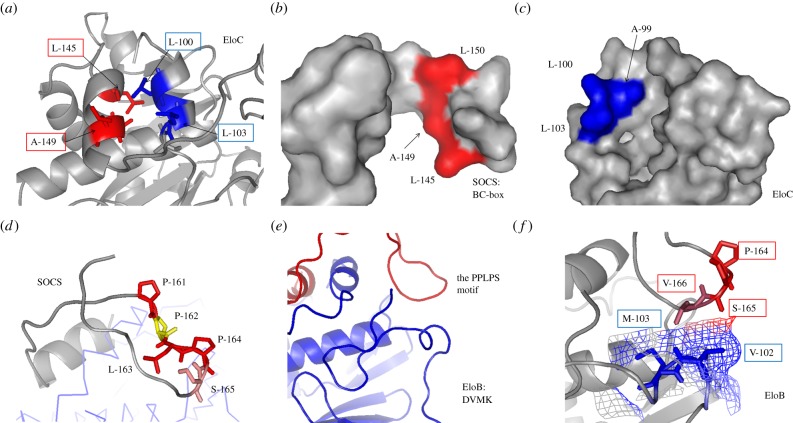
The structural model provides confirmation that the SOCS-box crosses the EloC carboxyl α-helix and binds to the flexible EloB carboxyl terminus. Residues L145, A149, L150 of the Vif SOCS-box and A99, L103 of EloC bind to each other (figure 5a) by forming hydrophobic interfaces that drive the SOCS–EloC interactions (figure 5b,c) typical of a high-affinity protein complex [58,59]. Hydrophobic interaction is the major force driving the formation of biological complexes [22,39]. The structure of this portion of the complex very closely matches that of the crystal structure (PDB ID: 3DCG) [38]. The studies on the PPLPS motif show that 164PS165 plus V166 interacts with the EloB DVMK stretch rather than the entire PPLPS motif (figure 5d,e). This proline–serine loop is stabilized by an antiparallel β-sheet-like structure. Within EloB, the C-terminus is flexible in solution, as shown by the narrow NMR line widths for this region [37], whereas in the presence of SOCS-box, the DVMK stretch experiences a conformational change and becomes partially helical (figure 5e). The interface between 164PS165 and the DVMK stretch is formed by close spatial positioning of the residues and are bound by weak van der Waals forces (figure 5f), implying that this interaction may not be a strong interaction although it was observed from the NMR perturbation experiments and ITC studies of SOCS-box and its mutants [37].
Figure 5.
Interaction between the SOCS-box and EloBC. (a) The interface of the BC-box and EloC binding site. Active side-chains are presented and labelled. (b) The binding surface of BC-box. Resides are coloured in red. (c) The binding surface of EloC C-terminal α-helix. Resides on the interface are coloured in blue. (d) The proline-rich motif downstream from the SOCS-box is coloured. Residues are highlighted in various colours. The second proline (residue 162) in yellow is buried in the complex. (e) The C-terminus of EloB, including the DVMK stretch. Blue, EloB; red, SOCS Peptide. (f) The interface of EloB DVMK stretch and the Vif proline-rich motif. The van der Waals radii of V102 and M103 are presented. The interacted regions by the PPLPS motif are coloured differently.
4.4. The interaction between the PPLPS motif and the DVMK stretch
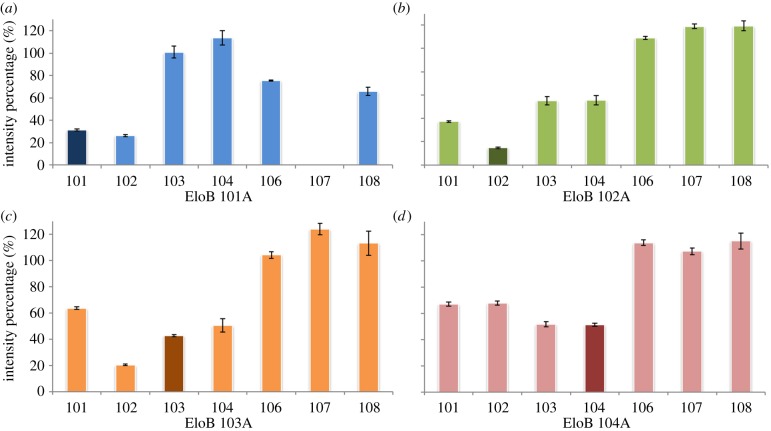
In order to define further the SOCS–EloB interface, we subsequently mutated the four residues at the C-terminus of EloB, respectively, and measured NMR perturbations to the line widths of distal amino acids. Interestingly, peaks from this domain follow a distinct decrease in intensity irrespective of which residue is mutated (figure 6). As for the EloB-D101A mutant, A101 is still perturbed upon binding, although residues M103 and K104 are not impacted. The NMR spectrum recorded for this mutant displayed peak shifts for residues 103–118, suggesting a conformational disruption by this mutation (data not shown). Profiles from the other three mutants match the same profile as for wild-type [37], indicating that the interaction between the PPLPS motif and the C-terminus of EloB is not specifically driven by these side-chains within the DVMK stretch.
Figure 6.
(a–d) The NMR perturbation studies on the Vif SOCS–EloBC complex. 1H–15N HSQC spectra were recorded on the labelled EloBC dimer in the absence and in the presence of SOCS-box peptide. The percentage represents the intensity ratio of bound peaks to unbound peaks. The numbers on the x-axis indicate the residues at the C-terminus of EloB.
We then asked the question whether individual residues in the PPLPS motif contribute more to the interaction with EloB. Several single-point mutants in the motif were made and ITC was used to quantify the thermodynamics of the interaction. Although sequence analysis of Vif and EloB amino acids indicates that the PPLPS motif of HIV-1 Vif and residues 101–104 (DVMK stretch) of EloB are both highly conserved (see electronic supplementary material, figure S2 and table S1), results from ITC reveal that the mutation of any residue within the PPLPS motif does not affect the binding affinity regardless of the entropy change (table 3; electronic supplementary material, figure S3), which is thought to be caused by internal conformational changes owing to the single-site mutations [60]. It can therefore be concluded that the second interface between the PPLPS motif and the DVMK stretch is driven by weak van der Waals forces.
Table 3.
Thermodynamic characterization of SOCS–EloBC interaction.
| Kd (μM) | Ka (μM−1) | ΔHobs (kcal mol–1) | ΔG (kcal mol–1) | –TΔS (kcal mol–1) | |
|---|---|---|---|---|---|
| WT | 1.225 | 0.82±0.06 | −10.2±1.6 | −7.9 | 2.3 |
| SPLPS | 2.14 | 0.52±0.18 | −8.3±2.3 | −7.6 | 0.7 |
| PSLPS | 1.24 | 0.86±0.24 | −8.8±2.0 | −7.9 | 0.9 |
| PPLSS | 1.36 | 0.77±0.19 | −10.7±2.8 | −7.8 | 2.9 |
| PPLPA | 1.18 | 0.89±0.24 | −8.6±0.3 | −7.9 | 0.7 |
| APLAS | 1.10 | 0.97±0.28 | −8.8±0.2 | −8.0 | 0.8 |
| AAAPS | 1.50 | 0.68±0.11 | −9.8±1.7 | −7.9 | 1.9 |
5. Discussion
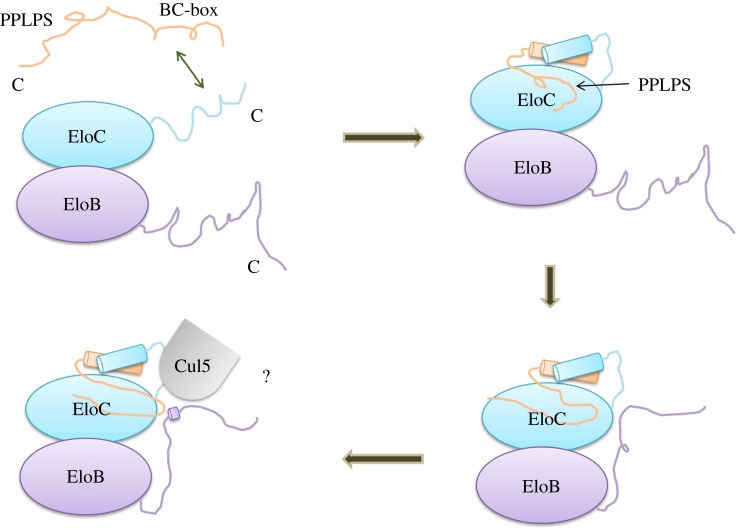
Here, we present the first structure of the HIV-1 Vif SOCS–EloBC complex including the conserved proline-rich motif of Vif. Interestingly, the side-chains of P161, L163 and P164 are exposed in the solution, whereas the side-chain of the second proline (P162) is buried within the protein, suggesting that this proline is less important compared with the other residues. This result agrees with previous work showing that mutations at the first and third proline decrease Vif function in cells and result in a lack of formation of the ubiquitination complex [37,40]. In our previous study [37], we reported that in the absence of SOCS-box the carboxyl terminus of EloB is flexible in solution and that the binding to SOCS-box induces structural changes in the disordered DVMK stretch according to the T1/T2 relaxation ratio [37]. This small helix in the EloB C-terminus is also observed in the SOCS2–EloBC complex [61]. Therefore, it can be concluded that the DVMK stretch forms a helix upon binding to SOCS-box. Considering the weak binding between the PPLPS motif and the DVMK stretch, which is not essential for SOCS–EloBC binding, though it is required for HIV-1 Vif function in cells, we further suggest that it is the α-helix of EloC that first drives the induced folding of Vif, followed by the interaction between the PPLPS motif and EloB. The final interface formed by the PPLPS motif and the induced folding of the DVMK stretch may be required to form an E3 ubiquitin ligase complex, perhaps specifically to recruit Cul5 (figure 7).
Figure 7.
Schematic of the proposed induced-folding mechanism. The SOCS-box peptide includes an α-helix domain (represented as SLQ) and a proline-rich domain (represented as PPLP). The formation of the Vif SOCS–EloBC complex is mainly driven by hydrophobic interactions between Vif BC-box and EloC C-terminus via a conformational change process. Then, the proline-rich motif induces the EloB C-terminal tail to fold, forming a common interface to recruit cellular factors, perhaps Cul5. The various interaction events are presented in succession. The C-terminus of each subunit is indicated.
As we have demonstrated here, the interaction between Vif's PPLPS motif and EloB's DVMK has low affinity and is mainly a weak van der Waals interaction. We also show that it is coupled with significant structural rearrangements in both Vif and EloB. Yet it is a critical interaction for the recruitment of a functional E3 ubiquitin ligase, CBFβ binding [62] and degradation of A3G in vivo [63]. It is therefore tempting to speculate that this interaction could be targeted by small molecule inhibitors for the design of a new class of anti-HIV drugs. Furthermore, the NMR spectra reported here could form the basis for an assay to screen small molecules libraries. Yet, as the interaction is based on flexible regions of these proteins, there is presumably no obvious binding pocket for a small molecule to bind to. In that regard, a small peptide inhibitor, mimicking the PPLPS motif engaging EloB upon Vif binding, would be a more likely candidate for interrupting this interaction.
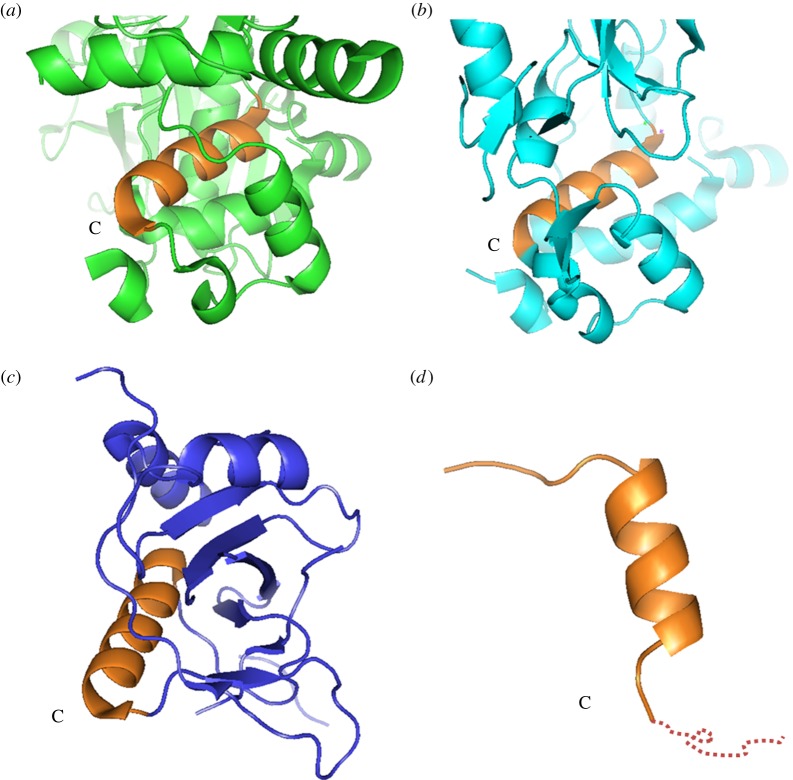
The SOCS family (SOCS1–7) proteins are indispensable regulators, functioning in many pathways, including ubiquitination and transcription, and have high sequence similarity. Published structures show that they all share a common α-helical structure. It is of note, however, that the downstream sequences have a different spatial portfolio depending on the structural family. Domains from SOCS2 and SOCS4 (PDB ID: 2C9W, 2IZV) contain three small α-helixes that are also known to interact with the EloB C-terminus (figure 8a,b) [61,64], whereas SOCS6 (PDB ID: 2VIF) adopts a partially folded structure (figure 8c), and the SOCS3 (PDB ID: 3DCG) downstream sequence cannot be identified in the X-ray crystallographic analysis (figure 8d) [38,65], which suggests that the downstream structure is flexible in solution and can be only observed by NMR. Interestingly, the SOCS-boxes of Vif proteins from other retroviruses, such as HIV-2, simian immunodeficiency virus from mandrill (SIVMND) or bovine immunodeficiency virus (BIV), contain the BC-box, but not a proline-rich motif [63,66,67]. Our results report, for the first time, structural insight into the whole HIV-1 Vif SOCS domain, including a BC-box and the proline-rich motif in the presence of EloBC, and its dynamic behaviour in the SOCS–EloBC interaction.
Figure 8.
Structures of SOCS family proteins. Close-up of the SOCS domains from published structures of different SOCS families. The C-terminus of each SOCS α-helical domain is labelled. (a) SOCS2 domain (2C9W). (b) SOCS4 domain (2IZV). (c) SOCS6 domain (2VIF). (d) SOCS3 domain (3DCG). The unobserved downstream sequence of SOCS3 is presented by a dotted line. The SOCS domain on each structure is shown in brown.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank Dr Geoff Kelly at MRC Biomedical NMR Centre, NIMR, Mill Hill, London, for the help with NMR data acquisition and processing. At King's College London, we thank Dr Luigi Martino for help with ITC, Dr Ivan Laponogov for help with NMR resonance assignment using his program ASSIGNER, and Daoud Akhtar and Bhavik Patel for help with protein preparation. NMR experiments were conducted at King's College London, and all ITC experiments were produced using the facilities of the Centre for Biomolecular Spectroscopy, King's College London, established with a Capital Award from the Wellcome Trust. Z.L. thanks the King's–China Scholarship Council PhD Scholarship Programme for a PhD studentship.
Data accessibility
The DNA accession information is listed in the electronic supplementary material, table S1.
Funding statement
Partial financial support for consumables was provided by grants from the Wellcome Trust (M.R.S., M.H.M.) and the Medical Research Council (M.H.M.).
References
- 1.LaRue RS, et al. 2009. Guidelines for naming nonprimate APOBEC3 genes and proteins. J. Virol. 83, 494–497 (doi:10.1128/JVI.01976-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger MS. 2005. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol. Biol. Evol. 22, 367–377 (doi:10.1093/molbev/msi026) [DOI] [PubMed] [Google Scholar]
- 3.Petersen-Mahrt SK, Harris RS, Neuberger MS. 2002. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature 418, 99–103 (doi:10.1038/nature00862) [DOI] [PubMed] [Google Scholar]
- 4.Pham P, Bransteitter R, Petruska J, Goodman MF. 2003. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature 424, 103–107 (doi:10.1038/nature01760) [DOI] [PubMed] [Google Scholar]
- 5.Wissing S, Galloway NL, Greene WC. 2010. HIV-1 Vif versus the APOBEC3 cytidine deaminases: an intracellular duel between pathogen and host restriction factors. Mol. Aspects Med. 31, 383–397 (doi:10.1016/j.mam.2010.06.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Albin JS, Harris RS. 2010. Interactions of host APOBEC3 restriction factors with HIV-1 in vivo: implications for therapeutics. Expert Rev. Mol. Med. 12, e4 (doi:10.1017/S1462399409001343) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirchhoff F. 2010. Immune evasion and counteraction of restriction factors by HIV-1 and other primate lentiviruses. Cell Host Microbe 8, 55–67 (doi:10.1016/j.chom.2010.06.004) [DOI] [PubMed] [Google Scholar]
- 8.Imahashi M, Nakashima M, Iwatani Y. 2012. Antiviral mechanism and biochemical basis of the human APOBEC3 family. Front Microbiol. 3, 250 (doi:10.3389/fmicb.2012.00250) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dang Y, Wang X, Esselman WJ, Zheng YH. 2006. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J. Virol. 80, 10 522–10 533 (doi:10.1128/JVI.01123-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiegand HL, Doehle BP, Bogerd HP, Cullen BR. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 23, 2451–2458 (doi:10.1038/sj.emboj.7600246) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng YH, Irwin D, Kurosu T, Tokunaga K, Sata T, Peterlin BM. 2004. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 78, 6073–6076 (doi:10.1128/JVI.78.11.6073-6076.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 14, 1392–1396 (doi:10.1016/j.cub.2004.06.057) [DOI] [PubMed] [Google Scholar]
- 13.Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302, 1056–1060 (doi:10.1126/science.1089591) [DOI] [PubMed] [Google Scholar]
- 14.Mehle A, Strack B, Ancuta P, Zhang C, McPike M, Gabuzda D. 2004. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 279, 7792–7798 (doi:10.1074/jbc.M313093200) [DOI] [PubMed] [Google Scholar]
- 15.Sheehy AM, Gaddis NC, Choi JD, Malim MH. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418, 646–650 (doi:10.1038/nature00939) [DOI] [PubMed] [Google Scholar]
- 16.Lecossier D, Bouchonnet F, Clavel F, Hance AJ. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300, 1112 (doi:10.1126/science.1083338) [DOI] [PubMed] [Google Scholar]
- 17.Sheehy AM, Gaddis NC, Malim MH. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9, 1404–1407 (doi:10.1038/nm945) [DOI] [PubMed] [Google Scholar]
- 18.OhAinle M, Kerns JA, Li MM, Malik HS, Emerman M. 2008. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe 4, 249–259 (doi:10.1016/j.chom.2008.07.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan L, Sarkis PT, Wang T, Tian C, Yu XF. 2009. Sole copy of Z2-type human cytidine deaminase APOBEC3H has inhibitory activity against retrotransposons and HIV-1. FASEB J. 23, 279–287 (doi:10.1096/fj.07-088781) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao K, Ishida Y, Oleksyk TK, Winkler CA, Roca AL. 2012. Evidence for selection at HIV host susceptibility genes in a West Central African human population. BMC Evol. Biol. 12, 237 (doi:10.1186/1471-2148-12-237) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahrour N, et al. 2008. Characterization of Cullin-box sequences that direct recruitment of Cul2-Rbx1 and Cul5-Rbx2 modules to Elongin BC-based ubiquitin ligases. J. Biol. Chem. 283, 8005–8013 (doi:10.1074/jbc.M706987200) [DOI] [PubMed] [Google Scholar]
- 22.Yu Y, Xiao Z, Ehrlich ES, Yu X, Yu XF. 2004. Selective assembly of HIV-1 Vif-Cul5-ElonginB-ElonginC E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes Dev. 18, 2867–2872 (doi:10.1101/gad.1250204) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. 2008. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell 134, 995–1006 (doi:10.1016/j.cell.2008.07.022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng N, et al. 2002. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416, 703–709 (doi:10.1038/416703a) [DOI] [PubMed] [Google Scholar]
- 25.Kamura T, Maenaka K, Kotoshiba S, Matsumoto M, Kohda D, Conaway RC, Conaway JW, Nakayama KI. 2004. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 18, 3055–3065 (doi:10.1101/gad.1252404) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou X, Evans SL, Han X, Liu Y, Yu XF. 2012. Characterization of the interaction of full-length HIV-1 Vif protein with its key regulator CBFbeta and CRL5 E3 ubiquitin ligase components. PLoS ONE 7, e33495 (doi:10.1371/journal.pone.0033495) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jager S, et al. 2012. Vif hijacks CBF-beta to degrade APOBEC3G and promote HIV-1 infection. Nature 481, 371–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim DY, Kwon E, Hartley PD, Crosby DC, Mann S, Krogan NJ, Gross JD. 2013. CBFbeta stabilizes HIV Vif to counteract APOBEC3 at the expense of RUNX1 target gene expression. Mol. Cell 49, 632–644 (doi:10.1016/j.molcel.2012.12.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang W, Du J, Evans SL, Yu Y, Yu XF. 2012. T-cell differentiation factor CBF-beta regulates HIV-1 Vif-mediated evasion of host restriction. Nature 481, 376–379 [DOI] [PubMed] [Google Scholar]
- 30.Du J, Zhao K, Rui Y, Li P, Zhou X, Zhang W, Yu XF. 2013. Differential requirements for HIV-1 Vif-mediated APOBEC3G degradation and RUNX1-mediated transcription by core binding factor beta. J. Virol. 87, 1906–1911 (doi:10.1128/JVI.02199-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He Z, Zhang W, Chen G, Xu R, Yu XF. 2008. Characterization of conserved motifs in HIV-1 Vif required for APOBEC3G and APOBEC3F interaction. J. Mol. Biol. 381, 1000–1011 (doi:10.1016/j.jmb.2008.06.061) [DOI] [PubMed] [Google Scholar]
- 32.Russell RA, Pathak VK. 2007. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J. Virol. 81, 8201–8210 (doi:10.1128/JVI.00395-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paul I, Cui J, Maynard EL. 2006. Zinc binding to the HCCH motif of HIV-1 virion infectivity factor induces a conformational change that mediates protein-protein interactions. Proc. Natl Acad. Sci. USA 103, 18 475–18 480 (doi:10.1073/pnas.0604150103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao Z, Xiong Y, Zhang W, Tan L, Ehrlich E, Guo D, Yu XF. 2007. Characterization of a novel Cullin5 binding domain in HIV-1 Vif. J. Mol. Biol. 373, 541–550 (doi:10.1016/j.jmb.2007.07.029) [DOI] [PubMed] [Google Scholar]
- 35.Mehle A, Thomas ER, Rajendran KS, Gabuzda D. 2006. A zinc-binding region in Vif binds Cul5 and determines cullin selection. J. Biol. Chem. 281, 17 259–17 265 (doi:10.1074/jbc.M602413200) [DOI] [PubMed] [Google Scholar]
- 36.Kobayashi M, Takaori-Kondo A, Miyauchi Y, Iwai K, Uchiyama T. 2005. Ubiquitination of APOBEC3G by an HIV-1 Vif-Cullin5-Elongin B-Elongin C complex is essential for Vif function. J. Biol. Chem. 280, 18 573–18 578 (doi:10.1073/pnas.0604150103) [DOI] [PubMed] [Google Scholar]
- 37.Bergeron JR, Huthoff H, Veselkov DA, Beavil RL, Simpson PJ, Matthews SJ, Malim MH, Sanderson MR. 2010. The SOCS-box of HIV-1 Vif interacts with ElonginBC by induced-folding to recruit its Cul5-containing ubiquitin ligase complex. PLoS Pathog. 6, e1000925 (doi:10.1371/journal.ppat.1000925) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stanley BJ, Ehrlich ES, Short L, Yu Y, Xiao Z, Yu XF, Xiong Y. 2008. Structural insight into the human immunodeficiency virus Vif SOCS box and its role in human E3 ubiquitin ligase assembly. J. Virol. 82, 8656–8663 (doi:10.1128/JVI.00767-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mehle A, Goncalves J, Santa-Marta M, McPike M, Gabuzda D. 2004. Phosphorylation of a novel SOCS-box regulates assembly of the HIV-1 Vif-Cul5 complex that promotes APOBEC3G degradation. Genes Dev. 18, 2861–2866 (doi:10.1101/gad.1249904) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simon JH, Sheehy AM, Carpenter EA, Fouchier RA, Malim MH. 1999. Mutational analysis of the human immunodeficiency virus type 1 Vif protein. J. Virol. 73, 2675–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Donahue JP, Vetter ML, Mukhtar NA, D'Aquila RT. 2008. The HIV-1 Vif PPLP motif is necessary for human APOBEC3G binding and degradation. Virology 377, 49–53 (doi:10.1016/j.virol.2008.04.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walker RC, Jr, Khan MA, Kao S, Goila-Gaur R, Miyagi E, Strebel K. 2010. Identification of dominant negative human immunodeficiency virus type 1 Vif mutants that interfere with the functional inactivation of APOBEC3G by virus-encoded Vif. J. Virol. 84, 5201–5211 (doi:10.1128/JVI.02318-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang B, Gao L, Li L, Lu Z, Fan X, Patel CA, Pomerantz RJ, DuBois GC, Zhang H. 2003. Potent suppression of viral infectivity by the peptides that inhibit multimerization of human immunodeficiency virus type 1 (HIV-1) Vif proteins. J. Biol. Chem. 278, 6596–6602 (doi:10.1074/jbc.M210164200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Techtmann SM, Ghirlando R, Kao S, Strebel K, Maynard EL. 2012. Hydrodynamic and functional analysis of HIV-1 Vif oligomerization. Biochemistry 51, 2078–2086 (doi:10.1021/bi201738a) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Engelman A, Cherepanov P. 2012. The structural biology of HIV-1: mechanistic and therapeutic insights. Nat. Rev. Microbiol. 10, 279–290 (doi:10.1038/nrmicro2747) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barraud P, Paillart JC, Marquet R, Tisne C. 2008. Advances in the structural understanding of Vif proteins. Curr. HIV Res. 6, 91–99 (doi:10.2174/157016208783885056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Babon JJ, Sabo JK, Soetopo A, Yao S, Bailey MF, Zhang JG, Nicola NA, Norton RS. 2008. The SOCS box domain of SOCS3: structure and interaction with the elonginBC-cullin5 ubiquitin ligase. J. Mol. Biol. 381, 928–940 (doi:10.1016/j.jmb.2008.06.038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (doi:10.1007/BF00197809) [DOI] [PubMed] [Google Scholar]
- 49.Vranken WF, et al. 2005. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59, 687–696 (doi:10.1002/prot.20449) [DOI] [PubMed] [Google Scholar]
- 50.Fogh RH, Boucher W, Vranken WF, Pajon A, Stevens TJ, Bhat TN, Westbrook J, Ionides JM, Laue ED. 2005. A framework for scientific data modeling and automated software development. Bioinformatics 21, 1678–1684 (doi:10.1093/bioinformatics/bti234) [DOI] [PubMed] [Google Scholar]
- 51.Korotkova N, et al. 2008. Binding of Dr adhesins of Escherichia coli to carcinoembryonic antigen triggers receptor dissociation. Mol. Microbiol. 67, 420–434 (doi:10.1111/j.1365-2958.2007.06054.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Battiste JL, Wagner G. 2000. Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear overhauser effect data. Biochemistry 39, 5355–5365 (doi:10.1021/bi000060h) [DOI] [PubMed] [Google Scholar]
- 53.Shen Y, Vernon R, Baker D, Bax A. 2009. De novo protein structure generation from incomplete chemical shift assignments. J. Biomol. NMR 43, 63–78 (doi:10.1007/s10858-008-9288-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramelot TA, et al. 2009. Improving NMR protein structure quality by Rosetta refinement: a molecular replacement study. Proteins 75, 147–167 (doi:10.1002/prot.22229) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dominguez C, Boelens R, Bonvin AM. 2003. HADDOCK: a protein–protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 125, 1731–1737 (doi:10.1021/ja026939x) [DOI] [PubMed] [Google Scholar]
- 56.de Vries SJ, van Dijk M, Bonvin AM. 2010. The HADDOCK web server for data-driven biomolecular docking. Nat. Protocol 5, 883–897 (doi:10.1038/nprot.2010.32) [DOI] [PubMed] [Google Scholar]
- 57.Zhou P, Lugovskoy AA, Wagner G. 2001. A solubility-enhancement tag (SET) for NMR studies of poorly behaving proteins. J. Biomol. NMR 20, 11–14 (doi:10.1023/A:1011258906244) [DOI] [PubMed] [Google Scholar]
- 58.Schreiber G. 2002. Kinetic studies of protein–protein interactions. Curr. Opin. Struct. Biol. 12, 41–47 (doi:10.1016/S0959-440X(02)00287-7) [DOI] [PubMed] [Google Scholar]
- 59.Wolfe LS, Stanley BJ, Liu C, Eliason WK, Xiong Y. 2010. Dissection of the HIVVif interaction with human E3 ubiquitin ligase. J. Virol. 84, 7135–7139 (doi:10.1128/JVI.00031-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tzeng SR, Kalodimos CG. 2012. Protein activity regulation by conformational entropy. Nature 488, 236–240 (doi:10.1038/nature11271) [DOI] [PubMed] [Google Scholar]
- 61.Bullock AN, Debreczeni JE, Edwards AM, Sundstrom M, Knapp S. 2006. Crystal structure of the SOCS2-elongin C-elongin B complex defines a prototypical SOCS box ubiquitin ligase. Proc. Natl Acad. Sci. USA 103, 7637–7642 (doi:10.1073/pnas.0601638103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang X, et al. 2013. Interactions between HIV-1 Vif and human ElonginB-ElonginC are important for CBF-beta binding to Vif. Retrovirology 10, 94 (doi:10.1186/1742-4690-10-94) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garvey KJ, Oberste MS, Elser JE, Braun MJ, Gonda MA. 1990. Nucleotide sequence and genome organization of biologically active proviruses of the bovine immunodeficiency-like virus. Virology 175, 391–409 (doi:10.1016/0042-6822(90)90424-P) [DOI] [PubMed] [Google Scholar]
- 64.Bullock AN, Rodriguez MC, Debreczeni JE, Songyang Z, Knapp S. 2007. Structure of the SOCS4-ElonginB/C complex reveals a distinct SOCS box interface and the molecular basis for SOCS-dependent EGFR degradation. Structure 15, 1493–1504 (doi:10.1016/j.str.2007.09.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zadjali F, et al. 2011. Structural basis for c-KIT inhibition by the suppressor of cytokine signaling 6 (SOCS6) ubiquitin ligase. J. Biol. Chem. 286, 480–490 (doi:10.1074/jbc.M110.173526) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ribeiro AC, Maia e Silva A, Santa-Marta M, Pombo A, Moniz-Pereira J, Goncalves J, Barahona I. 2005. Functional analysis of Vif protein shows less restriction of human immunodeficiency virus type 2 by APOBEC3G. J. Virol. 79, 823–833 (doi:10.1128/JVI.79.2.823-833.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oberste MS, Gonda MA. 1992. Conservation of amino-acid sequence motifs in lentivirus Vif proteins. Virus Genes 6, 95–102 (doi:10.1007/BF01703760) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The DNA accession information is listed in the electronic supplementary material, table S1.
Funding statement
Partial financial support for consumables was provided by grants from the Wellcome Trust (M.R.S., M.H.M.) and the Medical Research Council (M.H.M.).